Abstract
Novel treatments for Huntington’s disease (HD), a progressive neurodegenerative disorder, include selective targeting of the mutant allele of the huntingtin gene (mHTT) carrying the abnormally expanded disease-causing cytosine-adenine-guanine (CAG) repeat. WVE-120101 and WVE-120102 are investigational stereopure antisense oligonucleotides that enable selective suppression of mHTT by targeting single-nucleotide polymorphisms (SNPs) that are in haplotype phase with the CAG repeat expansion. Recently developed long-read sequencing technologies can capture CAG expansions and distant SNPs of interest and potentially facilitate haplotype-based identification of patients for clinical trials of oligonucleotide therapies. However, improved methods are needed to phase SNPs with CAG repeat expansions directly and reliably without need for familial genotype/haplotype data. Our haplotype phasing method uses single-molecule real-time sequencing and a custom algorithm to determine with confidence bases at SNPs on mutant alleles, even without familial data. Herein, we summarize this methodology and validate the approach using patient-derived samples with known phasing results. Comparison of experimentally measured CAG repeat lengths, heterozygosity, and phasing with previously determined results showed improved performance. Our methodology enables the haplotype phasing of SNPs of interest and the disease-causing, expanded CAG repeat of the huntingtin gene, enabling accurate identification of patients with HD eligible for allele-selective clinical studies.
Keywords: antisense oligonucleotide, Huntington’s disease, long-read sequencing, mRNA, single-nucleotide polymorphism
Graphical Abstract

Svrzikapa et al. describe a method using single-molecule real-time long-read sequencing and a custom algorithm to determine bases at SNPs on mutant alleles. Accurate haplotype phasing of SNPs in relationship to the expanded CAG repeat of the huntingtin gene enables identification of patients with Huntington’s disease eligible for allele-selective clinical studies.
Introduction
Huntington’s disease (HD) is a progressive neurodegenerative disorder that presents as a triad of motor, psychiatric, and cognitive dysfunction.1,2 HD is caused by a cytosine-adenine-guanine (CAG) trinucleotide repeat expansion in the huntingtin (HTT) gene, which results in production of a mutated form of the huntingtin protein (mHTT).1,3 Multiple studies have demonstrated an inverse correlation between CAG repeat length and age of symptom onset in patients with HD, clearly identifying the mHTT allele as a potential therapeutic target.1,4, 5, 6 Currently, there are no approved disease-modifying therapies for HD.7
Wild-type HTT protein (wtHTT) is ubiquitously expressed and has numerous physiologic roles, particularly in neurons and glia.8, 9, 10 In healthy neurons, wtHTT is required for neurogenesis11 and is involved in both retrograde and anterograde transport in axons and dendrites.12, 13, 14 Overexpression of wtHTT has been shown to protect against N-methyl-d-aspartate (NMDA)-mediated excitotoxicity,15 whereas expression of mHTT has been shown to increase NMDA receptor-mediated activation and cell death.16 In mice, reduced wtHTT levels result in deterioration of overall health, abnormal neuronal development, brain atrophy, and motor deficits;17,18 knockout of Hdh, the murine homolog of human HTT, leads to death at approximately embryonic day 7.5.19 Reduced expression of HTT has also been observed in invasive cells in patients with breast cancer and was associated with poor prognosis and development of metastasis.20,21 The long-term consequences of wtHTT reduction are currently unknown.
Various therapeutic strategies are in development to reduce HTT expression in patients with HD.22 Given the potentially deleterious effects associated with reduced wtHTT levels, and that few patients are homozygous carriers,23 selective suppression of mHTT with preservation of wtHTT expression may be a more beneficial targeting strategy than indiscriminate knockdown of both HTT alleles. One way to achieve selective suppression of mHTT is to design oligonucleotides that target heterozygous loci and preferentially bind mRNA transcripts containing single-nucleotide polymorphisms (SNPs) that are on the same allele (in haplotype phase) as the expanded CAG repeats in the mHTT allele.24,25 Although variable results have been reported in the literature, in HD, approximately two-thirds of patients with European ancestry have either SNP rs362307 (SNP1), SNP rs362331 (SNP2), or both SNPs in phase with the expanded CAG repeats, making these SNP variants promising therapeutic targets for most patients with HD.25,26
Wave Life Sciences Ltd. (Cambridge, MA, USA) has developed investigational stereopure oligonucleotides, WVE-120101 and WVE-120102, which selectively suppress mHTT production by specifically targeting the U variant of SNP1 and SNP2, respectively, in mHTT mRNA transcripts containing the disease-causing CAG expansion. These stereopure oligonucleotides have precisely controlled stereochemistry at phosphorothioate linkages, an approach that may lead to increased potency and improved target engagement.27 Accurate and efficient phasing, which involves determination of whether the U variant of SNP1 or SNP2 is on the same allele as the CAG expansion within mHTT, is imperative for identifying patients who could potentially benefit from these stereopure oligonucleotides that selectively suppress mHTT.
Identification of SNPs in phase with CAG expansions has been a challenge owing to the long distances between SNPs and CAG repeat regions within the HTT locus. The HTT gene is 180 kb long,28 and the targeted SNPs of interest are distal to the CAG repeat expansion. For instance, SNP1 and the CAG repeat expansion are approximately 165 kb apart in genomic DNA and 9.5 kb apart29 in HTT mRNA. Until recently, it was not possible to capture distal variants together with the long CAG expansions in HD on the same sequencing read. Long-read sequencing technologies can now facilitate resolution of bases at distant positions in a single read,30 enabling classification of an allele as targetable or not targetable with allele-selective treatment strategies.
