Abstract
Because of the complexity of cancer‐immune system interactions, combinations of biomarkers will be required for predicting individual patient responses to treatment and for monitoring combination strategies to overcome treatment resistance. To this end, the “immunogram” has been proposed as a comprehensive framework to capture all relevant immunological variables. Here, we developed a method to convert transcriptomic data into immunogram scores (IGS). This immunogram includes 10 molecular profiles, consisting of innate immunity, priming and activation, T cell response, interferon γ (IFNG) response, inhibitory molecules, regulatory T cells, myeloid‐derived suppressor cells (MDSCs), recognition of tumor cells, proliferation, and glycolysis. Using genes related to these 10 parameters, we applied single‐sample gene set enrichment analysis (ssGSEA) to 9417 bulk RNA‐Seq data from 9362 cancer patients with 29 different solid cancers in The Cancer Genome Atlas (TCGA). Enrichment scores were z‐score normalized (Z) for each cancer type or the entire TCGA cohort. The IGS was defined by the formula IGS = 3 + 1.5 × Z so that patients would be well distributed over a range of scores from 1 to 5. The immunograms constructed in this way for all individual patients in the entire TCGA cohort can be accessed at “The RNA‐Seq based Cancer Immunogram Web” (https://yamashige33.shinyapps.io/immunogram/).
Keywords: GSEA, immunogram, RNA‐Seq, TCGA, tumor immunity
We propose a novel scoring and visualization method for “immunogram” to assess the cancer immunity status of individual patients using the large TCGA dataset and RNA‐Seq of each patient. Immunograms for each individual will be a useful tool for precision immuno‐oncology, although their application needs to be validated in clinical trials.

Abbreviations
- ACC
adrenocortical carcinoma
- BLCA
bladder urothelial carcinoma
- BRCA
breast invasive carcinoma
- CESC
cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
cholangiocarcinoma
- COAD
colon adenocarcinoma
- CTS
cancer type–specific
- ESCA
esophageal carcinoma
- fpkm
fragments per kilobase of exon per million reads mapped
- GBM
glioblastoma multiforme
- HNSC
head and neck squamous cell carcinoma
- ICI
immune checkpoint inhibitors
- IGS
immunogram scores
- KICP
kidney chromophobe
- KIRC
kidney renal clear cell carcinoma,
- KIRP
kidney renal papillary cell carcinoma
- LGG
brain lower grade glioma
- LIHC
liver hepatocellular carcinoma
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MDSC
myeloid‐derived suppressor cell
- MESO
mesothelioma
- MSigDB
Molecular Signatures Database
- OV
ovarian serous cystadenocarcinoma
- PAAD
pancreatic adenocarcinoma
- PAN
pan‐cancer
- PCPG
pheochromocytoma and paraganglioma
- PRAD
prostate adenocarcinoma
- READ
rectum adenocarcinoma
- SARC
sarcoma
- SKCM
skin cutaneous melanoma
- ssGSEA
single‐sample gene set enrichment analysis
- STAD
stomach adenocarcinoma
- TCGA
The Cancer Genome Atlas
- THCA
thyroid carcinoma
- TME
tumor microenvironment
- UCEC
uterine corpus endometrial carcinoma
- UCS
uterine carcinosarcoma
- UVM
uveal melanoma
1. INTRODUCTION
Immunotherapy with immune checkpoint inhibitors (ICIs) has completely changed the therapeutic landscape for many types of solid tumors. 1 Several combinations of immunotherapy with chemotherapy, 2 , 3 , 4 molecular targeted drugs, 5 or combinations of different ICIs 6 , 7 are now approved for the first‐line treatment of various cancers. However, not all types of cancer respond equally well to ICIs, and even in responsive cancers, only a subset of patients experiences durable responses and favorable long‐term outcomes. This is because primary and acquired resistance occurs in a considerable proportion of patients across different cancer types. 8
Therefore, it is crucial to establish reliable predictive biomarkers to distinguish ICI responders from nonresponders, who may suffer unnecessary costs and toxicities, and to identify candidates for rational combination therapies. 9 Currently, tumor mutational burden 10 , 11 , 12 and PD‐L1 expression 13 , 14 are the two major variables used as biomarkers that have been validated in phase III clinical trials. Additionally, several other factors associated with response or resistance to ICIs across cancer types have been proposed as biomarkers, based on molecular profiling of cancers treated with different immunotherapies. These include an immune‐inflamed phenotype, 15 , 16 expression of T cell signaling pathway genes such as IFNγ, 17 microsatellite instability, 18 somatic copy‐number alterations, 19 human leukocyte antigen (HLA) class I diversity, 20 T cell repertoire clonality change, 21 WNT‐β‐catenin signaling, 22 TGFβ expression, 23 and even commensal microbiota. 24
However, as single biomarkers, none of the above is sufficient to identify individual patients who will likely benefit from immunotherapy. Unlike conventional cancer therapies, immunotherapies, including ICIs, do not directly target tumor cells; instead, they affect tumor cells through the patient's immune system or the tumor microenvironment (TME). 25 Therefore, the different components that affect tumor‐immune interactions need to be taken into account when developing predictive biomarkers for immunotherapy. Comprehensive analysis of multiple different functional pathways and molecular networks that reveal integrated mechanisms of tumor‐immune interactions are crucial for this purpose. General and local cancer immunity status in each patient needs to be taken into consideration. To this end, Blank et al proposed the concept of the cancer immunogram that integrates multiparameter biomarkers to visualize the immunological status of an individual patient. 26 We have applied this concept to lung cancer patients and developed an immunogram reflecting the cancer‐immunity cycle. 27 Since then, van Dijk et al have reported an immunogram informative specifically for urothelial cancer patients. 28
Although immunograms may be useful for visualizing the landscape of the tumor microenvironment and the compromised steps of antitumor immunity in each patient, both Blank et al and van Dijk et al had only theorized that they could be useful to patients but had not tested the concept in clinical practice. In contrast, we analyzed real‐world lung cancer patient data to generate immunograms with potential application for personalized immunotherapy. However, in the previous version of our immunogram, the parameters were normalized and scored within the cohort; this approach could, therefore, not be applied to other cohorts. Thus, in the present study, we utilized RNA‐Seq data from The Cancer Genome Atlas (TCGA) cohort as a standard and set up a scoring scale to quantify parameters incorporated in the immunogram. Here, we propose a novel versatile scoring method for constructing such individual immunograms.
