Abstract
Bromodomain-containing protein 4 (BRD4), the core component of transcriptional regulatory elements, plays a significant role in tumorigenesis and aggressiveness. However, the mechanisms regulating the functions of BRD4 in bladder cancer (BC) still remain elusive. Herein, we identify one exonic circular RNA (circRNA) generated from NR3C1 gene (circNR3C1) as a regulator of BRD4/C-myc complex. Our previous study indicated that BRD4 and C-myc promoter region form a complex, allowing C-myc to function as a transcription factor for BC progression. In the present study, mechanism studies reveal that circNR3C1 could interact with BRD4 protein, dissociating the formation of BRD4/C-myc complex. In vivo, ectopic expression of C-myc partly reverses the tumorigenesis of xenografts circNR3C1-induced in nude mice. Conclusively, these results demonstrate that circNR3C1 inhibits BC progression through acting as endogenous blocker of BRD4/C-myc complex.
Keywords: bladder cancer, circNR3C1, BRD4, endogenous blocker
Graphical Abstract

circNR3C1 is identified as a novel regulator of BRD4/C-myc complex. Mechanistically, circNR3C1 could interact with BRD4 to inhibit the tumorigenesis of BC through restraining the formation of the BRD4/C-myc complex and transcriptional alteration of target genes associated with C-myc activation.
Introduction
Bladder cancer (BC) is one of the most prevalent malignancies of the urinary system, with 429,000 new cases diagnosed and 165,000 deaths per year worldwide.1 Based on different prognosis, BC is divided into non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC).2,3 However, the recurrence and distant metastasis remains approximately to 50% and 5-year survival rate is only 50%–65%, even when the MIBC patients received radical cystectomy.4 Hence, these restrictions prompted us to search new diagnostic and therapeutic targets to improve the clinical effects of treatment for patients with BC.5,6
Bromodomain-4 protein (BRD4) is a member of the BET (bromodomain and extra-terminal domain) family of proteins, characterized by two tandem bromodomains (BD1 and BD2).7 BET proteins play pivotal roles during embryogenesis and cancer development by reading hyper-acetylated histone regions along the chromatin, regulating gene transcription both at initiation and elongation step.8 Deregulation of BRD4 protein has been widely accepted in hematological and solid malignancies.9, 10, 11 For example, BRD4 has been proved to cooperate with hematopoietic TFs in conjunction with the lysine acetyltransferase p300/CBP to support lineage-specific transcriptional circuits in acute myeloid leukemia.12 BRD4 promotes progression and metastasis of gastric cancer through stabilization of Snail at post-translational levels.13 Our previous work revealed that individual silencing of BRD4 or the BET inhibitor JQ1 strikingly diminished the recruitment of C-myc to EZH2 promoter in BC.14 However, the underlying mechanism of BRD4 recognition of hyper-acetylated chromatin sites (like enhancers, double-stranded breaks [DSBs], or telomeres) in BC remains largely unclear.
Circular RNAs (circRNAs), a new and large class of non-coding RNAs characterized by covalently closed continuous loops without 5′ to 3′ polar or polyadenylation tails, are formed by the back-splicing of exons or introns of parent genes.15,16 circRNAs are tissues or cell lines specifically expressed, evolutionarily conserved, and more stable than the corresponding linear mRNA.17 Exonic circRNAs are predominantly detected in the cytoplasm,18 and some of them regulate the expression of other genes by serving as microRNA (miRNA) sponges to arrest miRNA activities.19 Our previous studies demonstrated that circHIPK3 inhibited migration and invasion of BC cell via acting as miR-558 sponge, and BCRC-3 functioned as a tumor inhibitor to suppress BC cell proliferation through miR-182-5p/p27 axis.20,21 Besides the miRNA sponge mode, circRNAs may interact and sequester with RNA-binding proteins (RBPs) to regulate RNA-protein complex activities. Dysregulated circRNAs have been confirmed to be associated with both normal and pathological conditions, including neurodegenerative diseases, cardiovascular disease, and many types of cancer.22,23 However, the roles of circRNAs with RBP binding potential in BC still remain elusive.
In this study, based on the results of our RNA sequencing (RNA-seq) data and mass spectrometry (MS) analysis, we identify circNR3C1 as a novel regulator of BRD4/C-myc complex. Mechanistically, circNR3C1 physically binds BRD4 to suppress its competitive enrichment on promoter region of C-myc, resulting in transcriptional inhibition of C-myc and subsequent repression of BRD4/C-myc-mediated gene expression. These results indicate the endogenous blocker role of circNR3C1 in BC progression.
Results
BRD4 Is Upregulated in BC Tissues and Cell Lines and Associated with the Expression of circNR3C1 in BC
A previous study evaluated that both the mRNA and protein level of BRD4 were significantly higher in urothelial carcinoma of bladder tissues as compared with that in corresponding normal bladder tissues in our single-center.14 In this study, we analyzed the data from TCGA, including 408 BC tissues and 19 non-cancerous bladder tissues. However, the expression of BRD4 (p = 0.16) between normal and primary tumor had no statistical significance (Figure 1A). Higher levels of BRD4 (p = 0.01) were only observed in neuronal group of bladder urothelial carcinoma (BLCA), while there were no significant differences between other molecular subtypes of BLCA and the normal group (Figure 1B). But Kaplan-Meier survival analysis of this series of 406 BC cases indicated that high BRD4 expression was associated with poor overall survival of patients (Figure 1C). Furthermore, the overexpression of BRD4 was confirmed in BC tissues from GEO database (GSE48276), in which a similar number of normal and tumor cases were included (Figure 1D).24 These results suggest that BRD4 may play an important role in BC progression and its clinical significance needs larger and matched samples in the future.
Figure 1.
