Abstract
Background
miRNAs are considered important players in oncogenesis, serving either as oncomiRs or suppressormiRs. Although the accumulation of somatic alterations is an intrinsic aspect of cancer development and many important cancer-driving mutations have been identified in protein-coding genes, the area of functional somatic mutations in miRNA genes is heavily understudied.
Methods
Here, based on the analysis of large genomic datasets, mostly the whole-exome sequencing of over 10,000 cancer/normal sample pairs deposited within the TCGA repository, we undertook an analysis of somatic mutations in miRNA genes.
Findings
We identified and characterized over 10,000 somatic mutations and showed that some of the miRNA genes are overmutated in Pan-Cancer and/or specific cancers. Nonrandom occurrence of the identified mutations was confirmed by a strong association of overmutated miRNA genes with KEGG pathways, most of which were related to specific cancer types or cancer-related processes. Additionally, we showed that mutations in some of the overmutated genes correlate with miRNA expression, cancer staging, and patient survival.
Interpretation
Our study is the first comprehensive Pan-Cancer study of cancer somatic mutations in miRNA genes. It may help to understand the consequences of mutations in miRNA genes and the identification of miRNA functional mutations. The results may also be the first step (form the basis and provide the resources) in the development of computational and/or statistical approaches/tools dedicated to the identification of cancer-driver miRNA genes.
Funding
This work was supported by research grants from the Polish National Science Centre 2016/22/A/NZ2/00184 and 2015/17/N/NZ3/03629.
Keywords: miRNA, Somatic mutations, Pan-Cancer, TCGA, Non-coding
Research in Context Section.
Evidence before this study
Cancer genome analyses supported by data generated in large projects such as The Cancer Genome Atlas (TCGA) have led to the identification of hundreds of cancer-driving genes and thousands of cancer-driving mutations in protein-coding regions, which encompass barely 2% of the genome. Some of these genes/mutations, serve as important biomarkers for cancer-targeted therapies. However, very little (close to nothing) is known about genetic alterations in regions that encode non-coding RNAs, including miRNAs that are considered important players in oncogenesis, serving either as oncomiRs or suppressormiRs.
Added value of this study
With the use of sequencing datasets generated within the large cancer-genome projects, mostly TCGA, we identified and characterized >10,000 miRNA gene mutations in 33 types of cancers. Among the identified variants were mutations in well-known oncomiRs and miRNA suppressors, including let-7, miR-21, and miR-205. We identified and characterized dozens of significantly overmutated miRNA genes and hotspot nucleotide positions that are recurrently mutated in particular cancer types or the overall Pan-Cancer dataset and showed that some of these mutations affect miRNA expression, cancer staging, and patient survival as well as occur more frequently in miRNA genes playing role in cancer.
Implication of all the available evidence
This is the first comprehensive analysis of somatic mutations in miRNA genes in cancer. It may serve as the first step in the identification of driver mutations in miRNA genes and may help in understanding the role of particular miRNAs in cancer. After further functional validation, some of the mutations may be utilized as cancer biomarkers.
Alt-text: Unlabelled box
1. Introduction
Cancer encompasses a broad spectrum of heterogeneous diseases whose development (i.e., initiation, promotion, and progression) is associated with the accumulation of numerous genetic alterations in the cancer genome, which is the hallmark of all cancers. Although most of these alterations are neutral, some of the randomly occurring mutations are functional, providing a growth advantage to a neoplastic cell [1]. As a result of clonal selection of the fastest dividing cells, functional (driver) mutations often recur in genes playing an important role in cancer development (driver genes) and therefore may serve as indicators of such genes. Numerous large cancer genome sequencing studies (mostly whole-exome sequencing, WES) have been performed, and hundreds of cancer-driving genes and thousands of cancer-driving mutations have been detected. Some of these genes/mutations, e.g., EGFR, BRAF, and JAK2, serve as important biomarkers for cancer-targeted therapies. As the overwhelming majority of the cancer genome studies have focused on protein-coding genes, the most identified cancer-driver mutations are in protein-coding sequences, which encompass barely 2% of the genome. The spectacular exception are TERT promoter mutations, which occur most frequently in melanoma, brain, and bladder cancers but have also been identified in other cancers [2], [3], [4].
On the other hand, a growing body of evidence indicates that miRNAs, a class of short (~21 nt long) single-stranded non-coding RNAs, play an important role in cancer, and it was shown that particular miRNAs can either drive (oncomiRs, often upregulated in cancer) or suppress (suppressormiRs, often downregulated in cancer) oncogenesis. It was also proposed that miRNAs have great potential as cancer biomarkers and/or targets of cancer therapies [5], [6], [7], [8].
Among the most intensively studied miRNAs whose function in cancer is best documented are the let-7 family, miR-17-92 cluster (oncomiR-1), miR-21, and miR-205 (reviewed in [9]). Although the global level of miRNA is generally downregulated in cancer, many miRNAs are consistently either upregulated or downregulated in particular cancer types or specific cancer conditions. It was also shown that miRNA genes frequently show copy number alterations (either amplification or deletion) in cancer [10,11]. The cancer-related processes that are regulated by miRNAs include cell proliferation, epithelial-mesenchymal transformation (EMT), migration, angiogenesis, inflammation, apoptosis, and response to cancer treatment (reviewed in [12], [13], [14], [15]).
Despite the great interest in the role of miRNA in cancer, very little (close to nothing) is known about somatic mutations in miRNA genes (defined here as sequences coding for the most crucial part of miRNA precursors) occurring in cancer. Considering subsequent steps of miRNA biogenesis and the mechanism of miRNA posttranscriptional gene regulation, mutations may be expected to affect different attributes of miRNA genes. In addition to the most obvious consequences of mutations in seed sequences that affect the pivotal function of miRNAs, i.e., the ability to recognize and downregulate their specific targets, mutations in any part of the miRNA precursor may affect the effectiveness or precision of miRNA biogenesis by altering/destabilizing the hairpin structure of the miRNA precursor, by altering DROSHA or DICER1 cleavage sites, or by altering protein-interacting or other regulatory sequences/structure motifs [16], [17], [18], [19]. Additionally, mutations destabilizing one of the miRNA duplex ends may alter 5p/3p miRNA preference. Despite the scarcity of identified miRNA gene mutations, the individual examples of SNPs, germline or somatic mutations provide proof, at least for some of the scenarios listed above. Examples include (i) the mutation in the seed sequence of miR-204-5p, affecting target recognition, that causes inherited retinal dystrophy [20]; (ii) the mutation in the passenger strand of hsa-miR-96 that destabilizes the structure of the miRNA precursor, affects its processing, and decreases the miR-96-5p level, eventually resulting in the same phenotypic effect as mutations in the seed sequence of the guide strand, i.e., nonsyndromic inherited hearing loss [21,22]; (iii) the G>C substitution (SNP rs138166791) in the penultimate position of the 3p passenger strand of hsa-miR-890 that significantly lowers the cleavage efficiency by DROSHA and consequently decreases the levels of both mature miR-890-5p and passenger miR-890-3p [23]; (iv) the G>C substitution (SNP rs2910164) located in the 3p passenger strand of hsa-miR-146a that is associated with an increased risk of papillary thyroid carcinoma, where it was shown that the C allele of the SNP alters the structure of the precursor, decreases expression of the mature miRNA and activates the passenger strand, which becomes the second mature miRNA modulating many genes involved in the regulation of apoptosis [24]; (v) interesting example is a mutation in the seed sequence of miR-184-3p, causing familial keratoconus, whose effect is not the disruption of miR-184-3p target recognition but the inability to mask overlapping targets for miR-205 in INPPL1 and ITGB4 [25]; (vi) mutations in hsa-miR-30c-1 and hsa-miR-17 that affect the precursor structure and thereby increase the levels of mature miRNAs, downregulating BRCA1 in familial breast cancer cases without BRCA1/2 mutations [26]; and finally, (vii) two different somatic mutations in the seed sequence of miR-142-3p, found in acute myeloid leukemia (AML) samples, that were shown to decrease miR-142-5p and reverse the miR-5p/3p ratio (in favor of miR-3p) [27] (more details and references on mutations in hsa-miR-142 in the subsequent sections). An additional indication of miRNA gene sensitivity to genetic alteration is their general high conservation and the decreased density of common SNPs in miRNA hairpin sequences [28], [29], [30]. The SNP occurring in miRNA genes and other non-coding RNA sequences are collected in curated databases (e.g., MSDD and lincSNP 2.0), whose use may facilitate identification of functionally relevant SNP or mutations [31,32]. In our recent analysis of somatic mutations in lung cancers, we confirmed that seed mutations affect the vast majority of the predicted targets and showed that mutations in miRNA genes often alter the predicted structure of miRNA precursors [33]. miRNAs in cancers were also considered in the context of somatic mutations found in mRNAs that affect mRNA-miRNA and competing endogenous RNA (ceRNA)-miRNA interactions [34,35]. As we consider sequence variations in mRNAs that may affect miRNA function, the same should apply to the sequence variations in the miRNA genes themselves. The more general effect of sequence variants, including cancer somatic mutations on RNA structure, was recently demonstrated with the use of high-throughput combined experimental and computational approaches, that have led to the identification of many RNA structure motifs (termed riboSNitches) whose functionalities were affected by the variants detected in both coding and non-coding sequences [36,37].
Several multicenter projects have led to gathering data on somatic mutations from hundreds of cancers. One of the projects, The Cancer Genome Atlas (TCGA), covers over 10,000 samples from 33 types of cancers, including the most common human cancers. Importantly, the TCGA consortium works on standardized pipelines of data analysis [38], enabling comparisons across different cancer types (within the so-called Pan-Cancer set).
In the current study, we took advantage of data gathered by the TCGA consortium to analyze somatic mutations occurring in miRNA genes. As a result, we identified thousands of mutations in all subregions of miRNA genes and identified many Pan-Cancer or cancer-specific overmutated miRNA genes. We showed that mutations in some of the overmutated genes correlate with miRNA expression, cancer staging, and patient survival. Although the functionality of individual mutations or groups of mutations needs to be verified in independent functional studies, the strong association of the overmutated miRNA genes with cancer-related pathways indicates that miRNA gene mutations are not only random events and that at least some of them play a role in cancer.
2. Methods
2.1. Data resources
We used molecular and clinical data (Level 2) generated and deposited in the TCGA repository (http://cancergenome.nih.gov). These data included the results of somatic mutation calls in WES datasets preprocessed through the standard TCGA pipeline. We took advantage of somatic mutation data generated with four mutation caller algorithms (Mutect2, Muse, Varscan, and SomaticSnipper) and deposited as vcf.gz files. We analyzed the annotated somatic mutations with corresponding clinical information [39], miRNA and mRNA expression data [40].
2.2. Data processing
We analyzed somatic mutations in 1918 miRNA gene regions (Supplementary Table S1) annotated in the miRBase v.22.1 database. The miRNA genes were defined as pre-miRNA-coding sequences, extended upstream and downstream by 25 nucleotides. The pre-miRNA-coding sequences were reconstructed based on 5p and 3p mature miRNA sequences defined in miRBase (in cases when only one miRNA strand was indicated, the other pre-miRNA end was reconstructed assuming the pre-miRNA hairpin structure with a 2-nt 3p overhang). According to the number of reads reported for the particular pre-miRNA arm (miRBase), the analyzed precursors were classified into one of 3 categories: (i) generating mature miRNA predominantly from the 5p arm (≥90% of reads from the 5p arm); (ii) generating mature miRNA predominantly from the 3p arm (≥90% of reads from the 3p arm); and (iii) balanced (>10% of reads from each arm). As high-confidence miRNA genes, we considered genes coding for miRNA precursors annotated as “high confidence” in miRBase and/or deposited in MiRGeneDB v2.0. The precursors deposited in MiRGeneDB are defined based on criteria that include careful annotation of the mature versus passenger miRNA strands and evaluation of evolutionary hierarchy; therefore, they are much more credible than those in miRBase [41,42].
From the vcf.gz files generated by four algorithms (VarScan2, SomaticSniper, MuSE, and MuTect2) gathered from TCGA repository, we extracted somatic mutation calls with PASS annotation within defined gene regions (Supplementary Table S1). The extraction was performed with a set of in-house Python scripts (https://github.com/martynaut/pancancer_mirnome), an updated version of the scripts used in our earlier study [33]. To avoid duplicating mutations detected in multiple sequencing experiments in the same cancer patient (e.g., due to the presence of two cancer samples sequenced from a single cancer patient), we combined files summing reads associated with particular mutations. Next, the lists of mutations detected by different algorithms were merged, removing multiple calls of the same mutations. To further increase the reliability of the identified mutations, we removed mutations that did not fulfil the following criteria: (i) at least two mutation-supporting reads in a tumour sample (if no mutation-supporting read was detected in the corresponding normal sample); (ii) at least 5 × higher frequency of mutation-supporting reads in the tumour sample than in the corresponding normal sample; (iii) somatic score parameter (SSC) >30 (for VarScan2 and SomaticSniper); and (iv) base quality (BQ) parameter for mutation-supporting reads in the tumour sample >20 (for MuSE and MuTect2). We excluded mutations occurring in hypermutated samples, defined as samples with >10,000 mutations in the exome. Additionally, we checked if any of the analyzed miRNA genes are localized within DAC blacklisted regions wgEncodeDacMapabilityConsensusExcludable.bed.gz from Genome Browser for hg19) and found 7 miRNA genes at least partially overlapping with the blacklisted region (hsa-mir-12136, hsa-mir-4485, hsa-mir-9901, hsa-mir-3648-2, hsa-mir-10396a, hsa-mir-4477a-1, hsa-mir-4477b-1). None of the miRNA genes was considered significant in the performed analyses. Due to the used statistical approach, both ordinary binomial analysis and functionally-weighted analysis (see below) induced threshold of a minimal number of mutations assigned to relevant miRNA genes and only genes with at least 3 mutations (ordinary binomial) or 4 weighted points (functionally-weighted) were considered as overmutated. SNP coinciding mutations were not excluded from the analyses, because of their very low population frequency that does not justify the recurrence of the mutations.
Target prediction was performed with the use of TargetScan Custom (release 4.2) [43], which allows target prediction for both wild-type and mutant seed sequences. Secondary structure prediction was performed with the use of mfold software (default parameters) [44]. 3D pre-miRNA structures were predicted using RNAComposer software with default parameters [45] and visualized with PyMOL (Schrödinger, LLC, New York, NY, USA). Changes in motifs within miRNA precursors recognized by RNA-binding proteins were analyzed with a Python script based on miRNAmotif software [46].
The level of miRNA expression was classified to the following categories based on average reads_per_million_miRNA_mapped (RPM) values calculated for particular cancer types: <1 RPM, not expressed; 1-20 RPM, low level (low); 21-100 RPM medium level (medium); >101 RPM high level (high). At Pan-Cancer level miRNA genes were classified as either expressed (>1 RPM across all cancer samples) or not expressed. Expression data was gathered from FireBrowse (illuminahiseq_mirnaseq-miR_gene_expression).