However, this requires effective and reliable haplotype phasing methods, and current methods for phasing bases at a distant SNP on the mutant allele have limitations.31,32 Rather than directly phasing variants, these methods rely on familial data and statistical inference.30, 31, 32, 33, 34 When the linkage between CAG repeats and distant SNPs are ambiguous, they require consultation of genotypes and haplotypes of relatives, or, when such information is unavailable or uninformative for phase determination, they rely on probabilistic assignment of phase based on population data.31,35 A direct approach to phasing that is not reliant on population data would be more suitable for adoption in the clinic. Notably, several haplotype calling tools suitable for long reads were published during the development of our assay, and their potential utility is addressed in the Discussion.36,37
We sought to build off the promise of long-read sequencing technology and develop a haplotype phasing approach that could directly identify patients with expanded CAG repeats in phase with the U variant of a targeting SNP of interest without familial genotype or haplotype data, enabling reliable haplotype phasing determination of candidates for allele-selective clinical trials. This phasing approach couples single-molecule real-time (SMRT) DNA sequencing technology using the PacBio RS II system (Pacific Biosciences, Menlo Park, CA, USA), which produces read lengths exceeding 60 kb,38 with a custom algorithm developed to process the long-read sequencing data. With this approach, we can create long-range complementary (cDNA) templates of the HTT gene mRNA and amplify ∼9.8-kb regions that encapsulate the CAG repeat and SNP1 and SNP2 regions. During library preparation, a closed circular DNA template, termed a SMRTbell, is created by ligating flanking hairpins to the end of the polymerase chain reaction (PCR) product insert, which allows for circular polymerization of the insert, forming a continuous long read that may contain multiple passes of the insert.38 In our case, due to the length of the insert, this typically results in one complete pass over the amplified HTT sequence on the PacBio RS II instrument. However, recent advancements in the technology enable generation of longer reads and may further provide high-quality circular consensus sequencing (CCS) reads resulting from multiple sequencing passes of long inserts. When generating the DNA insert by long-range amplification of the HTT mRNA, the evaluated reads are of sufficient length to capture both the CAG repeat and exonic SNP regions of interest; thus, we developed an algorithm that can determine the phase of the SNP with the CAG expansion region.
PRECISION-HD1 (ClinicalTrials.gov: NCT03225833) and PRECISION-HD2 (ClinicalTrials.gov: NCT03225846), phase 1b/2a multicenter, randomized, double-blind, placebo-controlled studies, are currently evaluating the safety and tolerability of single and multiple doses of WVE-120101 and WVE-120102, respectively, in patients with HD. To be eligible for the PRECISION-HD studies, patients are screened to confirm the presence of CAG repeat expansions and heterozygosity of SNP1 and SNP2 and to determine whether they have the targeted SNP U variant on the same allele as the pathogenic CAG expansion (Figure 1).
Figure 1.
Prescreening and Screening Process for PRECISION-HD Clinical Studies
Prescreening included confirmation of CAG expansion ≥36 repeats and heterozygosity of the SNPs of interest. For confirmed samples, the long-read sequencing assay was used to determine haplotype phasing. CAG, cytosine-adenine-guanine; HD, Huntington’s disease; NGS, next-generation sequencing; PCR, polymerase chain reaction; SNP, single-nucleotide polymorphism.
Herein, we summarize our methodology for phasing determination between distant SNPs and CAG repeat expansions in HTT mRNA, which can be used to qualify patients for allele-selective clinical trials. We report validation of the approach using samples with known phasing results that were obtained using genome-wide human SNP array data33 and phasing of SNPs using the Markov chain haplotyping (MaCH) algorithm.31,35
Results
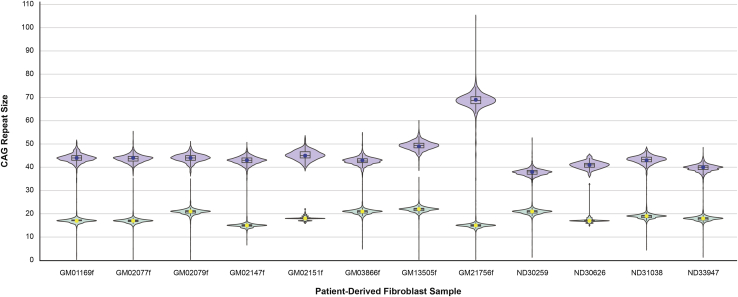
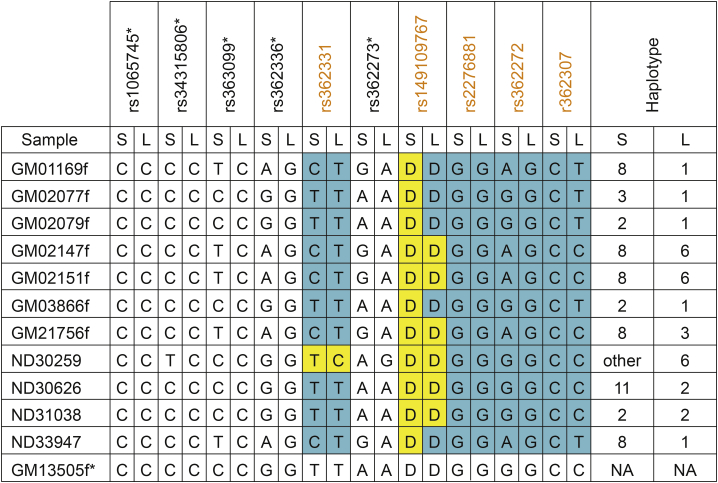
To assess the assay protocol, 12 HD patient-derived cell lines from the Coriell Institute were processed to determine whether the CAG repeat size, SNP heterozygosity, and haplotype phasing results generated using our methodology were comparable to the known CAG repeat size, SNP heterozygosity, and phasing data of the historical Coriell Institute samples. Following blinded sample analysis, Coriell Institute samples were unblinded and compared with the blinded sample data. Correlation analysis showed perfect concordance between CAG repeat data generated with our method and known repeat data (Figure 2), with results being within the ranges for CAG repeat quantitation given in the American College of Medical Genetics and Genomics technical standards and guidelines for HD.39 Similarly, heterozygosity and phasing generated with our method showed near-perfect concordance with known historical results (Figure 3) and high degrees of confidence (Table S1). We observed a difference between historical results and our phasing data for SNP2 on the mutant allele (T versus C, respectively) in sample ND30259. An additional discrepancy was found for rs149109767, in which, across all samples, we only observed the deletion.
Figure 2.
Concordance of CAG Sizing Data from the Coriell Institute and BioRep
Violin plot shows distribution of CAG repeat lengths as determined using the haplotype phasing assay. Yellow and blue dots indicate known CAG repeat size for short and long repeats, respectively. Boxes show interquartile range (IQR) and median.