The Expression of BRD4 in BC Tissues and Its Correlation with circRNA
(A) The relative expression level of BRD4 was determined in OMICS data (TCGA and MET500). (B) BRD4 expression was evaluated in molecular subtypes of BC. (C) Kaplan-Meier survival plots of 406 well-defined BC cases derived from TCGA database analysis showing the survival probability of patients with high or low expression of BRD4. (D) The relative expression level of BRD4 was determined in GEO database (GSE48276). (E) The correlation between the transcript levels of BRD4 and circHIPK3 by qRT-PCR in BC tissues (n = 54). (F) The correlation between the transcript levels of BRD4 and BCRC-3 by qRT-PCR in BC tissues (n = 54). (G) The correlation between the transcript levels of BRD4 and circNR3C1 by qRT-PCR in BC tissues (n = 54). Data information: Student’s t test and analysis of variance (ANOVA) compared the difference in (A) and (B). Log-rank test for survival comparison in (C). One-way ANOVA for expression comparison in (D). Pearson’s correlation coefficient analysis in (E)–(G). ∗p < 0.05 versus normal group.
In our previous studies, we identified a series of circRNAs according to high-throughput sequence in BC,20 while a few of them, including circNR3C1 (BCRC-1),25 circHIPK3 (BCRC-2),20 and BCRC-3,21 were confirmed to be significantly aberrant expressed in BC tissues compared with normal tissues. We thus investigated the correlation between BRD4 expression and our reported circRNAs (circHIPK3, BCRC-3, circNR3C1) by qRT-PCR in our series of 54 BC cases. As shown in Figures 1E–1G, there is a negative correlation between BRD4 and circNR3C1 (R = −0.2830, p < 0.05), while the expression of other circRNAs had no relationship with BRD4 protein. Together, the relationship between BRD4 and circNR3C1 may contribute to the progress of BC patients.
circNR3C1 Predominantly Interacts with BRD4 in the Nucleus
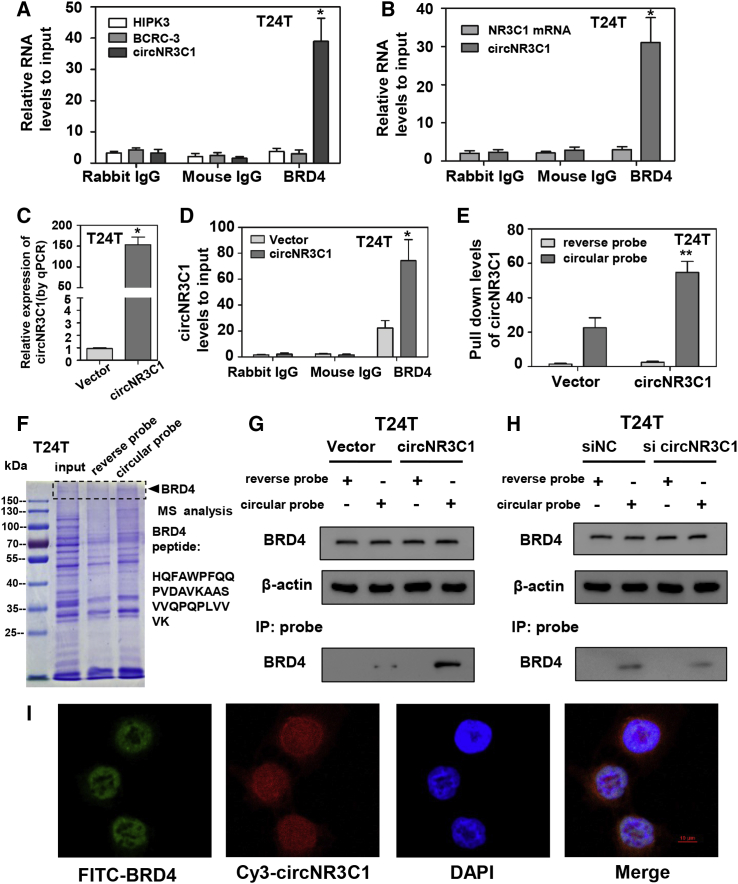
Previously, it was demonstrated that the expressions of circNR3C1 in EJ and T24T cells were the most significantly downregulated in BC cells compared with human immortalized uroepithelium cells (SV-HUC-1).25 To identify the relationship between BRD4 and circRNAs, we prepared cell lysis from T24T and EJ cells. The cell lysates were subject to immunoprecipitation (IP) with anti-rabbit immunoglobulin G (IgG), mouse IgG, and BRD4 (Figure S3A), followed by qRT-PCR with primers specific for the three alternative circRNAs (circHIPK3, BCRC-3, and circNR3C1). The experiment showed that circNR3C1 was pulled down by IP experiments with antibodies against BRD4, which did not pull down other circRNAs (Figure 2A; Figure S1A). In addition, the precipitated mixture was subject to qRT-PCR using primers specific for the linear NR3C1 mRNA or circNR3C1. The results suggested that circNR3C1 was pulled down by antibodies against BRD4, but the linear NR3C1 mRNA was not (Figure 2B; Figure S1B). circNR3C1 overexpression plasmid was stably transfected into T24T or EJ cells with G418 antibiotic selection (Figure 2C; Figure S1C). Cell lysates prepared from the vector- and circNR3C1-transfected cells were subject to IP assay with antibodies. Similarly, we found that only antibodies against BRD4 were able to pull down significantly higher levels of circNR3C1 from the circNR3C1-transfected cells than those from the control cells (Figure 2D; Figure S1D). These observations indicate the interactions between BRD4 and circNR3C1 in BC cells.
