Abstract
Multiple myeloma (MM) is an incurable hematopoietic neoplasm derived from plasma cells, and existing in the bone marrow. Recent developments in the field of myeloma onco‐biology have enabled the use of proteasome inhibitors (PIs) as key drugs for MM. PIs can increase cell sensitivity to endoplasmic reticulum stress, leading to apoptosis of myeloma cells. PI cannot kill all myeloma cells, however; one reason of this might be activation of autophagy via hypoxic stress in the bone marrow microenvironment. Hypoxia‐inducible gene(s) that regulate autophagy may be novel therapeutic target(s) for PI‐resistant myeloma cells. Here, a hypoxia‐inducible glycolytic enzyme hexokinase‐2 (HK2) was demonstrated to contribute by autophagy activation to the acquisition of an anti‐apoptotic phenotype in myeloma cells. We found that hypoxic stress led to autophagy activation accompanied by HK2 upregulation in myeloma cells. Under hypoxic conditions, HK2 knockdown inhibited glycolysis and impaired autophagy, inducing apoptosis. The cooperative effects of a PI (bortezomib) against immunodeficient mice inoculated with HK2‐knocked down myeloma cells were examined and significant tumor reduction was observed. An HK2 inhibitor, 3‐bromopyruvate (3‐BrPA), also induced apoptosis under hypoxic rather than normoxic conditions. Further examination of the cooperative effects between 3‐BrPA and bortezomib on myeloma cells revealed a significant increase in apoptotic myeloma cells. These results strongly suggested that HK2 regulates the activation of autophagy in hypoxic myeloma cells. Cooperative treatment using PI against a dominant fraction, and HK2 inhibitor against a minor fraction, adapted to the bone marrow microenvironment, may lead to deeper remission for refractory MM.
Keywords: autophagy, HK2, hypoxia, microenvironment, multiple myeloma
Contribution of the HK2‐autophagy pathway to cell survival of myeloma in an hypoxic microenvironment and complementary effects of conventional and hypoxia‐targeting therapies.
Abbreviations
- 3‐BrPA
3‐bromopyruvate
- BTZ
bortezomib
- ER
endoplasmic reticulum
- HIF
hypoxia‐inducible factor
- HK
hexokinase
- IL‐6
interleukin‐6
- IRF4
interferon regulatory factor 4
- KDM3A
lysine demethylase 3A
- MALAT1
metastasis associated in lung adenocarcinoma transcript‐1
- MM
multiple myeloma
- PI
proteasome inhibitor
- SQSTM1
sequestosome 1
1. INTRODUCTION
Multiple myeloma is a refractory plasma cell tumor that accumulates in the bone marrow. In recent years, the prognosis of MM has improved markedly with the development of various new agents such as immunomodulatory drugs, PIs, and monoclonal antibodies. However, all cases of MM remain incurable because they acquire drug resistance. 1 The drug‐resistant fraction of myeloma cells is thought to be protected by multiple factors in the bone marrow microenvironment. These factors are found in 2 compartments in the bone marrow microenvironment: 1 that contains cells and 1 that does not. The former includes blood cells, osteoclasts, osteoblasts, stromal cells, fibroblasts, and vascular endothelial cells, whereas the latter includes the extracellular matrix, hypoxia, starvation, low pH, and soluble factors such as cytokines and growth factors. 2 Among these factors, hypoxic stress in the bone marrow microenvironment is thought to play a crucial role in drug resistance in hematologic malignancies, including MM. 3 , 4
Drug‐resistant myeloma cells show an anti‐apoptotic phenotype. 5 , 6 This phenotype can occur either in normoxia or hypoxia, however the responsible genes are distinct between each condition. Hypoxic stress leads to the induction of distinct gene expression in myeloma cells. 7 , 8 For instance, the transcription factor IRF4, which plays crucial roles in acquisition of the anti‐apoptotic phenotype of MM, is typically upregulated in MM. 9 However, IRF4, and even its target c‐Myc, are downregulated under hypoxic stress. 10 Instead, accumulation of the transcription factor hypoxia‐inducible factor‐1α (HIF‐1α) upregulates anti‐apoptotic proteins, increases glycolysis, and enhances neovascularization, leading to the anti‐apoptotic phenotype of MM. 11 , 12
Moreover, recent studies have suggested that activation of autophagy is another factor in the acquisition of the anti‐apoptotic phenotype of MM. 13 , 14 Autophagy is an intracellular process that involves the encapsulation of cytoplasmic components and their targeting to the lysosome for degradation. This process helps cells to survive under nutrient‐starved or hypoxic conditions by inducing protein synthesis, removing damaged intracellular organelles, and promoting energy production. 15 , 16 , 17 , 18 , 19 This process is also closely related to cancer metabolism and tumor growth. 20 Autophagy may function in an oncogenic or tumor‐suppressive manner depending on the type of cancer. 21 In MM, both autophagy activation and proteasomal degradation are thought to contribute to protecting cells from excessive ER stress induced by misfolded proteins. 22
Based on these findings, we predicted that activation of autophagy in the hypoxic microenvironment increased the resistance capability to apoptosis in MM. This study was conducted to identify factors that induce autophagy in the hypoxic bone marrow microenvironment for use as therapeutic targets for treatment‐resistant MM.
2. MATERIALS AND METHODS
2.1. Primary MM samples
The study included 6 cases of primary samples from Akita University Hospital (Akita, Japan) and was conducted with written informed consent of the study participants and the approval of the Institutional Review Boards (approval no. 1313), according to the Declaration of Helsinki, before collection of specimens.
2.2. Myeloma cell lines and culture
Six well known MM cell lines (KMS‐12‐PE, KMS‐11, H929, RPMI‐8226, U266, and MM.1S) were used, and obtained from the American Type Culture Collection. A multi‐gas incubator MCO‐5M‐PJ (PHC) was used for hypoxic culture (1% O2), An F‐51 (Horiba) apparatus was used to measure the pH of culture media. The method of co‐culture with a human stromal cell line HS‐5 has been described previously. 7
2.3. DNA microarray
Gene expression was analyzed using Agilent DNA microarrays (Agilent) and an Agilent G2600A SureScan Microarray Scanner System. The experiment protocol was in accordance with Agilent Protocol v.6.7. Data were analyzed using GeneSpring software (Agilent), plus computed normalized signal intensity with 75%‐tile normalization. Data were uploaded to the Gene Expression Omnibus (GEO) database under identification numbers GSE80140 (KMS‐11, KMS‐12‐BM, MM.1S, and RPMI‐8226), GSE80545 (patient samples), and GSE96858 (U266).
2.4. Quantitative reverse transcription‐PCR (qRT‐PCR) analysis
qRT‐PCR was performed using TaqMan methodology (Applied Biosystems). TaqMan probes for GAPDH (Hs02758991_g1), HK1 (Hs00175976_m1), HK2 (Hs00606086_m1), HK3 (Hs01092850_m1), and glucokinase (GCK) (Hs01564555_m1) were purchased from Applied Biosystems. Expression levels were normalized separately against GAPDH and the relative expression levels of specific mRNA were presented as . qRT‐PCR was performed using a Light Cycler Nano instrument (Roche). Total RNA was extracted using TRIzol reagent (Life Technologies). Reverse transcription was performed using a Transcriptor First Strand cDNA Synthesis Kit (product no. 04379012001, Roche).