Figure 3.
Heterozygosity and Phasing Data from Patient-Derived Fibroblasts
rsIDs represent evaluated exonic SNPs, and orange denotes a SNP evaluated in Chao et al.31; rs149109767 is an indel, where D represents an AGG>delAGG observed deletion. Blue denotes bases concordant with previously published results, and yellow denotes bases discordant with previously published results. Haplotypes are reported as determined in Chao et al.31 “Other” is defined as a rare or uncertain haplotype. L, long (expanded) CAG repeat allele; NA, not reported; S, short CAG repeat allele. ∗SNP across the huntingtin mRNA sequence.
To further evaluate the method and reagents used for the phasing protocol, a validation study was undertaken with Q2 Solutions | EA Genomics (Morrisville, NC, USA). Specificity was initially assessed using a no template control (NTC). Measurements of long-range PCR products using the NTC were below the limit of detection of the assay, leading to the conclusion that the NTC was truly negative. Four cell lines, 9 whole-blood samples from 6 patients with HD, and 15 whole-blood samples from 12 healthy control donors were used for assay validation.
Accuracy of the assay for phase determination was analyzed using whole-blood samples from healthy controls and samples with known CAG repeat length and haplotype phasing. The assay produced the correct phasing results for all samples, leading to a total of 15 true positives, 26 true negatives, 0 false negatives, and 0 false positives (accuracy, 41/41 [100%]; Table S2). Both analytical sensitivity (15 true positives of 15 previously known in-phase samples) and specificity (15 true positives of all positives [15 true + 0 false positives]) were determined to be 100%. Intra-run precision, assessed by performing technical replicates within the same run, was 100% (6/6 samples with concordant phasing determinations). Inter-run precision was assessed by performing technical replicates across three runs; the assay led to concordant phasing results in 16 of 16 cases (100%; Table S3). Reproducibility was assessed using different lots of critical reagents; consistent results were observed for all replicates. However, one cell line, which was not included in the assay validation, failed to pass the Bayes factor threshold in the initial run, indicating possible run-to-run variability. Stability of RNA and cDNA was assessed after two freeze-thaw cycles. Freeze-thaw stability of long-range PCR amplicons (DNA) from downstream reactions was not tested since DNA is known to be more stable than RNA. Analysis of results demonstrated that phasing determination was not affected by freeze-thaw (Table S2).
Despite our expectation to observe two distinct haplotypes, we observed additional chimeric reads likely resulting from incomplete PCR products. This observation motivated the use of a statistical method that used the read counts of the two main haplotypes and conservatively included the counts of the chimeric products. This method allowed for determination of phase with Bayes factor values >9.19 × 1010, suggesting high degrees of confidence (Figure 4; Figure S1).
Figure 4.
Representative Results from the Allelic Phasing and SNP-CAG Repeat Association With Long-Read Sequencing
Inset shows prior (red), likelihood (yellow), and posterior (blue) probability for the long CAG repeat allele being in haplotype phase with the SNP U variant. Sample GM02077f was heterozygous for SNP1 and homozygous for SNP2. CAG repeat length was 17 and 44 for the short and long CAG repeats, respectively. SNP1 and SNP2 contained the U variant on the long CAG repeat; however, only SNP1 was heterozygous and therefore would qualify the patients for an allele-selective clinical trial. See Figure S1 for results for the 12 samples used in assay determination.
Thirty-one cycle PCR yielded an insufficient quantity of PCR product for library preparation. However, 34-cycle PCR amplification yielded a sufficient quantity of PCR product and significantly reduced the proportion of reads resulting from chimera artifacts compared with 38-cycle PCR amplification (8.3% versus 25.3%; p < 0.0001; Figure 5).
Figure 5.
Chimera Production as Percentage of Total Reads after 34 (Circles) or 38 (Triangles) PCR Amplification Cycles
Lines show mean and SD. ∗p < 0.0001.
We observed a marginal, but statistically significant enrichment of short CAG-containing reads in comparison with expanded CAG-containing reads (Figure S2). We applied our methodology to phase all exonic SNPs mentioned in Chao et al.31 located within the amplified region, with the exception of rs362267, rs115335747, and rs2530595, which are located upstream of our reverse primer (Figure 3; Figure S3; Table S4). In addition, we analyzed several other HTT SNPs distributed across the amplified region. We found an additional discrepancy with rs149109767, where we did not observe the insertion. Namely, in all samples, including the reference mRNA sequence GenBank: NM_002001, we only observed the deletion (Figure 3).
Discussion
Current haplotype phasing methods are limited by the inability to directly and definitively phase distant features in the absence of familial genotype or haplotype data,31,32 which impedes the identification of patients with HD eligible for allele-selective clinical studies. We took advantage of the availability of long-read sequencing technologies, which can capture SNPs of interest and CAG repeats from HD samples in one read, to develop a new phasing method for HD that can assess with confidence the base at a distant SNP on the mutant allele and enable appropriate identification of candidate patients with HD for clinical studies. Our assay produces single long-read sequences that allow visualization of distant SNP variants and the CAG repeat expansion on the mHTT allele, thus enabling haplotype phasing determination in patients with HD. This strategy can be utilized in multiple genes where a structural variant of variable size needs to be phased with a SNP of interest for allele-selective targeting.
Notably, during the development of our method, several haplotype callers suitable for long-read sequencing technologies were published. Solutions such as WhatsHap37 and, more recently, SHAPEIT436 rely on high-quality standard variant call format (VCF) files that must be generated directly from high-quality CCS reads on the more current Sequel II platform, or orthogonal short read sequencing of the same sample. While our sequencing reads generated on the RSII were not suitable for testing, it would be useful to evaluate the performance of these tools for phasing the long CAG repeat expansion and how the chimeric reads discussed below would affect their performance.