Figure 2.
circNR3C1 Interacts with BRD4 in the Nucleus
(A) Cell lysates prepared from T24T cells were incubated with antibodies against rabbit IgG, mouse IgG, and BRD4. The precipitated products were subjected to qRT-PCR with primers amplifying circHIPK3, BCRC-3, circNR3C1. (B) Cell lysates prepared from T24T cells were incubated with antibodies against rabbit IgG, mouse IgG, and BRD4. The precipitated products were subjected to qRT-PCR with primers amplifying circNR3C1 and NR3C1 mRNA. (C) qRT-PCR analysis verified the effective overexpression of circNR3C1 after transfection of the vector for 48 h in T24T cells. (D) Lysates were prepared from T24T cells transfected with circNR3C1 or the vector and incubated with antibodies as above followed by precipitation and qRT-PCR with primers amplifying circNR3C1. (E) Cell lysates prepared from T24T cells transfected with circNR3C1 or the vector were hybridized with circNR3C1 probe or reverse probe for RNA pull-down assays. (F) Coomassie bright blue staining (left) and mass spectrometry (MS, right) assays revealed the proteins pulled down by biotin-labeled probe from the lysates of T24T cells. (G) Cell lysates prepared from T24T cells transfected with circNR3C1 or the vector were subject to immunoprecipitation with BRD4 and were also hybridized with circNR3C1 probe or reverse probe for IP assays with anti-BRD4 antibody, followed by western blotting. (H) Cell lysates prepared from T24T cells transfected with si-circNR3C1 or the mock were subject to immunoprecipitation with BRD4 and were also hybridized with circNR3C1 probe or reverse probe for IP assays with anti-BRD4 antibody, followed by western blotting. (I) RNA-FISH and immunofluorescence staining assay showing the co-localization of circNR3C1 (red) and BRD4 (green) in T24T cells, with the nuclei staining with DAPI (blue). Scale bar, 10 μm. Data information: all values are expressed as mean ± SEM of three experiments. Student’s t test compared the difference. ∗p < 0.01 versus vector, ∗∗p < 0.01 versus reverse probe.
To explore the protein partner of circNR3C1, we subjected cell lysates prepared from BC cells transfected with circNR3C1 or the vector to the RNA pull-down assay. We found that the circular probe significantly pulled down more circNR3C1 than the reverse probe (Figure 2E; Figure S1E). In addition, we performed RNA pull-down assay followed by a proteomic analysis of RNA associated protein complex in T24T cells. Coomassie bright blue staining and MS assays reveal the proteins pulled down by the probe. We found that BRD4 was pulled down by the circular probe but not by the reverse probe (Figure 2F; Figure S1F). In the circNR3C1-transfected cells, the circular probe could pull down more of BRD4 while the equal levels of BRD4 were confirmed by western blotting in these two groups (Figure 2G; Figure S2A). Knockdown of circNR3C1 did not change the expression of BRD4 in T24T cells, while the circular probe could pull down decreased levels of BRD4 in the circNR3C1 small interfering RNA (siRNA)-transfected cells (Figure 2H).
Our previous study suggested that circNR3C1 suppressed cell-cycle progression by directly sponging miR-27a-3p. In order to figure out the relationship between miRNA sponge effect and BDR4 regulatory activity, we extracted cell lysis from T24T cells transfected with anti-miR-27a-3p or the inhibitor-NC. The cell lysates were subject to the RNA pull-down assay. Inhibition of miR-27a-3p did not change the expression of BRD4, while the circular probe pulled down the same level of BRD4 in anti-miR-27a-3p or inhibitor-negative control (NC) transfected cells (Figure S2B). The results indicated that the miRNA sponge effect did not influence BRD4 activity regulated by circNR3C1. Consistently, dual RNA-fluorescence in situ hybridization (FISH) and immunofluorescence assay revealed abundant signals and enrichment of circNR3C1-BRD4 complex in the nucleus of T24T cells (Figure 2I). We used ImageJ to make a quantification analysis of the dual RNA-FISH and immunofluorescence assay26 (Figure S2C). Together, these findings indicated that circNR3C1 could interact with BRD4 in the nucleus.
circNR3C1 Suppresses BRD4-Mediated Transactivation of C-myc
Our previous work has demonstrated that C-myc is an essential downstream regulator of BRD4 in BC cells (Figures S3C, S3D, and S3F).14 To investigate whether circNR3C1 influences the expression level of C-myc, we first detected the expression level of C-myc. As shown in Figures 3A and 3B, the mRNA and protein levels of C-myc were significantly reduced when the circNR3C1 plasmid was transfected into BC cells, but there was no significant effect on the expression of BRD4.
Figure 3.
circNR3C1 Leads to the Suppression of C-myc Transcription
(A) The proteins extracted from circNR3C1 or vector transfected cells were subjected to western blotting to determine the expression of BRD4 and C-myc proteins. β-actin protein was used as loading control. (B) The BRD4 and C-myc mRNA expression were detected by qRT-PCR as shown. β-actin mRNA was regarded as negative control. (C) Dual-luciferase assay indicating the activity of C-myc promoter in T24T cells transfected with shNC, sh-BRD4, vector, or circNR3C1. (D) Cell lysates prepared from T24T cells transfected with shBRD4 or circNR3C1 were subject to immunoprecipitation with BRD4 or rabbit IgG, followed by ChIP assay with primers amplifying C-myc promoter region. (E) The luciferase activities in BRD4 overexpression vector and circNR3C1 co-transfected cells were presented by dual-luciferase assay. (F) The negative correlation between the transcript levels of C-myc and circNR3C1 in BC tissues (n = 54). Data information: all values are expressed as mean ± SEM of three experiments. Student’s t test compared the difference. ∗p < 0.01 versus vector, # p < 0.01 versus shNC, ∗∗p < 0.01 versus empty+ vector, ## p < 0.01 versus empty+circNR3C1. Pearson’s correlation coefficient analysis in (F).
