Knowledge of the T4SS is derived from model systems, such as Agrobacterium tumefaciens. The structure of the T4SS in Rickettsiales differs from the classical arrangement. These differences include missing and duplicated components with structural alterations. Particularly, two sequenced virB6-4 genes encode unusual C-terminal structural extensions resulting in proteins of 4,322 (GenBank accession number AGR79286.1) and 9,935 (GenBank accession number ANC34101.1) amino acids. To understand how the T4SS is used in A. phagocytophilum, we describe the expression of the virB6 paralogs and explore their role as the bacteria replicate within its host cell. Conclusions about the importance of these paralogs for colonization of human and tick cells are supported by the deficient phenotype of an A. phagocytophilum mutant isolated from a sequence-defined transposon insertion library.
KEYWORDS: Anaplasma phagocytophilum, obligate intracellular pathogens, Rickettsiales, tick-borne bacteria, transposon mutagenesis, VirB6, virulence, attenuated growth, type IV secretion
ABSTRACT
Many pathogenic bacteria translocate virulence factors into their eukaryotic hosts by means of type IV secretion systems (T4SS) spanning the inner and outer membranes. Genes encoding components of these systems have been identified within the order Rickettsiales based upon their sequence similarities to other prototypical systems. Anaplasma phagocytophilum strains are obligate intracellular, tick-borne bacteria that are members of this order. The organization of these components at the genomic level was determined in several Anaplasma phagocytophilum strains, showing overall conservation, with the exceptions of the virB2 and virB6 genes. The virB6 loci are characterized by the presence of four virB6 copies (virB6-1 through virB6-4) arranged in tandem within a gene cluster known as the sodB-virB operon. Interestingly, the virB6-4 gene varies significantly in length among different strains due to extensive tandem repeats at the 3′ end. To gain an understanding of how these enigmatic virB6 genes function in A. phagocytophilum, we investigated their expression in infected human and tick cells. Our results show that these genes are expressed by A. phagocytophilum replicating in both cell types and that VirB6-3 and VirB6-4 proteins are surface exposed. Analysis of an A. phagocytophilum mutant carrying the Himar1 transposon within the virB6-4 gene demonstrated that the insertion not only disrupted its expression but also exerted a polar effect on the sodB-virB operon. Moreover, the altered expression of genes within this operon was associated with the attenuated in vitro growth of A. phagocytophilum in human and tick cells, indicating the importance of these genes in the physiology of this obligate intracellular bacterium in such different environments.
IMPORTANCE Knowledge of the T4SS is derived from model systems, such as Agrobacterium tumefaciens. The structure of the T4SS in Rickettsiales differs from the classical arrangement. These differences include missing and duplicated components with structural alterations. Particularly, two sequenced virB6-4 genes encode unusual C-terminal structural extensions resulting in proteins of 4,322 (GenBank accession number AGR79286.1) and 9,935 (GenBank accession number ANC34101.1) amino acids. To understand how the T4SS is used in A. phagocytophilum, we describe the expression of the virB6 paralogs and explore their role as the bacteria replicate within its host cell. Conclusions about the importance of these paralogs for colonization of human and tick cells are supported by the deficient phenotype of an A. phagocytophilum mutant isolated from a sequence-defined transposon insertion library.
INTRODUCTION
The type IV secretion system (T4SS) is a macromolecular protein complex of Gram-negative and -positive bacteria that forms a channel across the cell envelope, allowing the secretion of substrates. Depending on the bacterial species, this multimeric channel is a conduit for direct transfer of DNA, proteins, or effectors into eukaryotic host cells, enabling virulence and survival of bacteria (1–3). In some instances, this system is also involved in DNA uptake from the extracellular milieu (4) or interbacterial killing (5, 6). The T4SS is of major medical relevance, as it is used by several multidrug-resistant bacterial species to spread antibiotic resistance genes (7). The VirB/VirD4 system encoded by the pTi plasmid of Agrobacterium tumefaciens represents the canonical P-T4SS, consisting of 12 subunits, VirB1 throughout VirB11 and VirD4, which serve to deliver oncogenic nucleoprotein particles into plant cells, resulting in the development of crown gall tumors (8).
Anaplasma phagocytophilum (order Rickettsiales; family Anaplasmataceae) is an obligate intracellular tick-borne bacterium and the causative agent of human granulocytic anaplasmosis (HGA), a notifiable disease since 1998. This pathogen is of increasing concern in the United States and internationally as an emerging agent that causes disease in both humans and animals. Members of this species have a tropism for neutrophils and granulocytes, and infection is characterized by a nonspecific febrile illness with clinical manifestations that range from asymptomatic to fatal disease if left untreated (9–11). Moreover, all members of the order Rickettsiales sequenced to date encode components of a T4SS, prototypical to the VirB/VirD system of Agrobacterium tumefaciens (9, 12, 13).
Although the T4SS in A. phagocytophilum superficially resembles that of A. tumefaciens, significant differences exist. For example, in A. tumefaciens the genes that encode the transmembrane channel and coupling proteins are organized in a single locus that contains the virB and the virD operons (8, 14). However, in A. phagocytophilum these components are distributed in three distal clusters. The first includes virB8-1, virB9-1, virB10, virB11, and virD4, the second includes virB2s and virB4-2, and the third comprises virB3, virB4-1, and four virB6 paralogs (12, 15). Coinciding with the lack of genes required for peptidoglycan synthesis, A. phagocytophilum lacks virB1, a gene than encodes a murein-degrading transglycosylase required for channel assembly across the cell wall (16–19), and virB5, which encodes a pilus-associated protein, but has duplicate copies of genes encoding secretion channel-associated proteins (virB4, virB8, and virB9 genes), the four copies of virB6, and multiple copies (8 to 15) of virB2, which vary in number and sequence between A. phagocytophilum strains (15). These differences in organization suggest alternate apparatus assembly, regulation, and substrate transfer (12, 20) and perhaps even assembly of variable surface structures that may modulate host-pathogen interactions such as attachment to different host cells or evasion of host immune responses (21, 22).
Comparative genomics of these components in several A. phagocytophilum strains showed that they are highly conserved among strains, with the exception of virB2 and virB6. The virB6 loci, the focus of this study, are characterized by the presence of four copies in tandem (virB6-1 through virB6-4) arranged within an operon that contains sodB, virB3, and virB4-1. Interestingly, virB6-4 varies among strains due to the presence of an extensive repeat domain with various numbers of repeats found at the 3′ end (15). Although VirB6 is essential for substrate secretion and stability of the T4SS (23–26), very little is known about the expression of the four virB6 paralogs and their role in A. phagocytophilum infection and pathogenesis.
In this study, molecular and biochemical assays led us to conclude that the four virB6 paralogs are expressed in A. phagocytophilum during in vitro infection of human and tick cells and that VirB6-3 and VirB6-4 proteins are surface exposed. Transposon mutagenesis of A. phagocytophilum using the Himar1 system resulted in the isolation of a mutant carrying transposon (Tn) sequences within the virB6-4 gene. Insertion of Tn sequences within this gene not only disrupted its expression but also had a polar effect on the sodB-virB operon. Moreover, altered expression of genes within this operon was linked to attenuated growth of A. phagocytophilum in human and tick cells, indicating the importance of these genes during intracellular replication.
RESULTS
The virB6 loci are transcribed in A. phagocytophilum during infection of human and tick cells.
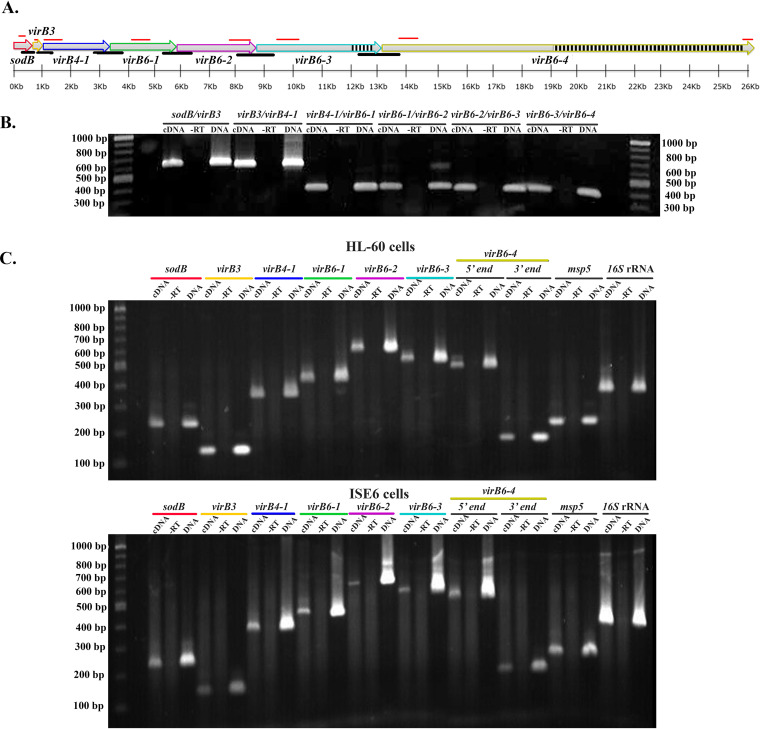
The virB6 loci are organized within an operon structure (sodB-virBs) comprised of sodB, virB3, virB4-1, virB6-1, virB6-2, virB6-3, and virB6-4 in tandem (Fig. 1A). Prior work failed to clarify the polycistronic nature or transcription status of each of these genes (15, 27, 28). Therefore, to help us understand the contribution of this locus to A. phagocytophilum virulence, we determined the cistronic organization and transcription of the sodB-virBs locus in both human and tick cells. Moreover, since the peptide used to raise antibodies against VirB6-4 represents sequences from the C-terminal repeat region, we also queried whether the full length of this gene is transcribed in both cell types by targeting transcripts from the 5′ and 3′ ends. Total RNA isolated from A. phagocytophilum-infected cells was reverse transcribed with random hexamer primers and the cDNA template used for PCR amplification with primers targeting sequences from intergenic regions and each gene within the operon (Fig. 1A). Reverse transcription-PCR (RT-PCR) products of appropriate size from the intergenic regions between sodB-virB3, virB3–virB4-1, virB4-1–virB6-1, virB6-1–virB6-2, virB6-2–virB6-3, and virB6-3–virB6-4 genes were detected (Fig. 1B), indicating that these genes are polycistronically transcribed. Moreover, transcripts from each gene and full-length virB6-4 were detected in both HL-60 and tick ISE6 cells (Fig. 1C).
FIG 1.
A. phagocytophilum virB6 paralogs are cotranscribed in mammalian and tick cells. (A) Organization of the sodB-virB operon in Anaplasma phagocytophilum. Black lines indicate locations of primers designed to amplify transcripts from intergenic regions sodB-virB3, virB3–virB4-1, virB4-1–virB6-1, virB6-1–virB6-2, virB6-2–virB6-3, and virB6-3–virB6-4. Red lines indicate binding sites for primers designed to amplify transcripts from sodB, virB3, virB4-1, virB6-1, virB6-2, virB6-3, and the 5′ and 3′ ends of virB6-4. Black and white boxes depict the repeats region of virB6-3 and virB6-4. The sodB-virB operon was drawn to scale using SnapGene software (from GSL Biotech). (B) Agarose gel analysis of cDNA products from sodB-virB operon intergenic regions. RT-PCR was performed on total RNA isolated from A. phagocytophilum-infected HL-60 cells. (C) Agarose gel analysis of cDNA products from sodB through virB6-4. RT-PCR was performed on total RNA isolated from A. phagocytophilum-infected HL-60 and ISE6 cells. Genomic DNA was used as a positive control, and reactions without reverse transcriptase (-RT) were used as negative controls. A 100-bp/1-kb DNA ladder was used. The msp5 and 16S rRNA genes were used as internal controls to ensure integrity of RNA/cDNA. Data were obtained from two independent experiments.