2.3. OncodriveFML analysis
OncordriveFML was run using CADD score (hg38, version 1.6) against the mutations in Pan-Cancer and individual cancer types. The maf file was generated based on Supplementary Table S2. The signature method was set as a complement with cancer type as a classifier, the statistical method was set to “amean” and indels were included using a max method (max_consecutive was set to 7 as default).
2.4. Analysis of mutations in the PCAWG-ICGC cohort
For validation purposes, we took advantage of a publicly available Pan-Cancer Analysis of Whole Genomes dataset generated by International Cancer Genome Consortium (maf file, version from Nov 25, 2019; PCAWG-ICGC dataset). For extraction/analysis of the mutations, we used a pipeline similar to that described above for the TCGA mutations, with minor modifications resulting from differences in input files (the code is available in the github repository on icgc_hg19 branch https://github.com/martynaut/pancancer_mirnome/tree/icgc_hg19). As the hg19 genome does not include regions covering 4 genes (i.e., hsa-mir-6859-2, hsa-mir-1234, hsa-mir-4477b, and hsa-mir-4477a) the analysis was performed on 1914 miRNA genes.
2.5. Statistics
Unless stated otherwise, all statistical analyses were performed with statistical functions from the Python module scipy.stats. Particular statistical tests are indicated in the text, and unless stated otherwise, a p-value <0.05 was considered significant. If necessary, p-values were corrected for multiple tests with the Benjamini-Hochberg procedure.
Hotspot miRNA genes were identified based on the probability of occurrence of the observed number of mutations in particular miRNA gene, which was calculated with the use of the 2-tailed binomial distribution, assuming a random occurrence of mutations in all analyzed miRNA genes and considering the miRNA gene length (based on the coordinates used in the study), although it should be noted that occurrence of mutations is not completely random and there are regions with increased and decreased background mutation rate [47,48]. The Benjamini-Hochberg procedure was used for multiple testing. To further evaluate the reliability of the identified hotspot miRNA genes, we recalculated the mutation enrichment significance, weighting the mutation occurrences by the following factors: 2 ×, mutations in seeds (guide strand only); 1.5 ×, mutations in miRNAs (miRNA duplex), mutations affecting the functional motifs (identified by miRNAmotif) or +/-1 positions of DROSHA/DICER1 cleavage sites; and 1 ×, other mutations. Weight correction was not used to search for hotspot positions within miRNA genes.
For patient survival analyses, we used a log-rank test (from lifelines library [49]) for specific cancers or a stratified version of the test for the Pan-Cancer cohort (survdiff function from statsmodels library [50]). To determine the direction of mutation effects on survival, we used Cox's proportional hazard model. Survival plots were created using KaplanMeierFitter from the lifelines library.
2.6. Ethics
Ethical approval was unnecessary because this work is a meta-analysis of previously published data. The use of the data was approved by TCGA (project ID: 16565).
2.7. Role of the funding source
The funders had no role in the study design, data analysis, interpretation, preparation of the manuscript, and any aspect of the study.
3. Results
3.1. Overview of miRNome mutations in TCGA cancers
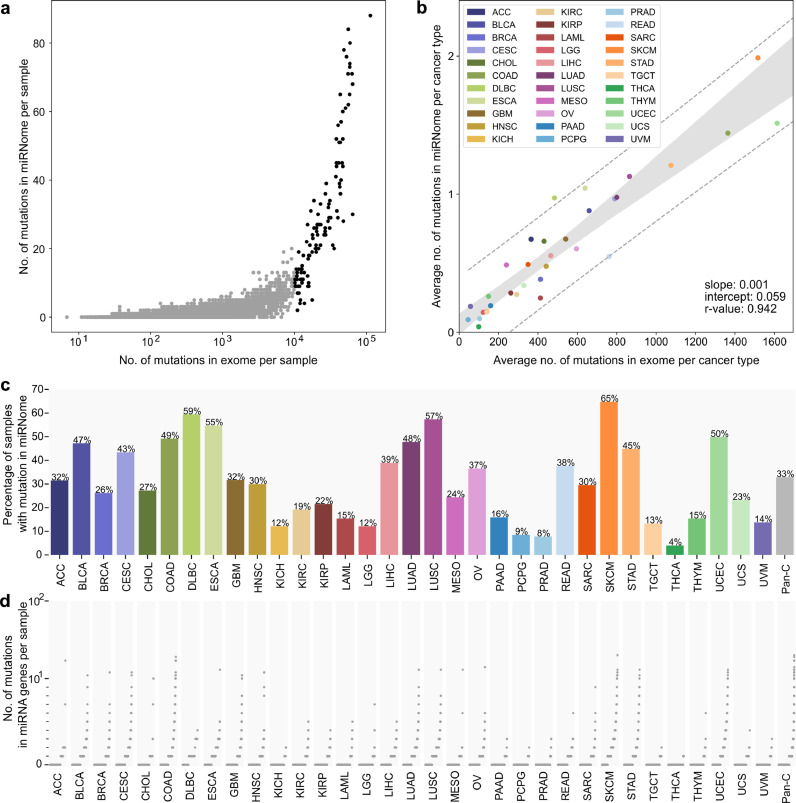
To investigate the occurrence of somatic mutations in miRNA genes (miRNome), we took advantage of the WES datasets of 10,369 tumour/control sample pairs representing 33 different cancer types collected and analyzed by the TCGA project. The list of all cancer types and their abbreviations is provided in Table 1 [to avoid confusion, we will use the abbreviations only for the TCGA sample sets but not generally for particular types of cancer; in the latter case, we will use full cancer type names or alternative abbreviations indicated in the text]. We defined miRNome as 1918 miRNA genes (Supplementary Table S1) encompassing ~100 nt long fragments of genomic DNA coding for all pre-miRNAs (with 25 nt flanks) defined in miRBase v22.1, including 537 high-confidence pre-miRNAs annotated by miRBase and 466 knowledge-based expert-curated pre-miRNAs annotated in MirGeneDB v.2.0. It should be noted, however, that not all miRNA genes were covered by TCGA WES. In total, we found 10,588 mutations in miRNA genes; however, as the number of miRNome mutations in hypermutated samples (samples with >10,000 mutations in the whole exome) was highly correlated with the general mutation burden in these samples (Fig. 1a, Supplementary Fig. S1) and therefore is likely highly enriched in randomly occurring nonfunctional mutations, we decided to remove the hypermutated samples (114, ~1% samples; 3,478, ~33% mutations) from further analysis. The remaining 7,110 mutations (Supplementary Table S2), including 6,312 substitutions, 198 insertions, and 600 deletions, were found in 1179 distinct miRNA genes. At the Pan-Cancer level, 3,370/10,255 (33%) samples had at least one miRNome mutation. This number was the highest in SKCM (298/460, 65%), DLBC (22/37, 59%), LUSC (285/497, 57%), and ESCA (99/181, 55%) and the lowest in PCPG (14/164, 9%), PRAD (39/497, 8%), and THCA (20/495, 4%) (Fig. 1c and Table 1). It should be noted, however, that the occurrence of mutations in miRNome is consistent with the general burden of mutations in particular cancer types (Fig. 1a and b, Supplementary Fig. S1). Additionally, as shown in Fig. 1d, there is a substantial fraction of samples with more than one mutation in miRNome. It is also noteworthy that some cancers, including COAD, STAD, and UCEC, have substantially heightened numbers of indel mutations (Table 1), which is consistent with known cancerous mechanisms, including DNA repair defects, associated with those cancers [51], [52], [53].
Table 1.
Summary of patients and mutations in miRNome per cancer type within the Pan-Cancer cohort.
| TCGA project (cancer type) | Full TCGA project name | No. of samples without (and with) hypermutated samples | No. (%) of samples with at least one mutation in miRNome | No. of mutated miRNA genes | No. of mutations | No. (%) of substitutions | No. (%) of insertions | No. (%) of deletions |
|---|---|---|---|---|---|---|---|---|
| ACC | Adrenocortical carcinoma | 92 (92) | 29 (31.5) | 49 | 63 | 55 (87.3) | 3 (4.8) | 5 (7.9) |
| BLCA | Bladder Urothelial Carcinoma | 411 (412) | 194 (47.2) | 283 | 372 | 354 (95.2) | 5 (1.3) | 13 (3.5) |
| BRCA | Breast invasive carcinoma | 1040 (1044) | 272 (26.1) | 297 | 415 | 371 (89.4) | 16 (3.9) | 28 (6.7) |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma | 304 (305) | 132 (43.4) | 251 | 299 | 277 (92.6) | 6 (2.0) | 16 (5.4) |
| CHOL | Cholangiocarcinoma | 44 (44) | 12 (27.3) | 27 | 29 | 22 (75.9) | 0 (0.0) | 7 (24.1) |
| COAD | Colon adenocarcinoma | 411 (432) | 202 (49.1) | 390 | 604 | 482 (79.8) | 21 (3.5) | 101 (16.7) |
| DLBC | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma | 37 (37) | 22 (59.5) | 22 | 36 | 35 (97.2) | 1 (2.8) | 0 (0.0) |
| ESCA | Esophageal carcinoma | 181 (182) | 99 (54.7) | 158 | 190 | 178 (93.7) | 0 (0.0) | 12 (6.3) |
| GBM | Glioblastoma multiforme | 394 (396) | 125 (31.7) | 207 | 269 | 246 (91.4) | 12 (4.5) | 11 (4.1) |
| HNSC | Head and Neck Squamous Cell Carcinoma | 510 (510) | 153 (30.0) | 199 | 248 | 228 (91.9) | 7 (2.8) | 13 (5.2) |
| KICH | Kidney Chromophobe | 66 (66) | 8 (12.1) | 6 | 10 | 9 (90.0) | 1 (10.0) | 0 (0.0) |
| KIRC | Kidney renal clear cell carcinoma | 339 (339) | 65 (19.2) | 85 | 95 | 81 (85.3) | 6 (6.3) | 8 (8.4) |
| KIRP | Kidney renal papillary cell carcinoma | 288 (288) | 62 (21.5) | 78 | 85 | 73 (85.9) | 5 (5.9) | 7 (8.2) |
| LAML | Acute Myeloid Leukemia | 149 (149) | 23 (15.4) | 36 | 39 | 33 (84.6) | 5 (12.8) | 1 (2.6) |
| LGG | Brain Lower Grade Glioma | 512 (513) | 62 (12.1) | 69 | 77 | 69 (89.6) | 3 (3.9) | 5 (6.5) |
| LIHC | Liver Hepatocellular Carcinoma | 375 (375) | 146 (38.9) | 172 | 214 | 198 (92.5) | 7 (3.3) | 9 (4.2) |
| LUAD | Lung adenocarcinoma | 567 (569) | 271 (47.8) | 356 | 561 | 536 (95.5) | 8 (1.4) | 17 (3.0) |
| LUSC | Lung squamous cell carcinoma | 497 (497) | 285 (57.3) | 365 | 576 | 554 (96.2) | 9 (1.6) | 13 (2.3) |
| MESO | Mesothelioma | 82 (82) | 20 (24.4) | 38 | 41 | 36 (87.8) | 0 (0.0) | 5 (12.2) |
| OV | Ovarian serous cystadenocarcinoma | 443 (443) | 162 (36.6) | 191 | 274 | 243 (88.7) | 18 (6.6) | 13 (4.7) |
| PAAD | Pancreatic adenocarcinoma | 182 (183) | 29 (15.9) | 30 | 36 | 34 (94.4) | 2 (5.6) | 0 (0.0) |
| PCPG | Pheochromocytoma and Paraganglioma | 164 (164) | 14 (8.5) | 15 | 15 | 15 (100.0) | 0 (0.0) | 0 (0.0) |
| PRAD | Prostate adenocarcinoma | 497 (498) | 39 (7.8) | 48 | 50 | 47 (94.0) | 0 (0.0) | 3 (6.0) |
| READ | Rectum adenocarcinoma | 146 (150) | 55 (37.7) | 69 | 83 | 77 (92.8) | 4 (4.8) | 2 (2.4) |
| SARC | Sarcoma | 240 (240) | 71 (29.6) | 98 | 120 | 112 (93.3) | 3 (2.5) | 5 (4.2) |
| SKCM | Skin Cutaneous Melanoma | 460 (470) | 298 (64.8) | 453 | 925 | 912 (98.6) | 7 (0.8) | 6 (0.6) |
| STAD | Stomach adenocarcinoma | 435 (441) | 195 (44.8) | 327 | 528 | 396 (75.0) | 16 (3.0) | 116 (22.0) |
| TGCT | Testicular Germ Cell Tumors | 150 (150) | 20 (13.3) | 19 | 22 | 21 (95.5) | 0 (0.0) | 1 (4.5) |
| THCA | Thyroid carcinoma | 495 (496) | 20 (4.0) | 20 | 21 | 19 (90.5) | 1 (4.8) | 1 (4.8) |
| THYM | Thymoma | 123 (123) | 19 (15.4) | 20 | 32 | 26 (81.2) | 1 (3.1) | 5 (15.6) |
| UCEC | Uterine Corpus Endometrial Carcinoma | 485 (542) | 242 (49.9) | 469 | 746 | 541 (72.5) | 31 (4.2) | 174 (23.3) |
| UCS | Uterine Carcinosarcoma | 56 (57) | 13 (23.2) | 18 | 19 | 17 (89.5) | 0 (0.0) | 2 (10.5) |
| UVM | Uveal Melanoma | 80 (80) | 11 (13.7) | 14 | 16 | 15 (93.8) | 0 (0.0) | 1 (6.2) |
| Pan-Cancer | - | 10255 (10369) | 3370 (32.9) | 1179 | 7110 | 6312 (88.8) | 198 (2.8) | 600 (8.4) |
Fig. 1.
Summary of mutations identified in miRNome in TCGA cancer types. (a) The number of mutations in the exome (x-axis, log10 scale) and miRNome (y-axis, linear scale) per sample in Pan-Cancer. Each dot represents a single sample. Black dots represent hypermutated (>10k somatic mutations in exome) samples. (b) The average number of mutations in the exome (x-axis, linear scale) and miRNome per cancer type (y-axis, linear scale). Each dot represents a single cancer type. Hypermutated samples were excluded prior to the analysis. (c) Percentage of patients with at least one somatic mutation detected in miRNA genes. (d) The number of mutations in miRNA genes per patient. Each dot represents a single patient. Patients are ranked by the number of mutations in miRNA genes. Scale is linear (values 0-10) and log10 (values 10+).