Our assay takes advantage of PacBio technology to produce longer read lengths than those that can be achieved using standard sequencing and other next-generation sequencing (NGS) methods.38 Thus, in every read, which represents a single allele, we are able to determine the base at the SNP of interest and the CAG repeat length, which can be up to approximately 10 kb apart in mHTT mRNA.40 In fact, in the current study, our approach produced an average read length of 15,951 bases, with a maximum read length of 21,460 bases, allowing for colocalization of SNPs and CAG repeats on the same amplicon. Superior performance is now achievable with the PacBio Sequel II platform, which is reported to provide maximum subread lengths of >200 kb, an insert (subread) length N50 of 22.3 kb, and a polymerase read length N50 of 41.7 kb, yielding 131.6 Gb of total sequence.41
The phasing of any structural variant with a SNP colocalized on the same read is a straightforward task. However, we observed above-background anomalous reads with an unexpected switched phase. One of the inherent problems of long-range PCR is the potential generation of chimeric PCR products during amplification.42 These chimeras are PCR artifacts that likely result from premature termination of an amplicon followed by reannealing of the amplicon to another template.43
A two-prong strategy was applied to mitigate the occurrence of chimeric reads in our sequencing data. We experimentally demonstrated that the number of chimeras can be reduced by decreasing the number of PCR amplification cycles and that this can be accomplished without significantly reducing the number of qualified reads. This supports development of future algorithms for the detection and elimination of chimeras. More critically, we conservatively accounted for the presence of chimeric products using our algorithm, and their numbers are built into our statistical model. As with previous HD datasets (R. Mouro Pinto, 2017, PacBio User Group, conference), we observed anomalous long CAG repeat regions and some degree of insertions and deletions in the CAG region, which could be polymerase errors possibly arising from their highly GC-rich nature, resulting in a distribution of CAG repeat lengths rather than one discrete length value (Figure S1) (R. Mouro Pinto, 2017, PacBio User Group, conference).
In our validation study, correlation analysis of blinded haplotype phasing results from patient-derived cell lines and whole-blood samples with known phasing results (obtained using our method at a separate facility)44 showed 100% accuracy, precision, sensitivity, and specificity (Table S2). CAG repeat sizing, SNP heterozygosity, and phasing data were directly and reliably obtained without the need for familial data or probabilistic phasing, demonstrating an advantage for this approach over existing haplotype phasing methods for HD.
The identified discrepancies between our findings and historical results confirm the potential for errors when using an indirect phasing approach and show the advantage of our strategy over previous phasing methods. Long-read sequencing can accurately capture CAG repeat expansions of various lengths and SNPs at distant locations within the same read, offering clear visibility to determine directly a SNP and its association with either the mHTT or the wtHTT allele, even for samples that may be challenging to phase using previous methods. The algorithm we developed can determine the phase of the targeted SNPs of interest, as well as accurately estimate the CAG repeat length and recapitulate zygosity at any position of the amplified insert. The initial generation of Bowtie indices facilitates future computationally efficient analysis of other sites of interest within the amplified insert region. We think that our method provides a conservative approach for determining the phase of the SNPs of interest with the CAG repeat type in both the mHTT allele and wtHTT alleles. Furthermore, we can phase any exonic SNPs within amplified regions with the CAG repeat region, enabling identification of a substantially greater number of SNPs than used previously.31
Direct phasing using our methodology allows for the possibility of allele-selective targeting of mHTT via SNPs that are in phase with CAG repeat expansions in patients with HD. Such targeting with stereopure oligonucleotides can reduce mHTT expression while preserving wtHTT expression and function. The investigational stereopure oligonucleotides WVE-120101 and WVE-120102 are currently being studied in clinical trials. These trials represent a novel clinical application of stereopure oligonucleotides that act specifically to degrade the disease-causing HTT mRNA by targeting a SNP in phase with the abnormal CAG repeat expansion. The haplotype phasing approach described herein represents the first long-read sequencing phasing method used to support a clinical trial. This approach is being used to screen patients for enrollment.44 While patients are prescreened to confirm CAG expansion of ≥36 repeats (i.e., confirm HD diagnosis) and heterozygosity at both SNPs by fluorescence PCR and Sanger sequencing, respectively, the algorithm we developed fully captures CAG repeat sizing, heterozygosity, and haplotype phasing.
This approach is also limited by the successful generation of a long-range PCR product and may not be applicable for phasing across longer distances, despite the ability of the sequencing technology to yield longer reads. Alternative strategies may be needed to phase events successfully across larger genomic distances. The 10x Genomics platform, for example, uses genomic DNA as a template to generate a library of approximately 50-kb fragments labeled with unique barcodes. The resulting barcoded library provides long-range information suitable for haplotype phasing and determination of SNPs of interest using existing short-range sequencing technologies. One limitation of this approach, related to the read length of these sequencing technologies, is the inability to resolve long CAG expansions. However, with prior determination of the CAG repeat lengths and Sanger sequencing to determine heterozygosity, this technology may be used to phase the short CAG expansion, providing sufficient information to infer the long CAG expansion haplotype. Another viable approach could use amplification of overlapping genomic regions followed by WhatsHap or SHAPEIT4 for haplotype calling.
In conclusion, our haplotype phasing assay offers benefits over current phasing methods by eliminating the need for familiar genotype or haplotype data or use of probabilistic population phasing. Our assay enabled the identification of patients with HD who have targeted SNPs on the disease-causing mHTT allele. Blinded assessment of haplotype phasing showed near-complete concordance with previously determined phasing data from Coriell Institute samples. The few observed discrepancies show the advantages of using long-read technology for haplotype phasing. Our method specifically treats the CAG structural variant and accounts for the occurrence of chimeras, both of which are likely to be problematic for general haplotype callers. This approach highlights a strategy for phasing that enables enrollment of patients with HD for allele-selective suppression of mHTT in clinical studies. More generally, our methodology can be readily used for phasing in other diseases for which allele-selective treatment may be desirable.
Materials and Methods
Samples
To evaluate allele-phasing capabilities in a blinded experiment, we analyzed patient-derived cell lines from the Coriell Institute (Camden, NJ, USA) with known determinations of SNPs in phase with CAG repeat expansions (Figure 3).31 To assess the reproducibility of the assay, we analyzed patient-derived cell lines from the Coriell Institute (Camden, NJ, USA) and whole-blood samples from patients with HD collected as part of an observational study designed to define SNP frequencies, each with known determinations of SNPs in phase with CAG repeat expansions,44 as well as whole-blood samples from healthy donors. The latter were obtained from Folio Conversant (now Discovery Life Sciences; Los Osos, CA, USA) in PAXgene blood RNA tubes (PreAnalytiX, Hombrechtikon, Switzerland). The blinded reproducibility study was performed at EA Genomics (Morrisville, NC, USA) with samples previously analyzed at HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA) using our method. The study was conducted in accordance with the Declaration of Helsinki and approved by an Institutional Review Board; all participants provided written informed consent prior to sample collection.