2. MATERIALS AND METHODS
2.1. Data and analysis
RNA‐Seq data for 29 solid tumors from TCGA were downloaded via the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/) (n = 9417). The dataset General Research Use in TCGA access as the project titled #12517: "Immunogram for personalized cancer immunotherapy" was approved by NIH (#49374‐7). For subjects with multiple RNA‐Seq data available, mean count data were converted into fragments per kilobase of exon per million reads mapped (fpkm) and used for gene set enrichment analysis (GSEA). RNA‐Seq data for 28 tumors from 27 melanoma patients who received anti‐PD‐1 treatment were obtained from Gene Expression Omnibus (Accession number: GSE78220). 29 The enrichment score is obtained using the single‐sample gene set enrichment analysis (ssGSEA) method 30 with R package ssGSEA 2.0 (https://github.com/broadinstitute/ssGSEA2.0) and R software version 3. 6. 0.
2.2. Availability of data and materials
All data analyzed during this study are included in this published article and its [Link], [Link], [Link], [Link] files. All data analyzed can be accessed at a web‐accessible database, “The RNA‐Seq based Cancer Immunogram Web” (https://yamashige33.shinyapps.io/immunogram/).
2.3. Statistical analyses
Spearman correlation coefficients between any pair of gene sets or axes for the immunogram were calculated. The results were interpreted according to the degree of association as strong (= 0.7‐1), moderate (= 0.5‐0.7), or low (<0.5) after taking significant correlation (or) values into consideration. Hierarchical clustering (Ward's method) was performed using JMP Pro15.0.0 (SAS Institute Inc).
3. RESULTS
3.1. Gene set selection for immunograms
An immunogram is a flexible system to illustrate the immunological status of each patient by adopting any and all appropriate parameters. Here, we depict an immunogram on a radar plot with ten axes to provide a useful snapshot of the immune landscape in the tumor microenvironment of an individual patient (Figure 1). These ten molecular profiles are innate immunity: natural killer (NK) cells (axis 1), priming and activation: dendritic cells (DCs) (axis 2), T cells: CD8+ T cell response (axis 3), interferon γ (IFNG) response (axis 4), inhibitory molecules (axis 5), inhibitory cells (regulatory T cells [Tregs], axis 6), inhibitory cells (myeloid‐derived suppressor cells [MDSCs], axis 7), recognition of tumor cells: antigen processing and presentation (axis 8), proliferation (axis 9), and glycolysis (axis 10). These pathways are all relevant for the development of antitumor immune responses. To quantify these molecular profiles, ssGSEA of bulk RNA‐Seq data was performed (Table 1, Table S1).
Figure 1.
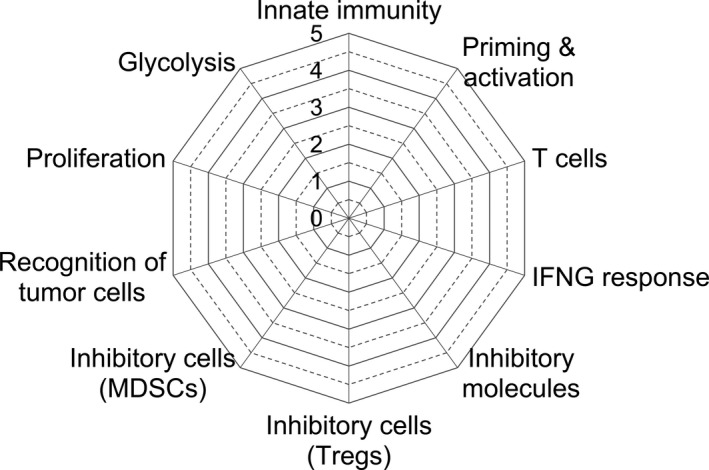
Immunogram radar plots. To depict the molecular profiles of the tumor microenvironment in each patient, 10 parameters related to this process were scored and plotted on the radar plot. These consist of innate immunity (axis 1), priming and activation (axis 2), T cell response (axis 3), IFNG response (axis 4), inhibitory molecules (axis 5), regulatory T cells (Treg, axis 6), myeloid‐derived suppressor cells (MDSCs, axis 7), recognition of tumor cells and presentation (axis 8), proliferation (axis 9), and glycolysis (axis 10), all relevant for the development of antitumor immune responses
Table 1.
Gene set for the 10‐axis immunogram
| Axis | Immunological parameter | Gene set_name |
|---|---|---|
| 1 | Innate immunity | LM22 NK cells activated (31) |
| 2 | Priming & activation | LM22 Dendritic cells activated (31) |
| 3 | T cells | LM22 T cells CD8 (31) |
| 4 | IFNG response | HALLMARK INTERFERON GAMMA RESPONSE (MSigDB) |
| 5 | Inhibitory molecules | Immune escape gene set (IEGS) immune escape (32) |
| 6 | Inhibitory cells (Tregs) | LM22 T cells regulatory (Tregs) (31) |
| 7 | Inhibitory cells (MDSC) | Angelova_MDSC (33) |
| 8 | Recognition of tumor cells | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION (MSigDB) |
| 9 | Proliferation | REACTOME DNA REPLICATION (MSigDB) |
| 10 | Glycolysis | HALLMARK GLYCOLYSIS (MSigDB) |
Molecular Signatures Database (MSigDB): https://www.gsea‐msigdb.org/gsea/msigdb/index.jsp.
