Abstract
Background
An artificial intelligence method could accelerate the clinical implementation of tumour-stroma ratio (TSR), which has prognostic relevance in colorectal cancer (CRC). We, therefore, developed a deep learning model for the fully automated TSR quantification on routine haematoxylin and eosin (HE) stained whole-slide images (WSI) and further investigated its prognostic validity for patient stratification.
Methods
We trained a convolutional neural network (CNN) model using transfer learning, with its nine-class tissue classification performance evaluated in two independent test sets. Patch-level segmentation on WSI HE slides was performed using the model, with TSR subsequently derived. A discovery (N=499) and validation cohort (N=315) were used to evaluate the prognostic value of TSR for overall survival (OS).
Findings
The CNN-quantified TSR was a prognostic factor, independently of other clinicopathologic characteristics, with stroma-high associated with reduced OS in the discovery (HR 1.72, 95% CI 1.24-2.37, P=0.001) and validation cohort (2.08, 1.26-3.42, 0.004). Integrating TSR into a Cox model with other risk factors showed improved prognostic capability.
Interpretation
We developed a deep learning model to quantify TSR based on histologic WSI of CRC and demonstrated its prognostic validity for patient stratification for OS in two independent CRC patient cohorts. This fully automatic approach allows for the objective and standardised application while reducing pathologists' workload. Thus, it can potentially be of significant aid in clinical prognosis prediction and decision-making.
Funding
National Key Research and Development Program of China, National Science Fund for Distinguished Young Scholar, and National Science Foundation for Young Scientists of China.
Keywords: Deep learning, Colorectal cancer, Tumour-stroma ratio, Whole-slide image, Prognosis prediction
Research in context.
Evidence before this study
Emerging evidence shows that tumour-stroma ratio (TSR) has an independent prognostic relevance in several oncologic diseases, including colorectal cancer (CRC). TSR is conventionally assessed on haematoxylin and eosin (HE) stained sections, under a microscope, by pathologists, visually. With inter-observer agreement ranging from 0.239 to 0.886 (Cohen's kappa), there exists the opportunity for improving the TSR evaluation. We screened MEDLINE, Web of Science for relevant articles on Aug 15, 2020, with the terms ("artificial intelligence" OR "deep learning") AND "whole-slide images" AND "tumour-stroma ratio" AND "colorectal cancer". And there was no study to develop a deep learning model for the fully automated tumour-stroma ratio quantification using whole-slide HE-stained images of CRC.
Added value of this study
We presented a deep learning model for the fully automated TSR quantification using whole-slide HE-stained images of CRC. We further showed the CNN-based TSR as a prognostic factor of overall survival in two independent CRC patient cohorts. Combined into a prediction model, TSR demonstrated its potential for integrating with the TNM staging system.
Implications of all the available evidence
This approach permits the standardisation and reproducibility of TSR assessment on ubiquitously available HE-stained histological images to eliminate variations documented with traditional visual assessment while reducing the pathologists' workload. This fully automatic workflow is well suited for its implementation in clinical practice and could accelerate the clinical implication of TSR for prognostication and decision making. The data sets and model are publicly available to facilitate further validation and use by other researchers and clinicians.
Alt-text: Unlabelled box
1. Introduction
Colorectal cancer (CRC), the third most common cancer worldwide, has a high mortality burden [1]. Although the tumour node metastasis (TNM) staging system serves as the basis for treatment decision for CRC patients, diverse prognosis observed within each stage calls for improved informative markers [2, 3, 4]. For decades, histopathology evaluation serves as the backbone for the definitive diagnosis of CRC, and routine haematoxylin and eosin (HE) stained tissue sections are indispensable to prognostic prediction [5]. Emerging evidence shows that tumour-stroma ratio (TSR), also known as tumour-stroma percentage, has an independent prognostic relevance in several oncologic diseases, including CRC [6, 7, 8, 9, 10]. TSR is conventionally assessed on HE-stained sections, under a microscope, by pathologists, visually [11]; with discrepancies reported among pathologists, and the process is not easily scalable [10–12]. With inter-observer agreement ranging from 0.239 to 0.886 (Cohen's kappa) [12], there exists the opportunity for improving the TSR evaluation. Since reliable assessment is crucial for subsequent patient stratification and follow-up, an automated method that could enable the objective and standardised TSR quantification, with optimal reproducibility, would have the scope of application.
On the other hand, the recent trend in digitalised pathology workflow also calls for automated evaluation methods with standardised protocol [10,13]. Automated histopathological analysis improves both efficiency and consistency compared with traditional evaluation [14,15]. For TSR, a few studies have applied computational image analysis to explore the possibility of classifying stroma and tumour epithelial cells using small parts of tissue samples [10,11,16]. However, fully automated TSR assessment that could be applied on whole-slide images (WSI) is yet unavailable [16–18]. The recent availability of digital WSI and the stunning success of convolutional neural networks (CNNs) in medical imaging, presents an opportunity for fully automatic pathologic assessment of CRC [19,20].
The deep learning model for digital whole-slide HE-stained image analysis could eliminate – or at least reduce – the variations in TSR assessment results, observed among pathologists. Therefore, we aimed to develop a deep learning model for the fully automated TSR assessment on HE-stained WSI and to validate its prognostic utility in independent patient cohorts with CRC.