It has been reported that BRD4 could bind directly to C-myc promoter region to facilitate its transcription.10 To further explore the role of circNR3C1 in mediating transcriptional regulation of C-myc by BRD4, we subsequently performed luciferase reporter assays and chromatin IP (ChIP) experiments. As shown in Figure 3C, transfection of circNR3C1 treatment resulted in decreased promoter activity of C-myc by luciferase reporter system, which was similar to the knockdown of BRD4. Besides, ChIP experiments also showed that transfection of circNR3C1 or knockdown of BRD4 lead to considerable loss of BRD4 recruitment to C-myc promoter region (Figure 3D). Overexpression of BRD4 could significantly facilitate C-myc promoter luciferase reporter activity, which was partly abolished by the ectopic expression of circNR3C1 (Figure 3E). In addition, the expression of circNR3C1 and C-myc were negatively correlated with each other by qRT-PCR in our series of 54 BC cases (Figure 3F). Since miRNA sponge activity often occurs in the cytoplasm and BRD4 interaction has been proposed to occur in the nucleus, we thus designed experiments about nucleus/cytoplasm translocation. We first determined the localization of circNR3C1 by preparing the cytoplasmic and nuclear fractions of T24T cells transfected with anti-miR-27a-3p or the inhibitor-NC. Real-time PCR analysis revealed that circNR3C1 was detected both in the nuclei and cytosol. More importantly, the inhibition of miR-27a-3p did not change the distribution of circNR3C1 in T24T cells. On western blotting, we found that suppression of miR-27a-3p had no impact on nucleus/cytoplasm translocation of BRD4 protein (Figure S3B). These results indicated that nucleus/cytoplasm translocation did not occur in course of regulating BRD4 controlled by circNR3C1. Collectively, endogenous interaction between circNR3C1 and BRD4 was noted in BC cells, and circNR3C1 restrained interaction between BRD4 and C-myc promoter region.
circNR3C1 Attenuates the Recruitment of C-myc to EZH2 Promoter in BC
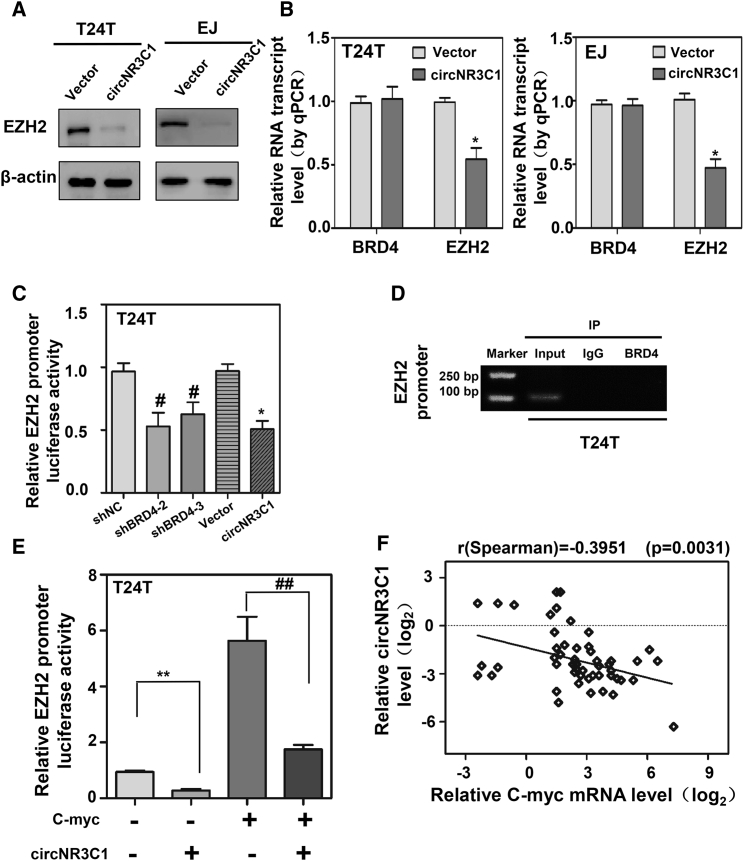
Our previous studies have reported that EZH2 plays an important role in regulating cell biological activity through BRD4/C-myc/EZH2 axis (Figures S3E and S3F). Next, we sought to explore the underlying mechanism of EZH2 regulated by circNR3C1. As evaluated by western blot and qRT-PCR (Figures 4A and 4B), the expression levels of EZH2 mRNA and protein were decreased in BC cells upon transfection of circNR3C1, while little change was found on BRD4.
Figure 4.
circNR3C1 Attenuates the Recruitment of C-myc to EZH2 Promoter
(A) The proteins extracted from circNR3C1 or vector transfected cells were subjected to western blotting to determine the expression of EZH2. β-actin protein was used as loading control. (B) The BRD4 and EZH2 mRNA expression were detected by qRT-PCR as shown. β-actin mRNA was regarded as negative control. (C) Dual-luciferase assay indicating the activity of EZH2 promoter in T24T cells transfected with shNC, sh-BRD4, vector, or circNR3C1. (D) Cell lysates prepared from T24T cells transfected with shBRD4 were subject to immunoprecipitation with BRD4 or rabbit IgG, followed by ChIP assay with primers amplifying EZH2 promoter region. (E) The luciferase activities in C-myc overexpression vector and circNR3C1 co-transfected cells were presented by dual-luciferase assay. (F) The negative correlation between the transcript levels of EZH2 and circNR3C1 in BC tissues (n = 54). Data information: All values are expressed as mean ± SEM of three experiments. Student’s t test compared the difference. ∗p < 0.01 versus vector, # p < 0.01 versus shNC, ∗∗p < 0.01 versus empty+ vector, ## p < 0.01 versus C-myc+vector. Pearson’s correlation coefficient analysis in (F).