Required marker genes for each axis are listed in Table S1.
For the quantification of NK cells, DCs, CD8+ T cells, and Tregs, gene sets from LM22 were utilized. 31 Gene sets for IFNG response, recognition of tumor cells: antigen processing and presentation, proliferation, and glycolysis were selected from the Molecular Signatures Database (MSigDB, https://www.gsea‐msigdb.org/gsea/msigdb/index.jsp). Gene set for inhibitory molecules was utilized from immune escape gene set (IEGS) immune escape. 1 , 2 , 32 For MDSC, we followed the published gene set by Angelova et al. 33 We validated these selected gene sets by comparing them with similar gene sets available in the literature (Table S2). As a test sample, we exploited the RNA‐Seq data of 103 melanoma patients from the TCGA and ran ssGSEA to obtain enrichment scores. For example, we compared 12 gene sets that are related to NK cells by Spearman correlation analysis to validate the gene set for axis 1 (Figure 2). Most gene sets showed a strong linear correlation with each other except the one reported by Şenbabaoğlu et al. 34 The LM22 NK cell gene set correlated well with the other gene sets. Similarly, gene sets for other axes were evaluated by Spearman correlation analysis, and their validity was confirmed (Figure. S1).
Figure 2.

Scatter plot matrix for Spearman correlation analysis of gene sets for axis 1. The RNA‐Seq data of 103 melanoma patients from The Cancer Genome Atlas (TCGA) were subjected to single‐sample gene set enrichment analysis (ssGSEA) with 12 gene sets related to innate immunity: Abbas_NK_cells, 39 Becht_NK_cells, 40 GO:0045087_innate_immune_response (AmiGo2), Huntington_NK_cells, 41 Angelova_NK_cells, 33 ImSig_NK_cells, 42 LM22_NK_cells_activated, 31 LM22_NK_activated_resting, 31 LM7_NK_cells, 43 Charoentong_NK_cells, 44 Schelker_NK_cells, 45 and Şenbabaoğlu_NK_cells. 34 The correlations of these gene sets were analyzed by Spearman correlation analysis. Pairwise correlation analyses of 12 gene sets are represented in the scatter plot matrix that contains all the pairwise data of ssGSEA scores with the indicated gene sets. The bivariate correlations for the dataset are shown with a color coding as follows: dark red is associated with Spearman correlation coefficient, R, equal to 1 and dark blue is associated with R = −1
3.2. Converting ssGSEA enrichment scores to immunogram scores
To plot an immunogram, the scoring method depends on having reliable standard values for each axis. To this end, we utilized 9417 RNA‐Seq data from 9362 cancer patients with 29 different solid cancers in the TCGA dataset. These consist of breast invasive carcinoma (BRCA), uterine corpus endometrial carcinoma (UCEC), kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), brain lower grade glioma (LGG), thyroid carcinoma (THCA), head and neck squamous cell carcinoma (HNSC), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), skin cutaneous melanoma (SKCM), colon adenocarcinoma (COAD), bladder urothelial carcinoma (BLCA), stomach adenocarcinoma (STAD), ovarian serous cystadenocarcinoma (OV), liver hepatocellular carcinoma (LIHC), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), kidney renal papillary cell carcinoma (KIRP), sarcoma (SARC), pheochromocytoma and paraganglioma (PCPG), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), glioblastoma multiforme (GBM), esophageal carcinoma (ESCA), mesothelioma (MESO), uveal melanoma (UVM), kidney chromophobe (KICP), uterine carcinosarcoma (UCS), cholangiocarcinoma (CHOL), and adrenocortical carcinoma (ACC) (Table 2). The RNA‐Seq data were subjected to ssGSEA with the 10 gene sets mentioned above (Table 1).
Table 2.
Patients with 29 solid cancers from The Cancer Genome Atlas (TCGA)
| Cancer type | Abbreviation | Number of patients | Number of samples |
|---|---|---|---|
| Breast invasive carcinoma | BRCA | 1091 | 1097 |
| Uterine corpus endometrial carcinoma | UCEC | 543 | 547 |
| Kidney renal clear cell carcinoma | KIRC | 530 | 534 |
| Lung adenocarcinoma | LUAD | 513 | 524 |
| Brain lower grade glioma | LGG | 511 | 511 |
| Thyroid carcinoma | THCA | 502 | 502 |
| Head and neck squamous cell carcinoma | HNSC | 500 | 500 |
| Lung squamous cell carcinoma | LUSC | 501 | 501 |
| Prostate adenocarcinoma | PRAD | 495 | 498 |
| Skin cutaneous melanoma | SKCM | 103 | 103 |
| Colon adenocarcinoma | COAD | 456 | 469 |
| Bladder urothelial carcinoma | BLCA | 408 | 412 |
| Stomach adenocarcinoma | STAD | 375 | 375 |
| Ovarian serous cystadenocarcinoma | OV | 374 | 374 |
| Liver hepatocellular carcinoma | LIHC | 371 | 371 |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | CESC | 304 | 304 |
| Kidney renal papillary cell carcinoma | KIRP | 288 | 288 |
| Sarcoma | SARC | 259 | 259 |
| Pheochromocytoma and paraganglioma | PCPG | 178 | 178 |
| Pancreatic adenocarcinoma | PAAD | 177 | 177 |
| Rectum adenocarcinoma | READ | 166 | 166 |
| Glioblastoma multiforme | GBM | 154 | 155 |
| Esophageal carcinoma | ESCA | 161 | 161 |
| Mesothelioma | MESO | 86 | 86 |
| Uveal melanoma | UVM | 80 | 80 |
| Kidney chromophobe | KICH | 65 | 65 |
| Uterine carcinosarcoma | UCS | 56 | 56 |
| Cholangiocarcinoma | CHOL | 36 | 45 |
| Adrenocortical carcinoma | ACC | 79 | 79 |
| Total | 9362 | 9417 |
For scoring the immunogram, z‐score normalization was applied to ssGSEA enrichment scores. Because of the considerable differences in gene expression profiles between cancer types, normalization was performed in subgroups of patients with each cancer type or in the entire TCGA cohort. Hence, we constructed two immunograms for each patient: a cancer type–specific (CTS) and a pan‐cancer (PAN) immunogram. The mean (MCTS) and standard deviation (SDCTS) values of the enrichment scores were calculated for subgroups of patients with each cancer type or the MPAN and SDPAN values of enrichment scores of all TCGA patients with 29 cancer types for pan‐cancer analysis. The enrichment score (ESn) of axis n in each patient was converted into z‐score ZCTS, n or ZPAN, n and then converted into the immunogram score IGSCTS, n or IGSPAN, n by the following formula.