3.2. Localization of mutations within miRNA genes
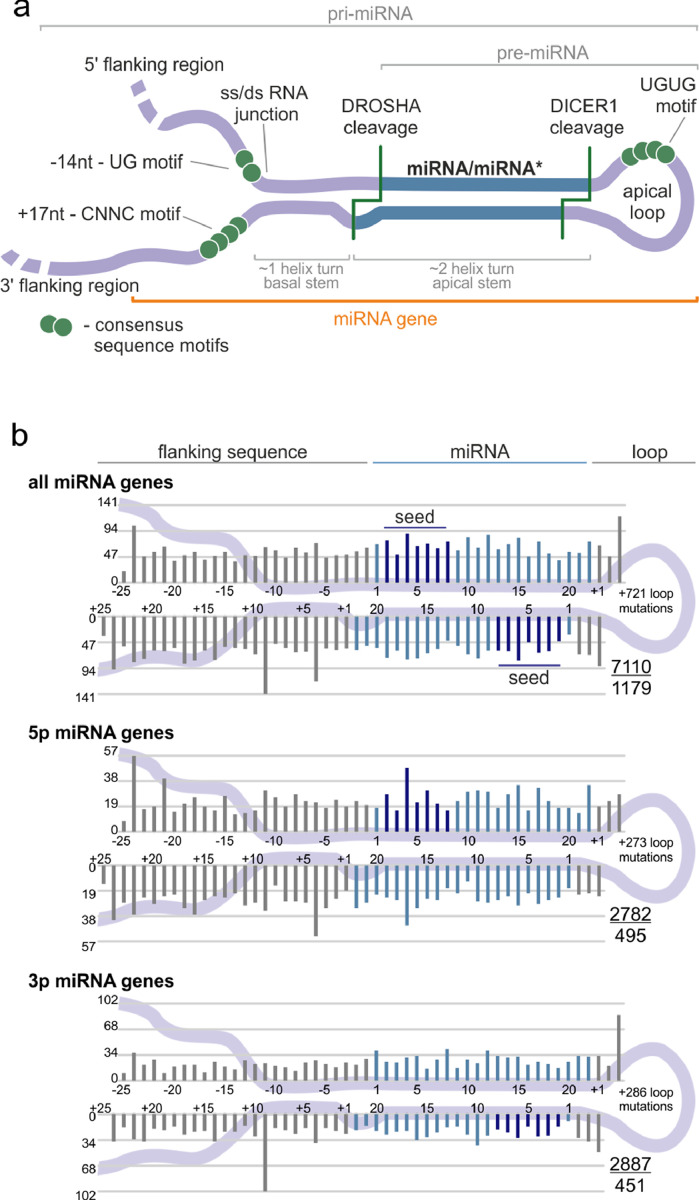
For a closer examination of the localization of sequence variants in subregions of miRNA precursors, we superimposed the identified variants on the consensus miRNA precursor structure and categorized them according to localization in the miRNA gene subregions (Fig. 2a). The analysis shows that mutations occur in all regions of the miRNA gene, and in general, there is no strong imbalance in mutation localization within the miRNA precursor (Fig. 2b). A similar mutation distribution was observed when precursors of predominantly 5p- and 3p-miRNAs were analyzed separately (Fig. 2b, lower panels) and when the analysis was narrowed only to the high-confidence miRNA genes defined either by miRBase or miRGeneDB (Supplementary Fig. S2) or performed separately for individual cancer types (data not shown). We observed only a slightly decreased mutation rate in the 5p flanking region (Table 2); however, this effect may result from lower sequencing coverage in the flanking regions.
Fig. 2.
Localization of somatic mutations in miRNA precursors in Pan-Cancer. (a) An overview of a primary miRNA transcript with the indicated subregions considered in the study. The miRNA duplex is indicated in blue, and representative sequence consensus motifs recognized as enhancers of miRNA biogenesis are represented by green circles. (b) Localization of all detected mutations in the Pan-Cancer cohort. miRNA duplex positions are indicated in blue, seed regions in dark blue, and flanking regions and terminal positions of the apical loop in grey. The numbers in the lower-right corner represent the number of plotted mutations (upper) and the number of mutated miRNA genes (lower). If present, sequence variants localized beyond position 22 in longer mature miRNAs are shown cumulatively at position 22. The plot shows mutations within six positions of the loop (first 3 and last 3 nucleotides). The number of remaining mutations is indicated within the loop. Analyses were also performed in narrowed groups of miRNAs that release the guide miRNA strand predominantly from the 5p or 3p arm (lower panels).
Table 2.
Distribution of somatic mutations within miRNA precursors.
| miRNA genes | Subregion | No. of mutations | Mut/Mbp | Fold change | P-value (binomial) |
|---|---|---|---|---|---|
| all | total | 7110 | 3.24 | 1.00 | |
| 5′flanking | 1355 | 2.76 | 0.85 | 4.26E−12 | |
| 3′flanking | 1746 | 3.55 | 1.09 | 2.12E−05 | |
| loop | 1187 | 3.30 | 1.02 | 5.01E−01 | |
| passenger strand | 1179 | 3.36 | 1.04 | 1.85E−01 | |
| guide | 1607 | 3.35 | 1.03 | 1.25E−01 | |
| seed | 536 | 3.45 | 1.06 | 1.33E−01 | |
| High Confidence (miRBase) | total | 3508 | 4.66 | 1.00 | |
| 5′flanking | 687 | 4.07 | 0.87 | 3.65E−05 | |
| 3′flanking | 823 | 4.87 | 1.04 | 1.57E−01 | |
| loop | 519 | 4.41 | 0.95 | 1.78E−01 | |
| passenger strand | 527 | 5.02 | 1.08 | 6.76E−02 | |
| guide | 952 | 4.97 | 1.06 | 2.59E−02 | |
| seed | 325 | 5.32 | 1.14 | 1.46E−02 | |
| MirGeneDB | total | 3594 | 5.99 | 1.00 | |
| 5′flanking | 692 | 5.08 | 0.85 | 6.93E−07 | |
| 3′flanking | 857 | 6.30 | 1.05 | 9.43E−02 | |
| loop | 497 | 5.61 | 0.94 | 1.26E−01 | |
| passenger strand | 644 | 6.60 | 1.10 | 7.64E−03 | |
| guide | 904 | 6.37 | 1.06 | 3.40E−02 | |
| seed | 277 | 6.13 | 1.02 | 6.58E−01 |
3.3. Significantly overmutated miRNA genes
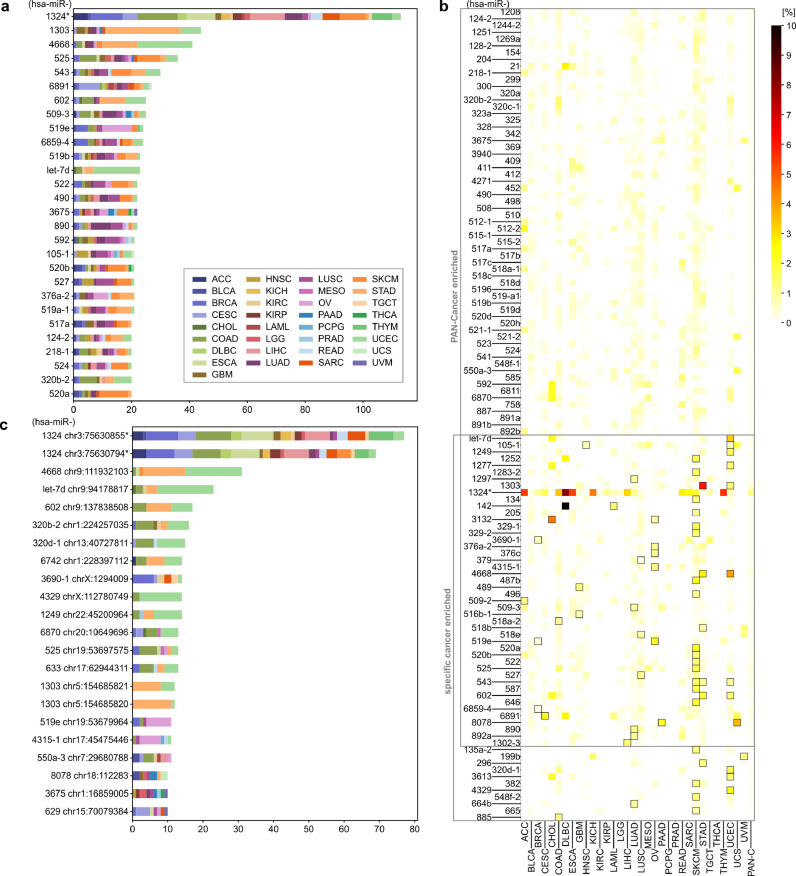
In the next step, we searched for miRNA genes overburdened with mutations. The most frequently mutated miRNA genes are presented in Fig. 3a. We examined the numbers of mutations occurring in particular miRNA genes and the overall frequency of mutations in miRNome with the use of a binomial distribution test (p < 0.01) and showed that 81 of the recurrently mutated genes are significantly overmutated in Pan-Cancer (Table 3, Supplementary Table S3A). As mutations may not be randomly distributed in the genome and to consider the enrichment of functional variants, we next performed functionally weighted analysis, increasing the value of mutations located in most likely functional sequences including seed regions, DROSHA/DICER1 cleavage sites, miRNA duplexes, and protein binding motifs (see Materials and Methods section). The weighted analysis revealed 108 significantly overmutated miRNA genes, of which a substantial fraction overlapped with the genes identified in the ordinary binomial analysis (Table 3, Supplementary Table S3B). Although the advantage of the weighted analysis cannot be formally tested due to lack of an appropriate list of credible cancer-related miRNA genes, hsa-miR-142, and hsa-miR-205, the most convincing examples of overmutated genes, see below), ranked substantially higher in the weighted vs. ordinary binomial Pan-Cancer analysis (rank 29th vs. 59th, and 52nd vs. 75th, respectively). Two main types of overmutated miRNA genes can be distinguished based on mutation occurrence in various cancer types: one shows a somewhat sample-number dependent distribution of mutations across various cancer types (e.g., hsa-miR-1324, hsa-miR-6891, hsa-miR-3675), and the other shows an overrepresentation of mutations in one or two cancer types (e.g., hsa-miR-1303 for STAD, hsa-miR-890 for LUAD, hsa-miR-519e for OV).
Fig. 3.
Most frequently mutated miRNA genes and hotspots in Pan-Cancer. (a) miRNA genes with at least 20 somatic mutations in Pan-Cancer. Each colour represents a distinct cancer type. (b) Heatmap showing the percentage of mutations in miRNA genes overmutated (according to functionally weighted analysis) in Pan-Cancer and in specific cancers. Framed squares indicate cancer types in which gene enrichment reached statistical significance (binomial distribution test, adjusted p < 0.01). Specific values of mutation frequencies are shown in Supplementary Table S4. (c) Hotspot positions in miRNA genes with at least 10 somatic mutations in Pan-Cancer (colour legend as in panel a). To simplify the figure, we omitted the prefix hsa-miR in the gene IDs; * note the comment on mutations in hsa-miR-1324 at the end of the section Examples of overmutated miRNA genes.
Table 3.
Summary of overmutated miRNA genes in Pan-Cancer and individual cancer types.
| Cancer type | Overmutated miRNA genes - ordinary binomial analysis (# mutations) | No. of genes | Overmutated miRNA genes - functionally weighted analysis (# mutations) | No. of genes |
|---|---|---|---|---|
| Pan-Cancer | let-7d (23); 105-1 (22); 1208 (18); 124-2 (20); 1249 (16); 1251 (17); 1269a (20); 1277 (15); 128-2 (16); 1283-2 (18); 1297 (15); 1303 (51); 1324! (166); 142 (16); 204 (15); 205 (15); 218-1 (20); 3132 (17); 320a (17); 320b-2 (20); 320c-1 (17);320d-1 (16); 323a (15); 329-1 (15); 329-2 (15); 3675 (22); 3690-1 (17); 376a-2 (26); 376c (22); 379 (20); 409 (19); 411 (19); 412 (18); 4271 (15); 4315-1 (20); 4329 (15); 4668 (41); 487b (16); 489 (16); 490 (22); 496 (15); 509-2 (17); 509-3 (25); 510 (15); 512-1 (15); 512-2 (17); 515-1 (17); 515-2 (16); 516b-1 (17); 517a (21); 517c (18); 518b (15); 518d (17); 5196 (16); 519a-1 (21); 519b (24); 519e (33); 520a (20); 520b (21); 522 (24); 524 (20); 525 (36); 527 (21); 541 (16); 543 (31); 548f-1 (18); 585 (17); 587 (16); 592 (21); 602 (25); 633 (15); 646 (16); 6742 (16); 6811 (16); 6859-4 (25); 6870 (20); 6891 (27); 758 (17); 887 (18); 890 (22); 892a (15) | 81 | let-7d (23); 105-1 (22); 1208 (18); 124-2 (20); 1244-2 (15); 1249 (16); 1251 (17); 1252 (13); 1269a (20); 1277 (15); 128-2 (16); 1283-2 (18); 1297 (15); 1303 (51); 1324! (166); 134 (14); 142 (16);154 (13); 204 (15); 205 (15);21 (14); 218-1 (20);299 (12); 300 (14); 3132 (17); 320a (17); 320b-2 (20); 320c-1 (17); 323a (15); 325 (14); 328 (13); 329-1 (15); 329-2 (15);342 (12); 3675 (22);369 (14); 3690-1 (17); 376a-2 (26); 376c (22); 379 (20);3940 (11); 409 (19); 411 (19); 412 (18); 4271 (15); 4315-1 (20);452 (13); 4668 (41); 487b (16); 489 (16); 490 (22); 496 (15);498 (13);508 (14); 509-2 (17); 509-3 (25); 510 (15); 512-1 (15); 512-2 (17); 515-1 (17); 515-2 (16); 516b-1 (17); 517a (21);517b (12); 517c (18);518a-1 (14);518a-2 (13); 518b (15);518c (13); 518d (17);518e (14); 5196 (16); 519a-1 (21); 519b (24);519d (14); 519e (33); 520a (20); 520b (21);520d (14);520h (14);521-1 (14); 521-2 (13); 522 (24);523 (14); 524 (20); 525 (36); 527 (21); 541 (16); 543 (31); 548f-1 (18); 550a-3 (14); 585 (17); 587 (16); 592 (21); 602 (25); 646 (16); 6811 (16); 6859-4 (25); 6870 (20); 6891 (27); 758 (17); 8078 (12); 887 (18); 890 (22);891a (14); 891b (14); 892a (15);892b (12) | 108 |
| ACC | 1324! (8) | 1 | 1324! (8);509-2 (3) | 2 |
| BRCA | 1324! (19); 3690-1 (7); 519e (6) | 3 | 1324! (19); 3690-1 (7); 519e (6); 6859-4 (5) | 4 |
| CESC | 1324! (10); 6891 (7) | 2 | 1324! (10); 6891 (7) | 2 |
| CHOL | 0 | 3132 (3) | 1 | |
| COAD | 1324! (19) | 1 | 1324! (19); 518a-2 (4); 885* (5) | 3 |
| DLBC | 1324! (6); 142 (10) | 2 | 1324! (6); 142 (10) | 2 |
| ESCA | 1324! (18) | 1 | 1324! (18) | 1 |
| GBM | 411 (5); 489 (5); 516b-1 (5) | 3 | 489 (5); 516b-1 (5) | 2 |
| HNSC | 0 | 105-1 (4) | 1 | |
| KICH | 1324! (5) | 1 | 1324! (5) | 1 |
| KIRP | 1324! (5) | 1 | 1324! (5) | 1 |
| LAML | 0 | 142 (2) | 1 | |
| LIHC | 1302-3*(5); 1324! (16) | 2 | 1302-3*(5); 1324! (16) | 2 |
| LUAD | 1297 (6); 379 (6); 664b*(6); 890 (7); 892a (6) | 5 | 1297 (6); 1324! (6); 379 (6);509-3 (5); 664b*(6); 890 (7); 892a (6) | 7 |
| LUSC | 0 | 518e (4); 527 (6) | 2 | |
| OV | 376a-2 (7); 376c (5); 4315-1 (6); 519e (16) | 4 | 3132 (4); 376a-2 (7); 376c (5); 4315-1 (6); 519e (16) | 5 |
| PAAD | 8078*(4) | 1 | 8078 (4) | 1 |
| READ | 1324! (5) | 1 | 1324! (5) | 1 |
| SARC | 1324! (9) | 1 | 1324! (9) | 1 |
| SKCM | 1252*(8); 1283-2 (7); 1324! (10); 135a-2*(8); 329-1 (8); 329-2 (7); 487b (10); 496 (7); 520a (10); 525 (8); 543 (7); 587 (8); 646 (8) | 13 | 1252 (8); 1283-2 (7); 1324! (10);134 (6); 135a-2*(8);205 (5); 329-1 (8); 329-2 (7);382* (6); 487b (10); 496 (7); 520a (10);520b (6); 522 (6); 525 (8); 543 (7); 548f-2* (6); 587 (8); 646 (8); 665* (5) | 20 |
| STAD | 1303 (32);454* (6); 4668 (12); 543 (6); 602 (9) | 5 | 1303 (32);296* (5); 4668 (12);518b (5); 543 (6); 602 (9) | 6 |
| THYM | 1324! (11) | 1 | 1324! (11) | 1 |
| UCEC | let-7d (16); 1249 (8); 1277 (7); 1303 (7); 320d-1 (9); 3613*(7); 4329 (12); 4668 (19); 602 (7) | 9 | let-7d (16);105-1 (4); 1249 (8); 1277 (7); 1303 (7); 320d-1*(9); 3613*(7); 4329*(12); 4668 (19); 543 (5); 602 (7) | 11 |
| UCS | 0 | 8078 (2) | 1 | |
| UVM | 199b*(3) | 1 | 199b*(3) | 1 |
cancer-specific miRNA gene (not overmutated in Pan-Cancer); underline indicates miRNAs expressed in particular cancers, for more information see Supplementary Table S3; bold indicates genes identified in both (ordinary binomial and functionally weighted) analyses; ! note the comment on mutations in hsa-miR-1324 at the end of the section Examples of overmutated miRNA genes. To simplify the table, we omitted the prefix hsa-miR in the gene IDs.