2. Methods
2.1. Patients
This retrospective study included a discovery and validation cohort, with follow-up information, to evaluate the prognostic value of a CNN-scored TSR (Supplementary Fig. 1a). Patients were recruited between March 2008 and May 2015. For the discovery cohort (Guangdong Provincial People's Hospital) and validation cohort (Yunnan Cancer Hospital), consecutive CRC patients who underwent surgery with curative intent, with available paraffin-embedded tumour samples, were enrolled. The Institutional Review Board (the Research Ethics Committee of Guangdong Provincial People's Hospital, the Institutional Review Boards of Yunnan Cancer Hospital) of each participating hospital approved the use of human tissues, with the need of informed consent waived for this retrospective study. Exclusion criteria were neo-adjuvant therapy (radiotherapy, chemotherapy) and death within 30 days of surgery. The outcome of interest was overall survival (OS), defined as the time from surgery to death due to any cause. Patients were followed-up using abdominal computed tomography every 6 to 12 months for the first two years, and then annually. The follow-up duration was measured from the time of surgery to the last follow-up date, with information regarding survival status at the last follow-up documented. Clinicopathological characteristics information (age at diagnosis, sex, TNM stage, and tumour anatomic site [colon or rectum]), were collected. TNM staging was performed according to the Union for International Cancer Control (UICC) guideline [3]. Patients with missing clinical information (on mortality or time) were excluded, and no imputation was used in this study. Image quality control was conducted, excluding blurry, artefacts, and over- or light-stained HE images.
2.2. Procedures
Routine HE-stained sections showing the most invasive part of the primary tumour were chosen from the tissue block for analysis by an experienced pathologist in each institute (S.Y. in Guangdong Provincial People's Hospital and L.W. in Yunnan Cancer Hospital) on conventional microscopy examination. The selection procedure was blinded to patient clinical information and outcome. The selected HE-stained tissue sections were imaged using digital Whole Slide Scanning (Leica, Aperio-AT2, USA) at 40 × magnification, and 0.252 μm/pixel resolution.
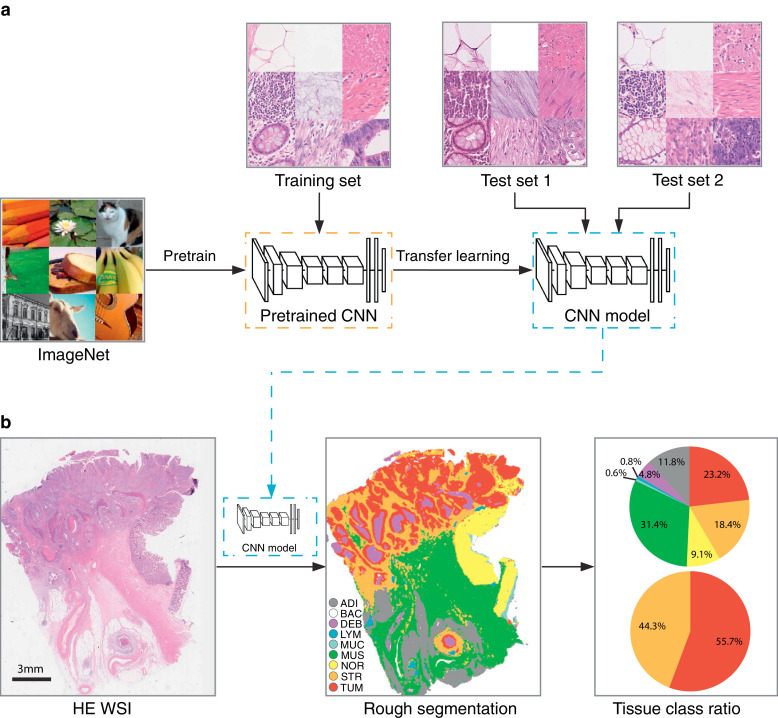
Inspired by the study conducted by Kather et al., CRC tissues were grouped into nine classes [20] (except BACK class, for different definition), including adipose (ADI), background (BAC), debris (DEB), lymphocytes (LYM), mucus (MUC), smooth muscle (MUS), normal mucosa (NOR), stroma (STR), and tumour epithelium (TUM). ImageScope (version 12.4.3, Leica, USA) was used for image annotation of the nine tissue class. Then image patches with size 224 × 224 pixel2 (20 × magnification) were extracted from annotation regions (Supplementary Fig. 2).
2.3. Data sets composition
The following four datasets (a training set, two test sets, and an evaluation set) were used for the training of the CNN model and the assessment of its tissue classification performance (Supplementary Fig. 1b). The training set used to train the CNN model, included 283.1k image patches, including HE slides of CRC tissue randomly selected from those retrieved from The Cancer Genome Atlas (training set part 1, 89.1k patches from 85 slides). The dataset was released with Kather et al. 's paper [19] (NCT-HE-100K dataset excluding BACK class, the training set part 2, 88.9k patches), and those randomly selected from the discovery cohort (training set part 3, 105.1k patches from 106 slides). Two independent test sets (test sets 1 and 2) were used to assess the classification performance of the trained CNN model. Dataset released with Kather et al. 's paper (CRC-VAL-HE-7K) was used to form the test set 1 (6.3k patches, excluding the BACK class), and those randomly chosen from validation cohort were used to establish the test set 2 (22.5k patches from 48 slides). The evaluation set, used for the TSR consistency analysis, included 126 image blocks (1 μm × 1 μm) from regions that only consisted of TUM and STR tissue classes. These image blocks were extracted from 42 randomly selected slides in the discovery cohort. Pixel-level annotation by a pathologist (S.Y.) was performed on these image blocks, as the ground truth.