2.5. Western blot analysis
Antibodies against HK2 (#2867), SQSTM1/p62 (#39749), and LC3A/B (#12741) were purchased from Cell Signaling Technology; tubulin (MS‐581‐P0) was purchased from NeoMarkers.
2.6. Transient siRNA transfection
The following Silencer Select siRNAs were purchased from Applied Biosystems: siHIF1A (s6539), siEPAS1 (s4698), siHK2 #1 (s6560), and siHK2 #2 (s6562). Information on the siRNA sequence is listed in Table S1. Negative control siRNA (scrambled siRNA) was designed and synthesized by Nippon Gene. Transfection of siRNA was performed using Nucleofector II and the Cell Line Nucleofector Kit V (VCA‐1003) (Amaxa) in accordance wiith the manufacturer's protocol. The assay protocol has been described previously. 7 , 10 In brief, cells were resuspended in nucleofector V solution, 100 μl cell suspension at a density of 1 × 107/mL was mixed with 1.0 μg siRNA, transferred to a cuvette and nucleofected using an Amaxa Nucleofector II apparatus with the S‐020 program for MM.1S, the G‐015 program for KMS‐11, and the X‐005 program for U266.
2.7. Stable knockdown constructs and lentivirus infection
The HK2 Human shRNA Plasmid Kit (TL312415) including control plasmid was purchased from OriGene (Rockville). Information on shRNA sequences is listed in Table S1. Together with 9.0 µg ViraPower Packaging mix (Invitrogen), 3.0 µg of vectors were transfected using Lipofectamine 3000 (Invitrogen) into the 293FT producer cells. After overnight culture, the medium was exchanged to remove transfection reagents. Viral supernatants were harvested the following day, at 48‐72 h after transfection. Then, 1 × 106 of KMS‐11 cells were prepared with changed medium, and medium containing virus was added. After 3‐d of culture, cells stably expressing siRNAs were sorted for GFP expression using a FACSAria III instrument (BD Biosciences).
2.8. ELISA assay
The CycLex Total p62 ELISA Kit (CY‐7055) was purchased from Medical & Biological Laboratories and the Glycolysis Cell‐Based Assay Kit (No. 600450) from Cayman Chemical. The assays were carried out in accordance with the manufacturer's protocol.
2.9. Autophagy detection on flow cytometry
DAPGreen‐Autophagy Detection (D676) was purchased from Dojindo Molecular Technologies. DAPGreen (0.1 or 0.05 µmol/L) was added after incubation of myeloma cells for 48 h under normoxic or hypoxic conditions. After a 1 h incubation, the double membranes of the autophagosome stained with the dye were assesssed using a FACSCanto instrument (BD Biosciences).
2.10. Apoptosis assay
APC‐Annexin V (550474), 7‐AAD (559925), and Annexin V binding buffer (556454) were purchased from BD Biosciences. The assays were carried out as described previously using a FACSCanto instrument (BD Biosciences). 7 , 10
2.11. Xenograft mouse model
KMS‐11 cells (1 × 106 each) were injected subcutaneously into the right or left side of the bodies of 6‐8‐wk‐old female NOD/Shi‐scid IL2rγnull (NOG) mice (Central Institute for Experimental Animals). The protocols for animal experimentation described in this paper were approved by the Animal Committee, Akita University (approval no. a‐1‐3133).
2.12. Reagents
3‐Bromopyruvate (3‐BrPA) (16490) was purchased from Sigma‐Aldrich. Bortezomib (021‐18901) was purchased from FUJIFILM Wako Pure Chemical. Recombinant human interleukin‐6 (IL‐6) (#8904SC) was purchased from Cell Signaling Technology. AZD8055 was purchased from ChemieTek.
2.13. Statistical analysis
Student t test (Figures 2, 3, 4, 5, 6) and two‐way ANOVA (Figure 5G) were used to test for significance. Bars represent the mean ± 95% confidence interval (CI) of 3 (Figures 2, 3, 4, 5, 6, except for Figure 4C) or 10 (Figure 4C) independent experiments. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001; NS, not significant.
FIGURE 2.

HK2 is upregulated through HIF activation in hypoxia‐stressed myeloma cells. A, Volcano plot showing the distribution of differentially expressed genes between primary MM samples (n = 4) cultured in hypoxia (1% O2) vs normoxia for 48 h (GSE80545). Red dots illustrate transcripts significantly upregulated by hypoxia (fold change >2.0, P < .05). B, Gene Ontology (GO) analysis of hypoxia‐induced genes (red dots in A). C, Heat map of 15 probes (13 genes) including GO “glycolytic process” of (B). D, Scheme of the glycolytic process. Red arrows: genes upregulated by hypoxia displayed in (C). E, Gene expression change of HK1, HK2, HK3, and HK4 in patient samples cultured in hypoxia (1% O2) vs normoxia for 48 h (GSE80545). F, qRT‐PCR of HK2 for cell lines (U266, KMS‐11, and MM.1S) cultured under hypoxia (1% O2), serum starvation, with recombinant IL‐6 (4 ng/mL), co‐cultured with a stromal cell line HS‐5 (only U266) for 24 h. G, qRT‐PCR of HK2 for cell lines (U266, KMS‐11, and MM.1S) transiently transduced with siHIF1A and/or siHIF2A and control scrambled siRNA and cultured in normoxia or hypoxia (1% O2) for 48 h. H, Western blot analysis of HK2 for primary samples (n = 2) and cell lines (KMS‐11, MM.1S, U266, RPMI‐8226, KMS‐12‐PE, and H929) cultured in normoxia or hypoxia (1% O2) for 48 h. H, hypoxia; N, normoxia. Asterisks indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001; NS, not significant
FIGURE 3.
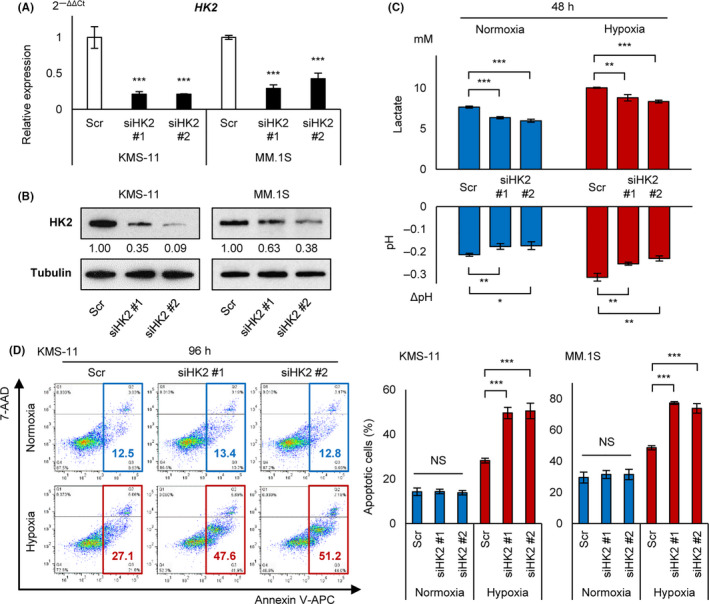
HK2 knockdown leads myeloma cells to induce apoptosis under chronic hypoxia. A, qRT‐PCR analysis of HK2 for KMS‐11 and MM.1S cell lines transiently transduced with siHK2 #1, siHK2 #2, and control scrambled siRNA (Scr). B, Western blot analysis of HK2 for KMS‐11 and MM.1S cell lines transiently transduced with siHK2 #1, siHK2 #2, and control scrambled siRNA (Scr). C, Lactate concentration by ELISA assay and pH measurement for KMS‐11 cell line transiently transduced with siHK2 #1, siHK2 #2, and control scrambled siRNA (Scr). Cells were cultured at 1 × 106/3 mL in normoxia or hypoxia (1% O2) for 48 h. D, Apoptosis assay of KMS‐11 and MM.1S cell lines transiently transduced with siHK2 #1, siHK2 #2, and control scrambled siRNA (Scr) and cultured in normoxia or hypoxia (1% O2) for 96 h. Left panel: x‐axis: Annexin V; y‐axis: 7‐AAD. Right panel: apoptotic cell rates of indicated cell lines were shown. Asterisks indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001; NS, not significant
FIGURE 4.