As some mutations were unevenly distributed across cancer types, we also performed mutation enrichment analysis for individual cancers. This analysis revealed 55 and 80 additional miRNA genes overmutated in individual cancers in the ordinary binomial and functionally weighted analyses, respectively (Table 3, Supplementary Table S3). These lists included 8 and 12 cancer-specific overmutated genes, respectively, i.e., genes enriched in mutations in one or more cancers but not in Pan-Cancer. Among the most striking examples of the cancer-specific overmutated genes are (i) hsa-miR-3613 with 7 mutations in UCEC but also with 1 mutation in CHOL and 1 in ESCA, (ii) hsa-miR-135a-2 with 8 mutations in SKCM and 2 in UCEC, and (iii) hsa-miR-664b with 6 mutations in LUAD and 1 or 2 in UCEC, STAD, SKCM, and CESC. The highest overlap of overmutated miRNAs was observed between STAD and UCEC, which shared 4 out of 6 and 11 miRNA genes, respectively, according to functionally weighted analysis. The highest number of overmutated miRNA genes per cancer was found for SKCM (20) and UCEC (11). Some cancer types had no overmutated miRNA genes. As cancer type groups consisted of very different numbers of samples, in Fig. 3b and Supplementary Table S4, we visualized the occurrence of mutations in overmutated genes as a percentage of patients in the individual cancers and Pan-Cancer. As shown in Fig. 3b, the highest frequency of mutations belonged to hsa-miR-142 in DLBC, but other overmutated genes often exceeded a frequency of 2% or even 5% in individual cancers, e.g., hsa-miR-1303 in STAD (6%) and hsa-miR-3132 in CHOL (4,5%). The localization of mutations in each of the overmutated genes is graphically illustrated in Supplementary Fig. S3. For reasons explained in the next section, we do not comment here on mutations in hsa-miR-1324. It should be noted, however, that the overmutation of particular genes is not a direct indicator of the functionality of mutations. Some overmutations may be accidental, among others resulting from increased background mutation rate or specific nucleotide composition in particular genomic regions [47,48]. Some indicator of the mutation functionality may be, however, the level of miRNA gene expression. Although the information on expression is not available for all tested miRNAs, we have indicated the level (not expressed/low/medium/high in individual cancers or not expressed/expressed in Pan-Cancer) of overmutated miRNAs in the corresponding cancers (Table 3, Supplementary Table S3).
The area of somatic mutations in miRNA genes is scarcely researched; therefore, it is difficult to compare our results directly to those of other studies. However, among the cancer-specific overmutated miRNA genes, we identified hsa-miR-142, in which somatic mutations were found before in several studies (for details see below). We also confirmed the recurrence of mutations in hsa-miR-21 as previously identified with the Annotative Database of miRNA Elements (ADmiRe) [54]. Not surprisingly, the current results overlap almost perfectly with our earlier results obtained for LUAD and LUSC [33]. Minor discrepancies result from some differences in the technical approach (see Materials and Methods).
As some miRNA coded in a cluster are not only simultaneously expressed but also are functionally related, e.g., may target different genes in the same pathway, we compared enrichment of the mutations in 155 clusters of miRNA genes defined as a group of miRNAs with genomic inter-miRNA distance <10,000 bp [55] (Supplementary Table S5). As shown in Supplementary Table S6, mutations are enriched (at adjusted p < 0.01, binomial distribution test) in 4 of the miRNA clusters in Pan-Cancer and up to 3 clusters in individual cancer types. Two of the clusters overmutated in Pan-Cancer and the most frequently overmutated in individual cancers are cluster 73 and cluster 33 (Supplementary Table S6), that are two biggest clusters with >40 miRNA genes in each of the clusters. An interesting example is cluster 148 known as miR-888 Cluster found overmutated in Pan-Cancer and LUAD. It was shown that this cluster plays a role in prostate cancer [56]. Another example can be cluster 131 overmutated in UCEC, comprising hsa-let-7a-1, hsa-let-7f-1, and hsa-let-7d, members of the let-7 family, whose role in cancer is well-known.
3.4. Significantly overrepresented recurring point mutations
In the next step, we tested which recurrently mutated nucleotide residues are significant hotspots, i.e., positions mutated more frequently than expected by chance, taking into account overall mutation frequency and the number of samples in a particular cancer or the Pan-Cancer dataset. As such analysis may be strongly affected by the uneven occurrence of mutations in different genomic regions and different sequence contexts [47,48], to minimize false-positive results, we set a very stringent threshold of significance, an adjusted p < 0.0001 (binomial distribution test). The analysis showed 62 hotspots in Pan-Cancer and 69 in individual cancers, including 5 cancer-specific hotspots, 1 for DLBC, 2 for OV, and 2 for SKCM. The list and characteristics of hotspot mutations along with the expression level of corresponding miRNAs are shown in Table 4 and Supplementary Table S7. The two most frequently recurring point mutations were found in hsa-miR-1324 (chr3:75630855T>C[+] and chr3:375630794C>G[+], Fig. 3c). Other interesting hotspot mutations include hsa-miR-142 (chr17:58331260A>G[-] in the seed sequence of miR-142-3p) found in DLBC (3 mutations) and hsa-miR-519e (chr19:53679964G>A/T[+] and chr19:53679965G>A[+]) found in Pan-Cancer and OV. Please note that mutation occurring in a miRNA gene encoded on the minus chromosome strand in the sequence of a miRNA precursor occurs in reverse/complementary orientation; therefore, to avoid, confusion, a [+] or [-] sign indicates the orientation of the affected gene. As the recurrence of some mutations may be artefacts of not efficient filtering of germline variants, we checked the overlap of hotspot positions with the positions of SNPs. Although, due to the very large number of currently annotated SNPs, some of the SNPs coincide with the detected mutations, the very low population frequency of the SNPs or their type preclude confusing the SNPs with the recurrent mutations (Supplementary Table S7).
Table 4.
Summary of hotspot positions found in miRNA genes in Pan-Cancer and individual cancer types.
| Cancer type | Significant hotspots (# mutations) | No. of hotspots |
| Pan-Cancer | let-7d chr9:94178817 (23); 1208 chr8:128150186 (7); 1244-2 chr5:118974595 (7); 1249 chr22:45200964 (14); 1277 chrX:118386402 (7); 1302-3 chr2:113583038 (5); 1302-7 chr8:141786250 (5); 1303 chr5:154685809 (6); 1303 chr5:154685819 (8); 1303 chr5:154685820 (12); 1303 chr5:154685821 (12); 1324 chr3:75630794! (69); 1324 chr3:75630855! (77); 1324 chr3:75630860! (5); 15a chr13:50049206 (5); 205 chr1:209432167 (5); 296 chr20:58817670 (6); 320b-2 chr1:224257035 (16); 320c-1 chr18:21683580 (8); 320d-1 chr13:40727811 (15); 320e chr19:46709336 (5); 328 chr16:67202382 (6); 3652 chr12:103930488 (8); 3658 chr1:165907957 (7); 3663 chr10:117167687 (5); 3675 chr1:16858942 (5); 3675 chr1:16859005 (10); 3690-1 chrX:1294009 (14); 4271 chr3:49274155 (8); 4313 chr15:75762211 (5); 4315-1 chr17:45475446 (11); 4329 chrX:112780749 (14); 454 chr17:59137873 (8); 4668 chr9:111932103 (31); 487b chr14:101046508 (5); 489 chr7:93483953 (6); 512-1 chr19:53666774 (6); 512-2 chr19:53669258 (6); 519e chr19:53679964 (11); 519e chr19:53679965 (7); 520b chr19:53701276 (5); 525 chr19:53697575 (13); 525 chr19:53697594 (5); 543 chr14:101032009 (6); 548f-1 chr10:54607963 (6); 550a-3 chr7:29680788 (11); 567 chr3:112112876 (6); 570 chr3:195699434 (5); 602 chr9:137838508 (17); 624 chr14:31014652 (8); 629 chr15:70079384 (10); 633 chr17:62944311 (13); 6742 chr1:228397112 (14); 6811 chr2:237510924 (5); 6821 chr22:49962899 (5); 6859-4 chr16:17058 (9); 6859-4 chr16:17089 (6); 6870 chr20:10649696 (13); 6875 chr7:100868123 (5); 6891 chr6:31355224 (5); 6891 chr6:31355262 (6); 8078 chr18:112283 (10) | 62 |
| ACC | 1324 chr3:75630794! (4); 1324 chr3:75630855! (3) | 2 |
| BRCA | 1324 chr3:75630794! (8); 1324 chr3:75630855! (9); 3690-1 chrX:1294009 (6) | 3 |
| CESC | 1324 chr3:75630794! (5); 1324 chr3:75630855! (5); 629 chr15:70079384 (4); 6891 chr6:31355224 (5) | 4 |
| COAD | 1324 chr3:75630794! (8); 1324 chr3:75630855! (10); 320b-2 chr1:224257035 (5); 320c-1 chr18:21683580 (4); 320d-1 chr13:40727811 (5); 525 chr19:53697575 (5); 602 chr9:137838508 (4); 633 chr17:62944311 (4) | 8 |
| DLBC | 1324 chr3:75630794! (3); 1324 chr3:75630855! (3); 142 chr17:58331260* (3) | 3 |
| ESCA | 1324 chr3:75630794! (8); 1324 chr3:75630855! (9); 624 chr14:31014652 (3) | 3 |
| KICH | 1324 chr3:75630855! (3) | 1 |
| KIRP | 1324 chr3:75630794! (3) | 1 |
| LIHC | 1302-3 chr2:113583038 (5); 1324 chr3:75630794! (7); 1324 chr3:75630855! (7) | 3 |
| OV | 1244-2 chr5:118974595 (3); 376a-2 chr14:101040083* (3); 376c chr14:101039694* (3); 4315-1 chr17:45475446 (6); 519e chr19:53679964 (7); 519e chr19:53679965 (4) | 6 |
| READ | 1324 chr3:75630855! (3) | 1 |
| SARC | 1324 chr3:75630794! (3); 1324 chr3:75630855! (5) | 2 |
| SKCM | 1324 chr3:75630794! (4); 329-1 chr14:101026832* (4); 487b chr14:101046508 (5); 524 chr19:53711049* (4); 525 chr19:53697594 (5) | 5 |
| STAD | 1208 chr8:128150186 (4); 1303 chr5:154685809 (4); 1303 chr5:154685819 (8); 1303 chr5:154685820 (11); 1303 chr5:154685821 (8); 296 chr20:58817670 (5); 454 chr17:59137873 (6); 4668 chr9:111932103 (12); 602 chr9:137838508 (7); 6742 chr1:228397112 (5) | 10 |
| THYM | 1324 chr3:75630794! (4); 1324 chr3:75630855! (7) | 2 |
| UCEC | let-7d chr9:94178817 (16); 1249 chr22:45200964 (8); 1277 chrX:118386402 (7); 1303 chr5:154685821 (4); 320b-2 chr1:224257035 (6); 320c-1 chr18:21683580 (4); 320d-1 chr13:40727811 (8); 3658 chr1:165907957 (5); 4271 chr3:49274155 (6); 4329 chrX:112780749 (12); 4668 chr9:111932103 (16); 602 chr9:137838508 (6); 633 chr17:62944311 (4); 6742 chr1:228397112 (5); 6870 chr20:10649696 (5) | 15 |
cancer-specific hotspots (not overmutated in Pan-Cancer); underline indicates miRNAs expressed in particular cancers, for more information see Supplementary Table S5; ! note the comment on mutations in hsa-miR-1324 at the end of the section Examples of overmutated miRNA genes. To simplify the table, we omitted the prefix hsa-miR in the gene IDs.