C-myc could increase the transcription of EZH2 and circNR3C1 indirectly reduced the expression of C-myc. Instinctively, we detected the relative EZH2 promoter luciferase activity in circNR3C1-overexpressed or BRD4-knockdown cells. As shown in Figure 4C, circNR3C1 gave rise to decreased promoter activity of EZH2 by luciferase reporter system which was same as BRD4-knockdown treatment. ChIP experiments demonstrated once again that BRD4 could not interact with EZH2 promoter region directly (Figure 4D). More importantly, ectopic expression of circNR3C1 inhibited promoter activity of EZH2, which was rescued by overexpression of C-myc (Figure 4E). In addition, qRT-PCR results suggested that the expression of circNR3C1 and EZH2 were negatively correlated in our series of 54 BC cases (Figure 4F). Together, these data indicated that circNR3C1 negatively regulated EZH2 transcription through downregulation of C-myc in BC cells.
circNR3C1 Inhibits Cell Proliferation through Targeting BRD4/C-myc/EZH2 Axis In Vitro and In Vivo
We further investigated the functional interplay between circNR3C1 and BRD4/C-myc/EZH2 axis in regulating the proliferation of BC cells. Flow cytometry indicated that overexpression of circNR3C1 inhibited cell-cycle progression in T24T cells (Figure 5A). Moreover, circNR3C1-induced cell-cycle arrest was partially reversed upon ectopic expression of C-myc and EZH2, respectively (Figure 5A). Cell Counting Kit-8 (CCK-8) results further validated this rescued experiment (Figure 5B; Figure S4A). Besides, transfection of C-myc or EZH2 could partly abolished circNR3C1-induced inhibition of C-myc or EZH2 expression (Figure 5C).
Figure 5.
circNR3C1 Restrains BC Proliferation via BRD4/C-myc/EZH2 Axis In Vitro and In Vivo
(A) Cell-cycle distributions in circNR3C1 overexpression vector and C-myc or EZH2 co-transfected T24T cells were presented by flow cytometry. (B) CCK-8 assay was performed in circNR3C1 overexpression vector and C-myc or EZH2 co-transfected T24T cells for 0 h, 12 h, 24 h, 48 h, 72 h, and 96 h. (C) The proteins extracted from circNR3C1 overexpression vector and C-myc or EZH2 co-transfected T24T cells were subjected to western blotting to determine the expression of C-myc or EZH2. β-actin protein was used as loading control. (D) Representative images in vivo of xenografts formed by subcutaneous injection of T24T cells stably transfected with circNR3C1 overexpression vector and C-myc plasmid into the dorsal flanks of nude mice (n = 5 for each group). Data information: all values are expressed as mean ± SEM of three experiments. Student’s t test compared the difference. ∗p < 0.01 versus empty+ vector, # p < 0.01 versus C-myc+ vector, ##p < 0.01 versus EZH2+ vector.
To further test the antitumor potential of circNR3C1 in vivo, we constructed T24T cells stably transfected with vector or circNR3C1 and those co-transfected with empty or C-myc overexpression plasmid. circNR3C1 injected mice showed a reduction in tumor weight and tumor volume compared with their control groups (p < 0.05, Figure 5D; Figures S4B–S4D). In addition, ectopic expression of C-myc partly abolished the antitumor effect of circNR3C1 in T24T cells (p < 0.05, Figure 5D; Figures S4B–S4D). Taken together, these results indicated that circNR3C1 suppressed BC progression through inhibiting BRD4/C-myc/EZH2 axis.
Discussion
Recent studies show that many circRNAs function as miRNA sponges or endogenous competing RNAs (ceRNAs) due to their miRNA binding sites.19 Previously, we reported that circNR3C1 possessed four targeting sites of miR-27a-3p and could effectively sponge miR-27a-3p to decrease the expression of cyclin D1.25 Alternatively, circRNAs may also exert functions in multiply ways, such as binding to proteins or translating into peptides. For example, oncogenic circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes.22 circRNA circ-Ccnb1 can inhibit the formation of the Ccnb1-Cdk1 complex and modulate nuclear translocation of these two molecules, thus inhibiting tumor progression.23 In this study, we identify a novel role for circNR3C1. On the basis of the abnormal high expression of BRD4, we applied RNA IP (RIP) and RNA pull-down assay to confirm the interaction between circNR3C1 and BRD4. Additionally, circNR3C1 hampers the formation of BRD4/C-myc promoter complex, affecting the expression of downstream gene. This role could be shared by other nuclear circRNAs. In this regard, our evidence demonstrates a novel action mode of circNR3C1 in BC progression.
BRD4, the most extensively studied member of the BET family, is an epigenetic regulator that recruits transcriptional regulatory complexes to acetylated chromatin.27 The BRD4/P-TEFb interaction is particularly important for rapid transcriptional induction, as occurs at mitotic exit and in response to signal-dependent activation of DNA-binding transcription factors (TFs).28 In addition, the ET domain of BRD4 has been related to transcriptional regulation by interacting with several histone modifiers, including the arginine demethylase Jmjd6 and the lysine methyltransferase Nsd3.29,30 Our previous work also demonstrated that BRD4 occupied similar sites across the C-myc genome and provided an interface between TFs and the transcriptional apparatus. Notably, EZH2 is not only a critical epigenetic repressor through histone methylation, but also an activator of gene expression through different pathways.31 Further, it can be regulated post-transcriptionally by the interaction with many miRNAs and long non-coding RNAs.32 Previous study demonstrated the therapeutic potential of targeting the antagonistic counterpart of EZH2 in BC.33 It is suggested that exogenous expression of lowly expressed circRNA can be performed by gene therapy where DNA cassettes designed for circRNA expression are delivered.34 Whether this method play a similar consequence in BC treatment and confer sensitivity toward EZH2 inhibition or EZH2 inhibitor-based combination therapy remains an area for further investigation. In this study, our results indicate that the competitive binding capacity of circNR3C1 dissociates BRD4/C-myc promoter complex. Nevertheless, our work will be more persuasive with the addition of molecular work, including circNR3C1-knockin and knockout studies, BRD4 or C-myc-knockin and knockout studies. The interactions between BRD4 and circNR3C1 probably change the structure of BRD4 protein and might affect non-transcriptional transversal function of BRD4, like DNA damage repair, checkpoint activation, or telomere homeostasis,8,35 with the underlying mechanisms need further exploration.