The four VirB6 paralogs were detected in A. phagocytophilum-infected human and tick cells.
The prototypical A. tumefaciens VirB6 is an integral membrane protein of ∼300 amino acid residues and four to five transmembrane domains (TMD1 through TMD5) (26, 29). Comparison of VirB6 protein sequences from A. tumefaciens with paralogs from A. phagocytophilum, Anaplasma marginale and Ehrlichia chaffeensis indicate sequence conservation restricted only toward the A. tumefaciens central DNA-transferring TrbL/VirB6 domain, specifically, three central TMDs and a cytoplasmic loop found between TMD3 and TMD4 (see Fig. S1A in the supplemental material). In A. tumefaciens, a tryptophan residue within the central cytoplasmic loop is essential for localization of this protein at the cell pole (30). This residue appears to be conserved among the VirB6 protein family (26, 30), including the five VirB6 paralogs from Rickettsia species (20, 31). Likewise, we found this residue to be invariant in VirB6 proteins from A. phagocytophilum, A. marginale, and E. chaffeensis (Fig. S1A), indicating conservation of this residue in Rickettsiales. Relative to A. tumefaciens, and similar to other Rickettsiales (12, 32), A. phagocytophilum encodes much larger VirB6 proteins characterized by extended N- or C-terminal hydrophilic regions flanking the TrbL/VirB6 domain (Fig. S1B), which place them within the “extended VirB6-like protein” subtype (4, 14, 16). In silico topology analysis indicated that in A. phagocytophilum, VirB6-1 and VirB6-2 have nine and seven TMDs, respectively, and a putative amino-terminal cleavable signal peptide (SP) (Fig. S1B). However, it is possible that SP residues from these proteins correspond to the first TMD, as it is known that prediction algorithms often confound TMDs with the SP due to similar biochemical characteristics (33). On the other hand, VirB6-3 and VirB6-4 have eight and six TMDs, respectively, and are predicted to be lipoproteins, as both contain highly conserved lipobox sequences (Fig. S1B). Common to these paralogs is the clustering of TMDs toward the TrbL/VirB6 hydrophobic region. VirB6-1, VirB6-2, and VirB6-3 carry the TrbL/VirB6 domain located toward the C terminus, while in VirB6-4 this domain is found toward the N terminus (Fig. S1B).
To determine expression of virB6 paralogs as proteins, specific antibodies against A. phagocytophilum strain HZ VirB6 proteins were prepared using synthetic peptides derived from unique predicted antigenic epitopes of each protein (Fig. S1B and Table 1). An enzyme-linked immunosorbent assay (ELISA) was performed to determine the titer of rabbit polyclonal antibodies specific to VirB6-1, -6-2, -6-3, and -6-4 peptides using conjugates of the VirB6 peptides with ovalbumin or ovalbumin alone as a coating agent. This assay showed that immunized animals developed high antibody titers against the peptides. In addition, there was a lack of reactivity of the preimmunization serum against the peptide (Fig. S2).
TABLE 1.
Peptides used to generate polyclonal antibodies against VirB6 proteins
| Peptide | Sequence | Length (residues) |
|---|---|---|
| VirB6-1 | C-SNYAWPKNRVSSSRYIEVCYRHPLGTVYMSPYVAARLGFAGRDPKEVLKESKYPRE | 56 |
| VirB6-2 | C-YPLYFSSKLGGGSGYYNSWYKVAKGEAPYITDGIPEYLHISGGIVKPSSMEKLDDEHE | 58 |
| VirB6-3 | C-REYRPGDGGVGEGDDRVYGSTTAGSGISGDAAGIDARGDHAREHQPADVGVGEGADRVSG | 60 |
| VirB6-4 | C-TELPREVVPEATEYGTKPDDQDGDKGDLRPERLDPDIGDGSAIEDEVEVRSSRSSESTDSVPSEVTERDA | 70 |
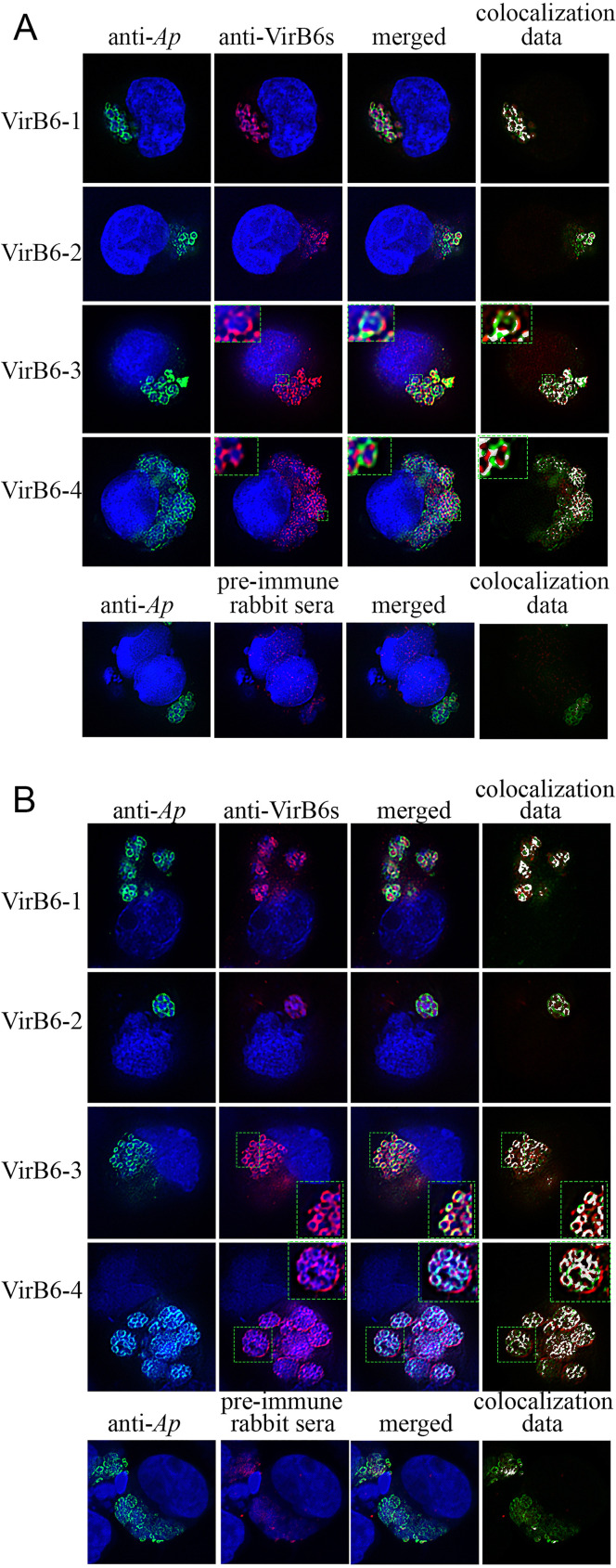
The expression of the individual VirB6 proteins in infected HL-60 and ISE6 cells was evaluated by immunofluorescence microscopy. As a localization control, antisera from mice infected with A. phagocytophilum strain HZ (with antibody responses mainly targeting the dominant surface protein MSP2/P44) (34, 35) were used. Binding of the infection sera to A. phagocytophilum generated a green fluorescent signal matching the known location of MSP2/P44 on the periphery of bacteria (Fig. 2). This allowed us to confidently identify the specific localization of rabbit anti-VirB6 peptides to A. phagocytophilum. Dual fluorescence indicated that the red fluorescent signals from VirB6-1, VirB6-2, VirB6-3, and VirB6-4 epitopes were specific to A. phagocytophilum organisms in infected HL-60 and ISE6 cells as shown by superimposed images and colocalization data (Fig. 2). Similar binding was not observed in infected cells reacted with preimmune rabbit sera. Interestingly, VirB6-3 and VirB6-4 appeared to be located predominantly at the periphery of A. phagocytophilum as multiple foci or punctate structures when growing in HL60 cells (Fig. 2A). In ISE6 cells, the red fluorescent signals from VirB6-3 and VirB6-4 were also located at the periphery of bacteria (Fig. 2B). Conversely, the signal from VirB6-4 was additionally observed in a location consistent with it being associated with the parasitophorous vacuole (PV), and the originating signal did not overlap the green fluorescence as indicated by colocalization data (Fig. 2B). However, as we did not use antibodies specific to PV associated proteins, we could not determine this localization unambiguously.
FIG 2.
A. phagocytophilum (Ap) VirB6 proteins are synthesized in infected HL-60 and ISE6 cells. Shown are representative dual immunofluorescence images of HL-60 (A) and ISE6 (B) cells infected with A. phagocytophilum. To indicate immunolabeling of VirB6-3 and VirB6-4, boxes delineated by the dotted lines are magnified in the insets. Infected cells were fixed and viewed by indirect immunofluorescence microscopy to determine immunoreactivity with A. phagocytophilum-infected mouse sera (green) and anti-VirB6-1, -VirB6-2, -VirB6-3, and -VirB6-4 sera (red). Host cell nuclei and bacterial DNA were stained with DAPI (blue). Preimmune rabbit serum was used as a negative control. The RG2B Colocalization ImageJ plug-in (C. P. Mauer, Northwestern University) was used to assess the overlap between the green and red channels. Representative images from two independent experiments are shown.
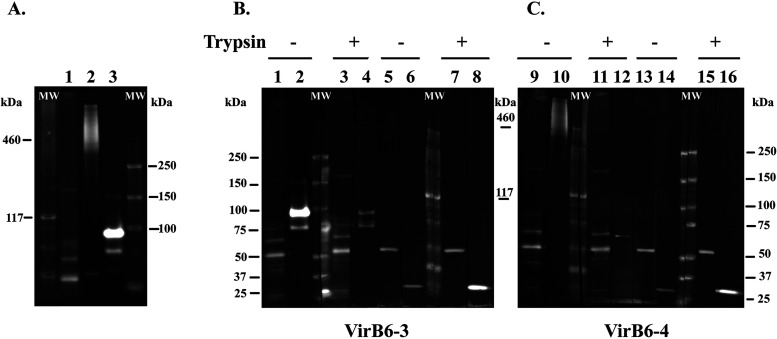
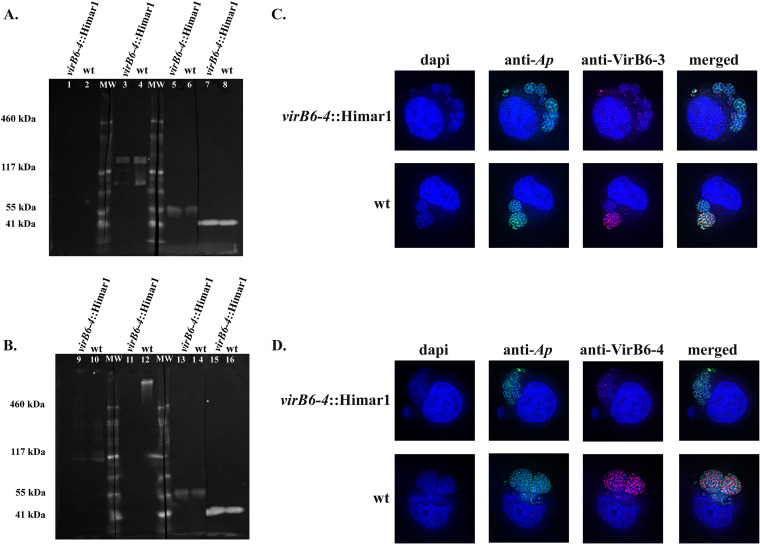
Due to the C-terminal hydrophilic repeat domains of VirB6-3 and VirB6-4, the focus of this work, it has been proposed that these proteins or their C-terminal domains are surface exposed or proteolytically released into the host cell (15). In addition, evidence of surface exposure of the extended VirB6 proteins has been presented for other Rickettsiales (36–38). Hence, we confirmed their expression in infected HL-60 cells by Western immunoblotting and determined whether these proteins are surface exposed by trypsin digestion of intact A. phagocytophilum organisms. Western immunoblotting showed specific protein bands in samples reacted with sera from VirB6-3 and VirB6-4 peptide-immunized rabbits that were not detected in samples reacted with preimmune sera (Fig. 3A). The sizes of the bands detected matched the predicted molecular weights of VirB6-3 (158.3 kDa) and VirB6-4 (predicted mass of 470 kDa, but can migrate anomalously because of larger repeat sequences). Additional protein bands of smaller size than predicted were detected in the sample reacted with sera against VirB6-3. It is possible that this protein undergoes proteolytic posttranslational processing that results in smaller molecules or represents nonspecific degradation products (Fig. 3A).