CAG Repeat Length and SNP Heterozygosity Determination
DNA was extracted from each sample, and CAG repeat length was analyzed at a commercial testing laboratory (BioRep, Milan, Italy) using fluorescence PCR. DNA samples were also analyzed for heterozygosity for HTT gene target SNPs (T or C) using Sanger sequencing at HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA; data not shown) (Figure 1).
Assay Parameters and Specifications
During the development of this methodology, we tested and determined several assay parameter specifications. In a prospective, observational study44 and the ongoing PRECISION-HD clinical studies (ClinicalTrials.gov: NCT03225833 and NCT03225846), a 2.5-mL whole blood sample was collected in each PAXgene RNA tube. The average amount of RNA obtained from each PAXgene whole-blood RNA tube was approximately 8.5 μg (range, 2.5–20 μg per PAXgene tube). The minimum amount of RNA required for a successful phasing assay was calculated to be 0.6 μg per reaction, and the minimum required RNA quality as estimated by the RNA integrity number (RIN) was 7. For the post-HTT gene-specific cDNA synthesis, a minimum of 295 and 325 ng of cDNA was used in a 12.5-μL reaction for cell-derived and blood-derived RNA, respectively, to generate the PCR product; 1 μg of PCR product was used to generate a sample library.
In a prospective, observational US study, the most frequently occurring genotype for HTT was a normal allele with 18 CAG repeats and a mutant allele with 43 repeats.44 In addition to PacBio sequencing for enumeration, CAG size was determined using a PAXgene blood DNA sample and a PCR-capillary electrophoresis (CE)-based orthogonal method that is widely used for HD diagnosis.45 In fibroblast cell lines, the longest CAG we have phased to date is 66 CAG repeats. Patients prescreened for the PRECISION-HD clinical studies were limited to early stage HD with an average repeat length of 44. The longest expansion tested so far was 62 CAG repeats; all repeats have been successfully enumerated. This is not the limit for the assay, but it is the longest we have encountered thus far. Longer CAG expansions have previously been sequenced and reported using the PacBio technology (R. Mouro Pinto, 2017, PacBio User Group, conference).
Haplotype Phasing
Methods and results for phasing determination of the Coriell Institute samples have been previously described.31 Briefly, phasing was previously determined using the MaCH algorithm.35 In this process, a pair of haplotypes compatible with observed genotypes is randomly generated. Initial haplotype estimates are refined through a series of iterations, and a consensus haplotype is constructed by merging the haplotypes sampled in each round. Phasing results for Coriell Institute samples that were previously determined through the MaCH method were blinded until completion of sample and data analysis.
Long-read sequencing technology was used to determine whether the targeted SNP U variant was present on the same allele as the pathogenic CAG expansion. These analyses were performed at HudsonAlpha Institute for Biotechnology and Q2 Solutions | EA Genomics, LLC (Morrisville, NC, USA) using methodology, algorithms, and analytic tools developed at Wave Life Sciences. Total RNA was extracted from patient-derived cells and whole-blood samples using the QIAGEN RNeasy Mini Kit (QIAGEN, Germantown, MD, USA) and the PAXgene IVD blood isolation kit (PreAnalytiX), respectively. Purified RNA was reverse transcribed into long cDNA with a gene-specific primer (5′-ATGCTGGGCTCTGTCACTAA-3′; Integrated DNA Technologies, Coralville, IA, USA) and the SuperScript IV first-strand synthesis system (Thermo Fisher Scientific, Waltham, MA, USA). Long-range PCR amplicons were generated from cDNA using gene-specific primers (forward, 5′-AGCTGATGAAGGCCTTCGAGTCCCTCAAGTC-3′; reverse: 5′-GTGTTCCCAAAGCCTGCTCACGGCAC-3′) and PCR TaKaRa LA Taq DNA polymerase with GC buffers (Clontech, Mountainview, CA, USA). For samples in which PCR produced 9- to 10-kb bands with no other bands >2 kb (see representative PCR results in Figure 6), the PCR reaction was purified and sequenced. For samples in which the reaction produced nonspecific bands >2 kb, amplicons were size selected using the BluePippin DNA size selection system (Sage Science, Beverly, MA, USA) before purification and sequencing. Only amplified DNA >6,000 bp was analyzed further. A library was prepared through ligation of blunt-ended double-stranded amplicons with PacBio SMRTbell templates and was subsequently sequenced on the PacBio RS II sequencer.
Figure 6.
Replicates of Long-Range PCR Products from Four Patient-Derived Cell Lines and 12 Whole-Blood Samples from Healthy Donors Run on an Agarose Gel
Long-range PCR produces 9- to 10-kb bands. “Directly to Library” Y indicates that the sample was directly analyzed, whereas N indicates that the PCR products underwent size selection before analysis. PF-HD, Huntington’s disease patient-derived fibroblasts.
The following parameters were used to validate the performance of the allele-phasing assay for determining SNP association with CAG repeat by reverse transcriptase PCR and NGS: specificity, sensitivity, lower limit of input, inter-run and intra-run precision, reproducibility, accuracy, and stability (Tables S1–S3).
Statistical Algorithm for Haplotype Phasing
Given the prevalence of SNP1 and SNP2 in the HD patient population, we developed allele-selective therapies targeting the U variant at these SNPs where the U variant of the U/C SNP was in phase with the disease-causing expanded CAG repeat. Thus, our assay and analytical methods were designed to evaluate the nature of the SNP and the length of the CAG repeat expansion. Both of these features were presented on one long read. Ideally, we expected to see two distinct haplotypes where in a heterozygous sample the long CAG repeat would be in phase with, for example, Z1 at the position of the Z1/Z2 SNP (Long | Z1), in which case the short/normal CAG repeat would be expected to be in phase with Z2 at the SNP position (Short | Z2). However, due to the observation of PCR chimeras, two additional switched haplotypes were observed (Long | Z2 and Short | Z1). As a result, we conservatively included the observation of the chimeric reads and used a statistical method in which we took the counts of each species in order to calculate the posterior probabilities of each potential phase with a high degree of confidence (Figure 7; Figure S4).