Given the multiple disease-related functions of BRD4 and well-proven concept of disrupting the BRD4−acetyl-lysine interactions as a therapeutic target, significant efforts have thus been made to develop BRD4 inhibitors from both pharmaceutical and academic settings. One of the BETi molecules was JQ1, which was initially described to be a potent suppressor of NUT midline carcinoma, B cell lineage malignancies, and other tumor entities.36,37 For example, a remarkable decrease of tumor size upon JQ1 administration was observed in a subcutaneous xenograft model of PDAC through blocking acinar-to-ductal metaplasia in the pancreas.38 In addition, JQ1 has also been demonstrated to suppress tumor cell growth specifically in colon cancers that are characterized by a CpG island methylator phenotype (CIMP).39 In our study, the interactions between BRD4 and circNR3C1 show inhibitory effects on BRD4 both in vitro and in vivo with selectivity reported at transcriptional level. Based on these findings, it is conceivable that circNR3C1 play its role in cancer progression as endogenous blocker, which shared the same effect as BETi molecules (JQ1). However, prospective clinical trials need to consider the promising effects of circRNA in preclinical BC models and have to validate whether this therapeutic strategy can be translated into the clinic.
In conclusion, it is demonstrated for the first time that circNR3C1 could interact with BRD4 to inhibit the tumorigenesis of BC through restraining the formation of the BRD4/C-myc complex and transcriptional alteration of target genes associated with C-myc activation. This study extends our knowledge about the regulation of BRD4/C-myc function by circRNA and proposes circNR3C1 as a potential endogenous blocker for curative management of BC.
Materials and Methods
Patient Tissue Specimens and Cell Lines
Tumor tissues and their adjacent normal tissues were obtained from patients suffering radical cystectomy for urothelial carcinomas of bladder at Union Hospital of Tongji Medical College. Patients with a history of preoperative chemotherapy or radiotherapy were excluded. The approval from the Institutional Review Board of Huazhong University of Science and Technology (Wuhan, China) was obtained before we collected the samples. All tissues were classified according to the 2004 World Health Organization Consensus Classification and Staging System for bladder neoplasms. The specimens were stored in liquid nitrogen. The human BC cell line T24T was provided by Dr. Dan Theodorescu (Departments of Urology, University of Virginia, Charlottesville, VA, USA) as described in our previous studies.21 Human BC cell lines EJ was purchased from American Type Culture Collection (ATCC, USA). Cells were cultured in RPMI-1640 medium (GIBCO) containing 10% fetal bovine serum (GIBCO) and 1% penicillin/streptomycin (GIBCO) at 37°C supplied with 5% CO2.
Compliance with Ethics Guidelines
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study. Additional informed consent was obtained from all patients for which identifying information is included in this article. All institutional and national guidelines for the care and use of laboratory animals were followed.
RNA Extraction, RNase R Treatment, and PCR Assays
Nuclear and cytoplasmic RNA was extracted according to the instruction of RNA Subcellular Isolation Kit (Active Motif, Carlsbad, CA, USA). Total RNA from tissues and cell lines was isolated using RNeasy Mini Kit (QIAGEN, Germany) following the manufacturer’s instructions. Reverse transcription and real-time PCR were performed using reverse transcription kit PrimeScript RT Master Mix (Takara, Dalian, China), the SYBR Premix Ex TaqTM kit (Takara), and primers shown in Table S1. The results of transcript levels were analyzed by 2−ΔΔCt method.
Western Blotting Analysis
Cellular protein was extracted in RIPA lysis buffer (Thermo Scientific) and determined using BCA Protein assay kit (Beyotime). Western blot was performed as previously described,40 with antibodies specific for β-actin (ab8226), BRD4 (ab128874), C-myc (ab32072), EZH2 (ab191080), PCNA (ab18197), α-Tubulin (ab7291).
Plasmids Construction and Stable Transfection
The human circNR3C1 plasmid was constructed and used as our previous study described.25 The C-myc and EZH2 promoter luciferase reporter vectors, ectopic vectors targeting BRD4, C-myc, and EZH2 were synthesized in our previous research.14 Oligonucleotides specific for short hairpin RNAs (shRNAs) against circNR3C1 or BRD4 (Table S1) were inserted into lentiviral vector GV298 (Genechem, Shanghai, China). Cell lines were transfected using Lipofectamine 2000 (Life Technologies, USA) according to the manufacturer’s instructions. After selection for neomycin or puromycin (Invitrogen) resistance, stable cell lines were obtained.
RIP
RIP assay was performed according to the instructions of Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore), with antibodies specific for BRD4 (ab128874), rabbit IgG (ab172730), and mouse IgG (ab190475). Co-precipitated RNAs were detected by qRT-PCR with specific primers (Table S1). Total RNAs (input) and antibody (rabbit IgG or mouse IgG) were applied as controls.
Biotin-Labeled RNA Pull-Down and MS Analysis
The biotinylated probe of circNR3C1 was synthesized by TSINGKE (Beijing, China). The sequence of the probe was listed in Table S1. In brief, lysates of 1 × 107 BC cells were incubated with biotin-labeled linear or circular probe for 4 h and treated with appropriate Streptavidin C1 magnetic beads (Invitrogen) for 30 min. After washed thoroughly with wash buffer for three times, the retrieved protein was detected by western blot or MS analysis at Wuhan Institute of Biotechnology (Wuhan, China).
FISH
Cy3-labeled circNR3C1 probes were obtained from RiboBio (Guangzhou, China). RNA hybridizations were carried out using FISH Kit (RiboBio, China) according to the manufacturer’s instructions. After that, cancer cells on coverslips were incubated with 5% milk for 1 h and treated with antibodies specific for BRD4 (ab128874, 1:100 dilution) at room temperature for 2 h. Then, coverslips were treated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (1:2,000 dilution) and 4',6-diamidino-2-phenylindole (DAPI) (300 nmol/L) staining. All fluorescence images were captured using Nikon A1Si Laser Scanning Confocal Microscope (Nikon Instruments, Japan).