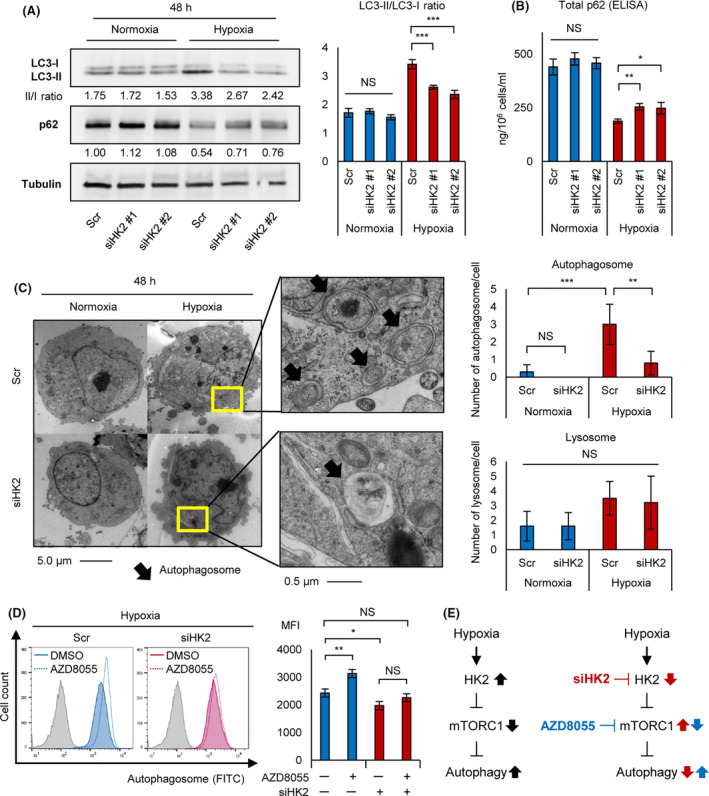
HK2 knockdown suppresses hypoxia‐induced autophagy. A, Western blot analysis of LC3‐I, LC3‐II, and p62 for KMS‐11 transiently transduced with siHK2 #1, siHK2 #2, and control scrambled siRNA (Scr) and cultured in normoxia or hypoxia (1% O2) for 48 h (left panel). LC3‐II/LC3‐I ratio of the western blot analysis is also shown (right panel). B, ELISA assay of p62 for KMS‐11 transiently transduced with siHK2 #1, siHK2 #2, and control scrambled siRNA (Scr) and cultured in normoxia or hypoxia (1% O2) for 48 h. Bars represent mean ± 95% CI of 3 replicates. C, Photograph of electron microscopy for KMS‐11 transiently transduced with siHK2 #1 or control scrambled siRNA (Scr) and cultured in normoxia or hypoxia (1% O2) for 48 h (left panel). Count of autophagosome/cell and lysosome/cell of siHK2‐ or control‐KMS‐11 (n = 10 each) is also shown (right panel). D, Flow cytometry assay of autophagosome stained with DAPGreen for KMS‐11 cell line cultured under hypoxia (1% O2) for 48 h with or without AZD8055 (100 nmol/L, 6 h). y‐axis: cell count, x‐axis: autophagosome‐FITC (left panel). Mean fluorescence intensity (MFI) of each samples are shown (right panel). E, Schematic illustrations of effects of hypoxic stress (black arrows), siHK2 (red arrows) and the mTOR inhibitor AZD8055 (blue arrows) on the HK2‐mTORC1‐autophagy axis. The left panel shows that hypoxia‐inducible HK2 activates autophagy via mTORC1 inhibition, and the right panels shows that siHK2 inhibits autophagy and that the mTORC1 inhibitor AZD8055 activates autophagy. Asterisks indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001; NS, not significant
FIGURE 5.
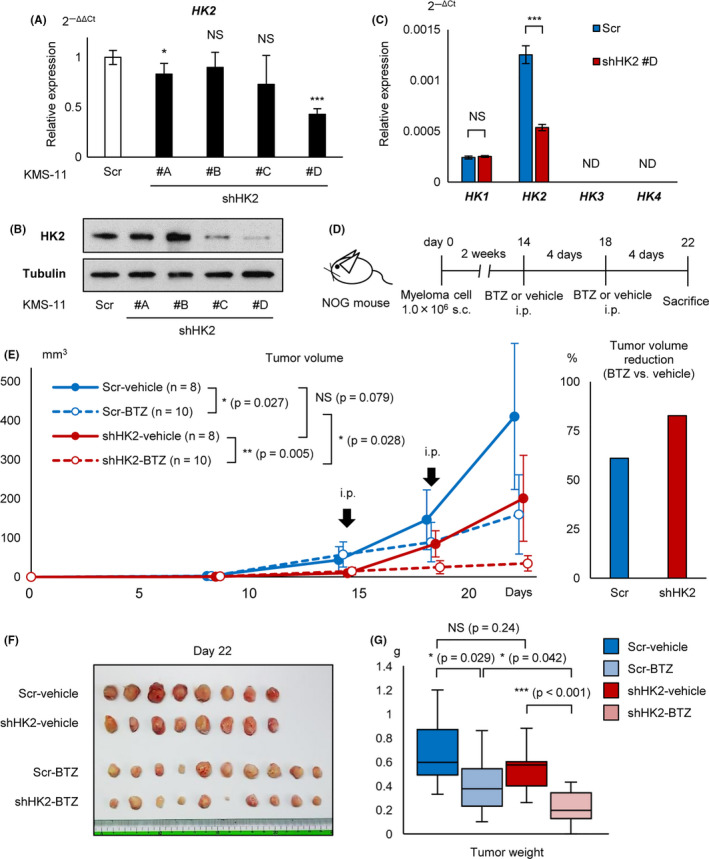
Cooperative effect of HK2 knockdown and proteasome inhibitor in vivo. A, qRT‐PCR analysis of HK2 for KMS‐11 stably transduced with shHK2 #A, #B, #C, #D, and control scrambled shRNA (Scr). B, Western blot analysis of HK2 for KMS‐11 stably transduced with shHK2 #A, #B, #C, #D, and control scrambled shRNA (Scr). C, qRT‐PCR analysis of HK1, HK2, HK3, and HK4 for KMS‐11 stably transduced with shHK2 #D or control scrambled shRNA (Scr). D, Illustration of the protocol of the in vivo transplantation and treatment; 1 × 106 of shHK2 #D or control shRNA stably transduced KMS‐11 were inoculated into NOG mice. Mice were treated with bortezomib (1.0 mg/kg) or phosphate‐buffered saline intraperitoneally. Scr‐vehicle; n = 8, Scr‐BTZ; n = 10, shHK2‐vehicle; n = 8, and shHK2‐BTZ; n = 10. BTZ, bortezomib. E, Tumor growth curves of each groups are shown. x‐axis, days after transplantation (days); y‐axis, tumor volume (mm3, major × minor2/2). Right panel: rates of tumor volume reduction by bortezomib (BTZ) administration for the Scr group and shHK2 group. F, Photograph of tumors from each group are shown. G, Tumor weights of each groups are shown. Asterisks indicate statistical significance: *.01 ≤ P < .05; ***P < .001; NS, not significant.