2.4. Training, testing, assessment of the neural network and TSR calculation
The full procedure is shown in Fig. 1. First, a CNN model (VGG-19) [21] was pre-trained on the ImageNet dataset (www.image-net.org). The final classification layer was replaced by a nine-category layer (corresponding to nine tissue classes). This model was then trained on the training dataset to classify different tissue types in pathologic CRC images using transfer learning with stochastic gradient descent with momentum (SGDM). We trained the network on a desktop workstation with two Nvidia 1080Ti GPUs, with a mini-batch size of 128 and a fixed learning rate of 3 × 10−4 for four epochs. The classification accuracy of the CNN model was assessed in two independent test sets (test sets 1 and 2) using metrics of accuracy and Cohen's kappa. The tSNE method was used for the visualisation of the CNN model deep layer activations (45-layer, fully connected layer).
Fig. 1.
Study design of the CNN model development and application. (a) A CNN model (VGG-19) was pre-trained on ImageNet dataset, and transfer learning was used to train the CNN model with the training set. Two independent image data sets were used to assess the classification accuracy of the model. (b) HE WSI image (20 × magnification) was segmented by the CNN model with sliding window methods. Eight tissue classes (excluding BAC class) ratio was calculated by counting each tissue area in the segmented result, and the tumour-stroma ratio was also obtained. CNN, convolutional neural network; HE, haematoxylin–eosin; WSI, whole-slide image; ADI, adipose; BAC, background; DEB, debris; LYM, lymphocyte aggregates; MUC, mucus; MUS, muscle; NOR, normal mucosa; STR, stroma; TUM, tumour epitheliu.
After testing the trained CNN model, we performed patch-level segmentation on WSI with the CNN model in two cohorts. A sliding window with size 224 × 224 pixel2 was used to extract partially overlapping tiles from WSI HE images at 20 × magnification. The step size of the sliding window was set at 84 pixels. As the sliding window traversed the entire WSI, each image tile was input into the CNN model to generate a prediction probability. The final classification result of the image tile was set as the tissue class with the maximum prediction probability. The entire process used GPUs for accelerated computation. Distinct colours for visualisation represented output nine classes. Lastly, tissue class ratio was calculated for each tissue types based on each tissue area. The TSR is defined as .
TSR consistency analysis was performed on the evaluation set by assessing the concordance in TSR estimation between the CNN model and pathologist annotation. The correlation coefficient (Pearson r) and intra-class correlation coefficient (ICC) were calculated. The agreement in TSR estimation between the CNN model and pathologist annotation was determined using the Bland–Altman plot.
2.5. Evaluation of the prognostic value of TSR
Potential cut-off of the TSR, to distinguish between stroma-high and stroma-low patients, associated with OS differences, were determined using maximally selected rank statistics [22] in the discovery cohort. Kaplan–Meier survival analysis was applied for the analysis of the survival curves, and log-rank statistics were used to test the differences in survival distributions. Stratified analyses were conducted to investigate the association between TSR and OS within subgroups of stage and clinicopathologic risk factors (age, sex, tumour site, and stage) on the entire discovery and validation cohort patients.
Uni- and multivariate survival analyses were performed using the Cox proportional hazard model, on the discovery and validation cohorts, for TSR and clinicopathological variables (age, sex, stage, and tumour site). We used Cox regression coefficients in multivariate analysis for the discovery cohort to generate a prediction model (TSR model). To investigate whether TSR could provide incremental value for clinicopathologic risk factors, we developed a reference model, which incorporating independent clinicopathologic factors only, except TSR.
The discrimination performance of the prediction models was assessed using the Harrell's C-statistics (C-index) and the integrated area under the ROC curve (iAUC). Time-dependent area under the curve (tAUC) was computed and plotted over time.
2.6. Statistical analysis
All statistical analyses were performed in R unless otherwise noted (R version 3.6.1) using the following R packages: survival, survminer, survcomp, rms, timeROC, MASS, BlandAltmanLeh, irr, and prodlim. P < 0.05 was considered statistically significant. The Student t test for dependent samples was used to compare two C-indices (and two time-dependent AUCs), and iAUCs comparison was conducted by a Wilcoxon rank sum test for dependent samples [23]. Neural network training and deployment were done in MATLAB (R2019a, MathWorks, USA).
2.7. Role of the funding source
The funder had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
3.1. Patients
Table 1 summarises the baseline clinicopathological characteristics of the discovery and validation cohorts. In the discovery cohort, of 499 patients, 158 deaths occurred, in a median follow-up of 89 (95% confidence interval [CI] 79–96) months, and a five-year survival rate of 74.3% (95% CI 70.6–78.3%). In the validation cohort, of 315 patients, 72 deaths occurred, in a median follow-up of 51 (95% CI 50–53) months, and a five-year survival rate of 72.5% (95% CI 66.1–79.5%). And significant differences were found between two cohorts on age, T-stage, N-stage, and TNM stage (P < 0.05, Table 1).
Table 1.
The distributions of demographic and clinicopathologic characteristics of colorectal cancer patients in the two cohorts.