In each patient, IGSs for all axes were determined by these formulae and plotted onto the radar chart to generate cancer type–specific or pan‐cancer immunograms. The lower and upper limits of the IGS were set at 1 and 5. By definition, IGS = 3 represents an ES equivalent to the mean ES of the TCGA cohort, and IGS = 4.5 or IGS = 1.5 represents ES equivalent to the mean plus or minus one SD. The IGS was defined in this way so that patients would be well distributed over the range from 1 to 5. We then developed a web‐accessible database designated “The RNA‐Seq based Cancer Immunogram Web with the results of the 9362 cancer patients of the TCGA cohort (https://yamashige33.shinyapps.io/immunogram/). Each patient has their own discrete immunogram, suggesting that the immune response and tumor microenvironment are unique to each individual.
3.3. Cancer type–specific and pan‐cancer immunograms
As an example, immunograms (IGPAN and IGCTS) for one patient in the TCGA with LUAD, KIC, SKCM, BRCA, PRAD, or LGG are depicted in Figure 3. For each patient, the shapes of the cancer type–specific (IGCTS) and pan‐cancer (IGPAN) are different. Some tumors are rich in T cell infiltration and designated as “hot,” while “cold” tumors lack such infiltrations. In the pan‐cancer analysis, all the data from patients with hot and cold tumors are pooled. Therefore, MPAN is higher than MCTS for tumor types in which cold tumors are dominant. The opposite is true for hot tumor–dominant types. As shown in Figure 3, the outer areas of IGPAN for LUAD and KIRC are more extended than those of the corresponding IGCTS, suggesting that these tumors are immunologically hot. In contrast, the IGPAN for PRAD and LGG appear compressed relative to those of the corresponding IGCTS, suggesting that these tumors are immunologically cold. IGPAN is suitable for comparing immunological status across the different cancer types, whereas by using IGCTS, we can compare subtle individual differences in intratumoral immune responses between patients with the same types of cancer.
Figure 3.
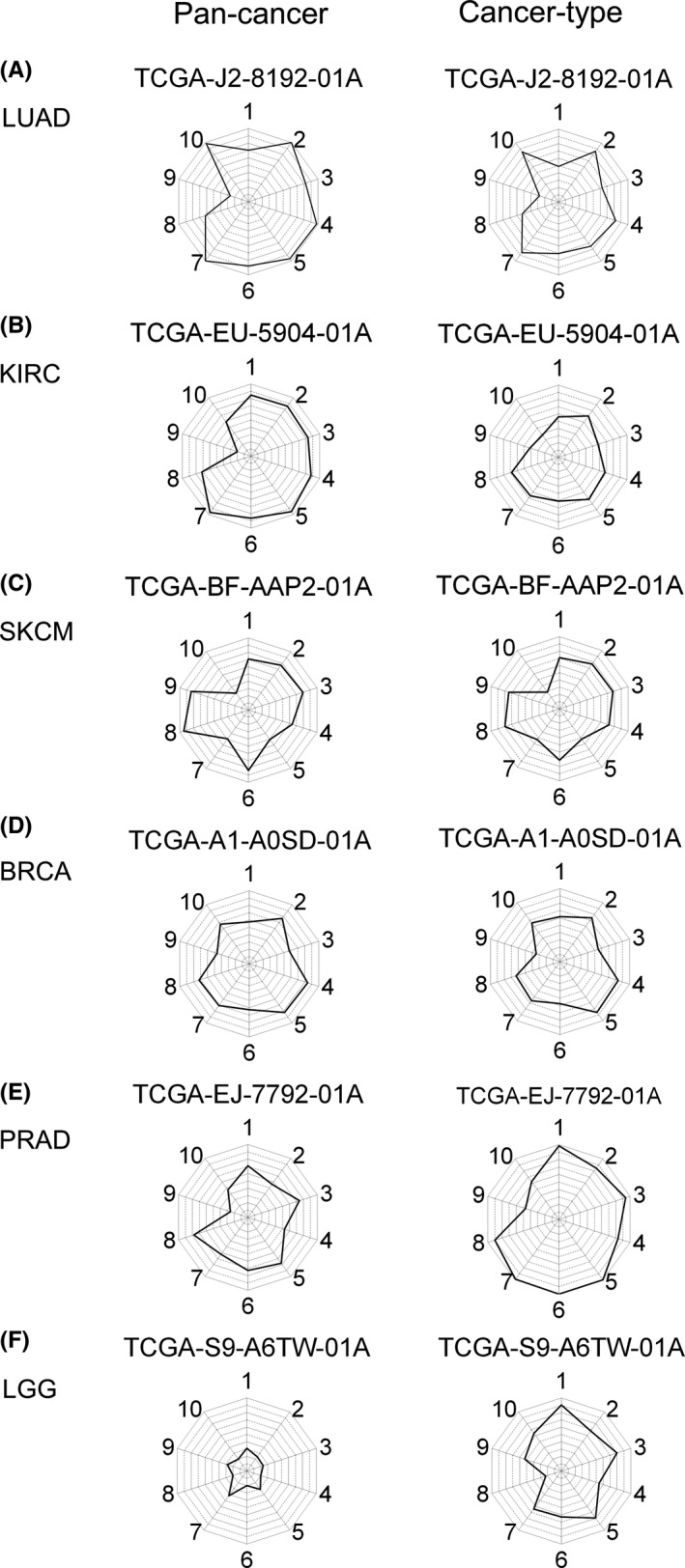
Pan‐cancer and cancer type–specific immunograms. In each patient, two immunograms can be depicted based on the different normalized scores, pan‐cancer and cancer type–specific. Immunograms for a randomly selected patient with LUAD (A), KIRC (B), SKCM (C), BRCA (D), PRAD (E), and LGG (F) from The Cancer Genome Atlas (TCGA) are shown. TCGA case ID is shown in the panel. 1. Innate immunity, 2. Priming & activation, 3. T cells, 4. IFNG response, 5. Inhibitory molecules, 6. Inhibitory cells (Tregs), 7. Inhibitory cells (MDSCs), 8. Recognition of tumor cells, 9. Proliferation, 10. Glycolysis. BRCA, breast invasive carcinoma; KIRC, kidney renal clear cell carcinoma; LGG, brain lower grade glioma; LUAD, lung adenocarcinoma; PRAD, prostate adenocarcinoma; SKCM, skin cutaneous melanoma