3.5. Examples of overmutated miRNA genes
3.5.1. Hsa-miR-142
Hsa-miR-142 is, to the best of our knowledge, the only miRNA gene convincingly shown to be recurrently mutated in several neoplasms, which include acute myeloid leukaemia (AML) [57,58] and different types of B-cell lymphoma [59,60], chronic lymphocytic leukaemia (CLL) [61] and diffuse large-cell B-cell lymphoma [62], [63], [64], [65]. Additionally, in our sequencing analysis performed within the framework of other projects, we found also one mutation in the seed region of hsa-miR-142 (chr17:58331263C>T[-]) in the Raji Burkitt lymphoma cell line (out of 5 Burkitt's lymphoma cell lines tested) (Fig. 4a). The occurrence of hsa-miR-142 mutations in haematological cancers may be consistent with the high abundance of miR-142-3p in mature hematologic cells [66,67] and with the observation that loss of the miRNA impairs the development and function of different hematologic lineages [68], [69], [70]. In the TCGA cohort, hsa-miR-142 is expressed at a high or very high level in most of the cancer types (~2000 RPM) with the highest level (>20,000 RPM) in LAML, DLBC, and THYM.
Fig. 4.
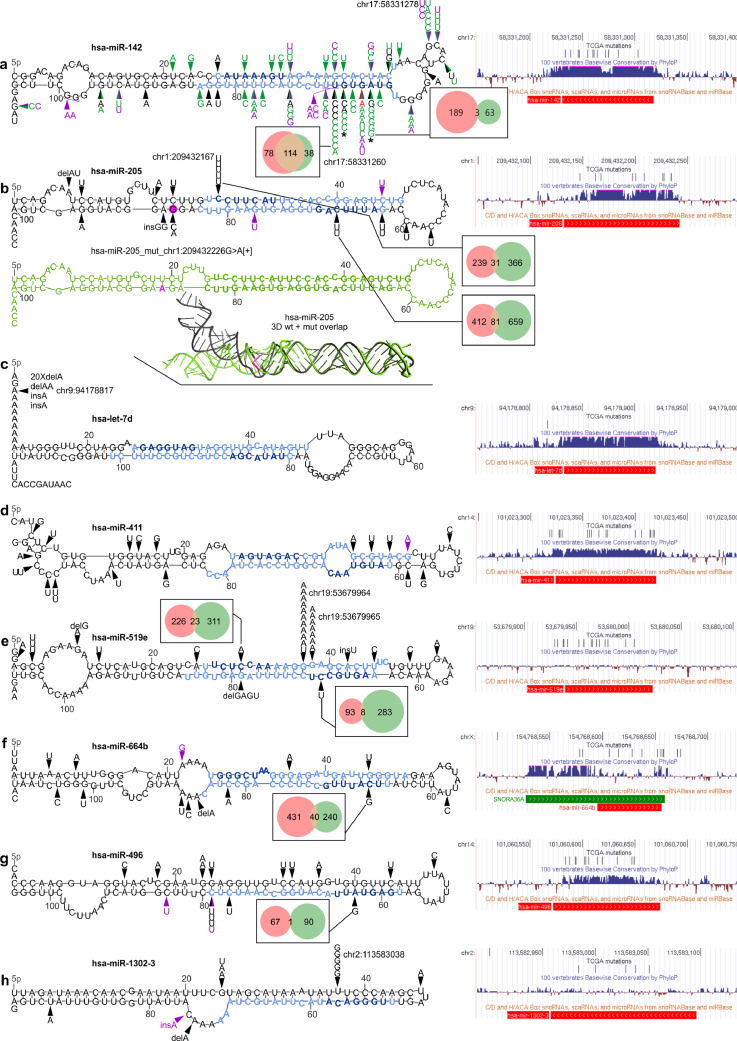
Localization of mutations in the selected overmutated miRNA genes. In each panel, on the left, mutations identified in the study in the TCGA samples (black arrowheads) and ICGC-PCAWG samples (purple arrowheads) are shown on mfold-predicted 2D structures of the miRNA precursors. Mature miRNA and seed sequences are indicated in light blue and dark blue, respectively. On the right, a screenshots from the UCSC genome browser, showing (from the top) a custom track with positions of the TCGA mutations detected in the study, conservation of the genomic region, and position of short non-coding RNAs, including pre-miRNAs (red bar; according to miRbase) and snoRNAs (green bar). For selected seed mutations, Venn-diagrams indicate the number of predicted targets for the wild-type (pink circle) and mutated (green circle) seeds. The effect of the seed mutations on the levels of the selected mutation-specific targets is shown in Supplementary Fig. S4. Genomic positions of the most significant hotspots are indicated next to mutations symbol. (a) Mutations found in hsa-miR-142. Green arrowheads indicate mutations detected in previous studies [[57], [58], [59], [60], [61],[63], [64], [65]] (for details see Supplementary Table S8); red arrowhead indicates the mutation identified by us in the Burkitt's lymphoma (Raji) cell line. * indicates the mutations tested functionally [27]. (b) Mutations found in hsa-miR-205 (above). Below: (i) the corresponding 2D structure of the precursor with the chr1:209432226G>A[+] mutation (drawn in green) and (ii) superimposed 3D structures of the wild-type (black) and mutant (green) precursors are shown. The position of the mutation and the mutant allele are marked in pink. (c-h) Mutations found in hsa-let-7d, hsa-miR-411, hsa-miR-519e, hsa-miR-664b, hsa-miR-496, and hsa-miR-1302-3, respectively.
In this study, we found 16 mutations in hsa-miR-142. Consistent with previous studies, we identified the highest number of mutations in DLBC (10 mutations, including 3 in one sample) and LAML (2 mutations), but we also found 4 mutations in solid tumours, i.e., in UCEC, BLCA, GBM, and BRCA, in which the hsa-miR-142 have not been found before. Five of the mutations in DLBC are located in the seed sequence of miR-142-3p, three in the 7th nucleotide (significantly recurring position chr17:58331260A>G[-]) and two in the 6th nucleotide (chr17:58331261C>G/T[-]) of the seed. Additionally, one mutation (chr17:58331264T>C[-]) was detected in the 3rd seed nucleotide in LAML. To better understand the distribution of mutations in hsa-miR-142, we combined the mutations detected in our study with the mutations detected previously (Fig. 4a, Supplementary Table S8). The distribution of mutations shows pronounced clustering of the mutations in the miR-142-3p seed region, with chr17:58331260A[-] being the most frequently mutated nucleotide, substituted with either G[-] (n=8) or T[-] (n=1). Nonetheless, a substantial fraction of the mutations is dispersed in other parts of the gene, including two recurring mutations in two subsequent positions of the loop. This result may suggest that the miRNA hairpin precursor is quite a fragile structure, and therefore, almost any mutation may be deleterious for the gene, either by disturbing the structure of the precursor or by disruption of the seed sequence. A recent functional study of two seed mutations, i.e., chr17:58331264T>C[-] and chr17:58331261C>G[-], showed that even though the mutations are located in miR-142-3p, they result in a decrease in both miR-142-3p and miR-142-5p levels and reverse the miR-5p:3p ratio (in favour of miR-3p) [27]. The functional consequences of the mutations are (i) aberration of hematopoietic differentiation, enhancing the myeloid and suppressing the lymphoid potential of hematopoietic progenitors, and (ii) inefficient repression of ASH1L, resulting in increased levels of HOXA9 and A10 (positively regulated by ASH1L) and ultimately leukemic transformation [27]. The question remains whether mutations in solid tumours, in which miR-142-3p acts predominantly as a tumour suppressor, among others targeting and downregulating TGFB1R and HMGB1 [71,72], may also have functional consequences.
3.5.2. Hsa-miR-205
Among the highly mutated miRNA genes, several encode miRNAs with an important and well-documented role in cancer. These genes include hsa-miR-205, whose miR-205-5p acts predominantly as a suppressormiR but also, depending on a tumour context and/or expression profile, as an oncomiR (reviewed in [73]). miR-205 is a highly conserved and well-validated miRNA. We found that hsa-miR-205 was overmutated in Pan-Cancer (in total, 15 mutations) with mutations in SKCM (5 mutations), CESC (3), LUSC (2), BLCA (2), COAD, ESCA, and THYM. The occurrence of mutations coincides with a very high expression of hsa-miR-205 in mutated cancers, exceeding 7,000 RPM in CESC, LUSC, BLCA, ESCA, and THYM. Five of the mutations are located in a single hotspot position (chr1:209432167C>T[+]) that is the first position of the seed sequence of miR-205-5p (guide miRNA). As we have shown before, the mutation may substantially affect target recognition, disrupting 250/288 (87%) predicted miR-205-5p targets and creating 471 new targets [33]. The disrupted targets include many validated miR-205-5p targets, including the oncogenes VEGFA (mediator of angiogenesis) and E2F1 (transcription factor controlling cell cycle) [74,75]. On the other hand, the recurrence of a specific mutation may suggest its gain-of-function character, such as the creation of a new seed/miRNA targeting the gene, whose downregulation may be beneficial for cancer. As shown in Supplementary Fig. S4 levels of some of the new target mRNAs are decreased in samples with the mutations. The other hsa-miR-205 mutations are dispersed alongside the miRNA duplex, hairpin loop, and flanking sequences. The mutations may affect the structure of the miRNA precursor and consequently its processing and effective miRNA biogenesis. An example of a mutation seriously affecting the structure is chr1:209432226G>A[+] transition, which disrupts the structure in the DROSHA cleavage site (Fig. 4b).
3.5.3. Hsa-let-7d
Another highly mutated miRNA gene playing a role in cancer is hsa-let-7d, which is located in a let-7a-1/let-7f-1/let-7d cluster and belonging to the let-7 family, one of the most extensively studied miRNA families in cancer. The let-7 miRNAs act as suppressormiRs and were found to be downregulated in many cancers. Hsa-let-7d is highly expressed in all tested samples. We found that hsa-let-7d was overmutated in Pan-Cancer (in total 23 mutations) and UCEC (16 mutations) but was also recurrently mutated in STAD (3) and COAD (2). All the mutations are indels of the poly-A10 tract (chr9:94178817[+] delA (20), delAA (1), and insA (2)) located in a 5p flanking sequence of the let-7d precursor (Fig. 4c). Although the mutation does not directly affect the sequence of mature let-7d, it may still affect miRNA processing. It should be noted, however, that indels in the polynucleotide tract, such as those observed in hsa-let-7d, may result from microsatellite instability (MSI). This is consistent with the overrepresentation of the hsa-let-7d indels in cancers such as UCEC, STAD, and COAD, in which MSI is especially frequent [53].
3.5.4. Hsa-miR-411
Hsa-miR-411 is overmutated in Pan-Cancer (19 mutations) and GBM (5 mutations) and is also recurrently mutated in SKCM (3) and ESCA (2). It is expressed in all but one tested cancers at a low to a high level. All mutations in hsa-miR-411 are substitutions and are generally dispersed over the gene without clustering in any specific region or hotspot (Fig. 4d), resembling a pattern of loss-of-function mutations, usually characteristic of tumour suppressor genes. This finding may be consistent with a predominantly tumour suppressor role and downregulation of miR-411-5p and miR-411-3p in different cancers [76], [77], [78], [79], [80]. Interestingly, miR-411-5p is posttranscriptionally modified (substitution A>I (inosine) in the 5th position of miR-411-5p seed) in both normal brain and glioblastoma multiforme tissues [80,81].
3.5.5. Hsa-miR-519e
With 33 identified mutations, hsa-miR-519e is overmutated in Pan-Cancer and two female-specific cancers, OV (16) and BRCA (6). The majority of the mutations are located in two subsequent hotspot positions, i.e., the 12th and 13th nucleotides (chr19:53679964G>A/T[+] and chr19:53679965G>A[+]) of the miR-519e-5p (passenger) strand (Fig. 4e), which are mutated predominantly in OV. Hsa-miR-519e belongs to the large (>50) miR-515 family coded in a cluster on 19q13.42. It is not a highly validated miRNA gene and is not well recognized as a cancer-related miRNA. In the TCGA cohort, hsa-miR-519e is expressed only in a few cancers (THYM, BLCA, and TGCT) on a low to medium level.
3.5.6. Hsa-miR-664b
Hsa-miR-664b is significantly overmutated in LUAD, with 6 mutations found in this cancer and 6 in other cancer types, including UCEC, STAD, SKCM, and CESC. The mutations were dispersed throughout the entire gene, with 5 mutations located in the mature miRNA sequences (Fig. 4f). Hsa-miR-664b is a highly validated (miRBase) and moderately conserved miRNA gene that substantially overlaps with the SNORA36A H/ACA box small nuclear RNA (snoRNA; snoRNABase) playing a role in the pseudouridylation of rRNAs and snRNAs; therefore, all mutations may also affect the function of snoRNA. miR-664b-5p was recently shown to act as a cancer suppressor in hepatocellular cancer cell lines [82]. The downregulated miR-664b-5p was associated with lower overall survival in cervical cancer [83], the proliferation of cutaneous malignant melanoma cells [84], and the progression of breast cancer [85]. The hsa-miR-664b is expressed in all of the TCGA cancer types in most of them including LUAD at a medium or high level.
3.5.7. Hsa-miR-496
Hsa-miR-496 was overmutated in the Pan-Cancer cohort with 15 mutations. Additionally, it was overmutated in SKCM with 7 mutations, 3 of which were located at a single position (chr14:101060649C>T[+]) in the DROSHA cleavage site (Fig. 4g), which may affect both the efficiency and precision of miRNA excision. Other mutated cancers include LUSC (2 mutations), HNC (2 mutations) and OV, UCEC, LUAD, and GBM (with single mutations). Hsa-miR-496 is expressed in most of the tested cancer types at low-medium levels. It is a conserved miRNA gene located in a large cluster (~40 miRNA genes) at ch14q31.31. It was shown that miR-496-3p plays a role in the regulation of the mTOR pathway [86] and Wnt pathway-mediated tumour metastasis in colorectal cancer [87].
3.5.8. Hsa-miR-1302-3
Hsa-miR-1302-3 is an example of a cancer-specific overmutated miRNA gene that is overmutated only in LIHC (5 mutations). Other mutated cancers include STAD (2 mutations) and ESCA, LUSC, CESC, and BRCA (with single mutations). Most of the mutations occur in two positions in the 5′ arm of the precursor, one of which is a hotspot mutation (chr2:113583038A>C[-]) significantly recurrent in LIHC and PAN-Cancer, localized within the passenger miRNA strand (Fig. 4h). The mutation replaces a U with G in a U:C mismatch and thus replaces the mismatch with a Watson-Crick pair G:C, greatly stabilizing the hairpin structure of the precursor (ddG=-6 kcal/mol; RNA mfold). miR-1302-3p has not been broadly researched, but its upregulation is associated with the recurrence and metastasis of prostate cancer [88]. The expression of hsa-miR-1302-3 was not observed in any of the TCGA cancer types, which argue against the functionality of the mutations, although, we cannot rule out that, in certain situations, some mutations may activate the expression or biogenesis of low- or not-expressed miRNAs.