FIGURE 6.
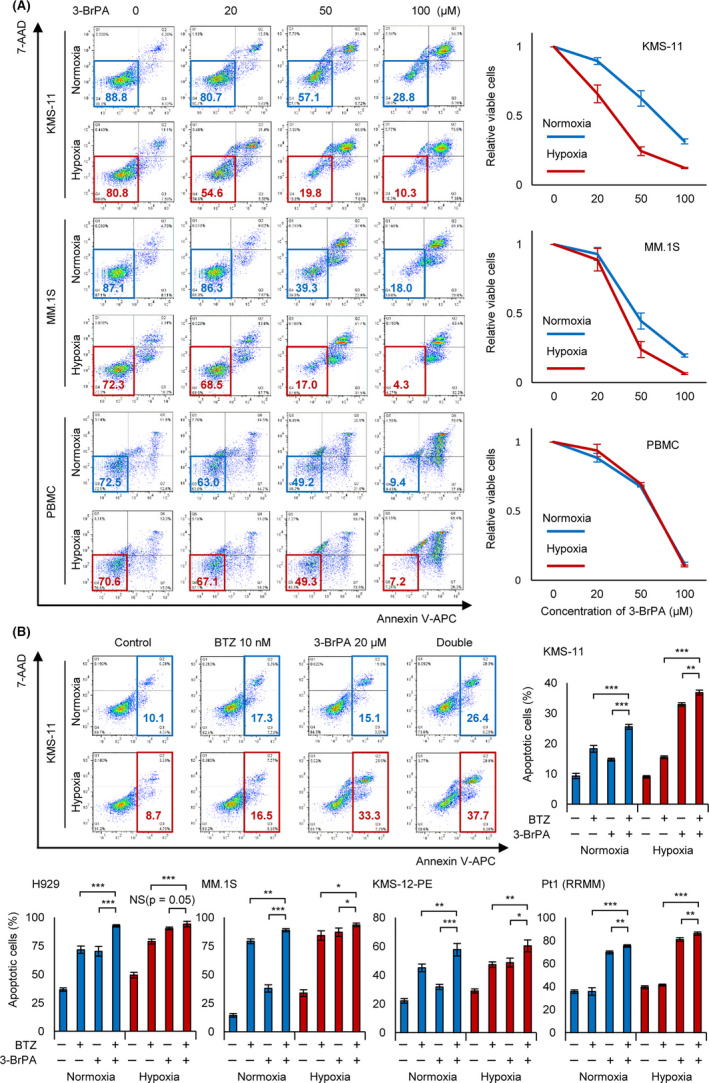
Complementary effect of bortezomib and 3‐BrPA under chronic hypoxia in vitro. A, Apoptosis assay of KMS‐11, MM.1S, and normal peripheral blood mononuclear cells (PBMC). Cells were cultured in normoxia or hypoxia (1% O2) for 48 h, and then 3‐bromopyruvate (3‐BrPA; 0, 20, 50, or 100 µmol/L) was added in the medium during 24 h. After the treatment, apoptosis assay was conducted. Left panel: x‐axis: Annexin V; y‐axis: 7‐AAD. B, Apoptosis assay of indicated cell lines and a refractory MM sample. Cells were cultured in normoxia or hypoxia (1% O2) for 24 h, and then bortezomib (for cell lines: 10 nmol/L, for patient sample: 50 nmol/L) and/or 3‐BrPA (20 µmol/L) were added in the medium during 24 h. After the treatment, apoptosis assay was conducted. Upper left panel: x‐axis: Annexin V; y‐axis: 7‐AAD. Asterisks indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001; NS, not significant
3. RESULTS
3.1. Hypoxic stress induces autophagy in MM cells
Intracellular organelles related to autophagy include autophagosomes, lysosomes, and autolysosomes. 23 Electron microscopy was performed to observe these organelles in patient samples (n = 2) and the myeloma cell line KMS‐11, which were incubated under normoxic or hypoxic conditions (1% O2) for 48 h. The number of organelles was markedly increased by hypoxia exposure (Figure 1A). To quantify autophagy activation in hypoxia‐subjected patient samples (n = 2) and cell lines (KMS‐11, H929, KMS‐12‐PE, and MM.1S), we conducted flow cytometry analysis. The fluorescent signal of the autophagosome was upregulated in all samples (Figure 1B). These results demonstrated that hypoxic stress activates autophagy in MM.
FIGURE 1.