Figure 7.
Long-Read Method for Haplotype Phasing of SNPs with the CAG Repeat Expansion in HD
(A) Each sequencing read contains information on the SNP of interest and a normal or expanded CAG repeat. (B) The CAG repeat length d is measured as the distance between two aligned flanking sequences, while the base at any SNP of interest is determined by progressive alignment of all potential bases at the SNP locus. (C) The counts of the two main read populations (|A| and |C|) and two minor read populations arising from PCR chimeras (|B| and |D|) are used by our statistical model to calculate the posterior probabilities of each potential phase.
Different approaches can be used to evaluate the counts of each potential phase. In order to enable quick analysis and allow the user to provide custom sequence queries in a FASTA format, our strategy uses the Bowtie software aligner (Bowtie software available at http://bowtie-bio.sourceforge.net/index.shtml) to resolve the base at both SNPs. To achieve this, the algorithm parameters were set to 0 allowable mismatches for each read. For SNP1 and SNP2, efforts were made to align the sequences 5′-GGAAGTCTGXGCCCTTGTGC-3′ and 5′-TCCCTCATCXACTGTGTGCA-3′, where all possibilities (A, C, G,T) were substituted for X, representing the position SNP1 (rs362307) and SNP2 (rs362331), respectively. More generally, we aligned a sequence (5′_flank – SNP – 3′_flank) and attempted to align all possibilities at the SNP position. Conversely, this can also be applied to indels, SNPs, or any multiple SNP permutations, where only one of the query permutations could possibly align on each read.
A similar approach was used to determine CAG repeat length but allowing two mismatches (tunable parameter) due to the high density of GC-rich regions. Namely, Bowtie was used to determine the alignment positions of the 5′ (CCCTCAAGTCCTTC) and 3′ (CAACAGCCGCCACC) sequences bordering the CAG repeat expansion. The CAG repeat category (normal, <36 repeats; expanded, ≥36 repeats) was determined by measuring the distance between the alignment positions of the CAG flanking regions after filtering reads to remove those with ≤75% CAG repeats content. Given that the CAG repeat lengths were previously determined by a fluorescence PCR assay, we developed an experimental algorithm to correct non-CAG artifacts occurring within the CAG region of each read. This correction recapitulates the fluorescence PCR estimates of the CAG repeat expansion lengths.
For each SNP, data were merged on the read ID, resulting in reads with either a long (≥36-repeat) or short (<36-repeat) CAG expansion and known base identities at the two SNPs of interest. Next, each observation was counted and binned by base identity at each SNP and CAG repeat type.
To assess the phase of a U variant with either a long or short CAG repeat at heterozygous SNP loci, a Bayesian model was applied to the counts data to assess the posterior of the joint probability that the long CAG repeat allele was in phase with the U variant and that the short CAG repeat was in phase with the C variant, denoted as
Individual probabilities of haplotype phase realizations were modeled using beta distributions with shape parameters determined by the numbers of reads that either supported or did not support the particular phase configuration; specifically:
The parameters were defined as follows: A indicates the number of reads with a long CAG with base identity of T at SNP, B indicates the number of reads with a short CAG with base identity of T at SNP, C indicates the number of reads with a short CAG with base identity of C at SNP, and D indicates the number of reads with a long CAG with base identity of C at SNP. Thus, the likelihood was estimated as:
An empirical prior was assigned that assumed completely ambiguous results based on the total number of counts containing the CAG repeat and SNP of interest regions:
The posterior was evaluated as the product of the prior and the likelihood and was used to provide two estimates. First, the 95% highest posterior density (HPD) was calculated as the 95% area under the curve of the posterior probability, where the lower and upper bounds were of equivalent probability density. Second, the Bayes factor of the posterior odds of the posterior probability over the prior probability was estimated according to the Savage-Dickey method46 at p = 0.5 or complete ambiguity.
Thus, to determine the phase of the expanded CAG repeat with the U variant with confidence, two conditions needed to be satisfied: (1) the 95% HPD interval must be p > 0.5, and (2) the posterior odds of the posterior probability over the prior probability must be >106:1.
Analysis of Chimera Production
During analysis, we frequently observed a small fraction of anomalous reads with a similar CAG repeat length but a SNP base call deviating from the majority read population. Suspecting that these reads were PCR chimera artifacts and were more likely to arise during later PCR cycles, we investigated the effect of reducing the number of PCR amplification cycles on their occurrence. cDNA from three different cell lines was amplified for 31, 34, or 38 cycles using a standard PCR protocol recommended by the manufacturer of the polymerase. For the standard protocol, reaction mixtures were incubated in a thermal cycler at 98°C for 3 min (denature), followed by the designated number of amplification cycles at 98°C for 25 s, 60°C for 15 s, and 68°C for 20 min, and holding at 4°C until analysis.
Analysis of CAG Repeat Imbalance
Percentages of short CAG-containing reads and long CAG-containing reads were estimated for 327 patient-derived samples. A marginal, but statistically significant, enrichment of the short CAG containing reads was observed (Figure S2). This is likely a PCR-introduced anomaly, as we expect both alleles to be produced at similar levels.
Data Availability
The haplotype phasing algorithm and an example FASTA file are available in the GitHub repository (https://github.com/WaveLifeSciences/hd_haplotype_phaser).
Author Contributions
N.S. conceptualized the study, performed experiments, designed and executed the haplotype phasing software, and analyzed the data. K.A.L., J.M.B., and C.V. conceptualized the study and analyzed the data. N.P., D.D., S.Y., J.P., and S.E.L. performed sequencing experiments and SNP determination. R.B., A.J.M., and J.G. conceptualized the study, performed validation experiments, and analyzed the data. All authors participated in manuscript preparation, provided critical review of each draft, and approved the final manuscript for submission.