3.5.9. Hsa-miR-1324
Finally, hsa-miR-1324 is the most commonly mutated gene, with a total of 166 mutations in Pan-Cancer, greatly exceeding the other highly mutated genes. The vast majority of the mutations (n = 146) were located in just two positions (i.e., chr3:75630855T>C[+] and chr3:375630794C>G[+]), which are also the two most highly mutated hotspots (Supplementary Fig. S3). We observed similar high frequency and a similar pattern of mutations in hsa-miR-1324 in a relatively small panel of diffuse large B-cell lymphoma and Hodgkin lymphoma cell lines sequenced in our laboratory with the conventional Sanger sequencing method (data not shown). However, the analysis of the genomic location of hsa-miR-1324 revealed that it is embedded in a large (>10 kb) segmentally duplicated region highly similar (>>95%) to at least 4 other sequences in the genome and likely variable in copy number [29,89,90]. The detailed comparison of the hsa-miR-1324 sequence with its paralog counterparts revealed that the substitutions differentiating paralogs correspond (position and type of substitution) with the identified mutations. This indicates that the hsa-miR-1324 mutations are most likely artefacts of the sequencing procedures and/or computational analyses (e.g., mapping). Additionally, miR-1324 is a low-confidence miRNA with only 48 confirming reads (miRBase, Mar 17, 2020) and is not annotated in MirGeneDB. It is also not detectable in any of the TCGA samples. In summary, based on the above facts, we concluded that the hsa-miR-1324 alterations are not credible somatic mutations, and therefore, we did not pursue further analysis of miR-1324.
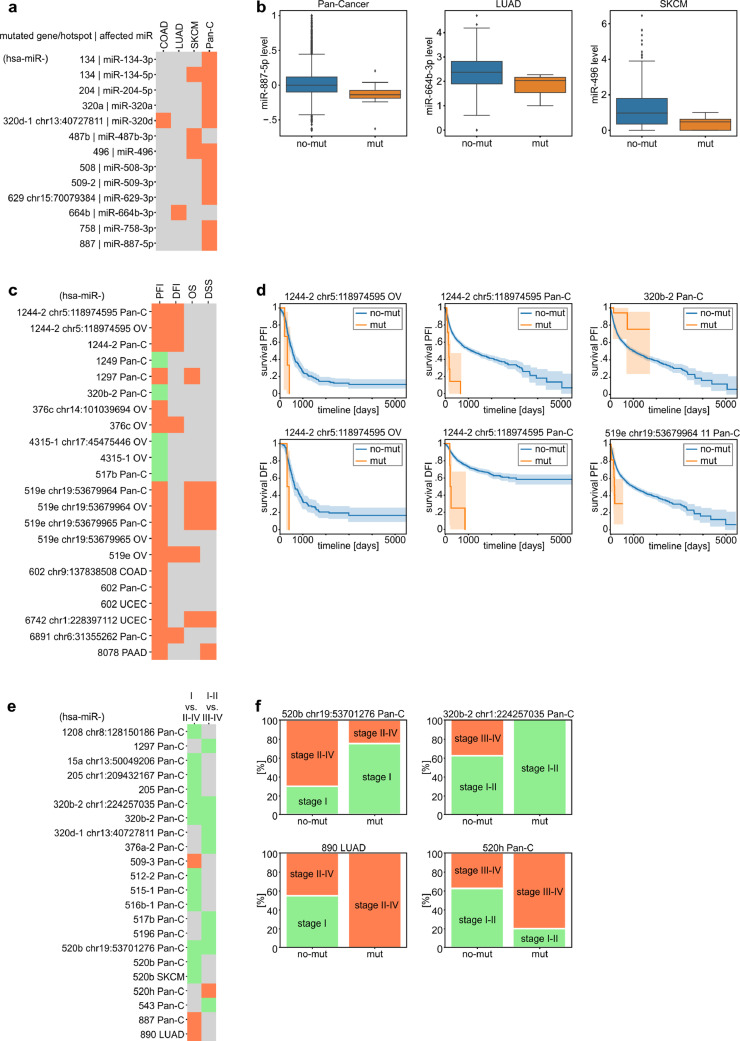
3.6. Effect of the mutations on the expression of the affected miRNA genes
To check whether mutations may affect miRNA expression, we compared the levels of miRNAs in samples with mutations vs. samples without mutations in genes either overmutated or with hotspot mutations in a particular cancer type or Pan-Cancer. To level the between-cancer expression differences, prior to Pan-Cancer analysis, we normalized the level of each miRNA to make its median level (equal to 0) and variation comparable between cancer types. We took into account only miRNAs whose level was >0 in at least 70% of the analyzed samples. Notably, not all miRNAs were covered in the TCGA miRNA expression data. As a result of the analysis, we identified 10 miRNA genes whose miRNA levels were downregulated in mutated samples, including hsa-miR-134, for which both miR-134-5p and miR-134-3p were downregulated in Pan-Cancer (Supplementary Table S9, and Fig. 5a and b). Additionally, we found 2 miRNAs whose levels were downregulated by particular hotspot mutations (Fig. 5a). No miRNA was upregulated by the mutations. The striking excess of downregulated miRNAs is consistent with the notion that most mutations are loss-of-function mutations for particular miRNA genes. It should be noted, however, that due to a low number of mutations, especially in the hotspots, the analysis is of relatively low statistical power, and most results are only nominally significant (Mann-Whitney or t-test, p<0.05; Supplementary Table S9), resulting in a relatively low number of miRNAs associated with the mutations in their genes. As some mature miRNAs are generated from more than one precursor (coded by groups of different miRNA genes), e.g., miR-320b (MIMAT0005792) is generated from hsa-miR-320b-1 and hsa-miR-320b-2, we compared the level of such miRNAs with mutations in the corresponding groups of miRNA genes. As shown in Supplementary Table S10 only mutations in groups of miRNA genes coding for miR-509-3p show borderline significant association with a decrease of the miRNA level. A similar trend for this miRNA is also visible in LUAD.
Fig. 5.
Association of miRNA gene mutations with miRNA expression, patient survival, and cancer stage. (a) Heatmap shows miRNAs whose levels are significantly changed in samples mutated in the indicated genes or hotspots. Orange indicates that all miRNAs were downregulated by mutations in their genes. (b) Box plots show representative examples of miRNAs whose levels were changed in samples with mutations (mut) vs. samples without mutations (no-mut) in the corresponding gene. For Pan-Cancer analysis, miRNA levels were normalized to allow comparison between cancer types. (c) Heatmap shows the miRNA genes or hotspot mutations significantly associated with PFI (green - positively, orange - negatively). In the next columns, associations of the genes/hotspots with the other survival metrics (DFI, DSS, and OS) are also shown. (d) Example survival plots comparing mut and no-mut samples (shown in c). (e) Heatmap shows miRNA genes or hotspot mutations associated with cancer stages. Orange and green colours indicate associations of samples bearing mutations with higher and lower cancer stages, respectively. (f) Examples of associations (shown in e) of mutation with the distribution of cancer stages. To simplify the figure, we omitted the prefix hsa-miR in the gene IDs.
3.7. Association of mutations in miRNA genes with patient survival and cancer aggressiveness
Changes in miRNA expression, processing, and target specificity may influence various cancer-related processes, including cell proliferation, metastasis, progression, and/or drug resistance. These changes may result in disease progression and treatment outcomes affecting patient survival. Multiple metrics associated with patient survival have been gathered within the TCGA project, including overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI), although not all are optimal for all cancers [39].
According to the recommendations in Liu et al. [39], we used PFI as a metric because it was permissible and most informative (had the highest statistical power) for the majority of TCGA cancer types. As survival metrics, including PFI, differ substantially between cancers, Pan-Cancer comparisons of survival in patients with mutations vs. patients without mutations may be affected by the fact that mutations are not equally distributed between cancer types. To overcome this effect, we used a stratified version of the log-rank test. We found 22 significant associations between mutations in the overmutated miRNA genes or hotspot positions and the PFI of cancer patients (either specific cancers or PAN-Cancer) (Supplementary Table S11). The associations were linked with mutations in 12 distinct miRNA genes (Fig. 5c). Interesting examples may be (i) hsa-miR-1244-2, in which hotspot mutations chr5:118974595C>T[+] are associated with decreased PFI in both OV and Pan-Cancer and total mutations decrease PFI in Pan-Cancer; (ii) hsa-miR-519e, in which total mutations are associated with decreased PFI in OV and hotspot mutations (chr19:53679964G>A/T[+], chr19:53679965G>A[+]) decrease PFI in OV and Pan-Cancer; and (iii) hsa-miR-602, in which hotspot mutations (chr9:137838508GC>G[+]) decrease PFI in COAD and total mutations decrease PFI in Pan-Cancer and UCEC (Fig. 5c and d). Additionally, we observed that mutations in hsa-miR-411 that was overmutated only in ordinary binomial analysis in GBM were associated with a decrease in PFI in GBM (log-rank test, p < 0.001, data not shown). As shown in the Fig. 5c, mutations of particular miRNA genes associated with PFI are also frequently associated with other measures of survival (DFI, OS, DSS), which were analyzed as appropriate for particular cancers [39]. A profound excess mutations associated with decreased survival may suggest a predominant tumour suppressor role of miRNAs, which is also consistent with the global decrease in miRNA levels observed in many cancers.
Next, we compared the occurrence of mutations in miRNA genes with cancer stages. The analysis showed 25 statistically significant associations of mutations, predominantly with lower cancer stages (Cochran–Mantel–Haenszel test for Pan-Cancer and Fisher exact test for specific cancers, p < 0.05, Supplementary Table S12, Fig. 5e and f). In two cases, i.e., hsa-miR-320b-2 and hsa-miR-517b, the association of the mutations with lower cancer stages corresponded with their positive effect on patient survival. However, due to the low number of identified mutations in particular miRNA genes or hotspots, the abovementioned associations with survival and cancer stages are of very low statistical power (not corrected for multiple comparisons) and therefore must be interpreted cautiously and cannot be generalized without further experimental validation.
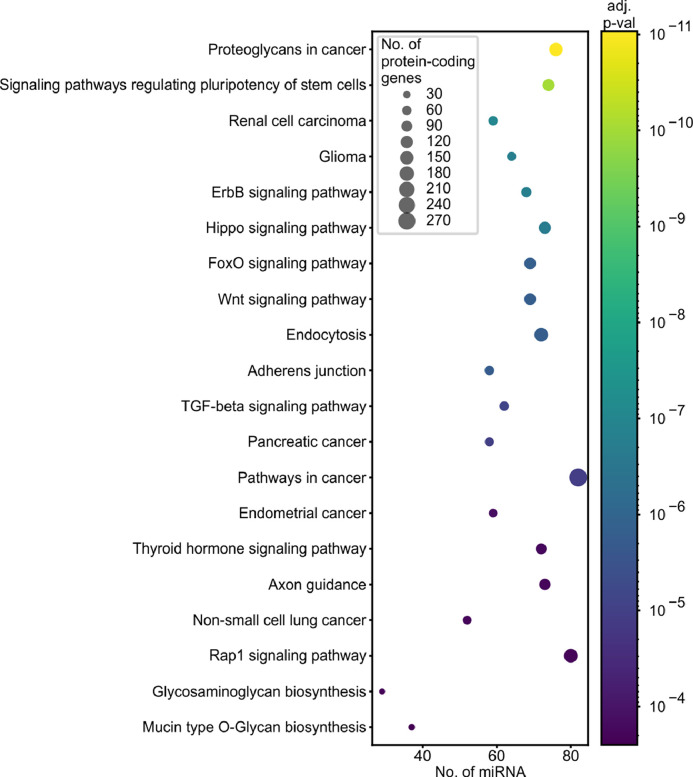
3.8. KEGG pathways associated with miRNA gene mutations
Finally, to identify pathways/processes enriched in the genes regulated by the most frequently mutated miRNA genes, we used miRPath v3.0 to perform KEGG pathway enrichment analysis. As shown in Supplementary Table S13 and Fig. 6, the vast majority of the associated (Fisher's combined probability method, adjusted p < 0.01) KEGG pathways are related to different cancers or cancer-related processes, such as the cell cycle, proliferation, or apoptosis. For example, the top ten most significant associations (Fisher's combined probability method, p < 0.000005) include the following terms: Proteoglycans in cancer, Signaling pathways regulating pluripotency of stem cells, Renal cell carcinoma, Glioma, ErbB signaling pathway, Hippo signaling pathway, FoxO signaling pathway, and Wnt signaling pathway (Supplementary Table S13, Fig. 6).
Fig. 6.
Functional association of overmutated miRNA genes with KEGG pathways. The graph shows the top 20 pathways (y-axis) enriched in protein-coding genes regulated by miRNAs (x-axis) encoded by overmutated miRNA genes. Dot size indicates the number of protein-coding genes; dot colour depicts an adjusted p-value (Fisher's combined probability method) of association. The enrichment analysis was performed with the use of miRPath v3.0 and encompassed the top 100 miRNA genes enriched in the functionally weighted test. The full list of enriched pathways is shown in Supplementary Table S13.
3.9. Identification of potential cancer drivers with the use of OncodriveFML
Although currently there is no software dedicated specifically to finding cancer driver miRNA genes and none of the currently available tools for non-coding regions take into account miRNA-specific characteristics (described above), we used OncodriveFML that predict potential cancer drivers based on CADD score of mutations deleteriousness that in non-coding regions is generally low. The results of the OncodriveFML analysis are shown in Supplementary Table S14. As expected, the analysis resulted in a smaller number of candidate driver genes with 8 miRNA genes identified in Pan-Cancer (hsa-miR-6891, hsa-miR-3918, hsa-miR-4726, hsa-miR-6728, hsa-miR-802, hsa-miR-1247, hsa-miR-623, and hsa-miR-939), 4 in UCEC, 3 in KIRP, 2 in LIHC, 2 in LUSC and 1 in PAAD, OV, LUAD, KIRC, GBM, DLBC, COAD, and CESC (at recommended Q-value <0.25, OncodriveFML). Overlapping miRNA genes between the weighted analysis and OncodriveFML analysis include hsa-miR-8078 in PAAD, hsa-miR-527 in LUSC, hsa-miR-142 in DLBC, and hsa-miR-6891 in CESC and PAN-Cancer.