3.4. A general overview of immune responses in each cancer type
To gain a general overview of cancer type–specific immune responses in the tumor, we created fictional patients representing the 29 cancer types in the TCGA cohort by providing MCTS, n for their ESn. When we depict IGCTS in these fictional patients, all immunograms show regular decagons with all axes equal to point 3.0. IGPANs for these patients are shown in Figure 4. Hierarchical clustering of these “patients” by their IGSs indicated that MESO, LUAD, KIRC, PAAD, LUSC, CESC, and HNSC are quite immunogenic, while UCS, GBM, ACC, PRAD, KICH, PCPG, UVM, and LGG are immunologically quiescent. These results are consistent with clinical experience as reflected in the approval of checkpoint inhibitors for these immunogenic cancer types (with the exception of PAAD).
Figure 4.
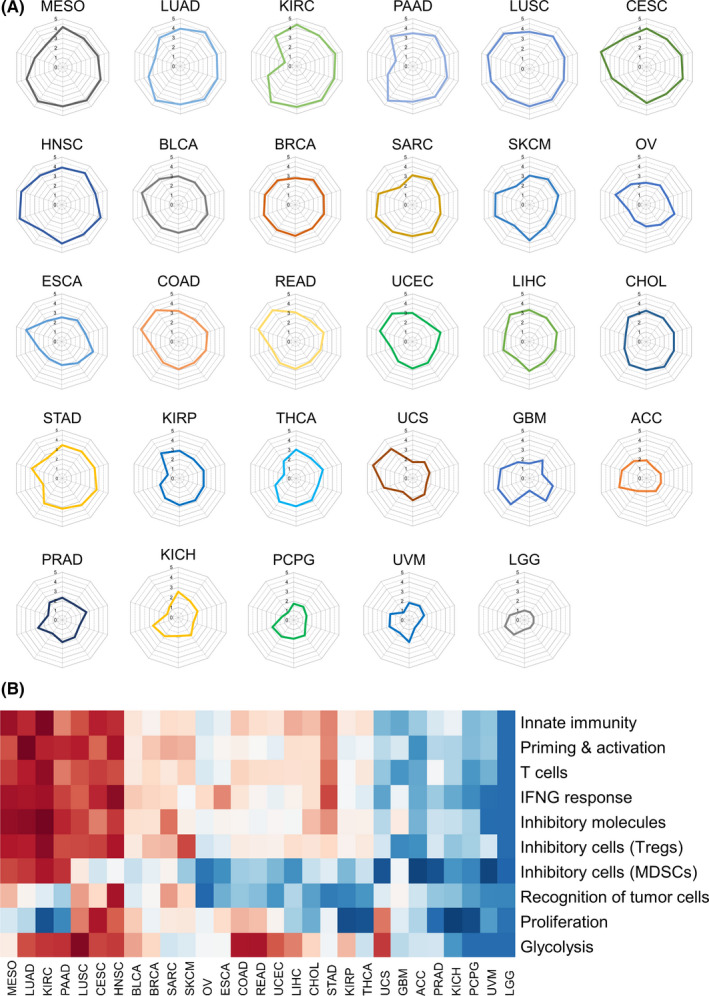
Pan‐cancer immunograms for fictional patients with 29 solid cancers. A, Pan‐cancer immunograms were generated for 29 fictional patients with the mean enrichment scores of the corresponding cancer type. B, Hierarchical clustering with Ward's method of these patients using the10 immunogram scores. Data are shown with a color coding as follows: dark red is associated with an immunogram score (IGS) equal to 5 and dark blue is associated with IGS = 1
3.5. Immunograms for personalized immuno‐oncology
Axes 1‐4 represent the series of dynamic processes involved in the induction of antitumor immune responses, and axis 8 is essential for the T cell recognition of tumor cells, whereas genes related to axes 5‐7 are seen as inhibitory counter‐regulators. Therefore, these axes often move together. To examine the correlation between all 10 axes of the immunogram, all IGSPANs of the 9417 patients were subjected to Spearman correlation analysis. As shown in Figure 5A, correlation coefficients among axes 1‐6 were >0.7. These results could be interpreted to imply that selecting only one parameter within axes 1‐6 would be sufficient for incorporation into the immunogram. This might be so if we evaluate the immune response as a whole. However, this might also be misleading if these parameters are not redundant and required for the evaluation of each patient's immunological status. Examples of lung cancer patients shown in Figure 5 demonstrate that each axis behaves differently. In the case of TCGA‐86‐8671‐01A, axes 1‐7 were equally high (Figure 5B), but in TCGA‐86‐8359‐01A, axes 1 and 3 were high, but axes 2, 4, and 5 were low. Axes 1, 3, and 6 were low and axes 2, 4, and 5 were high in TCGA‐97‐8175‐01A. The other three patients display additional different immunogram patterns. These results indicate that all these parameters are required to understand the antitumor immunity specifically in every single patient because combination immunotherapy could be selected to target each of the different detected impaired processes identified by the detailed immunogram.