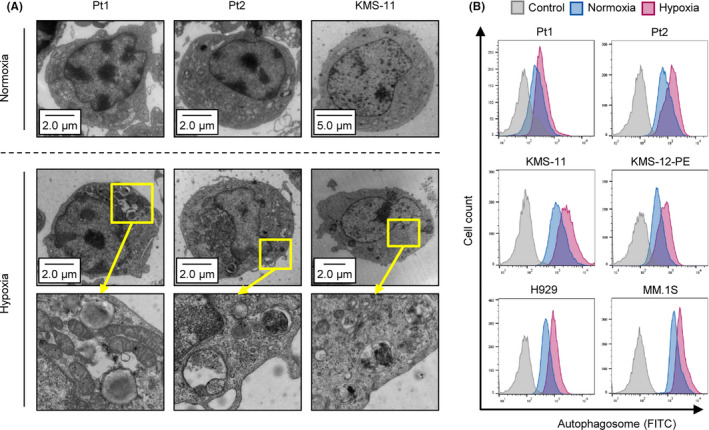
Hypoxia‐inducible autophagy in MM. A, Photograph of electron microscopy for 2 primary MM samples and a myeloma cell line KMS‐11 cultured under normoxia or hypoxia (1% O2) for 48 h. B, Flow cytometry assay of autophagosome stained with DAPGreen in patient samples (n = 2) and the indicated 4 myeloma cell lines cultured under normoxia or hypoxia (1% O2) for 48 h. y‐axis: cell count, x‐axis: autophagosome‐FITC
3.2. Hypoxic stress upregulates glycolytic genes including HK2 in samples from patients with MM
We hypothesized that activation of autophagy under hypoxic conditions was induced by hypoxia‐inducible genes in MM. To detect candidate gene(s) crucial to the acquisition of hypoxia‐inducible autophagy in MM, we performed volcano plot analysis using our comprehensive gene expression data (GSE80545) 10 obtained from patient samples (n = 4) that had been incubated under normoxic or hypoxic conditions (1% O2) for 48 h. In total, the levels of 546 probes were elevated significantly under hypoxic conditions (fold change (FC) > 2.0, P < .05; Figure 2A). Gene Ontology (GO) analysis of these genes revealed that all top 10 categories were energy metabolism related (Figure 2B). Thirteen genes were associated with the “glycolytic process,” including PFKFB4, ENO2, ALDOC, PFKFB3, HK2, PFKP, GPI, PGK1, LDHA, ALDOA, ENO1, PKM, and GAPDH (Figure 2C). Almost all of these genes were involved in glycolytic processes from glucose to the final metabolite product lactic acid (Figure 2D). Among these, an enzyme that metabolizes glucose to glucose‐6‐phosphate, hexokinase‐2 (HK2), was included in the top 5 (log2 FC ratio > 2.0), and was located mostly upstream in glycolysis. 24
HK2 is involved in autophagy induction under stress conditions via inhibiting mTORC1, 25 , 26 therefore we focused on the functions of HK2 under hypoxia in MM cells. HKs exist in 4 isoforms; our microarray data showed that only HK2 was elevated markedly in hypoxia‐exposed patient samples and cell lines (Figure 2E). Not only hypoxia but also serum starvation, IL‐6, and stromal cells are known as microenvironmental factors. 2 To determine which factors were the most important in HK2 upregulation, we examined HK2 expression in myeloma cell lines (U266, KMS‐11, and MM.1S) cultured either under hypoxic conditions, under serum starvation conditions, with IL‐6, or in co‐cultured with the stromal cell line HS‐5 (for floating cell line U266). qRT‐PCR showed that HK2 expression was increased significantly under hypoxic culture conditions compared with other microenvironmental factors (Figure 2F). qRT‐PCR analysis demonstrated that HIF‐1α and/or HIF‐2α knockdown significantly reduced HK2 expression in myeloma cell lines (U266, KMS‐11, and MM.1S) under hypoxic conditions (Figure 2G), indicating that HIFs ectopically transcripted HK2 under hypoxic conditions. Western blot analysis of patient samples (n = 2) and 6 myeloma cell lines revealed that HK2 was markedly upregulated under hypoxic conditions (Figure 2H). These results suggested that HK2/HK2 is upregulated by activation of HIFs under an hypoxic environment in MM.
3.3. Knockdown of HK2 leads to apoptosis of myeloma cells under hypoxic stress
To determine the role of HK2 in hypoxic stress, we performed transient knockdown of HK2 against myeloma cell lines (KMS‐11 and MM.1S). The siRNA sequences are listed in Table S1. These siRNAs (siHK2 #1 and #2) significantly reduced HK2 mRNA levels (80% and 60%‐70% reductions) and HK2 protein levels (60%‐90% and 40%‐60% reductions) in KMS‐11 and MM.1S cells, respectively (Figure 3A,B). Although hypoxic stress upregulated lactic acid production and acidification of the culture medium with activation of glycolysis, the siHK2 probes canceled these changes under both normoxic and hypoxic conditions (Figure 3C). This indicated that HK2 knockdown effectively inhibited glycolysis. An apoptosis assay against cell lines transduced with siHK2 revealed no difference in viable cells between control and siHK2‐treated cells under normoxic conditions, whereas a significant increase in the apoptotic cell rate was detected under chronic hypoxic conditions (Figure 3D). In addition, although long‐term chronic hypoxia for 96 h increased apoptotic cells, there was no apparent difference up to 48 h in acute hypoxia (data not shown). These results suggested that HK2 plays an important role in obtaining the anti‐apoptotic phenotype of MM in hypoxic environments such as in the bone marrow microenvironment.
3.4. HK2 knockdown inhibits hypoxia‐inducible autophagy in myeloma cells
We investigated whether HK2 controls autophagy against myeloma cells under hypoxic stress. Degradation of p62/SQSTM1 and an increased LC3‐II/LC3‐I ratio are considered useful for autophagy detection. 23 Therefore, we conducted western blot analysis of p62 and LC3 against siHK2‐KMS‐11 and control‐KMS‐11 cells, which were cultured under normoxic or hypoxic conditions. Hypoxic stress decreased p62 expression and increased the ratio LC3‐II/LC3‐I in control‐KMS‐11 cells (Figure 4A). However, these changes were decreased by HK2 knockdown, suggesting that HK2 knockdown inhibited hypoxia‐inducible autophagy. Furthermore, an ELISA assay to detect p62 revealed increased expression in siHK2‐KMS‐11 cells in hypoxic conditions, with no change under normoxic conditions (Figure 4B). We further conducted electron microscopy to confirm the accumulation of autophagosomes and lysosomes. Although we found increased autophagosome and lysosome formation in hypoxia‐treated myeloma cells, the number of autophagosomes was significantly decreased in siHK2‐KMS‐11 cells (Figure 4C). These results suggested that HK2 is required for hypoxia‐inducible autophagy in MM.
Next, we examined how HK2 regulates autophagy in a hypoxic environment. HK2 induces autophagy by inhibiting the activity of mTORC1 in normal murine tissues. 25 The mTOR inhibitor AZD8055 induces autophagy in myeloma cells. 27 We examined the effects of AZD8055 on autophagy in siHK2‐KMS‐11 or control‐KMS‐11 cells. As expected, flow cytometry analysis demonstrated that, under hypoxic conditions, AZD8055 treatment upregulated the fluorescence intensity of autophagosomes in control‐KMS‐11 cells, but had no significant effect on siHK2‐KMS‐11 cells compared with control cells (Figure 4D). Moreover, AZD8055 did not significantly upregulate the fluorescence intensity of autophagosomes in HK2 knockdown cells. This finding is consistent with the previous observation that HK2 regulates autophagy through inhibition of mTORC1. For a better understanding, these results are illustrated schematically in Figure 4E. Together, these data provide strong evidence that the HK2‐mTORC1 pathway plays a key role in autophagy activation under hypoxic conditions.
3.5. Cooperative effect of proteasome inhibitor on HK2‐knocked down myeloma cells inoculated into NOG mice
Because both autophagy and proteasomal degradation play essential roles in the processing of misfolded proteins, simultaneous inhibition of their activities has been reported to cause strong ER stress, inducing cell death in myeloma cells. 22 , 28 We examined the effect of cooperative inhibition of HK2 and proteasome. To determine the cooperative effects of both HK2 knockdown and PI in vivo, we established a stable HK2‐knocked down KMS‐11 cell line. Four sequences, shHK2 #A‐#D, against KMS‐11 cells were transfected using a lentiviral transfection system, and the knockdown effect of HK2 was examined by qRT‐PCR and western blot analysis. shRNA #D markedly reduced HK2 mRNA and protein levels by 60% and 90%, respectively (Figure 5A,B). Constitutive HK2 inhibition by lentivirus has been reported to induce compensatory upregulation of HK1, resulting in the promotion of glycolysis in some solid cancer cell lines, 29 therefore we examined the expression of other HK isoforms. There was no observation by qRT‐PCR of a compensatory change in HK1 expression in shHK2‐KMS‐11 cells. HK3 and HK4 expression was not detected (Figure 5C), therefore the possibility of compensatory effects of other HKs was excluded in subsequent experiments.