| Discovery cohort | Validation cohort | ||
|---|---|---|---|
| (158/499)* | (72/315)* | P | |
| Age | <0.001 | ||
| ≤ 60 year | 195 (39.1%) | 180 (57.1%) | |
| > 60 year | 304 (60.9%) | 135 (42.9%) | |
| Sex | 0.914 | ||
| Male | 301 (60.3%) | 188 (59.7%) | |
| Female | 198 (39.7%) | 127 (40.3%) | |
| T-stage | <0.001 | ||
| T1 | 14 (2.8%) | 0 (0%) | |
| T2 | 76 (15.2%) | 0 (0%) | |
| T3 | 360 (72.1%) | 261 (82.9%) | |
| T4 | 49 (9.8%) | 54 (17.1%) | |
| N-stage | 0.039 | ||
| N0 | 264 (52.9%) | 141 (44.8%) | |
| N1 | 145 (29.1%) | 117 (37.1%) | |
| N2 | 90 (18.0%) | 57 (18.1%) | |
| Stage | <0.001 | ||
| I | 71 (14.2%) | 0 (0%) | |
| II | 191 (38.3%) | 142 (45.1%) | |
| III | 228 (45.7%) | 173 (54.9%) | |
| IV | 9 (1.8%) | 0 (0%) | |
| Tumour site | 0.147 | ||
| Colon | 287 (57.5%) | 164 (52.1%) | |
| Rectum | 212 (42.5%) | 151 (47.9%) | |
| Median follow-up time (95% CI) | 89 (79-96) | 51 (50-53) | |
| 1-year survival rate (95% CI) | 93.2% (91.0%-95.4%) | 96.8% (94.9%-98.8%) | 0.038 |
| 3-year survival rate (95% CI) | 80.0% (76.5%-83.6%) | 83.5% (79.5%-87.7%) | 0.243 |
| 5-year survival rate (95% CI) | 74.3% (70.6%-78.3%) | 72.5% (66.1%-79.5%) | 0.590 |
Note: P value was performed by Kruskal–Wallis or χ2 test where appropriate.
Numbers in parentheses are number of events/total number of patients.
Abbreviation: CI, confidence interval.
3.2. TSR automated assessment
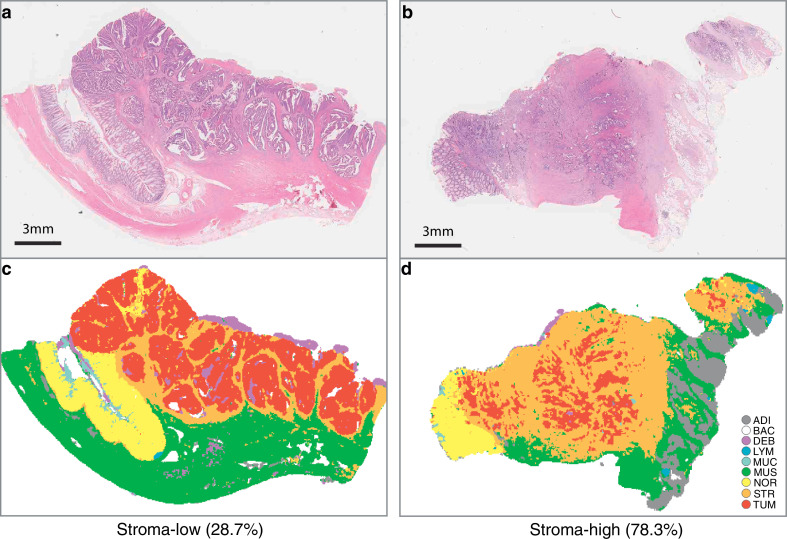
Supplementary Fig. 3 showed example images and the nine-class tissue distribution from the training dataset and two test sets. When we examined the internal features learned by the CNN using t-SNE, we observed that tissue classes naturally aggregated in separate clusters, especially in the test set 2 with more images (Supplementary Fig. 4). Representative examples of segmentation when the CNN model was applied to the stroma-low and stroma-high WSI are shown in Fig. 2.
Fig. 2.
Examples of stroma-low (a) and stroma-high (b) HE-stained WSI and corresponding segmented results (c, stroma-low; d, stroma-high). HE, haematoxylin–eosin; WSI, whole-slide image.
High CNN-based classification performance was achieved in all tissue classes (test set 1: 0.9572, 95% CI 0.9519–0.9621; test set 2: 0.9746, 0.9725–0.9766), which could be observed from the confusion matrixes of the CNN-based classification (Supplementary Fig. 5).
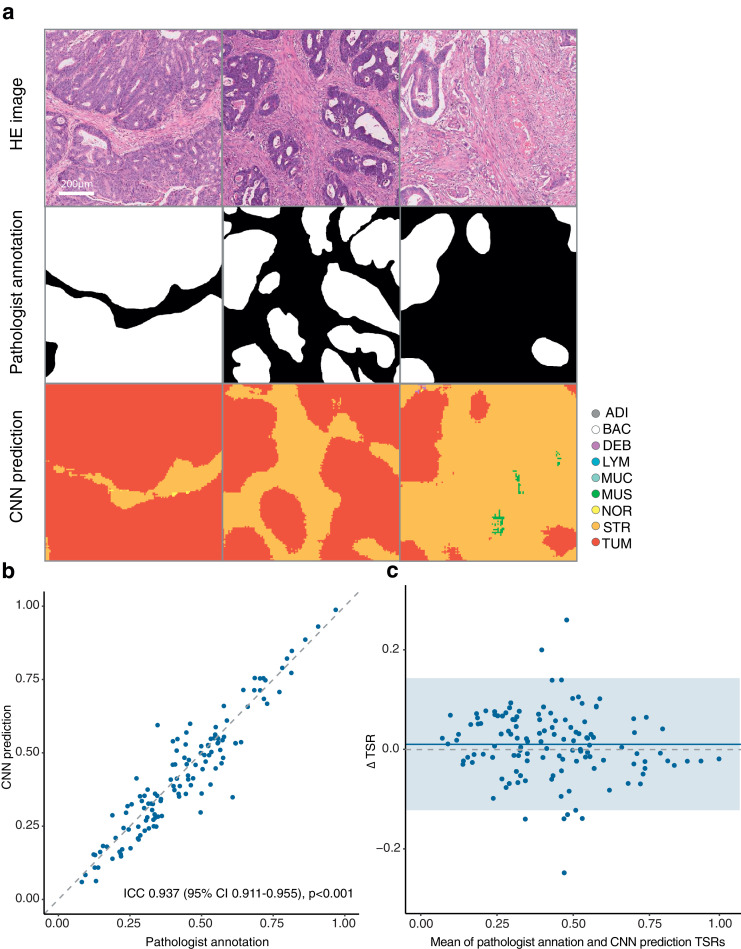
For TSR consistency analysis, Fig. 3a shows examples of manual pathologist annotation and automatic tissues segmentation by the CNN model. Good concordance was observed in the tissues' classification between the CNN model prediction and the pathologist annotation (Fig. 3b). Strong correlation occurred between TSR estimated by the CNN model and pathologist annotation (Pearson r = 0.939, 95% CI 0.914–0.957). A high agreement occurred between the annotated and predicted TSR (ICC = 0.937, 95% CI 0.911–0.955). Bland–Altman plot showed good agreement between TSR predicted by CNN model and that annotated by the pathologist (Fig. 3c). The mean difference of TSRs (annotation vs segmentation) was 0.01 (95% CI -0.12–0.14).
