Abstract
The draft genome sequence of Caldanaerobacter sp. strain 1523vc, a thermophilic bacterium, isolated from a hot spring of Uzon Caldera, (Kamchatka, Russia) is presented. The complete genome assembly was of 2 713 207 bp with predicted completeness of 99.38%. Genome structural annotation revealed 2674 protein-coding genes, 127 pseudogenes and 77 RNA genes. Pangenome analysis of 7 currently available high quality Caldanaerobacter spp. genomes including 1523vc revealed 4673 gene clusters. Of them, 1130 clusters formed a core genome of genus Caldanaerobacter. Of the rest 3543 Caldanaerobacter pangenome genes, 385 were exclusively represented in 1523vc genome. 101 of 2801 Caldanaerobacter CDS were found to be encoding carbohydrate-active enzymes (CAZymes). The majority of CAZymes were predicted to be involved in degradation of beta-linked polysaccharides as chitin, cellulose and hemicelluloses, reflecting the metabolism of strain 1523vc, isolated on cellulose. 5 of 101 CAZyme genes were found to be unique for the strain 1523vc and belonged to GH23, GT56, GH15 and two CE9 family proteins.
The draft genome of strain 1523vc was deposited at DBJ/EMBL/GenBank under the accessions JABEQB000000000, PRJNA629090 and SAMN14766777 for Genome, Bioproject and Biosample, respectively.
Keywords: Thermophiles, CAZymes, Caldanaerobacter, Genome, Extremophiles
Specifications Table
| Subject | Biology, Microbiology |
|---|---|
| Specific subject area | Microbial biotechnology |
| Type of data | Genomic sequence, predicted genes and functional analysis of respective proteins |
| How data was acquired | De novo whole genome sequencing Instrument: Illumina MiSeq |
| Data format | Raw data: annotated draft genome assembly; Secondary data: table of orthologous gene clusters of Caldanaerobacter representatives; table of average nucleotide indentity between Caldanaerobacter genomes |
| Parameters for data collection | Thermophilic anaerobic pure culture cultivation. Extraction of genomic DNA from a pure culture, fragment library preparation, Illumina sequencing, de novo assembly and annotation procedures |
| Description of data collection | Extraction of genomic DNA was performed with ISOLATE II Genome DNA kit (Bioline, UK); fragment library was prepared with NEBNext Ultra kit; sequencing was performed with Illumina MiSeq™ system. The genome was assembled using Unicycler and annotated with NCBI PGAP web server |
| Data source location | The culture of strain 1523vc is deposited in extremophiles metabolism laboratory collection at Federal Research Center “Biotechnology” RAS (Moscow, Russian Federation) |
| Data accessibility | Raw data is publicly available at NCBI Genbank.The Biosample, Bioproject and assembly/WGS accession numbers are: SAMN14766777 (https://www.ncbi.nlm.nih.gov/biosample/SAMN14766777) PRJNA629090 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA629090) https://www.ncbi.nlm.nih.gov/bioproject/PRJNA511806) and JABEQB000000000 RZHM00000000 (https://www.ncbi.nlm.nih.gov/nuccore/JABEQB000000000), https://www.ncbi.nlm.nih.gov/nuccore/RZHM00000000), respectively. Secondary data is available as Supplementary Table 1 and 2. |
| Related research article | Kublanov IV, Perevalova AA, Slobodkina GB, et al. Biodiversity of thermophilic prokaryotes with hydrolytic activities in hot springs of Uzon Caldera, Kamchatka (Russia). Appl Environ Microbiol. 2009;75(1):286‐291. doi:10.1128/AEM.00607-08 |
Value of the Data
-
•
Genome data for Caldanaerobacter sp. 1523vc can be used for genome-based phylogenetic and evolutionary analysis of Caldanaerobacter genus
-
•
385 of 3543 Caldanaerobacter pangenome genes were found to be represented exclusively in strain 1523vc genome. Among them are several carbohydrate-active enzymes (CAZymes, http://www.cazy.org) attributed to GH23, GT56 and GH15 and two CE9 family proteins, which can be further explored by biotechnologists using heterologous expression and activity analysis
-
•
The genome encodes a high number of CAZymes, participating in degradation of various beta-glucans, which could be relevant to various applications, including 2nd generation bioethanol production, as well as pulp and food industries. Genomic data, presented in this article unlock the coding potential of strain 1523vc for further biochemical analysis of its enzymes in the scope of biotechnological applications
1. Data Description
Caldanaerobacter is a genus of Firmicutes phylum, which was proposed by Fardeau et al., in 2004 upon isolation of two thermophilic bacterial strains and reclassification of three species, formerly representing the genus Thermoanaerobacter as well as Carboxydibrachium pacificum [1]. Later, a second species of the genus was proposed by Kozina and co-authors in 2010 [2]. The members of the genus are Gram-positive thermophilic strictly anaerobic chemoorganoheterotrophic bacteria, growing on carbohydrates and proteinaceous substrates. Among the biopolymers, known to be hydrolyzed by the genus members are xylan, starch and agarose [1], [2] as well as keratins [3], [4].