FIG 3.
VirB6-3 and VirB6-4 proteins are located on the surface of A. phagocytophilum. (A) Proteins from A. phagocytophilum isolated from HL-60 were separated by Tris acetate polyacrylamide gel electrophoresis. For immunoblotting, proteins were transferred to PVDF membranes and reacted with rabbit preimmune serum (lane 1) and rabbit anti-VirB6-4 or anti-VirB6-3 serum (lanes 2 and 3). (B and C) Intact A. phagocytophilum organisms were incubated with (+) or without (−) trypsin and solubilized, and equal amounts of proteins were separated by Tris acetate polyacrylamide gel electrophoresis. PVDF membranes with transferred proteins were processed for detection. (B) Rabbit anti-VirB6-3 serum (lanes 2 and 4), rabbit preimmune serum (lanes 1 and 3), normal mouse serum (NMS) (lanes 5 and 7), and polyclonal mouse anti-mCherry antibodies (lanes 6 and 8); (C) rabbit anti-VirB6-4 serum (lanes 10 and 12), rabbit preimmune serum (lanes 9 and 11), NMS (lanes 13 and 15), and polyclonal mouse anti-mCherry antibodies (lanes 14 and 16). “MW” indicates protein standards. Immunoblots are representative of those from three independent experiments.
To confirm that VirB6-3 and VirB6-4 are surface-exposed proteins in A. phagocytophilum, we performed trypsin digestion of intact A. phagocytophilum organisms. For these experiments, we used mCherry-tagged A. phagocytophilum organisms that carry the Himar1 Tn integrated within an intergenic region between the HGE1_00520 (phosphatidate cytidylyltransferase) and the HGE1_00525 (p44-66) genes and whose growth in HL-60 and ISE6 cells is comparabe to that of the parental strain A. phagocytophilum strain HGE1 (39). Intact A. phagocytophilum organisms were incubated with trypsin, followed by solubilization and Western blotting using rabbit anti-VirB6-3 and anti-VirB6-4 sera. Mouse anti-mCherry antibody was used as a control to show that trypsin would not breach the cell wall, as this protein localizes to the bacterial cytosol and is not surface exposed. Treatment of bacteria with trypsin completely digested the protein band of 158 kDa and dramatically reduced the detection of smaller protein bands in samples reacted with anti-VirB6-3 antisera relative to untreated samples (Fig. 3B). Likewise, anti-VirB6-4 sera did not detect VirB6-4 protein in samples digested with trypsin relative to untreated controls (Fig. 3C). In trypsin-treated samples, mCherry was not reduced or diminished in size, indicating that reduction of VirB6-3 and VirB6-4 was due to trypsin treatment of the cell surface and not to damaged organisms.
Isolation of a VirB6-4 mutant generated by transposon mutagenesis.
Anaplasma phagocytophilum strain HGE1, subsequently referred to here as the wild type (wt), was subjected to Himar1 Tn mutagenesis (40, 41). Tn sequencing (Tn-seq) using the Illumina platform detected a Tn insertion into the virB6-4 open reading frame. Cloning by limiting dilution allowed the isolation of a population of organisms isogenic for the Tn insertion within this gene. Therefore, these bacteria are designated here as virB6-4::Himar1 mutants.
Although the genome sequence of A. phagocytophilum strain HGE1 (GenBank accession number APHH00000000) is available, the 3′ end repeat region of virB6-4 is unresolved, containing gaps. Therefore, for our analysis we used the complete virB6-4 sequence from the A. phagocytophilum strain HGE1 mutant DU1 (GenBank accession number NZ_LASP01000002) as a reference. Alignment of Illumina reads to the reference genome mapped the Tn insertion at the TA dinucleotide coordinates (381514..381515) 5′ upstream from the extensive tandem repeat domain at the 3′ end (Fig. 4A and B). This analysis indicated that the mCherry and aadA genes from the Tn are integrated in the opposite orientation to virB6-4 (Fig. 4C). Orientation of the Himar1 transposon was also confirmed by Sanger sequencing.
FIG 4.
Himar1 Tn insertion site in the A. phagocytophilum virB6-4 gene. (A) Artemis (genome browser and annotation tool; Wellcome Sanger Institute, UK) window showing A. phagocytophilum wild-type virB6-4 gene used as a reference for the location of the Himar1 insertion site. The first panel shows a “zoomed out” view of the virB6-4 DNA sequence translated into six reading frames with black bars indicating stop codons. The open reading frame (ORF) for virB6-4 is shown in light blue. The black arrow indicates the Himar1 insertion location, 5′ upstream the 3′ end tandem repeats. The yellow box is “zoomed in” in the panel below to show in detail the virB6-4 sequences flanking the Himar1 Tn (yellow highlight), including the TA dinucleotide site required for successful transposition (red arrow). (B) Paired-end DNA sequencing reads indicating the same A. phagocytophilum sequences as described above (yellow highlight) and Himar1 Tn sequences (italics). (C) Artemis window of in silico cloning of the Himar1 Tn in the opposite orientation to virB6-4. Black bars within the virB6-4 ORF indicate putative stop codons.
Himar1 Tn insertion within virB6-4 abolished its expression and had a polar effect on the sodB-virBs operon.
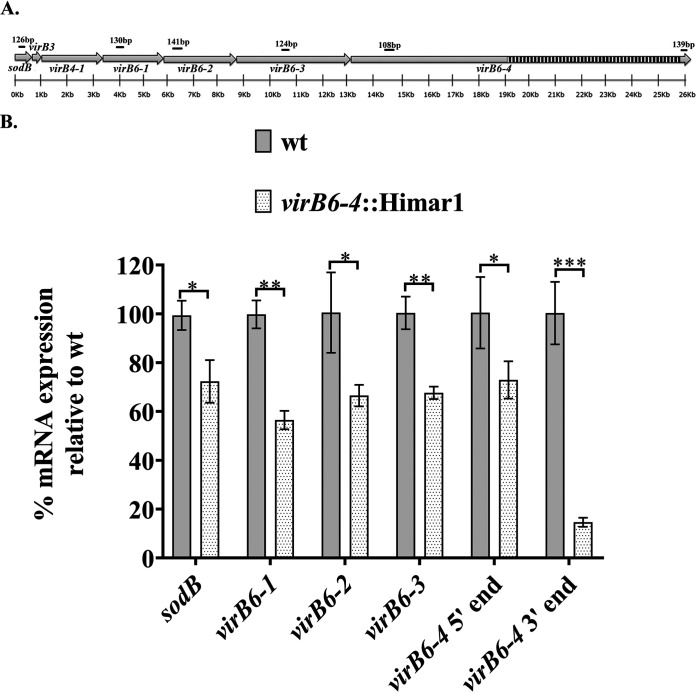
In silico cloning of Tn sequences within the virB6-4 gene suggested that its insertion resulted in a shift in the reading frame that was predicted to lead to premature termination of translation and the expression of a truncated protein (Fig. 4C). To test this assumption and determine whether the inserted transposon exerted polar effects on the expression of adjacent genes, we examined the expression of virB6-4 and upstream genes. Reverse transcription-quantitative PCR (RT-qPCR) was used to quantitatively determine differences in the expression of gene components of the sodB-virB operon in the A. phagocytophilum wt versus the virB6-4::Himar1 mutant. For this, total RNA from HL-60 cells infected with the A. phagocytophilum wt or the virB6-4::Himar1 mutant was reverse transcribed using random hexamer primers. The resulting cDNAs were quantified by real-time PCR amplification using primers targeting sodB, virB6-1, virB6-2, virB6-3, and the 3′ and 5′ ends of virB6-4 (Fig. 5A). Relative expression levels of these genes were normalized to the geometric mean of the reference genes rpoB and groEL. RT-qPCR analysis revealed significantly reduced transcripts from sodB (mean ± standard deviation [SD], 69.3% ± 6.83%), virB6-1 (54.26% ± 3.67%), virB6-2 (63.96% ± 5.8%), virB6-3 (64.97% ± 3.40%), the virB6-4 5′ end (70.14% ± 9.07%), and the virB6-4 3′ end (14.03% ± 1.49%) in the virB6-4::Himar1 mutant relative to the values for the wt, which were as follows: sodB, 99.38% ± 6.02%; virB6-1, 99.79% ± 5.71%; virB6-2, 100% ± 16.45%; virB6-3, 100% ± 6.67%; the virB6-4 5′ end, 100% ± 14.65%; the virB6-4 3′ end, 100% ± 12.79% (Fig. 5B). Initially, for gene expression data normalization, we also evaluated transcripts from the msp5 gene. However, during this analysis, we observed decreased transcription of this gene in the virB6-4::Himar1 mutant (mean ± SD, 78.44% ± 11.66%) relative to wt bacteria (99.91% ± 3.41%) (Fig. S3), suggesting that Himar1 insertion within virB6-4 may also have trans-acting effects on the expression of additional, more distant genes.
FIG 5.
Deletion of virB6-4 affects transcription of upstream genes. (A) Binding sites of primers designed to target transcripts from sodB, virB6-1, virB6-2, virB6-3, and the 5′ and 3′ ends of virB6-4. (B) Changes in expression of these genes were calculated based on geometric mean of the threshold cycle (ΔCT) values from reference genes, and the results were expressed as percentage of expression, with a 100% expression level being assigned to the control group, in this case, wt A. phagocytophilum. Transcripts from the rpoB and groEL genes were used as reference for data normalization. Bar lengths represent the percentage of expression of sodB, virB6-1, virB6-2, virB6-3, and the 5′ and 3′ ends of virB6-4 in A. phagocytophilum (wt) and the virB6-4::Himar1 mutant. *, P = 0.01 to 0.05; **, P = 0.001 to 0.01; ***, P < 0.001. Data were obtained from three independent experiments with two technical replicates.