Fig. 3.
The TSR consistency analysis. (a) Example of HE image blocks, pathologist annotations, and the CNN model prediction map. (b) Concordance between TSR estimated by CNN model and the pathologist. (c) Bland–Altman plot for TSR estimation. The solid horizontal line is the mean, the dashed line is zero, and the shaded regions are 95% CIs. TSR, tumour-stroma ratio; HE, haematoxylin–eosin; CNN, convolutional neural network; CI, confidence interval.
3.3. Evaluation of the prognostic value of TSR
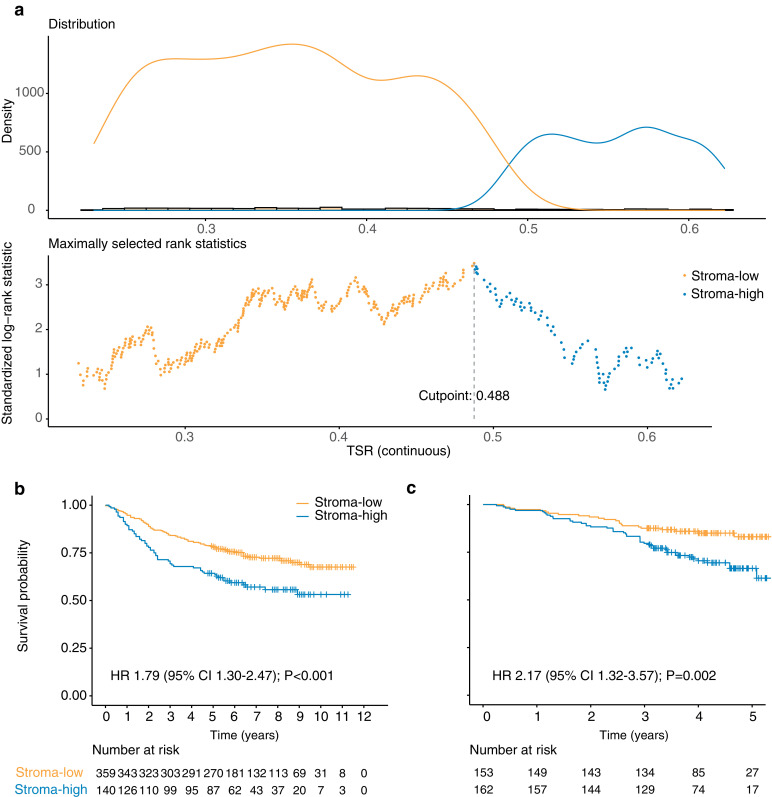
Patients were classified into stroma-low (TSR < 48.8%) or stroma-high (TSR ≥ 48.8%) groups based on the optimal cut-off point determined for the discovery cohort (Fig. 4a). Stroma-high was identified in 140 (28%) and 162 (51%) patients in the discovery and validation cohort, respectively.
Fig. 4.
The cut-off determination and Kaplan–Meier curves analysis. (a) The optimal cut-off to categorise TSR as stroma-low and stroma-high was determined by maximally selected rank statistics method. (b) Kaplan–Meier survival curves of overall survival of stroma-low vs stroma-high categories in the discovery and validation cohorts. TSR, tumour-stroma ratio; HR, hazard ratio; CI, confidence interval .
In discovery cohort, stroma-low and stroma-high groups’ median OS were 72 (interquartile range [IQR] 60-101) and 67 (28-99) months, respectively, with unadjusted hazard ratio (HR) of 1.79 (95% CI 1.30–2.47; log-rank test P < 0.001; Fig. 4b). In the validation cohort, these were 49 (IQR 40-58) and 46 (38-55) months, respectively, with unadjusted HR of 2.21 (1.35–3.63; P = 0.002; Fig. 4c). Furthermore, in both cohorts, stroma-low was associated with higher five-year survival rate (stroma-low vs stroma-high: 78.3% vs 64.3% in the discovery cohort; 80.5% vs 70.4% in the validation cohort). Furthermore, when we explored the feasibility of applying TSR to various patient subgroups, based on all discovery and validation cohort patients, the predictor remained a statistically significant prognostic factor when stratified by age, sex, and tumour site, and demonstrates marginal significance for the prediction when stratified by stage (Supplementary Fig. 6).
The univariate association of clinicopathological characteristics with OS is presented in Table 2. The prognostic association of TSR with OS was maintained in multivariate analysis, independently of TNM stage, age, sex, and tumour site, with stroma-high associated with reduced OS in the discovery (HR 1.72, 95% CI 1.24–2.37, P = 0.001) and validation cohort (2.08, 1.26–3.42, P = 0.004; Table 2).
Table 2.