Luciferase Reporter Assays
The C-myc and EZH2 promoter reporter vectors were constructed and used as our previous study.14 The reporter promoters were transiently transfected along with Renilla control plasmid, concomitantly with BRD4 shRNA, circNR3C1, C-myc, or BRD4 ectopic vectors. The luciferase activities were measured following dual luciferase reporter assay detection kit (Promega, WI, USA) as described after 24 h.21
ChIP Assay
The ChIP assay was performed using the EZ-ChIP kit (Upstate Biotechnology, Lake Placid, NY, USA). Chromatin DNA was immunoprecipitated with corresponding antibody or normal IgG washed, and the DNA-protein cross-links were subsequently reversed. PCR primers for the C-myc and EZH2 promoter were listed in Table S1. The recovered DNA was assessed by PCR analysis. The relative amount of immunoprecipitated DNA was evaluated by generating a standard and normalized against the negative control.
Flow Cytometry Assay for the Cell Cycle
T24T cells transfected with the plasmids were harvested and stained with propidium iodide buffer (BD PharMingen) for cell-cycle analysis. The results were analyzed by the ModFit LT software.
CCK-8 Assay
The cell viability was detected by CCK-8 assay (Dojin, Japan). Cells were starved in medium with 0.1% fetal bovine serum for 24 h. Next, approximately 2 × 103 cells were evenly seeded in 96-well plates in triplicate. The optical density (OD) value at 450 nm was measured using the SpectraMax M5 microplate reader (MD, USA) at 1, 2, 3, 4, and 5 days, respectively. CCK-8 solution (10 μL) was added into each well and incubated at 37°C for 2.5 h. The results were calculated using GraphPad Prism 6.0 software (La Jolla, USA) statistics analysis.
Tumor Xenografts
We chose 4-week-old female BALB/c nude mice for tumor xenografts experiments. T24T cell transfected plasmids were subcutaneously injected into the upper back of the nude mice (3 × 106, 200 μL). Mice were sacrificed and detected for tumor weight and gene expression after 1 month. All procedures were approved by the Animal Care Committee of Tongji Medical College.
Statistical Analysis
All data were indicated as means ± standard error of the mean (SEM) processed by GraphPad Prism 5.0 (La Jolla, USA). Student’s t test, chi-square, or ANOVA (p < 0.05) was used to evaluate the group difference. Pearson’s correlation coefficient assay was used to analyze the expression correlation. Log-rank test was used to assess survival difference.
Author Contributions
F.X. performed primers design, western blots experiments, data analysis, and circRNA pull-down. X.X. and D.T. contributed FISH, flow cytometry assay, CCK-8, and animal experiments. C.H. and L.W. collected and classified the human BC tissue samples. F.L. and H.Z. contributed to ChIP, RIP, and qRT-PCR. F.X., H.N., and G.J. wrote the paper. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 81874091, 81772724, 81702524, 81974396, and 81672529).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.09.016.
Contributor Information
Haitao Niu, Email: niuht0532@126.com.
Guosong Jiang, Email: jiangguosongdoc@hotmail.com.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Kamoun A., Reyniès A., Allory Y., Sjödahl G., Gordon Robertson A., Seiler R. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2019;77:420–433. doi: 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasad S.M., Decastro G.J., Steinberg G.D., Medscape Urothelial carcinoma of the bladder: definition, treatment and future efforts. Nat. Rev. Urol. 2011;8:631–642. doi: 10.1038/nrurol.2011.144. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z.L., Dong P., Li Y.H., Liu Z.W., Yao K., Han H., Qin Z.K., Zhou F.J. Radical cystectomy for bladder cancer: oncologic outcome in 271 Chinese patients. Chin. J. Cancer. 2014;33:165–171. doi: 10.5732/cjc.012.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriegmair M.C., Rother J., Grychtol B., Theuring M., Ritter M., Günes C. Multiparametric Cystoscopy for Detection of Bladder Cancer Using Real-time Multispectral Imaging. Eur. Urol. 2019;77:251–259. doi: 10.1016/j.eururo.2019.08.024. [DOI] [PubMed] [Google Scholar]
- 6.Lotan Y., Boorjian S.A., Zhang J., Bivalacqua T.J., Porten S.P., Wheeler T., Lerner S.P., Hutchinson R., Francis F., Davicioni E. Molecular Subtyping of Clinically Localized Urothelial Carcinoma Reveals Lower Rates of Pathological Upstaging at Radical Cystectomy Among Luminal Tumors. Eur. Urol. 2019;76:200–206. doi: 10.1016/j.eururo.2019.04.036. [DOI] [PubMed] [Google Scholar]
- 7.Donati B., Lorenzini E., Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol. Cancer. 2018;17:164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S., Pike A.M., Lee S.S., Strong M.A., Connelly C.J., Greider C.W. BRD4 inhibitors block telomere elongation. Nucleic Acids Res. 2017;45:8403–8410. doi: 10.1093/nar/gkx561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozer H.G., El-Gamal D., Powell B., Hing Z.A., Blachly J.S., Harrington B., Mitchell S., Grieselhuber N.R., Williams K., Lai T.H. BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor. Cancer Discov. 2018;8:458–477. doi: 10.1158/2159-8290.CD-17-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ba M., Long H., Yan Z., Wang S., Wu Y., Tu Y., Gong Y., Cui S. BRD4 promotes gastric cancer progression through the transcriptional and epigenetic regulation of c-MYC. J. Cell. Biochem. 2018;119:973–982. doi: 10.1002/jcb.26264. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y., Wang Y., Diao P., Zhang W., Li J., Ge H., Song Y., Li Z., Wang D., Liu L. Therapeutic Targeting of BRD4 in Head Neck Squamous Cell Carcinoma. Theranostics. 2019;9:1777–1793. doi: 10.7150/thno.31581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan S.H., Tang Y., Miao L., Darwich-Codore H., Vejnar C.E., Beaudoin J.D., Musaev D., Fernandez J.P., Benitez M.D.J., Bazzini A.A. Brd4 and P300 Confer Transcriptional Competency during Zygotic Genome Activation. Dev. Cell. 2019;49:867–881.e8. doi: 10.1016/j.devcel.2019.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin Z.-Y., Wang T., Su S., Shen L.-T., Zhu G.-X., Liu Q., Zhang L., Liu K.W., Zhang Y., Zhou Z.H. BRD4 Promotes Gastric Cancer Progression and Metastasis through Acetylation-Dependent Stabilization of Snail. Cancer Res. 2019;79:4869–4881. doi: 10.1158/0008-5472.CAN-19-0442. [DOI] [PubMed] [Google Scholar]