Figure 5.
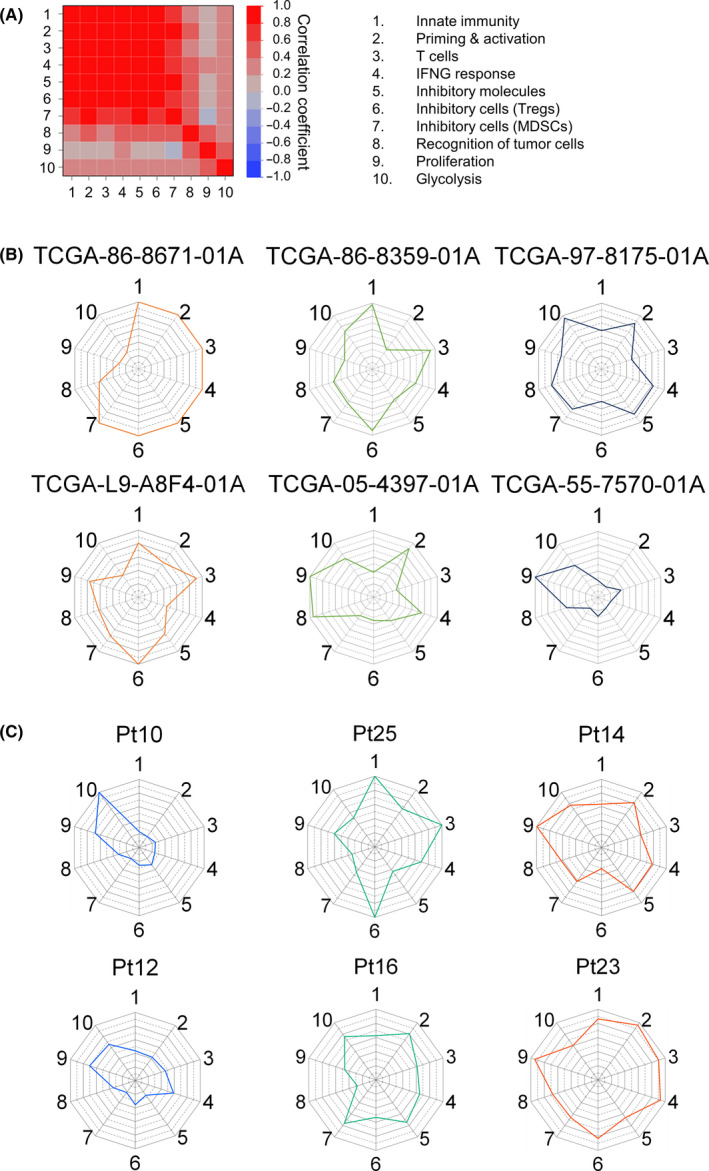
Immunograms for individual patients. A, Immunogram scores of each axis from 9417 patients’ data were subjected to Spearman correlation analysis. Pairwise correlation analyses of each axis of the immunogram are represented in the heatmap matrix that contains all the pairwise data of the indicated axes. Bivariate correlations for the dataset are shown with a color coding as follows: dark red is associated with Spearman correlation coefficient, R, equal to 1 and dark blue is associated with R = −1. B, Lung cancer–specific immunograms of six patients. The Cancer Genome Atlas (TCGA) case IDs are shown in the panel. C, Immunograms of melanoma patients who received anti‐PD‐1 therapy (cohort of Hugo et al 29 ). Six nonresponders were shown. See also Fig. S2. 1. Innate immunity, 2. Priming & activation, 3. T cells, 4. IFNG response, 5. Inhibitory molecules, 6. Inhibitory cells (Tregs), 7. Inhibitory cells (MDSCs), 8. Recognition of tumor cells, 9. Proliferation, 10. Glycolysis
We analyzed RNA‐Seq data of 28 pretreatment tumors from melanoma patients who received anti‐PD‐1 ICI. 29 Immunograms were depicted in these pretreatment tumors (Figure S2). Then, we focused on nonresponding (progressive disease) patients (Figure 5C) and examined whether we could recommend potential combination immunotherapy to these nonresponders using immunogram. The immunograms of Pt10 and Pt12 displayed a typical immunogram pattern of cold tumors. The strategies to induce T cell response in the tumor, for example, cancer vaccine or oncolytic virotherapy, might be recommended to combine with ICI. Besides, glycolysis stood out from the other axes in Pt10. T cell response is hampered by the high energy demand of tumor cells, that is a therapeutic target for combination immunotherapy. 35 Axis 6 (Treg) and axis 7 (MDSC) were high in Pt25 and Pt16, respectively, suggesting that the strategies to deplete Treg or MDSC might be recommended to these patients. In Pt14 and Pt23, axis 9 (proliferation) was high, suggesting that molecular targeted therapies or chemotherapy that suppress the proliferation of tumor cells might be combined with ICI. While these suggestions necessitate confirmation in a clinical trial, immunogram is considered to be an excellent platform for personalized immuno‐oncology.