To determine whether decreased mRNA levels from the virB6-3 and virB6-4 genes in the virB6-4::Himar1 mutant correlate with altered VirB6-3 and VirB6-4 protein levels, we performed Western blot analysis using polyclonal rabbit anti-VirB6-3 and anti-VirB6-4 sera. To be sure that observed changes in VirB6-3 and VirB6-4 protein levels in the virB6-4::Himar1 mutant relative to wt bacteria are due to altered protein expression rather than differences in sample loading, we used polyclonal dog serum against A. phagocytophilum, with antibody responses mainly targeting the dominant surface protein MSP2/P44 (42 to 44 kDa), as a loading control. Western blotting indicated a reduction in VirB6-3 expression in the virB6-4::Himar1 mutant compared to the wt (Fig. 6A). The ratio of the amount of VirB6-3 in the virB6-4::Himar1 mutant relative to the VirB6-3 amounts in the wt was determined by densitometry, normalized to the signal obtained from the MSP2/P44 band and converted to percentage. This indicated decreased levels of VirB6-3 expression in the virB6-4::Himar1 mutant (mean ± SD, 24.27% ± 14.82%) relative to the wt (100% ± 14.43%). VirB6-4 protein was detected only in protein samples from wt organisms and not in the virB6-4::Himar1 mutant (Fig. 6B). Bands of sizes corresponding to VirB6-3 and VirB6-4 proteins were not observed in samples reacted with the negative-control rabbit preimmune sera (Fig. 6A and B). Consistent with Western blot analysis, dual immunofluorescence of HL-60 cells infected with the virB6-4::Himar1 or wt strain indicated a dramatic reduction in the signals from VirB6-3 epitopes in the virB6-4::Himar1 mutant-infected relative to wt-infected cells (Fig. 6C). Likewise, fluorescent signals from VirB6-4 epitopes in cells infected with the virB6-4::Himar1 mutant were virtually absent compared to those in wt-infected cells (Fig. 6D).
FIG 6.
Synthesis of VirB6-4 and VirB6-3 is disrupted in the virB6-4::Himar1 mutant. Equal amounts of proteins from host cell purified virB6-4::Himar1 and wt A. phagocytophilum were separated by Tris acetate polyacrylamide gel electrophoresis. (A) PVDF membranes of transferred proteins were reacted with rabbit preimmune serum (lanes 1 and 2), rabbit anti-VirB6-3 serum (lanes 3 and 4), and dog preimmune serum (lanes 5 and 6). Polyclonal dog serum infected with A. phagocytophilum, with immune responses mainly targeting the MSP2/P44 surface protein, was used as a loading control to indicate equal loading of proteins from the virB6-4::Himar1 mutant and wt A. phagocytophilum (lanes 7 and 8). (B) PVDF membranes of transferred proteins were reacted with rabbit preimmune serum (lanes 9 and 10), rabbit anti-VirB6-4 serum (lanes 11 and 12), dog preimmune serum (lanes 13 and 14), and A. phagocytophilum-infected dog serum (loading control; lanes 15 and 16). (C and D) HL-60 cells infected with A. phagocytophilum virB6-4::Himar1 or the wt were fixed and viewed by indirect immunofluorescence microscopy to determine immunoreactivity with A. phagocytophilum-infected mouse serum (green) and rabbit anti-VirB6-3 (C, red) or anti-VirB6-4 (D, red) serum. Representative immunoblots were obtained from three independent experiments. Representative immunofluorescence assay images from several micrographs are shown.
The virB6-4::Himar1 mutant has a reduced proliferation in vitro.
We further evaluated the growth kinetics of the virB6-4::Himar1 mutant versus the wt by determining the numbers of A. phagocytophilum genome equivalents per cell (ApGE/cell) using duplex qPCR. For this, we used specific primers and probes targeting the A. phagocytophilum msp5 gene and the Toll-like receptor 9 (tlr9) gene in HL-60 cells or the calreticulin gene (crt) in ISE6 cells. During the course of infection in HL-60 cells, the virB6-4::Himar1 mutant maintained a reduced growth, as evidenced by significantly lower numbers of ApGE/cell (mean ± standard error of the mean [SEM], 28.82 ± 1.870 [P = 0.0317] at day 1 postinfection [p.i.] and 51.11 ± 5.966 [P = 0.0035] at day 6 p.i.) relative to ApGE/cell in cells infected with wt bacteria at the same time points (43.26 ± 4.05 and 164.2 ± 17.28 on days 1 and 6 p.i., respectively) (Fig. 7A). On the other hand, during the first 3 days p.i., there were no significant differences in the numbers of ApGE/cell in ISE6 cells infected with the virB6-4::Himar1 mutant relative to the wt. However, after day 3 p.i., the growth of the mutant declined, and by day 12 p.i., the numbers of ApGE/cell were significantly lower (mean ± SEM, 49.68 ± 15.6) (P = 0.0039) than in cells infected with wt bacteria (184.35 ± 4.43) (Fig. 7B).
FIG 7.
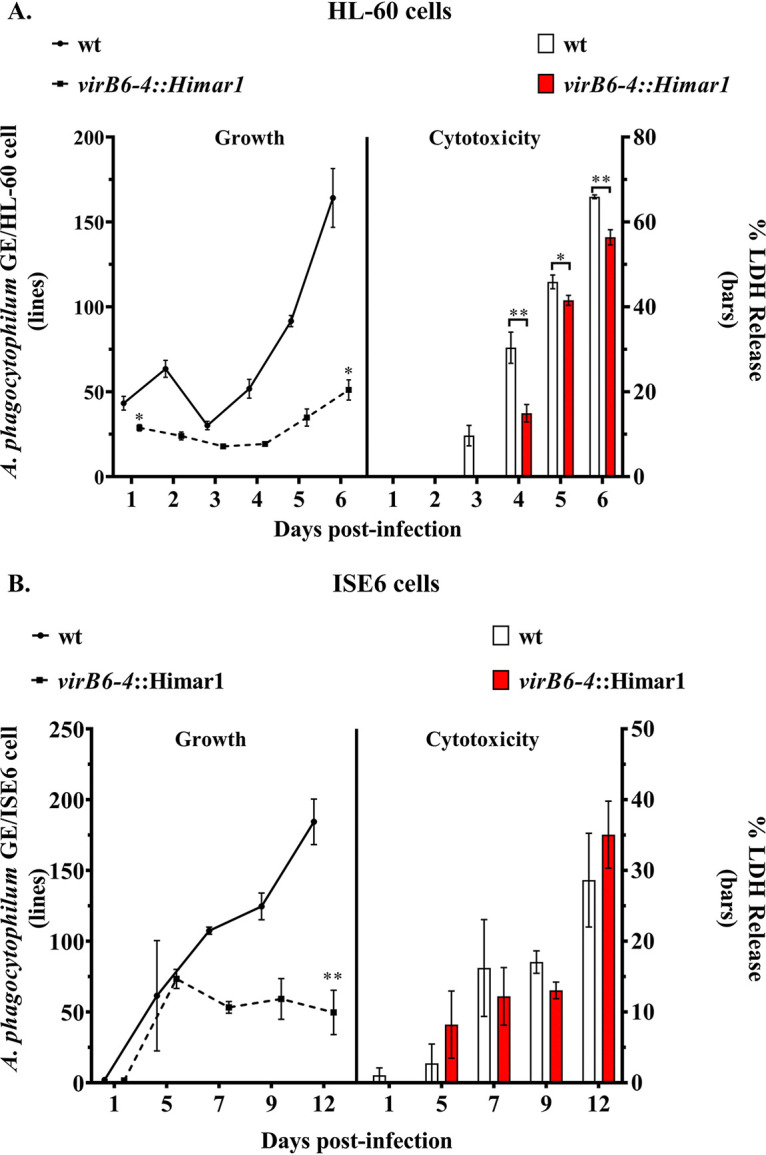
Growth and cytotoxicity of the virB6-4::Himar1 mutant. The growth of A. phagocytophilum virB6-4::Himar1 (dotted line) versus wild-type (wt) (black line) in infected HL-60 (A) and ISE6 (B) cells was followed by determining the number of A. phagocytophilum genome equivalents per host cell targeting the A. phagocytophilum single-copy gene msp5 and the tlr9 or crt genes from HL-60 and ISE6 cells, respectively. Bars indicate A. phagocytophilum virB6-4::Himar1 and wild-type cytotoxicity to host cells as indicated by the levels of LDH release into the cell culture supernatants. The culture medium was not changed at any time during the course of this experiment. *, P = 0.01 to 0.05; **, P = 0.001 to 0.01. Data were obtained from three independent experiments with three technical replicates.
Given the reduced growth exhibited by the virB6-4::Himar1 mutant, we expected decreased levels of host cell death. To determine this, aliquots of cell culture supernatants collected from virB6-4::Himar1- or wt-infected HL-60 and ISE6 cells were used to measure the levels of host cell lactate dehydrogenase (LDH cytotoxicity assay) released into the cell medium due to the pathogenic effects exerted by the bacteria. Effectively, the reduced burden of virB6-4::Himar1 organisms in HL-60 cells was associated with decreased host cell death, as there was a significantly reduced level of LDH in supernatants from virB6-4::Himar1-infected HL-60 cells relative to wt-infected cells at all tested times p.i. (Fig. 7A). In contrast, the reduced infection of ISE6 cells with the virB6-4::Himar1 mutant did not correlate with decreased host cell death, as the levels of LDH released in supernatants from infected cells were not significantly different from those in wt-infected cells at all tested times p.i. (Fig. 7B).
DISCUSSION
The role of the A. phagocytophilum T4SS during infection of human host and tick vector cells remains unclear. Here, we show that all four virB6 paralogs of A. phagocytophilum are cotranscribed into a polycistronic message that includes sodB, virB3, and virB4-1 during in vitro infection of mammalian and tick cells. VirB6-1, VirB6-2, VirB6-3, and, strikingly, a large VirB6-4 protein (∼470 kDa) were also detected in infected cultures. Given the unusual C-terminal structural extensions of VirB6-3 and VirB6-4 in A. phagocytophilum, we focused most of our work on characterization of the expression and potential subcellular localization of these two proteins. Evidence obtained from this work suggests that these proteins have evolved to serve a fundamentally different function in Rickettsiales than in A. tumefaciens. For example, dual labeling of A. phagocytophilum suggests that VirB6-3 and VirB6-4 are distributed throughout the bacterial cell membrane in infected HL-60 and ISE6 cells, whereas in A. tumefaciens, VirB6 is localized mainly to a single pole (30).
Surface trypsin digestion of host cell-free intact A. phagocytophilum provided evidence that major C-terminal domains of VirB6-3 and VirB6-4 are surface exposed. This also contrasts with A. tumefaciens, in which VirB6 consists of a periplasmic N terminus, four or five transmembrane domains, and a cytoplasmic C terminus (26, 29). Topology similar to that of A. tumefaciens was also reported for VirB6 in Brucella (42) and ComB6 (VirB6 homolog) in H. pylori (43). Although further experimental evidence is required, surface trypsin digestion data suggest that the C termini of A. phagocytophilum VirB6-3 and VirB6-4 are surface exposed on the outer membrane, as the peptide sequences used for the preparation of antibodies were derived from VirB6-3 and VirB6-4 C-terminal repeat sequences. It has been proposed that the extended C terminus may be cleaved and released into the target cell to effect regulatory functions (4, 15). This localization would optimize such a transfer. Hence, these data provide initial experimental evidence for an extracytoplasmic extended C terminus in polytopic “extended VirB6-like proteins” in bacteria of the P-T4SS group. Extended C-terminal domains have been found on the F-T4SS of Escherichia coli (TraG) and SXT-ICE (TraGSXT) of Vibrio cholerae. These C-terminal hydrophilic domains function during mating pair formation in association with TraS protein that is found in the inner membrane of the donor of the pair in a process known as entry exclusion to block transfer of redundant DNA (14).