Uni- and multivariate analyses including age, sex, stage, tumour site, and TSR for OS in the two cohort.
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Discovery cohort | Validation cohort | Discovery cohort | Validation cohort | |||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| Age | 1.02(1.01-1.04) | <0.001 | 1.01 (0.99-1.03) | 0.184 | 1.02 (1.01-1.04) | <0.001 | 1.02 (1.00-1.03) | 0.103 |
| Sex | ||||||||
| Male | 1 | |||||||
| Female | 0.93 (0.68-1.28) | 0.664 | 1.44 (0.90-2.28) | 0.125 | ||||
| Stage | ||||||||
| I | 1 | 1 | ||||||
| II | 2.70 (1.15-6.37) | 0.023 | 1 | 2.40 (1.02-5.68) | 0.046 | 1 | ||
| III | 7.20 (3.16-16.4) | <0.001 | 3.10 (1.80-5.35) | <0.001 | 6.47 (2.84-14.8) | <0.001 | 2.98 (1.73-5.15) | <0.001 |
| IV | 24.3 (8.40-70.0) | <0.001 | 24.7 (8.47-71.7) | <0.001 | ||||
| Tumour site | ||||||||
| Colon | 1 | 1 | ||||||
| Rectum | 0.99 (0.72-1.39) | 0.998 | 1.12 (0.71-1.78) | 0.625 | ||||
| TSR | ||||||||
| Stroma-low | 1 | 1 | 1 | 1 | ||||
| Stroma-high | 1.79 (1.30-2.47) | <0.001 | 2.17 (1.32-3.57) | 0.002 | 1.72 (1.24-2.37) | 0.001 | 2.08 (1.26-3.42) | 0.004 |
Abbreviations: OS, overall survival; HR, hazard ratio; CI, confidence interval; TSR, tumour-stroma ratio.
3.4. Construction and evaluation of prediction models
As TSR, stage, and age were identified as independent predictors of OS in multivariate analysis in the discovery cohort, we developed a prediction TSR model incorporating the above independent predictors, and a reference model with only stage and age incorporated.
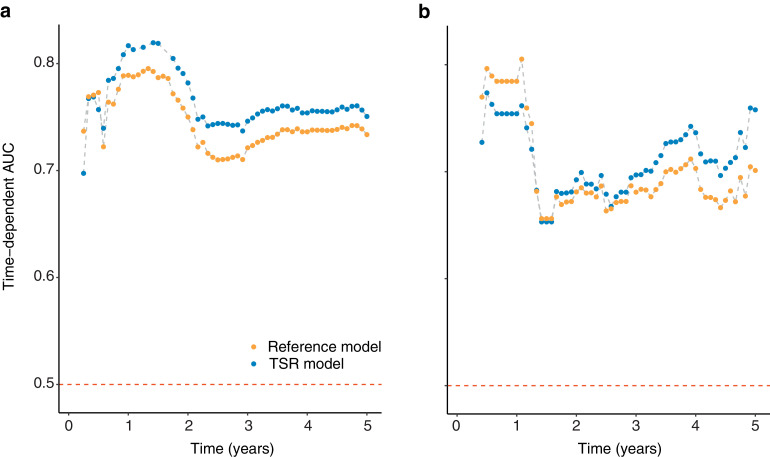
The TSR model showed better discrimination performance than the reference model (C-index: 0.721 [95% CI 0.684–0.759] vs 0.704 [0.667–0.742], P < 0.001; Supplementary Table S1). The resulting time-dependent AUC plotted over time are presented in Fig. 5. The TSR model showed higher AUC across the most time-points compared to the reference model.
Fig. 5.
Performance of the TSR model. Time-dependent AUC was measured from one month to five years at one-monthly intervals, reflecting the prediction performance at different time-points. The top-performing model (TSR) is shown compared with the reference model. (a) Discovery cohort. (b) Validation cohort. TSR, tumour-stroma ratio; AUC, area under curve.
4. Discussion
In this study, we presented a deep learning model for the fully automated TSR quantification using whole-slide HE-stained images of CRC. We further showed the CNN-based TSR as a prognostic factor of OS in two independent CRC patient cohorts. Combined into a prediction model, TSR demonstrated its potential for integrating with the TNM staging system. To the best of our knowledge, this is the first study to establish a deep learning model for the fully automated TSR quantification on WSI, with its prognostic utility validated in large multicentre patient cohorts. This approach permits the standardisation and reproducibility of TSR assessment on ubiquitously available HE-stained histological images to eliminate variations documented with traditional visual assessment while reducing the pathologists' workload. This fully automatic workflow is well suited for its implementation in clinical practice and could accelerate the clinical implication of TSR for prognostication and decision making.
TSR has gained increasing attention in cancer prognosis prediction fields [6,10,24,25]. By validating the prognostic relevance of TSR in two independent cohorts of patients, our study further elucidated TSR as a strong predictor of CRC patients' survival. Despite certain patient characteristic imbalance between the two patient cohorts, especially regarding TNM stage, results showed that TSR achieved comparable prognostic performance in two cohorts, which reflect the independent prognostic relevance of TSR. Multivariate analysis further confirmed that the derived TSR remained a stage-independent prognostic factor of reduced OS, which is concordant with previous studies [6–8,10]. The evolving knowledge that tumour-stroma plays an active role in cancer progression, since it interacts with tumour and non-malignant cells at different stages, from tumour onset to invasion and metastasis [26,27], explains the results of our work. That is, a high proportion of stroma corresponds to reduced OS in discovery and validation cohorts.