3.10. Analysis of the ICGC-PCAWG cohort
For comparison with the results obtained based on the TCGA analysis, we analyzed the ICGC-PCAWG dataset that currently covers 2793 genomes. In total, we extracted 1523 mutations (Supplementary Table S15) in 856 miRNA genes (from 1914 analyzed for hg19). Although the number of identified mutations was too small to perform formal mutation-enrichment analysis, especially in individual cancers, the identified mutations were significantly enriched in the miRNA genes identified as overmutated in the TCGA Pan-Cancer cohort (at least one mutation identified in 66% of overmutated miRNA genes vs. 43% of all other tested genes; Fisher exact test, p < 0.0001). Among the recurrently mutated genes were hsa-miR-142 with 19 mutations in B-cell non-Hodgkin lymphoma (Lymph-BNHL, n = 16) and chronic lymphocytic leukaemia (Lymph-CLL, n = 3) samples, hsa-miR-205 and hsa-miR-496 (Fig. 3), as well as 10 other Pan-Cancer overmutated genes with at least 4 mutations.
4. Discussion
Multiple functional somatic mutations with roles in cancer are known in the coding portion of the genome. In this study, we identified 7110 mutations in miRNA genes across 33 cancer types based on data available in the TCGA repository. Most of the mutations were substitutions (~89%), with indels overrepresented within a couple of analyzed cancer types (COAD, STAD, and UCEC). Overall, approximately 33% of Pan-Cancer samples have at least one mutation in miRNA genes, with percentages substantially differing among cancer types, similar to what is observed for mutations in other genomic regions. The mutations were in general evenly distributed across miRNA gene functional subregions. This could be attributed to the fact that the majority of detected sequence variants are spontaneous mutations randomly accumulating in the cancer genome.
Among the identified mutations, we found ones located in miRNA genes playing an important and well-recognized role in cancer (e.g., hsa-let-7 family, hsa-miR-205, and hsa-miR-142) as well as miRNAs that were not yet investigated broadly in relation to cancer. In total, we identified 108 overmutated miRNA genes within the Pan-Cancer cohort and 80 overmutated miRNA genes within individual cancer types. In particular, we found multiple mutations in hsa-miR-142, hsa-miR-205, hsa-let-7d, hsa-miR-411, hsa-miR-519e, hsa-miR-664b, hsa-miR-585, hsa-miR-496, and hsa-miR-1302-3. Although the frequency of mutations in overmutated miRNA genes is lower than in commonly mutated drivers such as TP53, CDKN2A, or KRAS, it is comparable to the cancer-specific frequencies of mutations in many other protein-coding driver genes that are generally much longer than miRNA genes, e.g., MET (7%), RB1 (4%), and RIT1 (2%) in LUAD [91], HRAS (4%), PTEN, RB1, NF1 (<1%) in PTC [92] or DRD5 (3%), and BRAF (2%) in GBM [93].
Additionally, we identified 62 hotspot positions in Pan-Cancer and 69 in individual cancers, including 5 cancer-specific hotspots. One group of recurring mutations covers primarily insertions and deletions in short repeats. The mutations were identified predominantly in STAD and UCEC cancers known to be associated with MSI. It was previously suggested that simple repeats in human miRNA genes are relatively rare and preserved from mutations due to MSI [94]. Only three such mutations were identified in hsa-miR-1303, hsa-miR-567, and hsa-miR-1273. In our study, we identified indels associated with MSI in numerous miRNA genes (e.g., hsa-miR-320c-1, hsa-miR-320b-2, and hsa-miR-1249), including previously observed ones. Many MSI-associated mutations are recurrently mutated hotspots; for example, hsa-let-7d (chr9:94178817delAA[+]) in UCEC, COAD, and Pan-Cancer (Fig. 4c), hsa-miR-1303 (chr5:154685821TTA>T[+]) in STAD, UCEC, and Pan-Cancer and hsa-miR-567 (chr3:112112876TA/TAAA>T[+]) in UCEC and Pan-Cancer. This result shows that the idea of the involvement of MSI in mutations within miRNA genes should be revisited, especially as those mutations may also be functional [95,96].
As mentioned earlier, depending on localization within the miRNA gene, mutations can have multiple effects on miRNA functionality, including changes in targets (mutations within seeds) or processing (mutations that change structure or are located in the DROSHA/DICER1 cleavage site), resulting in changed miRNA levels and/or strand balance. In our study, we detected 536 mutations and 7 recurrently mutated hotspots in seed sequences of different miRNAs, including miRNAs with defined roles in cancer. As shown before, in our previous study, such mutations affect the vast majority of predicted miRNA targets. An example of a seed hotspot mutation is chr1:209432167C>T[+] in miR-205-5p, which affects most of the predicted miRNA targets. Additionally, due to mutation occurrence in the seed region, mutated miRNAs may gain the ability to recognize and downregulate new targets (Supplementary Fig. S4). Consistent with the putative effect of mutations on the effectiveness of miRNA biogenesis, we identified associations of recurrently mutated genes with the level of the corresponding miRNA, predominantly resulting in decreased (e.g., miR-664b-3p, miR-134-5p) levels of the affected miRNAs. Although probably not all of the observed miRNA aberrations play any relevant functional role in cancer, a vast excess of downregulated miRNAs confirms that mutations in miRNA genes have mostly destructive effects on the structure or stability of the miRNA precursors, making them less optimal substrates for the miRNA biogenesis process. Changes in miRNA levels are a known aspect of cancer characteristics; however, they are usually attributed to other mechanisms, and the effect of somatic mutations on miRNA expression has not been systematically studied before.
Subsequent analyses of available clinical data, including patient survival and cancer stage, showed that mutations in 12 miRNA genes were associated with different metrics of patient survival (predominantly with decreases in survival), and mutations in 18 miRNA genes were associated with cancer stage. This observation further confirms the potential functionality of the miRNA gene mutations acting directly (e.g., a mutation in seed), by changes in miRNA levels, or by other secondary effects. Although we tested only the effects of overmutated miRNA genes, we cannot exclude the possibility that some of the individual mutations also affect miRNA biogenesis/function and/or cancer. On the other hand, the identified associations do not prove the functionality of the particular mutations or groups of mutations in cancer. To provide such proof, independent functional analyses are needed, in which the results presented in our study may serve as a starting point or support. Such analyses will often have to be performed in the context of a particular cancer type or condition. On the other hand, globally, the nonrandom character of the identified mutations was confirmed by a strong association of overmutated miRNA genes with KEGG pathways, of which the vast majority were specific to particular cancers or cancer-related processes.
Many approaches have been developed to discover and evaluate cancer-driver mutations in protein-coding sequences, e.g., MutSig2CV, HotSpot 3D, CLUMPS, and PARADIGM-SHIFT [47,[97], [98], [99]], and numerous cancer-driving mutations and genes have been identified by taking advantage of these tools. The majority of these approaches take into account (i) well-known and easy to predict consequences of mutations in protein-coding sequences, i.e., distinguishing frameshift, nonsense, missense, splicing or synonymous mutations, (ii) the predicted effects of the mutations on the AA properties and/or tolerance of AA change in particular protein domains and/or the effect of AA change on protein structure, and (iii) the conservation of the particular AA residue or particular protein. These factors allow estimation of the excess of deleterious functionally relevant mutations over neutral variants, which is one of the most important components of models identifying signals of cancer-driven selection. Unfortunately, such tools cannot be utilized for the identification of drivers in non-coding regions, including sequences encoding “non-coding” RNA. Recognizing this limitation, several approaches dedicated to the identification of drivers in non-coding sequences or with added functionalities for this purpose have been proposed (e.g., oncodriveFML, MutSigNC, ncDriver, and LARVA) [60,63,100,101]. It was also recognized that due to different roles and functionalities, e.g., promoters, 5′ and 3′ untranslated regions (5′ and 3′ UTRs), introns, long non-coding RNAs, and miRNAs, different non-coding elements have to be analyzed with separate approaches or different assumptions [63]. Nonetheless, among the available tools, the functionality of non-coding mutations is mostly recognized by two factors: impact on protein (e.g., transcription factor) binding properties and impact on RNA structure. Sometimes, for specific ncRNA regions, additional factors are taken into account, such as the impact on miRNA binding sites in 3’UTRs. Although the structure is an important factor of miRNA biogenesis/functionality, the impact on RNA structure (e.g., the RNAsnp score) is inferred based on structures predicted for standardized tailing RNA fragments not corresponding to the size and coordinates of miRNA precursors. Therefore, at present, there is no approach/algorithm dedicated to recognize driving selection signals in miRNA genes. To overcome this limitation, in addition to evaluating the excess of the mutation in particular genes, we also weighted the mutations based on our proposed functionally related factors, with higher scores for mutations within the seed sequence, mature miRNAs, DROSHA/DICER1 cleavage sites, and disrupting protein binding motifs. Our results, together with recently published insights on mutations occurring in non-coding regions [54,60], may provide a basis for the development of new tools focused on miRNA cancer drivers based on described potentially functional mutations.
Still, it has to be noted that the overmutated miRNA genes identified in our study cannot be simply interpreted as cancer drivers. Although in our analysis, we took into account many aforementioned miRNA-gene specific factors due to the limitations mentioned above the analysis did not consider important mutation distribution parameters such as sample-specific mutation signatures, sample and cancer type mutation burdens, and differences in background mutation rates that may affect the results [47,48]. In consequence, some of the identified overmutated genes may result from the spurious overrepresentation of mutations in particular regions. This underlines the need for the development of a tool dedicated to the identification of cancer-driver mutations in miRNA genes.
As there is no list of previously defined miRNA driver genes, we could not formally validate our approach; however, among the top-scored overmutated miRNA genes, we identified hsa-miR-142, which is the only miRNA gene in which mutations were identified in several hematologic neoplasms in several studies [[57], [58], [59], [60], [61],64,65], and their cancer relevance was functionally confirmed [27]. Our analysis confirmed the recurrence of hsa-miR-142 mutations in hematologic neoplasms, i.e., LAML, DLBC, and also the newly identified mutation in the Burkitt lymphoma Raji cell line, but also showed mutations in several solid tumours, i.e., UCEC, BLCA, GBM, and BRCA. Additionally, thanks to the large number of mutations identified in our study and the cumulative analysis of previously detected mutations, we could illustrate for the first time the distribution of mutations in the gene. This result showed that mutations may occur in any part of the gene, not only in the seed sequence, which indicates their loss-of-function character, acting most likely by destabilizing the precursor structure and impairing miRNA biogenesis. This observation may also have the more general intriguing implication that miRNA precursors are overall quite fragile structures that may be affected by almost any mutation within the hairpin-coding sequence. Such hypotheses may be tested by the functional analysis of a higher number of randomly selected mutations in different miRNA genes.
There are several limitations of computational analyses such as the one presented in our study. First, further functional analyses of the identified recurring mutations are needed to verify their role in particular cancers. Second, not all known (miRBase) miRNA genes were covered by TCGA WES experiments. Additionally, due to different versions of WES systems used in different TCGA projects, the sequencing of some miRNA genes may not be equal in all samples. Third, even working with over 10,000 samples, the statistical power of some analyses is not sufficient, and further analyses with even larger cohorts of particular cancers or groups of cancers are awaited. Fourth, some of the TCGA cancer type cohorts are quite heterogeneous, consisting of samples of different genetic backgrounds. Finally, the analyses of mutations involved in cancers would also benefit from a better understanding of the structure of miRNA genes, including more complete information about the full sequence of miRNA transcriptional units (full pri-miRNA sequences) and their regulatory elements [102] and better functional validation/annotation of the known miRNA genes, as proposed, e.g., in the miRGeneDB database [42].
In summary, we present the first comprehensive Pan-Cancer study of somatic mutations in miRNA genes in a large cohort of cancer samples. As a result, we detected thousands of different mutations located in different functionally relevant parts of miRNA genes, and many miRNA genes were overmutated either in Pan-Cancer or in specific cancer types. The frequency of the mutations in some of the overmutated miRNA genes corresponds to that observed in some validated protein-coding driver genes. Subsequent analyses (miRNA expression, survival analyses, and functional pathway associations) suggest that at least some of the overmutated miRNA genes or hotspots in miRNA genes may be driven by cancer-positive selection and therefore may play a role in cancer. Nonetheless, the functionality of particular mutations needs to be experimentally validated with the use of appropriate functional tests. Our results are also the first step (form the basis and provide the resources) for the development of computational and/or statistical approaches and tools dedicated to the identification of cancer-driver miRNA genes.
Data sharing
The study was based on the data available at TCGA https://www.cancer.gov/tcga. The code used for analysis is publicly available in the github repository under the links indicated in Methods.
Contributors
All authors have read and approved the final version of the manuscript. MOUT – participated in conceiving the study, wrote all the scripts, performed most of the computational and statistical analyses, drafted the manuscript, prepared figures, tables, and supplementary materials; PGM – participated in conceiving the study, discussed the study on all steps of analyses, participated in manuscript preparation, performed some analyses (3D structures and miRNA targets expression level); PMN – discussed the analyses on all steps of the study, performed some analyses, participated in manuscript preparation; EK – performed the sequencing experiments, critically read and corrected manuscript; SS – performed the sequencing experiments, critically read and corrected manuscript; MG – provided samples for the hsa-miR-1324 and hsa-miR-142 sequencing validation experiments, advised on hematological neoplasms and head and neck cancer, critically read and corrected manuscript; PK – received financing, participated in conceiving the study, supervised and coordinated the study, drafted the manuscript (with MOUT).
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgements
The results published here are based upon data generated by the TCGA project (project ID: 16565). This work was supported by research grants from the Polish National Science Centre [2016/22/A/NZ2/00184 (to P.K.) and 2015/17/N/NZ3/03629 (to M.O.U-T.)]
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103051.