Conforming to the case with other Rickettsiales (20), bioinformatics analysis indicated that sequence conservation of A. phagocytophilum VirB6 paralogs relative to VirB6 from A. tumefaciens is restricted to the TMDs and a central cytoplasmic loop. In addition, this analysis predicted SP sequences for VirB6-1 and VirB6-2 and identified VirB6-3 and VirB6-4 as putative lipoproteins given the presence of conserved lipobox signal sequences. Despite the importance of lipoproteins in physiology, virulence, and host immune evasion, very little is known about their role in A. phagocytophilum. In other Rickettsiales, such as E. chaffeensis (44) and the Wolbachia symbiont of filarial nematodes (38), VirB6-3, but not VirB6-4, has also been identified as a putative lipoprotein. Moreover, antibodies reacting with VirB6-3 proteins from E. chaffeensis and Wolbachia species were detected in sera from dogs infected with E. chaffeensis (44) and in sera from humans infected with the filarial nematode Wuchereria bancrofti (38). This is additional evidence that in Rickettsiales this protein not only is surface exposed but also induces immune responses. We noted that A. phagocytophilum contains genes that encode proteins required for lipoprotein biosynthesis, such as the prolipoprotein diacylglyceryl transferase (Lgt; GenBank accession number AGR79154) and the lipoprotein signal peptidase (LspA; GenBank accession number AGR79052.1) but not the apolipoprotein N-acyltransferase (Lnt/CutE). It is known that Wolbachia (45) synthesizes only diacyl-lipoproteins, which are involved in triggering of inflammatory responses (46). The fact that VirB6-3 and VirB6-4 are surface proteins suggests that they could be targeted as potential vaccine candidates against closely related A. phagocytophilum strains.
Transposon mutagenesis using the Himar1 system followed by high-throughput genome sequencing to identify Tn insertion loci in these transformants identified a population of mutants carrying the transposon sequences within the coding region of the virB6-4 gene. Relative gene expression experiments demonstrated that insertion of transposon sequences into virB6-4 not only resulted in the loss of its expression but also altered the expression of upstream genes, including sodB, virB6-1, virB6-2, and virB6-3, suggesting that gene expression from the entire operon (including virB3 and virB4-1) was disrupted. As the transposon sense strand is found in the opposite orientation to virB6-4, it is possible that transcriptional readthrough beyond Tn sequences generated antisense transcripts that reduced the expression of sequences upstream of virB6-4. Similar outcomes obtained by Himar1 insertion have been observed in several bacterial species, including tick-borne bacteria (47–49). Consistent with gene expression data, Western blotting and immunofluorescence indicated the absence of VirB6-4 and decreased levels of VirB6-3 in the virB6-4::Himar1 mutant relative to wt A. phagocytophilum. Importantly, our data show that the disruption of virB6-4 and the altered expression of upstream genes significantly decreased A. phagocytophilum infection in human and tick cells in vitro, indicating the important role of these T4SS components in the intracellular survival of A. phagocytophilum in these two different environments. Further work is required to determine whether reduced infection is due to a decreased replication or failure to evade host killing mechanisms such as avoidance of lysosomal degradation or failure to escape oxidative damage (50).
Given the polar effect exerted by the Himar1 insertion within the virB6-4 gene, it is not possible to assign the mutant phenotype to this gene alone, as expression of the other virB6 paralogs virB3, virB4-1, and sodB was also affected. However, it is possible that the altered expression levels of T4SS components present in this operon affect the abundance of other T4SS components, impairing the assembly and function of the secretion system and resulting in poor growth. A similar effect has been observed in other bacterial species, such as A. tumefaciens and a Brucella sp., carrying mutations in T4SS components, specifically virB6 mutations (25, 51). In Brucella species, VirB3 and VirB6 are essential for persistence in mice (25). Another important consequence of disruption of virB6-4 relates to the observed reduced expression of sodB in the virB6-4::Himar1 mutant as shown here. Expression of superoxide dismutase (SOD) in pathogenic bacteria contributes to virulence, as it helps them to escape killing by reactive oxygen species released by the host defense mechanisms (52, 53). In that context, it is worth noting that the virB6-4::Himar1 mutant showed reduced expression of sodB. It is known that A. phagocytophilum inhibits NADPH oxidase, thus avoiding killing by toxic oxygen intermediates (50, 54–57). However, it is not known which A. phagocytophilum effector proteins participate in this inhibitory process. It is possible that A. phagocytophilum uses SOD to protect itself from oxidative damage, and reduced expression of this protein as shown for the virB6-4::Himar1 mutant may contribute to impaired proliferation in infected cells.
The isolation of the virB6-4::Himar1 mutant with an altered expression pattern in the sodB-virB operon supports the contribution of these proteins to A. phagocytophilum infection. Reduced transcript levels from msp5 in the virB6-4::Himar1 mutant may reflect not only modification or remodeling of membrane protein components but also that genes from the sodB-virBs operon are possibly linked to regulation of additional genes. Therefore, comparative proteomics of virB6-4::Himar1 mutant and wt A. phagocytophilum will be of great significance to define differentially expressed proteins and pathways associated with the slow-growth phenotype of this mutant. Additionally, expression of a defective T4SS not only will help to understand how the T4SS functions but also will aid in the identification of T4SS substrates and will clarify the role of this operon in persistent infection.
The order Rickettsiales includes pathogens of high importance in human and veterinary health and, potentially, bioterrorism. Despite their genetic diversity, the different cellular niches they occupy, and the variety of hosts they infect, an element common to members of this order is the synteny of the virB3, virB4, and virB6 paralogs (4 copies in Anaplasmataceae and 5 copies in Rickettsiaceae) that are arranged in tandem within an operon unit (12). This genomic organization is most likely due to functional constrains. Hence, results obtained in working with A. phagocytophilum may also be applicable to other Rickettsiales. Our findings are of particular significance, as they provide the first evidence for surface exposure of the intriguing C-terminal repeat extensions of VirB6-3 and VirB6-4. The characterization of a mutant with an insertion into the virB6-4 gene offers insights about the key role that these T4SS components play in infection of mammalian and tick cells and will facilitate more detailed analysis of the T4SS structural components and the effectors they transport.
MATERIALS AND METHODS
Cultivation of Anaplasma phagocytophilum.
The A. phagocytophilum strains used for this work were the wt HZ (58), wt HGE1 (39), and the HGE1 virB6-4::Himar1 mutant and 120879::Himar1 mutant (used for trypsin digestion to confirm cell surface proteins). Two cell lines were used to propagate these bacteria, i.e., HL-60 human promyelocytic cells (ATCC CRL-240) and tick ISE6 cells (ATCC CRL-11974) derived from embryonated eggs of the black-legged tick, Ixodes scapularis. Uninfected and wt or mutant A. phagocytophilum-infected HL-60 cells were maintained in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (FBS; BenchMark, Gemini Bio-Products), 2 mM l-glutamine (Life Technologies), 0.25% NaHCO3 (Sigma-Aldrich), and 25 mM HEPES (Sigma-Aldrich) and were kept at 37°C in a 5% carbon dioxide (CO2) atmosphere. Uninfected ISE6 cell cultures were maintained in L-15B300 medium prepared as described previously (59) and supplemented with 5% FBS, 5% tryptose phosphate broth (TPB; Difco, Becton, Dickinson), and 0.1% bovine lipoprotein concentrate (LPC; MP-Biomedical). Infected ISE6 cells were maintained in L-15B300 medium additionally containing 0.25% NaHCO3 and 25 mM HEPES buffer.
RNA extraction and reverse transcription-PCR (RT-PCR).
Total RNA was isolated from infected HL-60 and ISE6 cells using the RNeasy kit (Qiagen) with an added “on-column” DNase I treatment (Qiagen) as per the manufacturer’s instructions and (1 μg) from each sample converted to cDNA by random hexamer priming using the Transcriptor first-strand cDNA synthesis kit (Roche Diagnostics) as per the manufacturer’s instructions. Fluorescence-based RNA concentrations were determined by using the Qubit RNA HS assay kit (Thermo Fisher Scientific) with a Qubit fluorometer (Thermo Fisher Scientific). Specific primers (Table 2) were designed to amplify transcripts from each gene component of the sodB-virB operon or intergenic regions. All reactions included primers targeting transcripts from the msp5 and 16S rRNA genes as internal controls and to ensure integrity of RNA/cDNA. Each reaction contained 200 ng of cDNA combined with 200 μM deoxynucleoside triphosphates (dNTPs), 0.25 μM forward and reverse primers, and 1.25 U of PrimeSTAR GXL DNA polymerase (TaKaRa). PCR amplification conditions were as follows: 94°C for 2 min, 30 cycles of denaturation 98°C for 10 s, annealing at 55°C for 15 s, and extension at 68°C for 1 min, and a final extension step at 68°C for 7 min. PCR products were electrophoretically separated using a 2% SeaKem LE (Lonza) agarose gel, and stained with SYBR gold nucleic acid gel stain (Thermo Fisher Scientific) for UV visualization. Genomic DNA and samples of reactions with no reverse transcriptase for each target were used as positive and negative controls, respectively.
TABLE 2.