Strain 1523vc was isolated from an in situ enrichment culture proliferating on a linen rope in a 70°C hot spring, and it is a first Caldanaerobacter representative, capable of growing on microcrystalline and carboxymethyl cellulose [4].
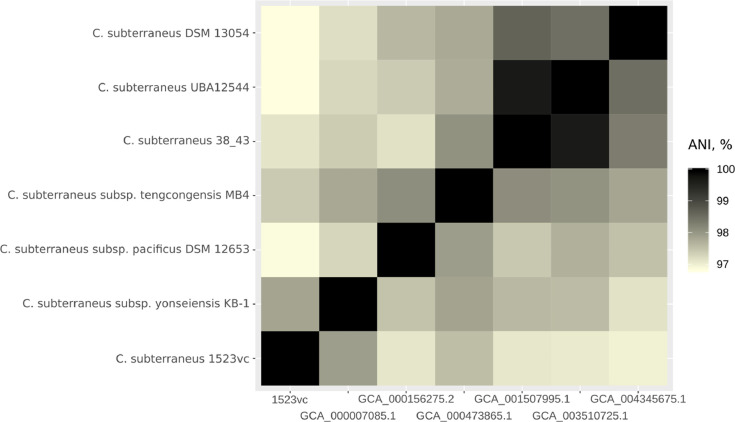
Strain 1523vc genome was sequenced using Illumina MiSeq™ platform. The complete genome assembly was of 2 713 207 bp with GC-content of 37.2 mol%. Completeness of the assembly was estimated to be 99.38%. Analysis of average nucleotide identity of 1523vc and genomes of Caldanaerobacter spp. (Fig. 1, Supplementary Table 2) showed that strain 1523vc is closely related to C.subterraneus subsp. yonseiensis, which was also isolated from a geothermal hot spring [1], [5].
Fig. 1.
Heatmap of average nucleotide identity values between Caldanaerobacter strains, calculated by ani.rb script of enveomics package [10]. Strain IDs are shown at vertical axis, corresponding NCBI Assembly IDs are shown on horizontal axis. ANI data, used for this heatmap are presented in Supplementary Table 2.
Genome annotation with NCBI Prokaryotic Genome Annotation Pipeline [6] revealed 2801 protein-coding sequences including 2674 CDSs, 127 pseudogenes, and 77 RNAs genes. Public genomic databases (NCBI, IMG) contain six high-quality Caldanaerobacter genome assemblies. Pangenome analysis of the seven Caldanaerobacter spp. genomes (including 1523vc) using ProteinOrtho [7] revealed 4673 gene clusters (Supplementary Table 1). Of them, 1130 clusters formed a Caldanaerobacter core genome. Of the rest 3543 Caldanaerobacter pangenome genes, 385 were exclusively represented in 1523vc genome. 92 of these genes were located in laterally acquired gene islands, detected by IslandViewer 4 [8].
101 of 2801 Caldanaerobacter CDS were found to be encoding CAZymes – the proteins, that degrade, modify, or create glycosidic bonds [9]. The most numerous families were CE9, CE14, CBM50, GH109, GT4. The majority of CAZymes were involved in degradation of beta-linked polysaccharides as chitin, cellulose and hemicelluloses reflecting the metabolism of strain 1523vc, isolated and growing on various cellulose substrates [4]. Of 101 CAZyme-related genes 5 glycozide hydrolazes and carbohydrate esterases were found to be unique for the strain 1523vc: HKI81_01480 and HKI81_01510 (CE9, adenine deaminase), HKI81_04210 (GH23, transglycosylase SLT domain-containing protein), HKI81_12285 (GT56, 4-alpha-L-fucosyltransferase) and HKI81_13925 (GH4, alpha-gluco/galactosidase). Thus, relatively small number of CAZymes, specific for strain 1523vc suggests consistent set of CAZymes within the Caldananaerobacter genus and hence, comparable capabilities to degrade polysaccharides within the genus members. Indeed, 30 CAZymes were found to be encoded by the Caldanaerobacter core genome genes, among which there were families with known cellulase (GH5), amylase (GH13), chitinase (GH18), lysozyme (GH23) and mannan phosphorylase (GH94) activities.
The draft genome of strain 1523vc was deposited at DBJ/EMBL/GenBank under the accessions JABEQB000000000, PRJNA629090 and SAMN14766777 for Genome, Bioproject and Biosample, respectively.
2. Experimental Design, Materials, and Methods
2.1. Strain isolation and deposition into collection
Strain 1523vc isolation procedure was described previously [4]. The strain is maintained in the extremophiles metabolism laboratory (Winogradsky Institute of Microbiology, now a part of FRC “Biotechnology”, RAS) collection by annual transfer on the medium, described previously [4]. For genomic sequencing one liter of the same medium was prepared, and strain 1523vc was cultivated in its optimal growth conditions. The grown cells were harvested by centrifugation at 12000 g.