Having assessed TSR prognostic validity, we present a prediction model to enable a step forward for individualised prognostic prediction. The proposed prediction model incorporates TSR, with independent risk factors (stage and age), and results showed that the model provides significant clinical values for patients' prognosis. TSR model provided adequate prediction, with high accuracy (iAUC 0.759 vs 0.728), and satisfied discriminative ability (C-index 0.721 vs 0.689), in discovery and validation cohorts, respectively. Furthermore, the finding that TSR integration into the prediction model showed improved prognostic capability compared with the reference model (Fig. 5) indicated that this biomarker could be a potential supplement to the TNM staging system while facilitating improved risk stratification. These results provided more evidence of recent discussion of the TNM Evaluation Committee (UICC) and the College of American Pathologists (CAP), on the potential of TSR integration in TNM staging system [10].
Despite the prognostic value of and consequently high interest of TSR in CRC, standardised, objective, and easily implemented assessment method is yet unavailable. Automated TSR assessment holds the potential to increase the reproducibility of this biomarker. Although recently, automatic discrimination of tissue image of epithelium and stroma in CRC was performed by traditional machine learning using handcrafted features of images [16,17], the vast amount of information in WSI remains a great computational challenge that lies ahead of the fully automated TSR assessment pipeline development [10]. Through making use of more abstract representation of input data, deep learning has achieved superior performance to traditional machine learning and holds promise in retrieving additional information from histopathology images, with continuous breakthroughs in medical image domain of CNNs [13,28,29].
Regarding efforts, leveraging deep learning, which aims for tumour "stroma" quantification based on WSI, two studies have yielded encouraging results [19,30]. Kather et al. trained a deep neural network to achieve tissue decomposition into tissue parts that could further be aggregated in a prognostic "deep stroma score" for CRC [19]. Their work supports the hypothesis that deep learning could be a significant aid in clinical prognostic settings. However, the clinical translation of their approach is still hampered by its semi-automatic nature (manual tumour region extraction form WSI was involved in their workflow), making it less objective and reproducible. It is noteworthy that they defined "stroma score" as the weighted sum of various non-tumour tissues (desmoplastic stroma, lymphocytes, and adipose tissue). Hence, being unable to provide clear biological insights into the computational "black boxes" is another weakness of their methodology. In Geessink et al. 's semi-automated TSR assessment method using deep learning, the derived TSR could serve as an independent prognosticator for patients with rectal cancer [30]. However, the study limitation was the need for human input for choosing user-provided hot-spot. Further automation at the WSI level, beyond the limited hot-spot area, is still warranted.
The strengthening of our work is that it fills the gap by providing a fully automated method that requires no manual input on HE-stained WSI to facilitate an objective and fast TSR assessment. By reducing pathologists' workload, this fully automatic workflow is well suited for its implementation in clinical practice. With success in recent studies that applied CNNs to reveal prognostic biomarkers directly from digitalised WSI, our work further supports the hypothesis that with deep-learning algorithms, conventional histopathology images could be better used to facilitate the extraction of prognostic information to enable more accurate clinical prediction and decision-making.
This work has the typical baggage inherent in retrospective studies; thus, prospective studies are warranted to validate the automatic scored TSR for routine clinical use. Secondly, one of the flaws in this study is that we only aimed at the OS, without analysing the prognostic value of the TSR in terms of disease-free survival (DFS), which also is an essential outcome-of-interest for CRC prognosis. Additional analysis on the CNN-quantified TSR for DFS prediction is among our further goals. Moreover, for fast annotation and given the vast availability of released open-source image data, the segmentation methodology in this work was based on image patch. However, this methodology could only achieve rough segmentation without fully-obtained tissue structure details. Hence, the pixel-wise methodology refinement to improve the classification accuracy is among our future research goals [31].
In summary, we present a deep learning model for the fully automated TSR quantification using whole-slide HE-stained images of CRC. The application to independent patient CRC cohorts confers prognostic relevance to our approach. The present study suggests that with deep learning, automated histopathology images analysis can potentially be of significant aid to clinical prognosis prediction and decision-making.
Data sharing
The data sets used for training and testing of the deep learning model are available (doi:10.5281/zenodo.4024676), including the training set part 1, the training set part 3, the test set 2, and the TSR evaluation set. The source code and the trained CNN model are also openly available online (doi: 10.5281/zenodo.4023999).
Declaration of Competing Interests
The authors have declared no conflicts of interest.
Acknowledgements
This work was supported by the National Key Research and Development Program of China [2017YFC130910002], the National Science Fund for Distinguished Young Scholars [81925023], National Natural Science Foundation of China [81771912] and the National Science Foundation for Young Scientists of China [81701782].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103054.
Contributor Information
Yanqi Huang, Email: yikiann@126.com.
Changhong Liang, Email: liangchanghong@gdph.org.cn.
Zaiyi Liu, Email: liuzaiyi@gdph.org.cn.