Appendix. Supplementary materials
References
- 1.Kumar S, Warrell J, Li S. Passenger mutations in more than 2500 cancer genomes: overall molecular functional impact and consequences. Cell. 2020;180:915–927. doi: 10.1016/j.cell.2020.01.032. .e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn S, Figl A, Rachakonda PS. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 3.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinagre J, Almeida A, Pópulo H. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 5.Hosseinahli N, Aghapour M, Duijf PHG, Baradaran B. Treating cancer with microRNA replacement therapy: a literature review. J Cell Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 6.Regouc M, Belge G, Lorch A, Dieckmann K-P, Pichler M. Non-coding microRNAs as novel potential tumor markers in testicular cancer. Cancers. 2020;12 doi: 10.3390/cancers12030749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neagu M, Constantin C, Cretoiu SM, Zurac S. miRNAs in the diagnosis and prognosis of skin cancer. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohel MMH. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117473. [DOI] [PubMed] [Google Scholar]
- 9.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Sevignani C, Dumitru CD. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czubak K, Lewandowska MA, Klonowska K. High copy number variation of cancer-related microRNA genes and frequent amplification of DICER1 and DROSHA in lung cancer. Oncotarget. 2015;6:23399–23416. doi: 10.18632/oncotarget.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florczuk M, Szpechcinski A, Chorostowska-Wynimko J. miRNAs as biomarkers and therapeutic targets in non-small cell lung cancer: current perspectives. Target Oncol. 2017;12:179–200. doi: 10.1007/s11523-017-0478-5. [DOI] [PubMed] [Google Scholar]
- 13.Krutovskikh VA, Herceg Z. Oncogenic microRNAs (OncomiRs) as a new class of cancer biomarkers. BioEssays. 2010;32:894–904. doi: 10.1002/bies.201000040. [DOI] [PubMed] [Google Scholar]
- 14.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 16.Vorozheykin PS, Titov II. How miRNA structure of animals influences their biogenesis. Russ J Genet. 2020;56:17–29. [Google Scholar]
- 17.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slezak-Prochazka I, Durmus S, Kroesen B-J, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA N Y N. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong J, Liu C, Liu W. An update of miRNASNP database for better SNP selection by GWAS data, miRNA expression and online tools. Database J Biol Databases Curation. 2015;2015 doi: 10.1093/database/bav029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conte I, Hadfield KD, Barbato S. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc Natl Acad Sci USA. 2015;112:E3236–E3245. doi: 10.1073/pnas.1401464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mencía A, Modamio-Høybjør S, Redshaw N. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 22.Soldà G, Robusto M, Primignani P. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum Mol Genet. 2012;21:577–585. doi: 10.1093/hmg/ddr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun G, Yan J, Noltner K. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jazdzewski K, Liyanarachchi S, Swierniak M. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci. 2009;106:1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes AE, Bradley DT, Campbell M. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89:628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J, Ambrosone CB, Zhao H. Novel genetic variants in microRNA genes and familial breast cancer. Int J Cancer. 2009;124:1178–1182. doi: 10.1002/ijc.24008. [DOI] [PubMed] [Google Scholar]
- 27.Trissal MC, Wong TN, Yao J-C. MIR142 loss-of-function mutations depreress ASH1L to increase HOXA gene expression and promote leukemogenesis. Cancer Res. 2018;78:3510–3521. doi: 10.1158/0008-5472.CAN-17-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders MA, Liang H, Li W-H. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcinkowska M, Szymanski M, Krzyzosiak WJ, Kozlowski P. Copy number variation of microRNA genes in the human genome. BMC Genom. 2011;12:183. doi: 10.1186/1471-2164-12-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quach H, Barreiro LB, Laval G. Signatures of purifying and local positive selection in human miRNAs. Am J Hum Genet. 2009;84:316–327. doi: 10.1016/j.ajhg.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning S, Yue M, Wang P. LincSNP 2.0: an updated database for linking disease-associated SNPs to human long non-coding RNAs and their TFBSs. Nucleic Acids Res. 2017;45:D74–D78. doi: 10.1093/nar/gkw945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue M, Zhou D, Zhi H. MSDD: a manually curated database of experimentally supported associations among miRNAs, SNPs and human diseases. Nucleic Acids Res. 2018;46:D181–D185. doi: 10.1093/nar/gkx1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galka-Marciniak P, Urbanek-Trzeciak MO, Nawrocka PM. Somatic mutations in miRNA genes in lung cancer—potential functional consequences of non-coding sequence variants. Cancers. 2019;11:793. doi: 10.3390/cancers11060793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharya A, Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 2016;44:D1005–D1010. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharya A, Ziebarth JD, Cui Y. SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013;41:D977–D982. doi: 10.1093/nar/gks1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lackey L, Coria A, Woods C, McArthur E, Laederach A. Allele-specific SHAPE-MaP assessment of the effects of somatic variation and protein binding on mRNA structure. RNA N Y N. 2018;24:513–528. doi: 10.1261/rna.064469.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellrott K, Bailey MH, Saksena G. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6:271–281. doi: 10.1016/j.cels.2018.03.002. .e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Lichtenberg T, Hoadley KA. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldman M, Craft B, Hastie M, et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv2019;: 326470.
- 41.Fromm B, Billipp T, Peck LE. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fromm B, Domanska D, Høye E. MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 2020;48:D132–D141. doi: 10.1093/nar/gkz885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antczak M, Popenda M, Zok T. New functionality of RNAComposer: an application to shape the axis of miR160 precursor structure. Acta Biochim Pol. 2016;63:737–744. doi: 10.18388/abp.2016_1329. [DOI] [PubMed] [Google Scholar]
- 46.Urbanek-Trzeciak MO, Jaworska E, Krzyzosiak WJ. miRNAmotif-A tool for the prediction of pre-miRNA−protein interactions. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19124075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence MS, Stojanov P, Polak P. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez-Perez A, Sabarinathan R, Lopez-Bigas N. Local determinants of the mutational landscape of the human genome. Cell. 2019;177:101–114. doi: 10.1016/j.cell.2019.02.051. [DOI] [PubMed] [Google Scholar]
- 49.Davidson-Pilon Cameron, Kalderstam Jonas, Jacobson Noah. CamDavidsonPilon/lifelines: 0.24.6. Zenodo. 2020 doi: 10.5281/zenodo.3787142. [DOI] [Google Scholar]
- 50.Seabold S, Perktold J. 2010. Statsmodels: Econometric and Statistical Modeling with Python; pp. 92–96. Austin, Texas. [Google Scholar]
- 51.Reilly NM, Novara L, Di Nicolantonio F, Bardelli A. Exploiting DNA repair defects in colorectal cancer. Mol Oncol. 2019;13:681–700. doi: 10.1002/1878-0261.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maruvka YE, Mouw KW, Karlic R. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat Biotechnol. 2017;35:951–959. doi: 10.1038/nbt.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cortes-Ciriano I, Lee S, Park W-Y, Kim T-M, Park PJ. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun. 2017;8:15180. doi: 10.1038/ncomms15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oak N, Ghosh R, Huang K-L, Wheeler DA, Ding L, Plon SE. Framework for microRNA variant annotation and prioritization using human population and disease datasets. Hum Mutat. 2019;40:73–89. doi: 10.1002/humu.23668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cantini L, Bertoli G, Cava C. Identification of microRNA clusters cooperatively acting on epithelial to mesenchymal transition in triple negative breast cancer. Nucleic Acids Res. 2019;47:2205–2215. doi: 10.1093/nar/gkz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hasegawa T, Glavich GJ, Pahuski M. Characterization and evidence of the miR-888 cluster as a novel cancer network in prostate. Mol Cancer Res MCR. 2018;16:669–681. doi: 10.1158/1541-7786.MCR-17-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thol F, Scherr M, Kirchner A. Clinical and functional implications of microRNA mutations in a cohort of 935 patients with myelodysplastic syndromes and acute myeloid leukemia. Haematologica. 2015;100:e122–e124. doi: 10.3324/haematol.2014.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cancer Genome Atlas Research Network. TJ Ley, Miller C. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouska A, Zhang W, Gong Q. Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia. 2017;31:83–91. doi: 10.1038/leu.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rheinbay E, Nielsen MM, Abascal F. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020;578:102–111. doi: 10.1038/s41586-020-1965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puente XS, Beà S, Valdés-Mas R. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 62.Morin RD, Assouline S, Alcaide M. Genetic landscapes of relapsed and refractory diffuse large B-Cell lymphomas. Clin Cancer Res Off J Am Assoc Cancer Res. 2016;22:2290–2300. doi: 10.1158/1078-0432.CCR-15-2123. [DOI] [PubMed] [Google Scholar]
- 63.Hornshøj H, Nielsen MM, Sinnott-Armstrong NA. Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. NPJ Genomic Med. 2018;3:1. doi: 10.1038/s41525-017-0040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hezaveh K, Kloetgen A, Bernhart SH. Alterations of miRNAs and miRNA-regulated mRNA expression in GC B cell lymphomas determined by integrative sequencing analysis. Haematologica. 2016 doi: 10.3324/haematol.2016.143891. published online July 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwanhian W, Lenze D, Alles J. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 2012;1:141–155. doi: 10.1002/cam4.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur J Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 67.Petriv OI, Kuchenbauer F, Delaney AD. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci. 2010 doi: 10.1073/pnas.1009320107. published online Aug 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mildner A, Chapnik E, Manor O. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood. 2013;121:1016–1027. doi: 10.1182/blood-2012-07-445999. [DOI] [PubMed] [Google Scholar]
- 69.Rivkin N, Chapnik E, Mildner A. Erythrocyte survival is controlled by microRNA-142. Haematologica. 2017;102:676–685. doi: 10.3324/haematol.2016.156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kramer NJ, Wang W-L, Reyes EY. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood. 2015;125:3720–3730. doi: 10.1182/blood-2014-10-603951. [DOI] [PubMed] [Google Scholar]
- 71.Xiao P, Liu W-L. MiR-142-3p functions as a potential tumor suppressor directly targeting HMGB1 in non-small-cell lung carcinoma. Int J Clin Exp Pathol. 2015;8:10800–10807. [PMC free article] [PubMed] [Google Scholar]
- 72.Lei Z, Xu G, Wang L. MiR-142-3p represses TGF-β-induced growth inhibition through repression of TGFβR1 in non-small cell lung cancer. FASEB J. 2014;28:2696–2704. doi: 10.1096/fj.13-247288. [DOI] [PubMed] [Google Scholar]
- 73.Qin A-Y, Zhang X-W, Liu L. MiR-205 in cancer: an angel or a devil. Eur J Cell Biol. 2013;92:54–60. doi: 10.1016/j.ejcb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 74.Vosgha H, Ariana A, Smith RA, Lam AK-Y. miR-205 targets angiogenesis and EMT concurrently in anaplastic thyroid carcinoma. Endocr Relat Cancer. 2018;25:323–337. doi: 10.1530/ERC-17-0497. [DOI] [PubMed] [Google Scholar]
- 75.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286:16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin H, Sun W, Zhang Y. MicroRNA-411 downregulation enhances tumor growth by upregulating MLLT11 expression in human bladder cancer. Mol Ther - Nucleic Acids. 2018;11:312–322. doi: 10.1016/j.omtn.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo L, Yuan J, Xie N. miRNA-411 acts as a potential tumor suppressor miRNA via the downregulation of specificity protein 1 in breast cancer. Mol Med Rep. 2016;14:2975–2982. doi: 10.3892/mmr.2016.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xia L-H, Yan Q-H, Sun Q-D, Gao Y-P. MiR-411-5p acts as a tumor suppressor in non-small cell lung cancer through targeting PUM1. Eur Rev Med Pharmacol Sci. 2018;22:5546–5553. doi: 10.26355/eurrev_201809_15816. [DOI] [PubMed] [Google Scholar]
- 79.Chen FD, Chen HH, Ke SC, Zheng LR, Zheng XY. SLC27A2 regulates miR-411 to affect chemo-resistance in ovarian cancer. Neoplasma. 2018;65:915–924. doi: 10.4149/neo_2018_180122N48. [DOI] [PubMed] [Google Scholar]
- 80.Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 2011;6:e24248. doi: 10.1371/journal.pone.0024248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paul D, Sinha AN, Ray A. A-to-I editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme. Sci Rep. 2017;7:2466. doi: 10.1038/s41598-017-02397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, Guo D, Zhang Y, Yang S, Zhang R. miR-664b-5p inhibits hepatocellular cancer cell proliferation through targeting oncogene AKT2. Cancer Biother Radiopharm. 2020 doi: 10.1089/cbr.2019.3043. published online Jan 20. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Y-X, Qin L-L, Yang S-Y. Down-regulation of miR-664 in cervical cancer is associated with lower overall survival. Eur Rev Med Pharmacol Sci. 2016;20:1740–1744. [PubMed] [Google Scholar]
- 84.Ding Z, Jian S, Peng X. Loss of MiR-664 Expression enhances cutaneous malignant melanoma proliferation by upregulating PLP2. Medicine. 2015;94:e1327. doi: 10.1097/MD.0000000000001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Song W, Tang L, Xu Y. PARP inhibitor increases chemosensitivity by upregulating miR-664b-5p in BRCA1-mutated triple-negative breast cancer. Sci Rep. 2017;7:42319. doi: 10.1038/srep42319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alqurashi N, Hashimi SM, Alowaidi F, Ivanovski S, Farag A, Wei MQ. miR-496, miR-1185, miR-654, miR-3183 and miR-495 are downregulated in colorectal cancer cells and have putative roles in the mTOR pathway. Oncol Lett. 2019;18:1657–1668. doi: 10.3892/ol.2019.10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang H, Yan B, Zhang P. MiR-496 promotes migration and epithelial-mesenchymal transition by targeting RASSF6 in colorectal cancer. J Cell Physiol. 2020;235:1469–1479. doi: 10.1002/jcp.29066. [DOI] [PubMed] [Google Scholar]
- 88.Nam RK, Amemiya Y, Benatar T. Identification and validation of a five MicroRNA signature predictive of prostate cancer recurrence and metastasis: a cohort study. J Cancer. 2015;6:1160–1171. doi: 10.7150/jca.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Conrad DF, Pinto D, Redon R. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Veerappa AM, N MM, Vishweswaraiah S. Copy number variations burden on miRNA genes reveals layers of complexities involved in the regulation of pathways and phenotypic expression. PLoS One. 2014;9:e90391. doi: 10.1371/journal.pone.0090391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brennan CW, Verhaak RGW, McKenna A. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.El-Murr N, Abidi Z, Wanherdrick K. MiRNA genes constitute new targets for microsatellite instability in colorectal cancer. PLoS One. 2012;7:e31862. doi: 10.1371/journal.pone.0031862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Woerner SM, Yuan YP, Benner A, Korff S, von Knebel Doeberitz M, Bork P. SelTarbase, a database of human mononucleotide-microsatellite mutations and their potential impact to tumorigenesis and immunology. Nucleic Acids Res. 2010;38:D682–D689. doi: 10.1093/nar/gkp839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Timmermann B, Kerick M, Roehr C. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PloS One. 2010;5:e15661. doi: 10.1371/journal.pone.0015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen S, He X, Li R, Duan X, Niu B. HotSpot3D web server: an integrated resource for mutation analysis in protein 3D structures. Bioinformatics DOI:10.1093/bioinformatics/btaa258. [DOI] [PubMed]
- 98.Kamburov A, Lawrence MS, Polak P. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci USA. 2015;112:E5486–E5495. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ng S, Collisson EA, Sokolov A. PARADIGM-SHIFT predicts the function of mutations in multiple cancers using pathway impact analysis. Bioinformatics. 2012;28:i640–i646. doi: 10.1093/bioinformatics/bts402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17:128. doi: 10.1186/s13059-016-0994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lochovsky L, Zhang J, Fu Y, Khurana E, Gerstein M. LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations. Nucleic Acids Res. 2015;43:8123–8134. doi: 10.1093/nar/gkv803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.SiamiGorji S, Jorjani I, Tahamtan A, Moradi A. Effects of microRNAs polymorphism in cancer progression. Med J Islam Repub Iran. 2020;34:3. doi: 10.34171/mjiri.34.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.