Conflicts of Interest
N.S., K.A.L., R.B., C.V., and J.G. are employees and stockholders of Wave Life Sciences Ltd. N.P. and D.D. were employees of HudsonAlpha Discovery, Discovery Life Sciences and Genomic Services Laboratory, HudsonAlpha Institute for Biotechnology at the time the work was conducted. J.M.B. was an employee of Wave Life Sciences Ltd. at the time the work was conducted. S.Y. is an employee of Q2 Solutions | EA Genomics, LLC; J.P. was an employee of Q2 Solutions | EA Genomics, LLC at the time the work was conducted. S.E.L. is an employee of HudsonAlpha Discovery, Discovery Life Sciences and Genomic Services Laboratory, HudsonAlpha Institute for Biotechnology. A.J.M. was an employee of Wave Life Sciences Ltd. at the time the work was conducted and is a current stockholder of Wave Life Sciences Ltd.
Acknowledgments
The authors would like to thank Alesia Antoine, MS, Andrew Hoss, PhD, Ravindra Kodihalli, PhD, Robert Sebra, PhD, Michael Weiand, MS, and Maria Zapp, PhD, for technical advice and support during the early stages of assay development, and Giulia Malferrari, PhD, for assistance with the CAG PCR assay. The authors would also like to thank CHDI for enabling this work by kindly providing access to the cell lines used in this study. This work was supported by Wave Life Sciences. Funding for open access charges was provided by Wave Life Sciences. Editorial support for development of this manuscript was provided by Krystina Neuman, PhD, at ICON plc (North Wales, PA, USA) and funded by Wave Life Sciences.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.09.003.
Contributor Information
Nenad Svrzikapa, Email: nsvrzikapa@wavelifesci.com.
Jaya Goyal, Email: jgoyal@wavelifesci.com.
Supplemental Information
References
- 1.Sturrock A., Leavitt B.R. The clinical and genetic features of Huntington disease. J. Geriatr. Psychiatry Neurol. 2010;23:243–259. doi: 10.1177/0891988710383573. [DOI] [PubMed] [Google Scholar]
- 2.Paulsen J.S. Cognitive impairment in Huntington disease: diagnosis and treatment. Curr. Neurol. Neurosci. Rep. 2011;11:474–483. doi: 10.1007/s11910-011-0215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 4.Keum J.W., Shin A., Gillis T., Mysore J.S., Abu Elneel K., Lucente D., Hadzi T., Holmans P., Jones L., Orth M. The HTT CAG-expansion mutation determines age at death but not disease duration in Huntington disease. Am. J. Hum. Genet. 2016;98:287–298. doi: 10.1016/j.ajhg.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrew S.E., Goldberg Y.P., Kremer B., Telenius H., Theilmann J., Adam S., Starr E., Squitieri F., Lin B., Kalchman M.A. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat. Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 6.Snell R.G., MacMillan J.C., Cheadle J.P., Fenton I., Lazarou L.P., Davies P., MacDonald M.E., Gusella J.F., Harper P.S., Shaw D.J. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nat. Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 7.Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R., Nance M., Ross C.A., Scahill R.I., Wetzel R. Huntington disease. Nat. Rev. Dis. Primers. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 8.Schulte J., Littleton J.T. The biological function of the Huntingtin protein and its relevance to Huntington’s disease pathology. Curr. Trends Neurol. 2011;5:65–78. [PMC free article] [PubMed] [Google Scholar]
- 9.Landles C., Bates G.P. Huntingtin and the molecular pathogenesis of Huntington’s disease. Fourth in molecular medicine review series. EMBO Rep. 2004;5:958–963. doi: 10.1038/sj.embor.7400250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saudou F., Humbert S. The biology of huntingtin. Neuron. 2016;89:910–926. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 11.White J.K., Auerbach W., Duyao M.P., Vonsattel J.P., Gusella J.F., Joyner A.L., MacDonald M.E. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat. Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 12.Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.J., Saudou F., Humbert S. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Twelvetrees A.E., Yuen E.Y., Arancibia-Carcamo I.L., MacAskill A.F., Rostaing P., Lumb M.J., Humbert S., Triller A., Saudou F., Yan Z., Kittler J.T. Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin. Neuron. 2010;65:53–65. doi: 10.1016/j.neuron.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma B., Savas J.N., Yu M.S., Culver B.P., Chao M.V., Tanese N. Huntingtin mediates dendritic transport of β-actin mRNA in rat neurons. Sci. Rep. 2011;1:140. doi: 10.1038/srep00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leavitt B.R., van Raamsdonk J.M., Shehadeh J., Fernandes H., Murphy Z., Graham R.K., Wellington C.L., Raymond L.A., Hayden M.R. Wild-type huntingtin protects neurons from excitotoxicity. J. Neurochem. 2006;96:1121–1129. doi: 10.1111/j.1471-4159.2005.03605.x. [DOI] [PubMed] [Google Scholar]
- 16.Zeron M.M., Hansson O., Chen N., Wellington C.L., Leavitt B.R., Brundin P., Hayden M.R., Raymond L.A. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington’s disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 17.Auerbach W., Hurlbert M.S., Hilditch-Maguire P., Wadghiri Y.Z., Wheeler V.C., Cohen S.I., Joyner A.L., MacDonald M.E., Turnbull D.H. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum. Mol. Genet. 2001;10:2515–2523. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- 18.Dietrich P., Johnson I.M., Alli S., Dragatsis I. Elimination of huntingtin in the adult mouse leads to progressive behavioral deficits, bilateral thalamic calcification, and altered brain iron homeostasis. PLoS Genet. 2017;13:e1006846. doi: 10.1371/journal.pgen.1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasir J., Floresco S.B., O’Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 20.Thion M.S., Humbert S. Cancer: from wild-type to mutant huntingtin. J. Huntingtons Dis. 2018;7:201–208. doi: 10.3233/JHD-180290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thion M.S., McGuire J.R., Sousa C.M., Fuhrmann L., Fitamant J., Leboucher S., Vacher S., du Montcel S.T., Bièche I., Bernet A. Unraveling the role of huntingtin in breast cancer metastasis. J. Natl. Cancer Inst. 2015;107:djv208. doi: 10.1093/jnci/djv208. [DOI] [PubMed] [Google Scholar]