4. DISCUSSION
The concept of the cancer immunogram has attracted a great deal of attention since Blank et al proposed using radar plots as frameworks for describing the diversity of cancer‐immune interactions in each individual patient. 26 Although their potential value is widely recognized, no versatile scoring method has been developed to construct immunograms in a real‐world setting. Here, we propose a novel scoring method based on RNA‐Seq data and the application of ssGSEA to quantify parameters related to antitumor immunity. We have constructed a web‐accessible database “The RNA‐Seq based Cancer Immunogram Web” (https://yamashige33.shinyapps.io/immunogram/) with the results of 9362 cancer patients (9417 RNA‐Seq data) from the TCGA cohort.
Although cancer type–specific immune responses can be appreciated and compared by immunogram analysis (Figure 4), the real value of using immunograms would be in personalized immuno‐oncology. Indeed, we can easily see that the immunogram patterns differ greatly between patients even with the same type of cancer, reflecting the heterogeneity of immune responses and TME in each individual (Figures 3 and 5, The RNA‐Seq based Cancer Immunogram Web). As shown in Figure 5B, for example, the immunogram for TCGA‐86‐8671‐01A suggests pre‐existing T cell responses but tumor‐induced immunosuppressive molecules and cells as counter‐regulators. For these patients, ICI might be expected to be effective. Low scores in priming and activation and high scores for Tregs observed in patient TCGA‐86‐8359‐01A, on the other hand, suggest that combination therapies with DC vaccines and Treg depletion therapy might be recommended. Once we can identify the impaired steps of the antitumor immune response in each patient individually, we might be better able to select some of the several drugs that have already been developed and approved for clinical use in treating certain forms of cancer to overcome these hurdles. 36 Using immunograms of nonresponding tumors to ICI (Figure 5C), we could point out the impaired step that might be a potential target for combination therapies in each patient. Immunograms for each individual will be a useful tool for precision immuno‐oncology, although their application needs to be validated in clinical trials.
Comprehensive assessment and integration of a myriad of potentially immune‐relevant factors is required to generate informative immunograms. To this end, the flexibility of approaches based on RNA‐seq is ideal. RNA‐Seq provides comprehensive transcriptome data, 37 and ssGSEA scoring provides a useful approach for quantifying selected molecular signatures in the sample transcriptome. 30 There is a collection of annotated gene sets available for GSEA, such as the Molecular Signatures Database (MSigDB). It is possible to supplement the immunogram with novel relevant biomarkers and easily test different combinations of gene sets by selecting other panels of genes to generate different immunograms. For example, immunograms for hallmarks of cancer could also be compiled by adopting gene sets for the eight hallmarks: sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, reprogramming of energy metabolism, and evading immune destruction. 38
In the present study, we described an immunogram with 10 axes just as an example to introduce our scoring method, but other parameters may need to be considered as well. These 10 parameters have not yet been optimized for developing predictive biomarkers for ICI and for tailoring personalized combination immunotherapy. Accumulating evidence indicates that ICI efficacy is affected by a combination of local and systemic factors involving tumor‐intrinsic, host‐related, and microenvironmental biomarkers. The immunogram described here lacks any data on tumor mutational burden or other genomic data. It also lacks information on systemic factors, environmental factors, and data from immunohistochemistry and flow cytometry. These data can be incorporated into the immunogram; however, methods for normalizing these modalities are set up differently and require considerable labor. We will add these parameters one by one to the immunogram in the future.
In conclusion, we propose a novel scoring and visualization method for assessing the cancer immunity status of individual patients using the large TCGA dataset and RNA‐Seq of each patient. This study is the first to describe a way of depicting immunograms using real‐world patient data. Immunograms generated in this way are flexible and can incorporate a myriad of gene sets available to the community. Further refinement and validation of such immunograms should contribute to understanding the immunological status of each individual patient for predicting the efficacy of ICI and tailoring optimal combination immunotherapies in a personalized manner.
CONFLICT OF INTEREST
Dr Kakimi reports grants from TAKARA BIO Inc, grants from MSD, outside the submitted work; the Department of Immunotherapeutics, The University of Tokyo Hospital, is endowed by TAKARA BIO Inc. Dr Yamaguchi is a founder of cBioinformatics. Other authors have no competing interests to disclose.
ETHICAL CONSIDERATION
This article does not contain any studies involving human participants performed by any of the authors. The dataset General Research Use in TCGA access as the project titled #12517: "Immunogram for personalized cancer immunotherapy" was approved by NIH (#49374‐7).
Supporting information
Fig S1
Fig S2
Table S1
Table S2
ACKNOWLEDGMENTS
The authors thank Dr Koji Nagaoka, Dr Akihiro Hosoi, Dr Taro Teshima, Ms Mikiko Shibuya, and Ms Jerena Arsenic for technical assistance. This work was conducted with the institutional support of RIKEN. The funder had no role in study design, data collection and analysis, interpretation, decision to publish, preparation of the manuscript, or any aspect of the study.