As a preliminary experiment, 1 × 106 of control‐KMS‐11 or shHK2‐KMS‐11 cells were injected into immunodeficient mice (NOG mice) subcutaneously at day 0 (Figure S1), and the tumor was found to have became palpable from day 15. The tumor size in the shHK2 group tended to be smaller than that in the control group. Intraperitoneal administration of 1.0 mg/kg bortezomib appeared to exert a tumor‐suppressive effect, although 0.2 mg/kg bortezomib showed no apparent anti‐myeloma effects.
Based on this result, we injected 1 × 106 of control‐KMS‐11 or shHK2‐KMS‐11 cells subcutaneously into NOG mice at day 0 and later administered 1.0 mg/kg bortezomib or phosphate‐buffered saline intraperitoneally every 4 d, starting on day 14 (Figure 5D). shHK2‐KMS‐11 xenografted mice treated with bortezomib showed significantly reduced tumor volumes; the percent inhibitions of control and shHK2 groups were 61% and 83%, respectively (Figure 5E). The mice were sacrificed and the tumors removed and weighed; the shHK2‐KMS‐11 xenografted mice treated with bortezomib showed significantly reduced tumor weights (Figure 5F,G). There was a positive correlation between the tumor volume calculated from the tumor size and measured tumor weight (Figure S2). Together, these results suggested that cooperative inhibition of HK2 and proteasome can further enhance their anti‐myeloma effects in vivo.
3.6. Apoptosis‐inducible capability of HK2 inhibitor (3‐BrPA) in hypoxia is greater than that in normoxia, and combination with bortezomib increased the effect
The HK2 inhibitor 3‐BrPA induces apoptosis in myeloma cells and other cancer cells by suppressing ATP production and inhibiting drug efflux transporters. 30 , 31 , 32 As HK2 knockdown increased apoptotic myeloma cells under hypoxic conditions (Figure 2), we hypothesized that its inhibitor 3‐BrPA would also strongly induce apoptosis of myeloma cells under hypoxic stress. As expected, an apoptosis assay demonstrated that the numbers of 3‐BrPA‐treated viable cells under hypoxic conditions were significantly lower than those under normoxic conditions (Figure 6A). We further performed the same experiment using normal peripheral blood mononuclear cells (PBMC) and found no apparent differences between hypoxic and normoxic conditions, suggesting that 3‐BrPA had greater effects on myeloma cells compared with normal cells under hypoxic conditions. These data indicated that HK2 inhibitors such as 3‐BrPA are useful for treating myeloma cells adapted to the hypoxic microenvironment.
Finally, we evaluated the combined effects of 3‐BrPA with bortezomib in vitro. Under normoxic conditions, administration of bortezomib alone or administration of 3‐BrPA alone induced significantly greater cell death than in the control; combined administration further increased the number of apoptotic cells (Figure 6B). Importantly, both bortezomib and 3‐BrPA administration under hypoxic conditions further increased apoptotic cells compared with under normoxic conditions. We conducted the same experiment using other myeloma cell lines (H929, MM.1S, and KMS‐12‐PE) and a PI‐resistant patient sample and found that 3‐BrPA enhanced the apoptotic effect of bortezomib (Figure 6C). Notably, 3‐BrPA was also effective against a bortezomib‐resistant patient sample that did not show apoptosis when treated with a high concentration of bortezomib (50 nmol/L). These results strongly suggested that an HK2 inhibition/inhibitor can induce apoptosis against PI‐resistant myeloma cells.
4. DISCUSSION
The average oxygen tension of human bone marrow aspirates is 52‐56 mm Hg in both healthy control subjects and patients with MM. 33 However, the endosteal niche, in which hematopoietic stem cells or “myeloma stem cells” are still present, is thought to have a much lower oxygen tension (~10 mm Hg) compared with other bone marrow regions. 11 , 34 , 35 In hypoxic environments such as the bone marrow microenvironment, the transcription factor HIF is upregulated, leading to upregulation of HIF‐dependent genes in myeloma cells. 11 Thus, the gene expression pattern and phenotypes of myeloma cells in the bone marrow microenvironment may be distinct from those in other bone marrow regions. Several studies including this study have been conducted to detect the effects of hypoxic signals on myeloma cells. Numerous studies have also attempted to identify useful HIF inhibitors. However, because HIF moderates various essential genes involved in the survival of not only malignant cells but also normal cells, HIF inhibitors have not been applied practically to treat various cancers including MM. Therefore, inhibition of downstream genes of HIF is considered a therapeutic target. For instance, Maiso et al found that HIF‐1α‐inducible glycolytic enzymes including lactate dehydrogenase induced anti‐apoptosis for myeloma cells. 8 Recently, we also found that miR‐210 and the H3K9 demethylase KDM3A were specifically upregulated in hypoxia‐exposed myeloma cells. In hypoxia, the HIF‐1α‐miR‐210 axis led myeloma cells to cellular quiescence via the suppression of IRF4, which is typically upregulated in MM and plays a crucial role in myeloma oncogenesis. 9 , 10 Moreover, we found that the HIF‐1α‐KDM3A axis regulates glycolytic enzymes by upregulating the long non‐coding RNA MALAT1, leading to the anti‐apoptotic phenotype of MM. 7 These findings indicated that glycolytic enzymes, but not oncogenes of MM such as IRF4, are important therapeutic targets for myeloma cells adapted to the hypoxic environment.
In the present study, we focused on the glycolytic gene HK2 among hypoxia‐inducible factors. The glycolytic enzyme HK2 phosphorylates glucose to form glucose‐6‐phosphate in the first step of the glycolytic system and also plays an important role in tumorigenesis. 36 , 37 HK has 4 isoforms: HK1, HK2, HK3, and HK4 (GCK). 24 HK1 is expressed ubiquitously in various tissues; HK2 is expressed in insulin‐sensitive tissues such as muscle and fat cells; HK3 is expressed in the lung and pancreas; and HK4 is expressed in the liver and pancreas. Among these HKs, HK2 is the most important isoform in various cancers. 38 , 39 , 40 Several groups including our group have suggested the clinical significance of HK2 in MM. For instance, Rasche et al 41 reported that HK2 expression was related to the positive rate of FDG‐PET in MM. Moreover, Abe et al reported that low HK2 expression‐related FDG‐PET false‐negative cases showed a good prognosis. 42 These reports strongly suggested that aberrant HK2 overexpression is associated with a worse prognosis of patients with MM. Moreover, activation of HK2 may be oncogenic. However, in previous studies, samples were likely to have been obtained from a normoxic bone marrow blood rather than an hypoxic bone marrow microenvironment. No studies have precisely examined the role of HK2 in myeloma cells adapted to hypoxic stress.
Glycolytic enzymes are promising targets for cancer therapy. 43 As HK2 is a critical enzyme that regulates the first step of glycolysis, we cannot completely rule out the possibility that apoptosis induction in HK2‐knocked down myeloma cells was due to the inhibitory effect of glycolysis. Additionally, we found that HK2 knockdown suppressed the production of lactic acid, the final glycolytic metabolite, under both normoxic and hypoxic conditions. Even in an aerobic environment, glycolysis is activated in cancer cells, a phenomenon known as the Warburg effect. Nevertheless, in our study, HK2 knockdown induced apoptosis under hypoxic conditions, but not under normoxic conditions. Hence, our data strongly suggested that, in a hypoxic environment, HK2 contributes to apoptosis inhibition by other mechanism rather than glycolysis. This is in accordance with a previous study that reported that HK2 exerts its cytoprotective effects by inducing not only glycolysis but also autophagy. 26 Thus, our findings revealed an autophagy‐inducing function of the glycolytic factor, HK2, which is ectopically expressed under hypoxic conditions.