- 22.Wild E.J., Tabrizi S.J. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017;16:837–847. doi: 10.1016/S1474-4422(17)30280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cubo E., Martinez-Horta S.I., Santalo F.S., Descalls A.M., Calvo S., Gil-Polo C., Muñoz I., Llano K., Mariscal N., Diaz D., European HD Network Clinical manifestations of homozygote allele carriers in Huntington disease. Neurology. 2019;92:e2101–e2108. doi: 10.1212/WNL.0000000000007147. [DOI] [PubMed] [Google Scholar]
- 24.Watts J.K., Corey D.R. Silencing disease genes in the laboratory and the clinic. J. Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kay C., Skotte N.H., Southwell A.L., Hayden M.R. Personalized gene silencing therapeutics for Huntington disease. Clin. Genet. 2014;86:29–36. doi: 10.1111/cge.12385. [DOI] [PubMed] [Google Scholar]
- 26.Lombardi M.S., Jaspers L., Spronkmans C., Gellera C., Taroni F., Di Maria E., Donato S.D., Kaemmerer W.F. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Iwamoto N., Butler D.C.D., Svrzikapa N., Mohapatra S., Zlatev I., Sah D.W.Y., Meena, Standley S.M., Lu G., Apponi L.H. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017;35:845–851. doi: 10.1038/nbt.3948. [DOI] [PubMed] [Google Scholar]
- 28.Ambrose C.M., Duyao M.P., Barnes G., Bates G.P., Lin C.S., Srinidhi J., Baxendale S., Hummerich H., Lehrach H., Altherr M. Structure and expression of the Huntington’s disease gene: evidence against simple inactivation due to an expanded CAG repeat. Somat. Cell Mol. Genet. 1994;20:27–38. doi: 10.1007/BF02257483. [DOI] [PubMed] [Google Scholar]
- 29.Pfister E.L., Kennington L., Straubhaar J., Wagh S., Liu W., DiFiglia M., Landwehrmeyer B., Vonsattel J.P., Zamore P.D., Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantere T., Kersten S., Hoischen A. Long-read sequencing emerging in medical genetics. Front. Genet. 2019;10:426. doi: 10.3389/fgene.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao M.J., Gillis T., Atwal R.S., Mysore J.S., Arjomand J., Harold D., Holmans P., Jones L., Orth M., Myers R.H. Haplotype-based stratification of Huntington’s disease. Eur. J. Hum. Genet. 2017;25:1202–1209. doi: 10.1038/ejhg.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kay C., Collins J.A., Skotte N.H., Southwell A.L., Warby S.C., Caron N.S., Doty C.N., Nguyen B., Griguoli A., Ross C.J. Huntingtin haplotypes provide prioritized target panels for allele-specific silencing in Huntington disease patients of European ancestry. Mol. Ther. 2015;23:1759–1771. doi: 10.1038/mt.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J.-M., Gillis T., Mysore J.S., Ramos E.M., Myers R.H., Hayden M.R., Morrison P.J., Nance M., Ross C.A., Margolis R.L. Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am. J. Hum. Genet. 2012;90:434–444. doi: 10.1016/j.ajhg.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warby S.C., Montpetit A., Hayden A.R., Carroll J.B., Butland S.L., Visscher H., Collins J.A., Semaka A., Hudson T.J., Hayden M.R. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaneau O., Zagury J.F., Robinson M.R., Marchini J.L., Dermitzakis E.T. Accurate, scalable and integrative haplotype estimation. Nat. Commun. 2019;10:5436. doi: 10.1038/s41467-019-13225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin M., Patterson M., Garg S., Fischer S.O., Pisanti N., Klau G.W., Schöenhuth A., Marschall T. WhatsHap: fast and accurate read-based phasing. bioRxiv. 2016 doi: 10.1101/085050. [DOI] [Google Scholar]
- 38.Rhoads A., Au K.F. PacBiol. sequencing and its applications. Genomics Proteomics Bioinformatics. 2015;13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bean L., Bayrak-Toydemir P. American College of Medical Genetics and Genomics Standards and Guidelines for Clinical Genetics Laboratories, 2014 edition: technical standards and guidelines for Huntington disease. Genet. Med. 2014;16:e2. doi: 10.1038/gim.2014.146. [DOI] [PubMed] [Google Scholar]
- 40.Liu W., Kennington L.A., Rosas H.D., Hersch S., Cha J.H., Zamore P.D., Aronin N. Linking SNPs to CAG repeat length in Huntington’s disease patients. Nat. Methods. 2008;5:951–953. doi: 10.1038/nmeth.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kingan S.B., Urban J., Lambert C.C., Baybayan P., Childers A.K., Coates B., Scheffler B., Hackett K., Korlach J., Geib S.M. A high-quality genome assembly from a single, field-collected spotted lanternfly (Lycorma delicatula) using the PacBio Sequel II system. Gigascience. 2019;8:giz122. doi: 10.1093/gigascience/giz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S.K., Sodergren E., Human Microbiome Consortium Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel R., Lin M., Laney M., Kurn N., Rose S., Ullman E.F. Formation of chimeric DNA primer extension products by template switching onto an annealed downstream oligonucleotide. Proc. Natl. Acad. Sci. USA. 1996;93:2969–2974. doi: 10.1073/pnas.93.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Claassen D.O., Corey-Bloom J., Dorsey E.R., Edmondson M., Kostyk S.K., LeDoux M.S., Reilmann R., Rosas H.D., Walker F., Wheelock V. Genotyping single nucleotide polymorphisms for allele-selective therapy in Huntington disease. Neurol. Genet. 2020;6:e430. doi: 10.1212/NXG.0000000000000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quarrell O.W., Handley O., O’Donovan K., Dumoulin C., Ramos-Arroyo M., Biunno I., Bauer P., Kline M., Landwehrmeyer G.B., European Huntington’s Disease Network Discrepancies in reporting the CAG repeat lengths for Huntington’s disease. Eur. J. Hum. Genet. 2012;20:20–26. doi: 10.1038/ejhg.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickey J.M., Lientz B.P. The weighted likelihood ratio, sharp hypothesis about chances, the order of a Markov chain. Ann. Math. Stat. 1970;41:214–226. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The haplotype phasing algorithm and an example FASTA file are available in the GitHub repository (https://github.com/WaveLifeSciences/hd_haplotype_phaser).