Oligonucleotides used in this study
| Purpose and oligonucleotide | Sequence | Amplicon size (bp) | Target |
|---|---|---|---|
| RT-PCR | |||
| AB1727 | TGGCAATCCATGAAGCCGAA | 230 | sodB |
| AB1728 | GGAAGCTGCCCCTGAGTAAG | ||
| AB1729 | TTAACCAGGCCCACCATGCT | 130 | virB3 |
| AB1730 | CGTGCATACCTGGCGCTAAA | ||
| AB1731 | AAGCTAGGTTGGTCTTCGGC | 347 | virB4-1 |
| AB1732 | CGCCACTGCATATTCACAGC | ||
| AB1733 | CCAGAGATCGGGTGTCAGAT | 414 | virB6-1 |
| AB1734 | AAGCTCCCCTGCACTTTGTA | ||
| AB1735 | TTTGCAGGGCCTATAGGTTG | 588 | virB6-2 |
| AB1736 | TGCTGTCGTACCCACTTCAG | ||
| AB1737 | TACATAGCTCCGGGTTCTGG | 512 | virB6-3 |
| AB1738 | TTCCCAGGTAACGCAGTAGG | ||
| AB1739 | TGTCTAGGACTGTCGCATCG | 471 | virB6-4 5′ end |
| AB1740 | TGCGTAAGTTCTGCATCACC | ||
| AB1741 | AATCCGGCAAAGAAGAGGAT | 132 | virB6-4 3′ end |
| AB1742 | GTGCCCGAAAACAGCTCTAA | ||
| AB1745 | TGTTCGGCTTTTCTTCATGC | 204 | msp5 |
| AB1746 | CTTCCTCATCCCCAGTCAGC | ||
| AB1747 | AAGAAGTCCCGGCAAACTCC | 342 | 16S |
| AB1748 | CCCACATTCAGCACTCATCG | ||
| AB1727 | Same as above | 633 | sodB-virB3 |
| AB1730 | Same as above | ||
| AB1729 | Same as above | 608 | virB3–virB4-1 |
| AB1732 | Same as above | ||
| AB1960 | CGAGACCGTTGCCATACTTC | 367 | virB4-1–virB6-1 |
| AB1961 | AAACCGAGAGGCCTAAATCC | ||
| AB1962 | TCCTCTGATAGTGGAGGTACAG | 369 | virB6-1–virB6-2 |
| AB1963 | TAGAGCTATCCTTCTGTCCCTTC | ||
| AB1964 | TGGAACAGCATCAGCATCAG | 357 | virB6-2–virB6-3 |
| AB1965 | TCCAGAATGCACCCAATACG | ||
| AB1966 | TGCCGTGAGGGATGATTTG | 362 | virB6-3–virB6-4 |
| AB1967 | CCCTCCACCTTTACTACTATCTC | ||
| RT-qPCR | |||
| AB2053 | CGGTAGTGTGGAAGGGTTTAAT | 126 | sodB |
| AB2054 | GCATTCGCTGTGCTAACAAC | ||
| AB2057 | GCCGCTGATGGAAGTAAGAT | 130 | virB6-1 |
| AB2058 | CCACAACCACCGACGTTATAG | ||
| AB2059 | TAGGGTTACCTGGACCGATATT | 141 | virB6-2 |
| AB2060 | CTGTCGTACCCACTTCAGAAAG | ||
| AB2061 | GCACGTACTGGTGAGGATATT | 124 | virB6-3 |
| AB2062 | GTAACGCAGTAGGAGGTGTATC | ||
| AB2063 | CTCACACACAGAGGCAGATTT | 108 | virB6-4 5′ end |
| AB2064 | AGAGCATCCAATGGCGAATAG | ||
| AB1741 | Same as above | 132 | virB6-4 3′ end |
| AB1742 | Same as above | ||
| AB2065 | TGCGGAACTTGGTATGGTATC | 118 | msp5 |
| AB2066 | CTCATTTAACCTTTCAACAGTGTCA | ||
| AB2121 | AGGGAGGTAGTACGCATCCTAGA | 102 | groEL |
| AB2122 | TGTGATCTCTGGCGACCCATAA | ||
| AB2125 | GGCCTATGGTGCTGCTTATAC | 115 | rpoB |
| AB2126 | CCACACTCGAAGTTGCTATCC | ||
| qPCR | |||
| AB1334 | AGATGCTGACTGGGGATGAG | 125 | msp5 (59) |
| AB1335 | TCGGCATCAACCAAGTACAA | ||
| AB1336a | CGTAGGTGAGTCTGATAGTGAAGG | ||
| AB2039 | GTCAAGTCCGGCACAATCT | 111 | crt |
| AB2040 | CATCTTCTTCTCGGCATCCTT | ||
| AB2041b | TTGCTGACTGACGACGAAGAGTATGC | ||
| AB2042 | CCCAGTCTTGGACTCAGAATTAG | 100 | tlr9 |
| AB2043 | GGTATAGCCAGGGATTGGTTAAG | ||
| AB2044b | TCTAGGTCTCAGTCCTGGTTCTGAAGC | ||
Probe labeled with hexachloro-fluorescein (HEX) at the 5′ end and tetramethylrhodamine (TAMRA) at the 3′ end.
Probe labeled with 6-carboxyfluorescein (6-FAM) at the 5′ end and black hole quencher-1 (BHQ-1) at the 3′ end.
RT-qPCR.
Transcript differences between sodB, virB6-1, virB6-2, virb6-3, and the virB6-4 5′ and 3′ ends in the A. phagocytophilum virB6-4::Himar1 mutant relative to the wild type were determined using RT-qPCR, and the results were based on the means of three biological replicates (individual RNA extracts). For SYBR green quantitative PCR, cDNA obtained from HL-60 cells infected with the A. phagocytophilum wt or the virB6-4::Himar1 mutant was used with primers (Table 2) designed to amplify sodB, virB6-1, virB6-2, virb6-3, the virB6-4 5′ or 3′ end, msp5, groEL, and rpoB sequences. Reactions were performed using the hot-start LightCycler-FastStart DNA Master SYBR Green I (Roche Diagnostics) in a LightCycler 96 instrument (Roche Diagnostics). Twenty microliters of reaction mixture contained 1× LightCycler FastStart DNA Master SYBR green I, 0.4 μM primers, and 2 μl of template (∼20 ng of cDNA). No-template and no-reverse transcriptase control samples to assess DNA contamination were included for all reactions. The amplification program included the following conditions: 95°C for 10 min and 45 cycles of 95°C for 10 s and 60°C for 30 s. After amplification, a melting curve was acquired by heating of the product at 4.4°C/s to 95°C for 10 s, cooling it at 2.2°C/s to 65°C for 60 s, and then slowly heating at 0.1°C/s to 97°C. After melting-curve acquisition, reaction mixtures were cooled at 37°C for 30 s. Significant differences between the A. phagocytophilum virB6-4::Himar1 mutant versus the wt were calculated using Student’s t test (P < 0.05), comparing threshold cycle (ΔCT) values (target gene − reference gene) of the virB6-4::Himar1 mutant and the wild type and calculated based on the Pfaffl formula (60), which yields the gene expression ratio. The expression ratio was then expressed as percent expression by multiplying the gene expression values by 100. The geometric means of ΔCT values from the reference genes rpoB and groEL were used for gene expression normalization. Transcript amplification efficiencies for each target were 87% for sodB, 99% for virB6-1, 97% for virB6-2, 100% for virB6-3, 96% for the virB6-4 5′ end, 100% for virB6-4, 93% for msp5, 100% for rpoB, and 91% for groEL.
VirB6 synthetic peptides and antibodies.
Specific antibodies were raised against A. phagocytophilum strain HZ VirB6 paralogs using synthetic peptides derived from predicted antigenic epitopes using the protein analysis and peptide prediction programs TMpred (61), Phobius (62), BcePred (B-Cell Epitope Prediction of Continuous B-Cell Epitopes in Antigenic Sequences Using Physicochemical Properties) (63), LEPS (Linear Epitope Based on Propensity Scale) (64), and the antigenicity predicting tools from EMBOSS (65). Peptide sequences (56–70 residues) (Table 1) were selected on the basis of their predicted hydrophilicity, antigenic propensity, flexibility, surface probability and lack of sequence identity with other A. phagocytophilum proteins based on BLASTP alignments (66).
VirB6-1 through VirB6-4 peptides were synthesized commercially by LifeTein, NJ. An amino-terminal cysteine was included for conjugation, and 4 mg of each peptide was prepared (2 mg was conjugated to keyhole limpet hemocyanin (KLH) and 2 mg to ovalbumin). Production of polyclonal antibodies against VirB6-1, VirB6-2, VirB6-3, and VirB6-4 was done by Pocono Rabbit Farm and Laboratory Inc., PA. Each rabbit was subcutaneously injected with 200 μg of KLH-conjugated peptide emulsified in complete Freund’s adjuvant. At days 14 and 28 postimmunization, the rabbits were boosted by subcutaneous injection of 200 μg of peptide conjugates in incomplete Freund’s adjuvant. Affinity-purified anti-VirB6 antibodies were prepared by immobilization of sulfhydryl-containing peptides using a commercially available coupling resin (SulfoLink coupling resin; Thermo Fisher Scientific) according to the manufacturer instructions (67). All animal work was performed in accordance with the standards and approval of the University of Florida Institutional Animal Care and Use Committee.
To determine titers and the specificity of the sera for the VirB6 paralogs, we used an enzyme-linked immunosorbent assay (ELISA) in an antibody capture format. For this, 96-well MaxiSorp microtiter plates (Nunc) were coated with 1 μg/well of ovalbumin-conjugated peptide or 2 μg/well of VirB6-ovalbumin-conjugated peptide. Pre- and postimmune sera from each rabbit were serially diluted 10-fold 102 to 105. All assays were performed in triplicate, and antibody binding was detected with recombinant protein A/G conjugated to alkaline phosphatase. Color was developed using 4-nitrophenol phosphate, and the optical densities were measured using an ELISA plate reader at 405 nm.
Detection of VirB6 paralogs by immunofluorescence.
Antigen slides were prepared from A. phagocytophilum-infected HL-60 and ISE6 cells essentially as described previously (68). Briefly, infected cells were washed twice in 1× phosphate-buffered saline (PBS), collected by centrifugation at 200 × g for 5 min at room temperature, fixed in 1 ml of 4% paraformaldehyde–0.0075% glutaraldehyde in PBS for 30 min at room temperature, then washed in PBS, and permeabilized in ice-cold 0.3% Nonidet P-40 (NP-40) for 10 min. After being washed with 1× PBS, the samples were blocked with 1 ml of blocking buffer (5% bovine serum albumin [BSA] and 1% normal goat serum in PBS) and incubated for 3 h at room temperature. After blocking, cells were aliquoted into five 1.5-ml centrifuge tubes and collected by centrifugation at 200 × g for 5 min at room temperature. Dual A. phagocytophilum staining was done with a mixture of two primary antibodies, mouse A. phagocytophilum-positive serum (1:320) and the appropriate anti-VirB6 serum, unpurified anti-VirB6-1 serum (1:640), unpurified anti-VirB6-2 serum (1:640), affinity-purified anti-VirB6-3 and anti-VirB6-4 antibodies at concentrations of 0.208 mg/ml and 0.232 mg/ml, respectively, or preimmune rabbit serum (1:640) or protein A-purified 0.768-mg/ml preimmune serum (negative control). All samples were incubated overnight at 4°C. After incubation with primary antibodies, the samples were collected by centrifugation as described above and washed 3 times with washing buffer (1% BSA and 0.1% Tween 20 in PBS), followed by incubation for 1.5 h at room temperature with goat anti-rabbit IgG Alexa Fluor 568-conjugated antibody (Thermo Fisher Scientific) and goat anti-mouse IgG Alexa Fluor 488-conjugated antibody (Thermo Fisher Scientific), both at a dilution of 1:800. The samples then were washed as described above, followed by a final wash with 1× PBS, and mounted with ProLong gold antifade reagent with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Thermo Fisher Scientific).
Immunoblotting.
A. phagocytophilum organisms were purified from infected HL-60 cells as described previously (69). Host cell free bacterial samples used for immunoblots were resuspended in 1× protein stabilizing cocktail (Thermo Fisher Scientific) and stored at −80°C.
To determine the expression of VirB6-3 and VirB6-4 proteins in wt A. phagocytophilum or the virB6-4::Himar1 mutant, host cell purified bacteria were solubilized with n-dodecyl-d-maltoside (DDM) lysis buffer (DDM at 1.50%, 1× cOmplete proteinase inhibitor [Roche Diagnostics], MgCl2 at 2 mM, NaCl at 150 mM, Tris at 25 mM, and lysozyme at 1 mg/ml) for 20 min at 4°C on a rotator, followed by sonication as described previously (60) using a Q-125A-110 sonicator (Qsonica, Newtown, CT). After sonication, lysates were centrifuged for 10 min at 12,000 × g and 4°C. Total protein concentration was determined using the noninterfering protein assay (NI protein assay) kit (G-Bioscience) as per the manufacturer’s instructions. Equal amounts of proteins were loaded per lane and separated on NuPAGE 3 to 8% Tris acetate gels (Thermo Fisher Scientific) and then transferred to polyvinylidene difluoride (PVDF) membranes for immunoblotting. Membranes were cut into strips and probed with either preimmune or postimmunization rabbit anti-VirB6-3, anti-VirB6-4, or A. phagocytophilum-infected dog serum diluted 1:103 fold, followed by incubation with 10 ng/ml of horseradish peroxidase (HRP)-conjugated goat anti-rabbit (SeraCare) or HRP-conjugated anti-dog (1:150,000) antibodies. Antibody reactions against VirB6-3, VirB6-4, or A. phagocytophilum MSP2/p44 were visualized with the chemiluminescent substrate SuperSignal West Femto (Thermo Fisher Scientific).
Surface trypsin digestion of intact A. phagocytophilum.