We found that HK2‐inducible autophagy is “oncogenic” in MM, particularly in myeloma cells in the hypoxic bone marrow microenvironment. In other hematopoietic tumors, such as diffuse large B‐cell lymphoma, autophagy can also act as a tumor suppressor. 44 In contrast, the oncogenic autophagy mechanism is likely to be myeloma specific because, unlike other cancers, myeloma cells are exposed to intense ER stress caused by large amounts of non‐functional monoclonal proteins. Therefore, the survival of myeloma cells depends on proteasomal protein degradation. Even when the proteasome was inhibited by PIs, the activation of autophagy led myeloma cells to acquire an apoptosis‐resistant phenotype, as autophagy can also reduce ER stress. 13 , 28 Considering the importance of autophagy of myeloma cells in the hypoxic environment, 18 , 19 targeting the HK2‐autophagy axis is a promising therapeutic strategy (Figure 7A). The development of a specific inhibitor of HK2 may improve the prognosis of patients with refractory MM. Indeed, Xu et al 45 recently demonstrated the effectiveness of antisense oligos of HK2 against myeloma xenografted mice. However, because inhibition of the glycolytic system is also likely to damage normal tissues, it may be necessary to develop a drug delivery system that allows a HK2 inhibitor to exert its effect only in the bone marrow microenvironment. The development of a novel therapeutic strategy including a drug delivery system to inhibit HK2 activity only under hypoxic conditions is promising for refractory MM.
FIGURE 7.
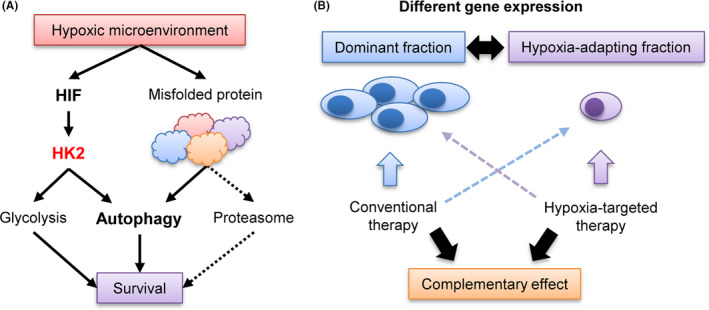
Schematic illustration of the role of autophagy and HK2 in myeloma pathogenesis and hypoxia‐targeting therapy. A, Contribution of HK2‐autophagy pathway for cell survival of myeloma in hypoxic microenvironment. B, Complementary effect of conventional therapy and hypoxia‐targeting therapy
Because PIs compensatively activate autophagy, a better therapeutic effect may be obtained by simultaneous inhibition of both autophagy and the proteasome. 22 , 28 Therefore, combinations of autophagy‐targeted drugs and PIs have been evaluated by several groups. 46 , 47 In our study, there was no apparent synergistic effect between HK2 inhibition and bortezomib in vitro and in vivo. However, when we used bortezomib against shHK2 inoculated mice, the mice showed the most anti‐tumor effect. The anti‐myeloma effect of PI under normoxia and that of HK2 inhibitor under hypoxia may have led to complementary therapeutic effects against myeloma cells. In fact, clinical trials of combination therapy with a hypoxia‐targeting drug evofosfamide (TH‐302) and bortezomib (in a Phase I/II trial) have been successfully conducted for refractory/relapsed MM. 48 , 49 , 50 Our results suggested that PIs and inhibitor of hypoxia‐inducible factors may be effective for each fractions with different gene expression, but not for a single fraction, leading to complementary deep responses (Figure 7B).
In conclusion, our data indicated that hypoxia‐inducible HK2 promotes autophagy and inhibits apoptosis in myeloma cells subjected to hypoxic stress. These data also strongly suggest that a combination of PIs (effective in the normoxic environment) and HK2 inhibition (effective in the hypoxic environment) may complementarily result in a greater response to treatment‐resistant/refractory MM.
DISCLOSURE
SI has received research funding from Nippon Shinyaku. NT received honoraria from Pfizer, Otsuka, and Novartis and research funding from Novartis.
Supporting information
Fig S1
Fig S2
Table S1
ACKNOWLEDGMENTS
We thank Jyunsuke Yamashita, Yukiko Abe, Yuko Chiba, and Hiromi Kataho for their outstanding technical assistance. This study was supported by Nippon Shinyaku Research Grant (Nippon Shinyaku, Kyoto, Japan) to SI.
Ikeda S, Abe F, Matsuda Y, Kitadate A, Takahashi N, Tagawa H. Hypoxia‐inducible hexokinase‐2 enhances anti‐apoptotic function via activating autophagy in multiple myeloma. Cancer Sci. 2020;111:4088–4101. 10.1111/cas.14614
REFERENCES
- 1. Mateos MV, Ludwig H, Bazarbachi A, et al. Insights on multiple myeloma treatment strategies. Hemasphere. 2018;3:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kawano Y, Moschetta M, Manier S, et al. Targeting the bone marrow microenvironment in multiple myeloma. Immunol Rev. 2015;263:160‐172. [DOI] [PubMed] [Google Scholar]
- 3. Muz B, de la Puente P, Azab F, Luderer M, Azab AK. The role of hypoxia and exploitation of the hypoxic environment in hematologic malignancies. Mol Cancer Res. 2014;12:1347‐1354. [DOI] [PubMed] [Google Scholar]
- 4. Gezer D, Vukovic M, Soga T, Pollard PJ, Kranc KR. Concise review: genetic dissection of hypoxia signaling pathways in normal and leukemic stem cells. Stem Cells. 2014;32:1390‐1397. [DOI] [PubMed] [Google Scholar]
- 5. Ri M. Endoplasmic‐reticulum stress pathway‐associated mechanisms of action of proteasome inhibitors in multiple myeloma. Int J Hematol. 2016;104(3):273‐280. [DOI] [PubMed] [Google Scholar]
- 6. Robak P, Drozdz I, Szemraj J, Robak T. Drug resistance in multiple myeloma. Cancer Treat Rev. 2018;70:199‐208. [DOI] [PubMed] [Google Scholar]
- 7. Ikeda S, Kitadate A, Abe F, Takahashi N, Tagawa H. Hypoxia‐inducible KDM3A addiction in multiple myeloma. Blood Adv. 2018;2:323‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maiso P, Huynh D, Moschetta M, et al. Metabolic signature identifies novel targets for drug resistance in multiple myeloma. Cancer Res. 2015;75(10):2071‐2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shaffer AL, Emre NC, Lamy L, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ikeda S, Kitadate A, Abe F, et al. Hypoxia‐inducible microRNA‐210 regulates the DIMT1‐IRF4 oncogenic axis in multiple myeloma. Cancer Sci. 2017;108:641‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin SK, Diamond P, Gronthos S, Peet DJ, Zannettino AC. The emerging role of hypoxia, HIF‐1 and HIF‐2 in multiple myeloma. Leukemia. 2011;25:1533‐1542. [DOI] [PubMed] [Google Scholar]
- 12. Borsi E, Terragna C, Brioli A, Tacchetti P, Martello M, Cavo M. Therapeutic targeting of hypoxia and hypoxia‐inducible factor 1 alpha in multiple myeloma. Transl Res. 2015;165:641‐650. [DOI] [PubMed] [Google Scholar]
- 13. Yun Z, Zhichao J, Hao Y, et al. Targeting autophagy in multiple myeloma. Leuk Res. 2017;59:97‐104. [DOI] [PubMed] [Google Scholar]