Kobayashi Y, Kushihara Y, Saito N, Yamaguchi S, Kakimi K. A novel scoring method based on RNA‐Seq immunograms describing individual cancer‐immunity interactions. Cancer Sci. 2020;111:4031–4040. 10.1111/cas.14621
REFERENCES
- 1. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350‐1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gandhi L, Rodríguez‐Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non–small‐cell lung cancer. N Engl J Med. 2018;378:2078‐2092. [DOI] [PubMed] [Google Scholar]
- 3. Paz‐Ares L, Luft A, Vicente D, et al. Pembrolizumab plus chemotherapy for squamous non–small‐cell lung cancer. N Engl J Med. 2018;379:2040‐2051. [DOI] [PubMed] [Google Scholar]
- 4. Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for first‐line treatment of metastatic nonsquamous NSCLC. N Engl J Med. 2018;378:2288‐2301. [DOI] [PubMed] [Google Scholar]
- 5. Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal‐cell carcinoma. N Engl J Med. 2019;380:1116‐1127. [DOI] [PubMed] [Google Scholar]
- 6. Hodi FS, Chiarion‐Sileni V, Gonzalez R, et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4‐year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 2018;19:1480‐1492. [DOI] [PubMed] [Google Scholar]
- 7. Motzer RJ, Tannir NM, McDermott DF, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal‐cell carcinoma. N Engl J Med. 2018;378:1277‐1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharma P, Hu‐Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA‐4 blockade in melanoma. N Engl J Med. 2014;371:2189‐2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science. 2015;348:124‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA‐4 blockade in metastatic melanoma. Science. 2015;350:207‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism‐driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor‐based immunotherapy. Lancet Oncol. 2016;17:e542‐e551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herbst RS, Soria J‐C, Kowanetz M, et al. Predictive correlates of response to the anti‐PD‐L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563‐567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tumeh PC, Harview CL, Yearley JH, et al. PD‐1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao J, Shi LZ, Zhao H, et al. Loss of IFN‐gamma pathway genes in tumor cells as a mechanism of resistance to anti‐CTLA‐4 therapy. Cell. 2016;167:397‐404.e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD‐1 blockade. Science. 2017;357:409‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355:eaaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chowell D, Morris LGT, Grigg CM, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Riaz N, Havel JJ, Makarov V, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171(4):934‐949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spranger S, Bao R, Gajewski TF. Melanoma‐intrinsic beta‐catenin signalling prevents anti‐tumour immunity. Nature. 2015;523:231‐235. [DOI] [PubMed] [Google Scholar]
- 23. Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD‐L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368‐1379. [DOI] [PubMed] [Google Scholar]
- 25. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blank CU, Haanen JB, Ribas A, Schumacher TN. The “cancer immunogram”. Science. 2016;352:658‐660. [DOI] [PubMed] [Google Scholar]
- 27. Karasaki T, Nagayama K, Kuwano H, et al. An immunogram for the cancer‐immunity cycle: towards personalized immunotherapy of lung cancer. J Thorac Oncol. 2017;12:791‐803. [DOI] [PubMed] [Google Scholar]
- 28. van Dijk N, Funt SA, Blank CU, Powles T, Rosenberg JE, van der Heijden MS. The cancer immunogram as a framework for personalized immunotherapy in urothelial cancer. Eur Urol. 2019;75:435‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hugo W, Zaretsky JM, Sun LU, et al. Genomic and transcriptomic features of response to anti‐PD‐1 therapy in metastatic melanoma. Cell. 2016;165:35‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS‐driven cancers require TBK1. Nature. 2009;462:108‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laurent C, Charmpi K, Gravelle P, et al. Several immune escape patterns in non‐Hodgkin's lymphomas. Oncoimmunology. 2015;4:e1026530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Angelova M, Charoentong P, Hackl H, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Şenbabaoğlu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kishton RJ, Sukumar M, Restifo NP. Metabolic regulation of T cell longevity and function in tumor immunotherapy. Cell Metab. 2017;26:94‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel SA, Minn AJ. Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity. 2018;48:417‐433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nagalakshmi U, Waern K, Snyder M. RNA‐Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol. 2010;Chapter 4:Unit 4.11.11‐13. [DOI] [PubMed] [Google Scholar]
- 38. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abbas AR, Baldwin D, Ma Y, et al. Immune response in silico (IRIS): immune‐specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319‐331. [DOI] [PubMed] [Google Scholar]
- 40. Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue‐infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cursons J, Souza‐Fonseca‐Guimaraes F, Foroutan M, et al. A gene signature predicting natural killer cell infiltration and improved survival in melanoma patients. Cancer Immunol Res. 2019;7:1162‐1174. [DOI] [PubMed] [Google Scholar]
- 42. Nirmal AJ, Regan T, Shih BB, Hume DA, Sims AH, Freeman TC. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol Res. 2018;6:1388‐1400. [DOI] [PubMed] [Google Scholar]
- 43. Tosolini M, Pont F, Poupot M, et al. Assessment of tumor‐infiltrating TCRVgamma9Vdelta2 gammadelta lymphocyte abundance by deconvolution of human cancers microarrays. Oncoimmunology. 2017;6:e1284723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Charoentong P, Finotello F, Angelova M, et al. Pan‐cancer immunogenomic analyses reveal genotype‐immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248‐262. [DOI] [PubMed] [Google Scholar]
- 45. Schelker M, Feau S, Du J, et al. Estimation of immune cell content in tumour tissue using single‐cell RNA‐seq data. Nat Commun. 2017;8:2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Table S1
Table S2
Data Availability Statement
All data analyzed during this study are included in this published article and its [Link], [Link], [Link], [Link] files. All data analyzed can be accessed at a web‐accessible database, “The RNA‐Seq based Cancer Immunogram Web” (https://yamashige33.shinyapps.io/immunogram/).