Freshly collected A. phagocytophilum strain HGE1 120879::Himar1 mutant bacteria, released from HL60 cells, were subjected to trypsinolysis (70), suspended in PBS, and split into aliquots. Sequencing-grade modified trypsin (Promega) and Tris (50 mM; pH 8), at final concentrations of 0.05 mg/ml 1.25 mM, respectively, were added to the trypsin-treated sample. A non-trypsin-treated sample received Tris (50 mM; pH 8) at a final concentration of 1.25 mM. Both samples were incubated at 37°C for 15 min with periodic gentle mixing. After trypsinolysis, the protease inhibitor phenylmethylsulfonyl fluoride (PMSF; 100 mM) (final concentration of 6.25 mM) was added to both samples and incubated for 10 min to inactivate trypsin. After incubation, bacteria from both samples were washed 3 times with 320 μl of wash solution (300 μl of Tris and 20 μl of PMSF) and pelleted by centrifugation at 12,000 × g for 10 min at room temperature. Final bacterial pellets were solubilized and proteins resolved as described above. Membranes were cut into strips and screened with anti-VirB6-3, anti-VirB6-4, and anti-mCherry antibodies (1:1,000 mouse anti-mCherry; Novus Biologicals).
Isolation of virB6-4::Himar1 mutant.
A library of mCherry-tagged A. phagocytophilum mutants was generated by Tn mutagenesis using the Himar1 system. The pHimarcisA7mCherry-SS plasmid (49) was introduced into A. phagocytophilum strain HGE1 by electroporation as described previously (40). Following electroporation, bacteria were incubated with HL-60 cells, diluted into three 96-well tissue culture plates, and kept under spectinomycin/streptomycin antibiotic selection (40). Wells containing cells infected with red fluorescent bacteria were expanded into 12-well tissue culture plates. A total of 900 mutants were obtained, and frozen stocks and DNA from each individual culture were archived. Transposon insertion sites were determined by high-throughput genome sequencing of pooled DNA. Briefly, DNA from 25 cultures was combined into one pool, for a total of 36 pools, barcoded, and sequenced on a single lane of an Illumina HiSeq 2000 instrument. Raw Illumina sequencing data were processed and analyzed as described previously (49) using the open-source GALAXY platform located at the University of Florida web site http://galaxy.hpc.ufl.edu. A total of 35,467 reads containing A. phagocytophilum genome-Tn junctions were identified using the A. phagocytophilum strain HGE1 (GenBank accession number APHH01000000) genome and the Himar1 inverted-repeat sequences as references. Through this analysis, a total of 1,200 transposon insertion sites were identified.
Since the DNA used for Tn-seq corresponds to pooled DNA, a PCR screen using primer pairs that bind upstream and downstream of the Tn insertion site within the virB6-4 gene was used to match this particular mutant to its frozen stock. Reactions used PrimeStar GXL DNA polymerase (TaKaRa) according to the manufacturer instructions, and PCR conditions were as follows: 94°C for 2 min, 30 cycles of denaturation at 98°C for 10 s, annealing at 60°C for 60 s, and extension at 68°C for 3 min, and a final extension step at 72°C for 5 min. PCR products were run on a 1.0% agarose gel and stained with SYBR gold nucleic acid stain. Agarose gel electrophoresis analysis of PCR products showed a PCR product consistent with the size (2,202 bp) of the Tn insertion within the targeted region in the DNA from transformant stock culture number 667 (data not shown). A DNA band of the size of the wild-type locus (368 bp) was also detected in the same stock, indicating the presence of a background mutant population with other Tn insertions. Cloning by limiting dilution resulted in the isolation of a clonal virB6-4::Himar1 mutant population.
Anaplasma phagocytophilum growth curves and cytotoxicity assay.
We compared the growth rates of the A. phagocytophilum virB6-4::Himar1 mutant and wild type in HL-60 and ISE6 cells. On the day of infection, 1 ml of uninfected HL-60 cells was seeded in two 24-well tissue culture plates at a density of 3 × 105 cells/ml. A. phagocytophilum wt and virB6-4::Himar1 bacteria were purified from heavily infected HL-60 cells (>80% cells contained morulae) and resuspended in 1.2 ml of RPMI 1640. To quantify isolated bacteria, we performed a LIVE/DEAD staining assay (Thermo Fisher Scientific). As the excitation/emission wavelengths from propidium iodide (PI) dye overlap those of mCherry, the PI was replaced with Sytox blue (Thermo Fisher Scientific) to calculate the number of dead cells. Each well was inoculated with host cell-free wt or virB6-4::Himar1 organisms at a multiplicity of infection (MOI) of 10 (∼3 × 106 organisms per well) and incubated at 37°C in the presence of CO2. After 24 h the inoculum was removed and replaced with 1 ml of fresh RPMI 1640 medium. Triplicate wells of infected cells with each A. phagocytophilum strain were harvested at different time points (1, 2, 3, 4, 5, and 6 days) postinfection (p.i.).
To evaluate growth of the virB6-4::Himar1 versus the wild type in ISE6 cell cultures, 2 days before infection, two 24-well tissue culture plates were seeded with 1 ml of uninfected ISE6 cells (3 × 105 cells/ml). On the day of infection, bacteria were purified and resuspended in 1.2 ml of L-15B3SE6 cells. Each well was inoculated with host cell-free wild-type or virB6-4::Himar1 bacteria at an MOI of 10. The next day, the medium was replaced and infected cells were collected at different time points (1, 3, 6, 9, and 12 days) p.i. HL-60 and ISE6 cells infected with the A. phagocytophilum wt or virB6-4::Himar1 mutant harvested at different times p.i. were processed for DNA extraction, using the Quick-gDNA kit (Zymo Research) as per the manufacturer’s instructions, and the supernatant was used for cytotoxicity assays.
Toxic damage in HL-60 and ISE6 cells due to A. phagocytophilum infection was measured by quantitatively determining the release of lactate dehydrogenase (LDH) into the medium using the Cyto Tox 96 nonradioactive cytotoxicity assay (Promega) following the manufacturer’s instructions. Supernatants from infected cultures were collected at the above-indicated time points p.i. All assays were performed in triplicate, and LDH activity was calculated by measuring optical densities at 490 nm using the Synergy HT plate reader (BioTek Instruments). Final absorbance values were determined after subtracting background values obtained from medium-only or no-cell control wells. To calculate LDH release, supernatants from wells containing uninfected cells (spontaneous LDH activity controls) and from wells containing uninfected cells lysed with 10× lysis buffer (maximum LDH activity controls) were added. Hence, cytotoxicity is expressed as 100× [(LDH activity from infected culture − spontaneous LDH activity controls)/(maximum LDH activity controls − spontaneous LDH activity controls)].
qPCR.
Duplex quantitative PCR (qPCR) was used to determine A. phagocytophilum GE per host cell as a measure of its growth in cell culture. Quantitation of A. phagocytophilum GE was performed by targeting the single-copy gene msp5 (Table 2). To normalize bacterial numbers, we used primers and probes targeting the single-copy gene for Toll like receptor 9 (tlr9) in HL-60 cells and the calreticulin gene (crt) in ISE6 cells. Triplicate reactions with DNA from three experimental wells were used. Reaction mixtures of 20 μl containing 2 μl of genomic DNA, 1× of LightCycler multiplex DNA master mix (Roche Diagnostics), 0.4 μM forward and reverse primers, and 0.2 μM probe were used for amplification in a LightCycler 96 instrument (Roche Diagnostics) with the following conditions: 95°C for 30 s and 45 cycles of 95°C for 5 s and 60°C for 30 s fluorescence detection. To allow for equimolar ratios of templates for standard curve preparation, a linear double-stranded DNA (dsDNA) fragment (Eurofins Genomics) carrying msp5, tlr9, and crt sequences was used. A. phagocytophilum and host cell GE number were calculated based on the standard curve.
Statistical analysis.
Data are expressed as means ± SEs. Unpaired two-tailed t test analysis was used for comparison of growth and cytotoxicity of the A. phagocytophilum wild type versus the virB6-4::Himar1 mutant. Statistical comparisons and graphics were made with Prism (GraphPad Software Inc., La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants R21AI109496 and RO1AI042792.
The funding agency had no role in the study, design, data collection and interpretation, or the decision to submit the work for publication.
A.F.B., U.G.M., and D.R.A. supervised this project, contributed to manuscript writing, and helped with critical revisions. M.J.H. generated the A. phagocytophilum Tn mutant library, and C.M.N. isolated the virB6-4::Himar1 mutant. A.F.B. performed bioinformatics analysis to identify Tn insertion sites. A.M.L. performed Western blotting, and Y.-P.X. provided technical assistance. F.L.C. conceived and authored the manuscript, carried out experimental design, acquired data, and interpreted results.