2.1. DNA extraction, library preparation and sequencing
Genomic DNA was isolated using ISOLATE II Genome DNA kit (Bioline, UK). Fragmentation of genomic DNA was performed with Bioruptor™ sonicator (Diagenode, Belgium) to achieve an average fragment length of 400 bp. Further steps of library preparation were performed with NEBNext® Ultra™ fragment library kit (New England BioLabs) according to the manufacturer's instructions. Bead-based size-selection was performed to get fragment sizes in the range of 300–500 bp. Sequencing was done with Illumina MiSeq™ platform (Illumina, USA) using 300 cycles paired-end sequencing reagents. 1,600,832 read pairs were obtained from the sequencing run.
2.2. De novo assembly
Raw sequencing reads were trimmed by quality with CLC Genomics Workbench v. 10.0.1 (Qiagen, Germany). Adapter sequences were trimmed with SeqPrep tool (https://github.com/jstjohn/SeqPrep). Finally, 1,462,277 read pairs were used for the assembly. Genome was assembled with Unicycler v.0.4.8 [11]. Genome completeness and contamination were assessed with CheckM [12] using Thermoanaerobacteraceae-specific marker set.
2.3. Genome annotation and analysis
Genome was annotated with NCBI PGAP [6].Average nucleotide identity (ANI) was calculated using ani.rb script (https://github.com/lmrodriguezr/enveomics) [10]. ANI heatmap was plotted using ggplot2 library for R [13].CAZymes [9] were searched using hmmscan [14] in dbCAN v. 2.0 [15] followed by manual verification using hmmscan and Pfam databases [16].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgements
Genome assembly and analysis was supported by the NRC “Kurchatov institute” (internal grant #1360 from 25.06.2019 “Genomes of industrially-relevant microorganisms”). Sequencing was supported by a grant from Ministry of Science and Higher Education of Russian Federation allocated to the Kurchatov Center for Genome Research (grant 075-15-2019-1659).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.106336.
Appendix. Supplementary materials
References
- 1.Fardeau M.L., Bonilla Salinas M., L'Haridon S., Jeanthon C., Verhé F., Cayol J.L., Patel B.K.C., Garcia J.L., Ollivier B. Isolation from oil reservoirs of novel thermophilic anaerobes phylogenetically related to Thermoanaerobacter subterraneus: reassignment of T. subterraneus, Thermoanaerobacter yonseiensis, Thermoanaerobacter tengcongensis and Carboxydibrachium pacificum toCaldanaerobacter subterraneus gen. nov., sp. nov., comb. nov. as four novel subspecies. Int. J. Syst. Evol. Microbiol. 2004;54:467–474. doi: 10.1099/ijs.0.02711-0. [DOI] [PubMed] [Google Scholar]
- 2.Kozina I.V, Kublanov I.V, Kolganova T.V, Chernyh N.A., Bonch-Osmolovskaya E.A. Caldanaerobacter uzonensis sp. nov., an anaerobic, thermophilic, heterotrophic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2010;60:1372–1375. doi: 10.1099/ijs.0.012328-0. [DOI] [PubMed] [Google Scholar]
- 3.Riessen S., Antranikian G. Isolation of Thermoanaerobacter keratinophilus sp. nov., a novel thermophilic, anaerobic bacterium with keratinolytic activity. Extremophiles. 2001;5:399–408. doi: 10.1007/s007920100209. [DOI] [PubMed] [Google Scholar]
- 4.Kublanov I.V, Perevalova A.A., Slobodkina G.B., Lebedinsky A.V, Bidzhieva S.K., Kolganova T.V, Kaliberda E.N., Rumsh L.D., Haertlé Thomas, Elizaveta A.B.O. Biodiversity of thermophilic prokaryotes with hydrolytic activities in hot springs of Uzon Сaldera, Kamchatka (Russia) Appl. Environ. Microbiol. 2009;75:286–291. doi: 10.1128/AEM.00607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim B.C., Grote R., Lee D.W., Antranikian G., Pyun Y.R. Thermoanaerobacter yonseiensis sp. nov., a novel extremely thermophilic, xylose-utilizing bacterium that grows at up to 85°C. Int. J. Syst. Evol. Microbiol. 2001;51:1539–1548. doi: 10.1099/00207713-51-4-1539. [DOI] [PubMed] [Google Scholar]
- 6.Tatusova T., Dicuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lechner M., Findeiß S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (Co-)orthologs in large-scale analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertelli C., Laird M.R., Williams K.P., Lau B.Y., Hoad G., Winsor G.L., Brinkman F.S.L. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.L. Rodriguez-R, K. Konstantinidis, The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes, (2016). 10.7287/peerj.preprints.1900v1. [DOI]
- 11.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. ISBN 978-3-319-24277-4. [Google Scholar]
- 14.Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H., Yohe T., Huang L., Entwistle S., Wu P., Yang Z., Busk P.K., Xu Y., Yin Y. DbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A., Sonnhammer E.L.L., Hirsh L., Paladin L., Piovesan D., Tosatto S.C.E., Finn R.D. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.