- 14. Desantis V, Saltarella I, Lamanuzzi A, et al. Autophagy: a new mechanism of prosurvival and drug resistance in multiple myeloma. Transl Oncol. 2018;11:1350‐1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814‐822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang Y, Tan J, Zhang Q. Signaling pathways and mechanisms of hypoxia‐induced autophagy in the animal cells. Cell Biol Int. 2015;39:891‐898. [DOI] [PubMed] [Google Scholar]
- 19. Daskalaki I, Gkikas I, Tavernarakis N. Hypoxia and selective autophagy in cancer development and therapy. Front Cell Dev Biol. 2018;6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037‐1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gewirtz DA. The four faces of autophagy: implications for cancer therapy. Cancer Res. 2014;74:647‐651. [DOI] [PubMed] [Google Scholar]
- 22. Hoang B, Benavides A, Shi Y, Frost P, Lichtenstein A. Effect of autophagy on multiple myeloma cell viability. Mol Cancer Ther. 2009;8:1974‐1984. [DOI] [PubMed] [Google Scholar]
- 23. Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12:1‐222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003;206:2049‐2057. [DOI] [PubMed] [Google Scholar]
- 25. Roberts DJ, Tan‐Sah VP, Ding EY, Smith JM, Miyamoto S. Hexokinase‐II positively regulates glucose starvation‐induced autophagy through TORC1 inhibition. Mol Cell. 2014;53:521‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan VP, Miyamoto S. HK2/hexokinase‐II integrates glycolysis and autophagy to confer cellular protection. Autophagy. 2015;11:963‐964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cirstea D, Santo L, Hideshima T, et al. Delineating the mTOR kinase pathway using a dual TORC1/2 inhibitor, AZD8055, in multiple myeloma. Mol Cancer Ther. 2014;13:2489‐2500. [DOI] [PubMed] [Google Scholar]
- 28. Hideshima T, Bradner JE, Wong J, et al. Small‐molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci USA. 2005;102:8567‐8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kudryavtseva AV, Fedorova MS, Zhavoronkov A, et al. Effect of lentivirus‐mediated shRNA inactivation of HK1, HK2, and HK3 genes in colorectal cancer and melanoma cells. BMC Genet. 2016;17(Suppl 3):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakano A, Tsuji D, Miki H, et al. Glycolysis inhibition inactivates ABC transporters to restore drug sensitivity in malignant cells. PLoS One. 2011;6:e27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakano A, Miki H, Nakamura S, et al. Up‐regulation of hexokinaseII in myeloma cells: targeting myeloma cells with 3‐bromopyruvate. J Bioenerg Biomembr. 2012;44:31‐38. [DOI] [PubMed] [Google Scholar]
- 32. Majkowska‐Skrobek G, Augustyniak D, Lis P, et al. Killing multiple myeloma cells with the small molecule 3‐bromopyruvate: implications for therapy. Anticancer Drugs. 2014;25:673‐682. [DOI] [PubMed] [Google Scholar]
- 33. Colla S, Storti P, Donofrio G, et al. Low bone marrow oxygen tension and hypoxia‐inducible factor‐1α overexpression characterize patients with multiple myeloma: role on the transcriptional and proangiogenic profiles of CD138(+) cells. Leukemia. 2010;24:1967‐1970. [DOI] [PubMed] [Google Scholar]
- 34. Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci USA. 2007;104:5431‐5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lévesque JP, Winkler IG, Hendy J, et al. Hematopoietic progenitor cell mobilization results in hypoxia with increased hypoxia‐inducible transcription factor‐1 alpha and vascular endothelial growth factor A in bone marrow. Stem Cells. 2007;25:1954‐1965. [DOI] [PubMed] [Google Scholar]
- 36. Mathupala SP, Rempel A, Pedersen PL. Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J Biol Chem. 2001;276:43407‐43412. [DOI] [PubMed] [Google Scholar]
- 37. Patra KC, Wang Q, Bhaskar PT, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24:213‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683‐4696. [DOI] [PubMed] [Google Scholar]
- 39. Shinohara Y, Ichihara J, Terada H. Remarkably enhanced expression of the type II hexokinase in rat hepatoma cell line AH130. FEBS Lett. 1991;291:55‐57. [DOI] [PubMed] [Google Scholar]
- 40. Mathupala SP, Rempel A, Pedersen PL. Glucose catabolism in cancer cells. Isolation, sequence, and activity of the promoter for type II hexokinase. J Biol Chem. 1995;270:16918‐16925. [DOI] [PubMed] [Google Scholar]
- 41. Rasche L, Angtuaco E, McDonald JE, et al. Low expression of hexokinase‐2 is associated with false‐negative FDG‐positron emission tomography in multiple myeloma. Blood. 2017;130:30‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abe Y, Ikeda S, Kitadate A, et al. Low hexokinase‐2 expression‐associated false‐negative 18F‐FDG PET/CT as a potential prognostic predictor in patients with multiple myeloma. Eur J Nucl Med Mol Imaging. 2019;46:1345‐1350. [DOI] [PubMed] [Google Scholar]
- 43. Abdel‐Wahab AF, Mahmoud W, Al‐Harizy RM. Targeting glucose metabolism to suppress cancer progression: prospective of anti‐glycolytic cancer therapy. Pharmacol Res. 2019;150:104511. [DOI] [PubMed] [Google Scholar]
- 44. Li M, Chiang YL, Lyssiotis CA, et al. Non‐oncogene addiction to SIRT3 plays a critical role in lymphomagenesis. Cancer Cell. 2019;35:916‐931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu S, Zhou T, Doh HM, et al. An HK2 antisense oligonucleotide induces synthetic lethality in HK1‐HK2+ multiple myeloma. Cancer Res. 2019;79:2748‐2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mishima Y, Santo L, Eda H, et al. Ricolinostat (ACY‐1215) induced inhibition of aggresome formation accelerates carfilzomib‐induced multiple myeloma cell death. Br J Haematol. 2015;169:423‐434. [DOI] [PubMed] [Google Scholar]
- 47. Jarauta V, Jaime P, Gonzalo O, et al. Inhibition of autophagy with chloroquine potentiates carfilzomib‐induced apoptosis in myeloma cells in vitro and in vivo. Cancer Lett. 2016;382:1‐10. [DOI] [PubMed] [Google Scholar]
- 48. Laubach JP, Liu CJ, Raje NS, et al. A phase I/II study of evofosfamide, a hypoxia‐activated prodrug with or without bortezomib in subjects with relapsed/refractory multiple myeloma. Clin Cancer Res. 2019;25:478‐486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu J, Handisides DR, Van Valckenborgh E, et al. Targeting the multiple myeloma hypoxic niche with TH‐302, a hypoxia‐activated prodrug. Blood. 2010;116:1524‐1527. [DOI] [PubMed] [Google Scholar]
- 50. Phillips RM. Targeting the hypoxic fraction of tumours using hypoxia‐activated prodrugs. Cancer Chemother Pharmacol. 2016;77:441‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Table S1