- 14.Wu X., Liu D., Tao D., Xiang W., Xiao X., Wang M., Wang L., Luo G., Li Y., Zeng F., Jiang G. BRD4 Regulates EZH2 Transcription through Upregulation of C-MYC and Represents a Novel Therapeutic Target in Bladder Cancer. Mol. Cancer Ther. 2016;15:1029–1042. doi: 10.1158/1535-7163.MCT-15-0750. [DOI] [PubMed] [Google Scholar]
- 15.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 17.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S., Hou J., Zhang H., Wu Y., Hu M., Zhang L., Xu J., Na R., Jiang H., Ding Q. The evaluation of the risk factors for non-muscle invasive bladder cancer (NMIBC) recurrence after transurethral resection (TURBt) in Chinese population. PLoS ONE. 2015;10:e0123617. doi: 10.1371/journal.pone.0123617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 20.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C., Liu D., Wang M., Wang L., Zeng F., Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie F., Li Y., Wang M., Huang C., Tao D., Zheng F., Zhang H., Zeng F., Xiao X., Jiang G. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol. Cancer. 2018;17:144. doi: 10.1186/s12943-018-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang L., Du W.W., Awan F.M., Dong J., Yang B.B. The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett. 2019;459:216–226. doi: 10.1016/j.canlet.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Choi W., Porten S., Kim S., Willis D., Plimack E.R., Hoffman-Censits J., Roth B., Cheng T., Tran M., Lee I.L. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng F., Wang M., Li Y., Huang C., Tao D., Xie F., Zhang H., Sun J., Zhang C., Gu C. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 2019;460:139–151. doi: 10.1016/j.canlet.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 26.Zinchuk V., Wu Y., Grossenbacher-Zinchuk O. Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci. Rep. 2013;3:1365. doi: 10.1038/srep01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gobbi G., Donati B., Do Valle I.F., Reggiani F., Torricelli F., Remondini D., Castellani G., Ambrosetti D.C., Ciarrocchi A., Sancisi V. The Hippo pathway modulates resistance to BET proteins inhibitors in lung cancer cells. Oncogene. 2019;38:6801–6817. doi: 10.1038/s41388-019-0924-1. [DOI] [PubMed] [Google Scholar]
- 28.Shi J., Wang Y., Zeng L., Wu Y., Deng J., Zhang Q., Lin Y., Li J., Kang T., Tao M. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A., Rosenfeld M.G. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q., Zeng L., Shen C., Ju Y., Konuma T., Zhao C., Vakoc C.R., Zhou M.M. Structural Mechanism of Transcriptional Regulator NSD3 Recognition by the ET Domain of BRD4. Structure. 2016;24:1201–1208. doi: 10.1016/j.str.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deb G., Singh A.K., Gupta S. EZH2: not EZHY (easy) to deal. Mol. Cancer Res. 2014;12:639–653. doi: 10.1158/1541-7786.MCR-13-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez-Fernández M., Rubio C., Segovia C., López-Calderón F.F., Dueñas M., Paramio J.M. EZH2 in Bladder Cancer, a Promising Therapeutic Target. Int. J. Mol. Sci. 2015;16:27107–27132. doi: 10.3390/ijms161126000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ler L.D., Ghosh S., Chai X., Thike A.A., Heng H.L., Siew E.Y., Dey S., Koh L.K., Lim J.Q., Lim W.K. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci. Transl. Med. 2017;9:eaai8312. doi: 10.1126/scitranslmed.aai8312. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y.G., Kim M.V., Chen X., Batista P.J., Aoyama S., Wilusz J.E., Iwasaki A., Chang H.Y. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol. Cell. 2017;67:228–238. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanlie A., Yousif A.S., Akiyama H., Honjo T., Begum N.A. Chromatin reader Brd4 functions in Ig class switching as a repair complex adaptor of nonhomologous end-joining. Mol. Cell. 2014;55:97–110. doi: 10.1016/j.molcel.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 36.Chapuy B., McKeown M.R., Lin C.Y., Monti S., Roemer M.G.M., Qi J., Rahl P.B., Sun H.H., Yeda K.T., Doench J.G. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stathis A., Zucca E., Bekradda M., Gomez-Roca C., Delord J.-P., de La Motte Rouge T., Uro-Coste E., de Braud F., Pelosi G., French C.A. Clinical Response of Carcinomas Harboring the BRD4-NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer Discov. 2016;6:492–500. doi: 10.1158/2159-8290.CD-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller A.L., Fehling S.C., Garcia P.L., Gamblin T.L., Council L.N., van Waardenburg R.C.A.M., Yang E.S., Bradner J.E., Yoon K.J. The BET inhibitor JQ1 attenuates double-strand break repair and sensitizes models of pancreatic ductal adenocarcinoma to PARP inhibitors. EBioMedicine. 2019;44:419–430. doi: 10.1016/j.ebiom.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCleland M.L., Mesh K., Lorenzana E., Chopra V.S., Segal E., Watanabe C., Haley B., Mayba O., Yaylaoglu M., Gnad F., Firestein R. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Invest. 2016;126:639–652. doi: 10.1172/JCI83265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo G., Liu D., Huang C., Wang M., Xiao X., Zeng F., Wang L., Jiang G. LncRNA GAS5 Inhibits Cellular Proliferation by Targeting P27Kip1. Mol. Cancer Res. 2017;15:789–799. doi: 10.1158/1541-7786.MCR-16-0331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.