All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Grohmann E, Christie PJ, Waksman G, Backert S. 2018. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol 107:455–471. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, Cavalcante NS, Alegria MC, Barbosa LR, Salinas RK, Guzzo CR, Farah CS. 2015. Bacterial killing via a type IV secretion system. Nat Commun 6:6453. doi: 10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- 6.Bayer-Santos E, Cenens W, Matsuyama BY, Oka GU, Di Sessa G, Mininel IDV, Alves TL, Farah CS. 2019. The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog 15:e1007651. doi: 10.1371/journal.ppat.1007651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudaher E, Shaffer CL. 2019. Inhibiting bacterial secretion systems in the fight against antibiotic resistance. Medchemcomm 10:682–692. doi: 10.1039/C9MD00076C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie PJ. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim Biophys Acta 1694:219–234. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rikihisa Y. 2011. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin Microbiol Rev 24:469–489. doi: 10.1128/CMR.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woldehiwet Z. 2010. The natural history of Anaplasma phagocytophilum. Vet Parasitol 167:108–122. doi: 10.1016/j.vetpar.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Jin H, Wei F, Liu Q, Qian J. 2012. Epidemiology and control of human granulocytic anaplasmosis: a systematic review. Vector Borne Zoonotic Dis 12:269–274. doi: 10.1089/vbz.2011.0753. [DOI] [PubMed] [Google Scholar]
- 12.Gillespie JJ, Brayton KA, Williams KP, Diaz MA, Brown WC, Azad AF, Sobral BW. 2010. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect Immun 78:1809–1823. doi: 10.1128/IAI.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Eisen J, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie PJ. 2016. The mosaic type IV secretion systems. EcoSal Plus 7:ESP-0020-2015. doi: 10.1128/ecosalplus.ESP-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Khedery B, Lundgren AM, Stuen S, Granquist EG, Munderloh UG, Nelson CM, Alleman AR, Mahan SM, Barbet AF. 2012. Structure of the type IV secretion system in different strains of Anaplasma phagocytophilum. BMC Genomics 13:678. doi: 10.1186/1471-2164-13-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatty M, Laverde Gomez JA, Christie PJ. 2013. The expanding bacterial type IV secretion lexicon. Res Microbiol 164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.den Hartigh AB, Sun YH, Sondervan D, Heuvelmans N, Reinders MO, Ficht TA, Tsolis RM. 2004. Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect Immun 72:5143–5149. doi: 10.1128/IAI.72.9.5143-5149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoppner C, Carle A, Sivanesan D, Hoeppner S, Baron C. 2005. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology 151:3469–3482. doi: 10.1099/mic.0.28326-0. [DOI] [PubMed] [Google Scholar]
- 19.Arends K, Celik EK, Probst I, Goessweiner-Mohr N, Fercher C, Grumet L, Soellue C, Abajy MY, Sakinc T, Broszat M, Schiwon K, Koraimann G, Keller W, Grohmann E. 2013. TraG encoded by the pIP501 type IV secretion system is a two-domain peptidoglycan-degrading enzyme essential for conjugative transfer. J Bacteriol 195:4436–4444. doi: 10.1128/JB.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillespie JJ, Phan IQ, Driscoll TP, Guillotte ML, Lehman SS, Rennoll-Bankert KE, Subramanian S, Beier-Sexton M, Myler PJ, Rahman MS, Azad AF. 2016. The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion. Pathog Dis 74:ftw058. doi: 10.1093/femspd/ftw058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Rivera C, Bhatty M, Christie PJ. 2016. Mechanism and function of type IV secretion during infection of the human host. Microbiol Spectr 4:VMBF-0024-2015. doi: 10.1128/microbiolspec.VMBF-0024-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christie PJ, Gomez Valero L, Buchrieser C. 2017. Biological diversity and evolution of type IV secretion systems. Curr Top Microbiol Immunol 413:1–30. doi: 10.1007/978-3-319-75241-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumari R, Shariq M, Sharma S, Kumar A, Mukhopadhyay G. 2019. CagW, a VirB6 homologue interacts with Cag-type IV secretion system substrate CagA in Helicobacter pylori. Biochem Biophys Res Commun 515:712–718. doi: 10.1016/j.bbrc.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Mary C, Fouillen A, Bessette B, Nanci A, Baron C. 2018. Interaction via the N terminus of the type IV secretion system (T4SS) protein VirB6 with VirB10 is required for VirB2 and VirB5 incorporation into T-pili and for T4SS function. J Biol Chem 293:13415–13426. doi: 10.1074/jbc.RA118.002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.den Hartigh AB, Rolán HG, de Jong MF, Tsolis RM. 2008. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J Bacteriol 190:4427–4436. doi: 10.1128/JB.00406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Judd PK, Mahli D, Das A. 2005. Molecular characterization of the Agrobacterium tumefaciens DNA transfer protein VirB6. Microbiology (Reading) 151:3483–3492. doi: 10.1099/mic.0.28337-0. [DOI] [PubMed] [Google Scholar]
- 27.Ohashi N, Zhi N, Lin Q, Rikihisa Y. 2002. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect Immun 70:2128–2138. doi: 10.1128/IAI.70.4.2128-2138.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson CM, Herron MJ, Felsheim RF, Schloeder BR, Grindle SM, Chavez AO, Kurtti TJ, Munderloh UG. 2008. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics 9:364. doi: 10.1186/1471-2164-9-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion System. J Mol Biol 341:961–977. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Judd PK, Kumar RB, Das A. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol Microbiol 55:115–124. doi: 10.1111/j.1365-2958.2004.04378.x. [DOI] [PubMed] [Google Scholar]
- 31.Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, Sobral BS, Azad AF. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4:e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao W, Kumagai Y, Niu H, Yamaguchi M, Miura K, Rikihisa Y. 2009. Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J Bacteriol 191:278–286. doi: 10.1128/JB.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai JS, Cheng CW, Sung TY, Hsu WL. 2012. Computational comparative study of tuberculosis proteomes using a model learned from signal peptide structures. PLoS One 7:e35018. doi: 10.1371/journal.pone.0035018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.IJdo JW, Zhang Y, Hodzic E, Magnarelli LA, Wilson ML, Telford SR III, Barthold SW, Fikrig E. 1997. The early humoral response in human granulocytic ehrlichiosis. J Infect Dis 176:687–692. doi: 10.1086/514091. [DOI] [PubMed] [Google Scholar]
- 35.Brown WC. 2012. Adaptive immunity to Anaplasma pathogens and immune dysregulation: implications for bacterial persistence. Comp Immunol Microbiol Infect Dis 35:241–252. doi: 10.1016/j.cimid.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rancès E, Voronin D, Tran-Van V, Mavingui P. 2008. Genetic and functional characterization of the type IV secretion system in Wolbachia. J Bacteriol 190:5020–5030. doi: 10.1128/JB.00377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sears KT, Ceraul SM, Gillespie JJ, Allen ED Jr, Popov VL, Ammerman NC, Rahman MS, Azad AF. 2012. Surface proteome analysis and characterization of surface cell antigen (Sca) or autotransporter family of Rickettsia typhi. PLoS Pathog 8:e1002856. doi: 10.1371/journal.ppat.1002856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voronin D, Guimaraes AF, Molyneux GR, Johnston KL, Ford L, Taylor MJ. 2014. Wolbachia lipoproteins: abundance, localisation and serology of Wolbachia peptidoglycan associated lipoprotein and the type IV secretion system component, VirB6 from Brugia malayi and Aedes albopictus. Parasit Vectors 7:462. doi: 10.1186/s13071-014-0462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman JL, Nelson C, Vitale B, Madigan JE, Dumler JS, Kurtti TJ, Munderloh UG. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N Engl J Med 334:209–215. doi: 10.1056/NEJM199601253340401. [DOI] [PubMed] [Google Scholar]
- 40.Felsheim RF, Herron MJ, Nelson CM, Burkhardt NY, Barbet AF, Kurtti TJ, Munderloh UG. 2006. Transformation of Anaplasma phagocytophilum. BMC Biotechnol 6:42. doi: 10.1186/1472-6750-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliva Chavez AS, Fairman JW, Felsheim RF, Nelson CM, Herron MJ, Higgins L, Burkhardt NY, Oliver JD, Markowski TW, Kurtti TJ, Edwards TE, Munderloh UG. 2015. An O-methyltransferase is required for infection of tick cells by Anaplasma phagocytophilum. PLoS Pathog 11:e1005248. doi: 10.1371/journal.ppat.1005248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villamil Giraldo AM, Mary C, Sivanesan D, Baron C. 2015. VirB6 and VirB10 from the Brucella type IV secretion system interact via the N-terminal periplasmic domain of VirB6. FEBS Lett 589:1883–1889. doi: 10.1016/j.febslet.2015.05.051. [DOI] [PubMed] [Google Scholar]
- 43.Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. 2006. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol 188:882–893. doi: 10.1128/JB.188.3.882-893.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H, Lin M, Wang X, Kikuchi T, Mottaz H, Norbeck A, Rikihisa Y. 2008. Proteomic analysis of and immune responses to Ehrlichia chaffeensis lipoproteins. Infect Immun 76:3405–3414. doi: 10.1128/IAI.00056-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnston KL, Wu B, Guimaraes A, Ford L, Slatko BE, Taylor MJ. 2010. Lipoprotein biosynthesis as a target for anti-Wolbachia treatment of filarial nematodes. Parasit Vectors 3:99. doi: 10.1186/1756-3305-3-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner JD, Langley RS, Johnston KL, Gentil K, Ford L, Wu B, Graham M, Sharpley F, Slatko B, Pearlman E, Taylor MJ. 2009. Wolbachia lipoprotein stimulates innate and adaptive immunity through Toll-like receptors 2 and 6 to induce disease manifestations of filariasis. J Biol Chem 284:22364–22378. doi: 10.1074/jbc.M901528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier TM, Casey MS, Becker RH, Dorsey CW, Glass EM, Maltsev N, Zahrt TC, Frank DW. 2007. Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect Immun 75:5376–5389. doi: 10.1128/IAI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng C, Nair AD, Jaworski DC, Ganta RR. 2015. Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PLoS One 10:e0132657. doi: 10.1371/journal.pone.0132657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crosby FL, Wamsley HL, Pate MG, Lundgren AM, Noh SM, Munderloh UG, Barbet AF. 2014. Knockout of an outer membrane protein operon of Anaplasma marginale by transposon mutagenesis. BMC Genomics 15:278. doi: 10.1186/1471-2164-15-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlyon JA, Fikrig E. 2003. Invasion and survival strategies of Anaplasma phagocytophilum. Cell Microbiol 5:743–754. doi: 10.1046/j.1462-5822.2003.00323.x. [DOI] [PubMed] [Google Scholar]
- 51.Jakubowski SJ, Krishnamoorthy V, Christie PJ. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J Bacteriol 185:2867–2878. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Broxton CN, Culotta VC. 2016. SOD enzymes and microbial pathogens: surviving the oxidative storm of infection. PLoS Pathog 12:e1005295. doi: 10.1371/journal.ppat.1005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynch M, Kuramitsu H. 2000. Expression and role of superoxide dismutases (SOD) in pathogenic bacteria. Microbes Infect 2:1245–1255. doi: 10.1016/S1286-4579(00)01278-8. [DOI] [PubMed] [Google Scholar]
- 54.Carlyon JA, Abdel-Latif D, Pypaert M, Lacy P, Fikrig E. 2004. Anaplasma phagocytophilum utilizes multiple host evasion mechanisms to thwart NADPH oxidase-mediated killing during neutrophil infection. Infect Immun 72:4772–4783. doi: 10.1128/IAI.72.8.4772-4783.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.IJdo JW, Mueller AC. 2004. Neutrophil NADPH oxidase is reduced at the Anaplasma phagocytophilum phagosome. Infect Immun 72:5392–5401. doi: 10.1128/IAI.72.9.5392-5401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlyon JA, Fikrig E. 2006. Mechanisms of evasion of neutrophil killing by Anaplasma phagocytophilum. Curr Opin Hematol 13:28–33. doi: 10.1097/01.moh.0000190109.00532.56. [DOI] [PubMed] [Google Scholar]
- 57.Rikihisa Y. 2010. Molecular events involved in cellular invasion by Ehrlichia chaffeensis and Anaplasma phagocytophilum. Vet Parasitol 167:155–166. doi: 10.1016/j.vetpar.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rikihisa Y, Zhi N, Wormser GP, Wen B, Horowitz HW, Hechemy KE. 1997. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York state. J Infect Dis 175:210–213. doi: 10.1093/infdis/175.1.210. [DOI] [PubMed] [Google Scholar]
- 59.Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, Greig B, Mellencamp MA, Goodman JL. 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J Clin Microbiol 37:2518–2524. doi: 10.1128/JCM.37.8.2518-2524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikeda M, Arai M, Okuno T, Shimizu T. 2003. TMPDB: a database of experimentally-characterized transmembrane topologies. Nucleic Acids Res 31:406–409. doi: 10.1093/nar/gkg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kall L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 63.Saha SR, Raghava GPS. 2004. BcePred: prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. Lect Notes in Comput Sci 3239:197–204. doi: 10.1007/978-3-540-30220-9_16. [DOI] [Google Scholar]
- 64.Wang HW, Lin YC, Pai TW, Chang HT. 2011. Prediction of B-cell linear epitopes with a combination of support vector machine classification and amino acid propensity identification. J Biomed Biotechnol 2011:432830. doi: 10.1155/2011/432830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 66.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma H, O’Kennedy R. 2015. The purification of natural and recombinant peptide antibodies by affinity chromatographic strategies. Methods Mol Biol 1348:153–165. doi: 10.1007/978-1-4939-2999-3_15. [DOI] [PubMed] [Google Scholar]
- 68.Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, Cowman AF, McFadden GI. 2004. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol 137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Crosby FL, Lundgren AM, Hoffman C, Pascual DW, Barbet AF. 2018. VirB10 vaccination for protection against Anaplasma phagocytophilum. BMC Microbiol 18:217. doi: 10.1186/s12866-018-1346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kahlon A, Ojogun N, Ragland SA, Seidman D, Troese MJ, Ottens AK, Mastronunzio JE, Truchan HK, Walker NJ, Borjesson DL, Fikrig E, Carlyon JA. 2013. Anaplasma phagocytophilum Asp14 is an invasin that interacts with mammalian host cells via its C terminus to facilitate infection. Infect Immun 81:65–79. doi: 10.1128/IAI.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.