Appendix. Supplementary materials
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Deschoolmeester V, Baay M, Specenier P, Lardon F, Vermorken JB. A review of the most promising biomarkers in colorectal cancer: one step closer to targeted therapy. Oncologist. 2010;15(7):699–731. doi: 10.1634/theoncologist.2010-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edge SB BD, Compton CC, Fritz AG, Greene FL, Trotti A. 7th ed. Springer; New York: 2010. AJCC cancer staging manual. [Google Scholar]
- 4.Nagtegaal ID, Quirke P, Schmoll HJ. Has the new TNM classification for colorectal cancer improved care? Nat Rev Clin Oncol. 2011;9(2):119–123. doi: 10.1038/nrclinonc.2011.157. [DOI] [PubMed] [Google Scholar]
- 5.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 6.Huijbers A, Tollenaar RA, V Pelt GW, Zeestraten EC, Dutton S, McConkey CC. The proportion of tumor-stroma as a strong prognosticator for stage II and III colon cancer patients: validation in the VICTOR trial. Ann Oncol. 2013;24(1):179–185. doi: 10.1093/annonc/mds246. [DOI] [PubMed] [Google Scholar]
- 7.Park JH, Richards CH, McMillan DC, Horgan PG, Roxburgh CS. The relationship between tumour stroma percentage, the tumour microenvironment and survival in patients with primary operable colorectal cancer. Ann Oncol. 2014;25(3):644–651. doi: 10.1093/annonc/mdt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JH, McMillan DC, Powell AG, Richards CH, Horgan PG, Edwards J. Evaluation of a tumor microenvironment-based prognostic score in primary operable colorectal cancer. Clin Cancer Res. 2015;21(4):882–888. doi: 10.1158/1078-0432.CCR-14-1686. [DOI] [PubMed] [Google Scholar]
- 9.Hansen TF, Kjaer-Frifeldt S, Lindebjerg J, Rafaelsen SR, Jensen LH, Jakobsen A. Tumor-stroma ratio predicts recurrence in patients with colon cancer treated with neoadjuvant chemotherapy. Acta Oncol. 2018;57(4):528–533. doi: 10.1080/0284186X.2017.1385841. [DOI] [PubMed] [Google Scholar]
- 10.van Pelt GW, Sandberg TP, Morreau H, Gelderblom H, van Krieken J, Tollenaar R. The tumour-stroma ratio in colon cancer: the biological role and its prognostic impact. Histopathology. 2018;73(2):197–206. doi: 10.1111/his.13489. [DOI] [PubMed] [Google Scholar]
- 11.van Pelt GW, Kjaer-Frifeldt S, van Krieken J, Al Dieri R, Morreau H, Tollenaar R. Scoring the tumor-stroma ratio in colon cancer: procedure and recommendations. Virchows Arch. 2018;473(4):405–412. doi: 10.1007/s00428-018-2408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courrech Staal EF, Smit VT, van Velthuysen ML, Spitzer-Naaykens JM, Wouters MW, Mesker WE. Reproducibility and validation of tumour stroma ratio scoring on oesophageal adenocarcinoma biopsies. Eur J Cancer. 2011;47(3):375–382. doi: 10.1016/j.ejca.2010.09.043. [DOI] [PubMed] [Google Scholar]
- 13.Yu KH, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2(10):719–731. doi: 10.1038/s41551-018-0305-z. [DOI] [PubMed] [Google Scholar]
- 14.Yu KH, Berry GJ, Rubin DL, Re C, Altman RB, Snyder M. Association of omics features with histopathology patterns in lung adenocarcinoma. Cell Syst. 2017;5(6):620–627. doi: 10.1016/j.cels.2017.10.014. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu KH, Zhang C, Berry GJ, Altman RB, Re C, Rubin DL. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francesco Bianconi, AAl-Ln, Fernández Antonio. Discrimination between tumour epithelium and stroma via perception-based features. Neurocomputing. 2015;154:119–126. [Google Scholar]
- 17.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3(108):108ra13. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 18.West NP, Dattani M, McShane P, Hutchins G, Grabsch J, Mueller W. The proportion of tumour cells is an independent predictor for survival in colorectal cancer patients. Br J Cancer. 2010;102(10):1519–1523. doi: 10.1038/sj.bjc.6605674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 2019;16(1) doi: 10.1371/journal.pmed.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kather JN, Pearson AT, Halama N, Jager D, Krause J, Loosen SH. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25(7):1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonyan KZ, Andrew. Very deep convolutional networks for large-scale image recognition. arXiv. 2014;abs/1409.1556.
- 22.Torsten Hothorn BL. On the exact distribution of maximally selected rank statistics. Comput Stat Data Anal. 2003;43(2):121–137. [Google Scholar]
- 23.Haibe-Kains B, Desmedt C, Sotiriou C, Bontempi G. A comparative study of survival models for breast cancer prognostication based on microarray data: does a single gene beat them all? Bioinformatics. 2008;24(19):2200–2208. doi: 10.1093/bioinformatics/btn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogelaar FJ, van Pelt GW, van Leeuwen AM, Willems JM, Tollenaar RA, Liefers GJ. Are disseminated tumor cells in bone marrow and tumor-stroma ratio clinically applicable for patients undergoing surgical resection of primary colorectal cancer? The Leiden MRD study. Cell Oncol (Dordr) 2016;39(6):537–544. doi: 10.1007/s13402-016-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandberg TP, Stuart M, Oosting J, Tollenaar R, Sier CFM, Mesker WE. Increased expression of cancer-associated fibroblast markers at the invasive front and its association with tumor-stroma ratio in colorectal cancer. BMC Cancer. 2019;19(1):284. doi: 10.1186/s12885-019-5462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 29.Jha S, Topol EJ. Adapting to artificial intelligence: radiologists and pathologists as information specialists. JAMA. 2016;316(22):2353–2354. doi: 10.1001/jama.2016.17438. [DOI] [PubMed] [Google Scholar]
- 30.Geessink OGF, Baidoshvili A, Klaase JM, Ehteshami Bejnordi B, Litjens GJS, van Pelt GW. Computer aided quantification of intratumoral stroma yields an independent prognosticator in rectal cancer. Cell Oncol (Dordr) 2019;42(3):331–341. doi: 10.1007/s13402-019-00429-z. [DOI] [PubMed] [Google Scholar]
- 31.Pinheiro PO, Collobert R. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2015. From image-level to pixel-level labeling with convolutional networks. IEEE:1713-21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.