Abstract
Background
As a highly malignant tumor, cholangiocarcinoma poses a serious threat to human life and health, so exploring the mechanisms of its development and progression at a molecular level is of great significance to the diagnosis and treatment of the disease.
Objective
This study was aimed at investigating the effects and related mechanisms of LncRNA TUG1 on cholangiocarcinoma cells.
Methods
Cholangiocarcinoma tissues and adjacent tissues (n=82 each), human cholangiocarcinoma cell lines (RBE, QBC939, HuH28), and a human normal biliary epithelial cell line (HIBE) were collected. miR-29a-mimics, miR-29a-inhibitor, miR-NC, si-TUG1, pcDNA3.1 TUG1, and NC were transfected into the cholangiocarcinoma cells. qRT-PCR was performed to detect TUG1 and miR-29a expression in the cholangiocarcinoma tissues and cells. Western blotting (WB) was conducted to detect the expression of Bax, Caspase-3, and Bcl-2 in the cells. CCK-8 assay, Transwell, and flow cytometry were carried out to detect cell proliferation, invasion, and apoptosis. Dual luciferase reporter gene assay (DLRGA) was performed to confirm the correlation of TUG1 with miR-29a.
Results
TUG1 was highly expressed while miR-29a was poorly expressed in cholangiocarcinoma cells. TUG1 expression was negatively correlated with miR-29a expression, and TUG1 had a relatively high diagnostic value for cholangiocarcinoma. Cell experiments showed that inhibiting TUG1 expression or up-regulating miR-29a expression could inhibit cholangiocarcinoma cells from proliferation and invasion, and promote their apoptosis, while up-regulating TUG1 or inhibiting miR-29a could promote the proliferation and invasion but inhibit the apoptosis. Rescue experiment showed that overexpressing miR-29a could reverse the effects of high TUG1 expression on cholangiocarcinoma cells. DLRGA confirmed that there was a regulatory relationship between TUG1 and miR-29a.
Conclusion
TUG1 is highly expressed in cholangiocarcinoma tissues. It can promote the growth and metastasis of cholangiocarcinoma cells by inhibiting miR-29a, so it may be a new target for diagnosing and treating cholangiocarcinoma.
Keywords: LncRNA TUG1, miR-29a, cholangiocarcinoma, proliferation, invasion, apoptosis
Introduction
As a highly heterogeneous malignant tumor of the biliary tract, cholangiocarcinoma appears anywhere in the biliary tree and its global incidence has been rising in recent years, so it is a great threat to human life and health.1,2 At present, surgical treatment is still the most effective method for treating patients with the disease. However, cholangiocarcinoma has unapparent symptoms in its early stage, and it has been usually in its advanced stage when diagnosed. As a result, many patients with the disease have lost their opportunities for operation.3,4 Cholangiocarcinoma cells are highly invasive and metastatic, and are easy to metastasize far away through blood, lymph, and other channels. Therefore, even after surgical treatment, some patients may have a poor prognosis due to tumor metastasis.5 For this reason, exploring the mechanisms of cholangiocarcinoma cells’ growth and metastasis has important significance for improving the therapeutic effect on cholangiocarcinoma and the prognosis of patients.
LncRNAs are non-coding RNAs over 200nt long. With the development of molecular biology in recent years, their roles in the development and progression of tumors and many others diseases have been gradually valued.6 Their roles in cholangiocarcinoma have also been reported more. For example, the high expression of LncRNA H19 in the disease indicates patients’ poor prognoses and promotes cancer cells’ migration and invasion.7 It has been confirmed that as a LncRNA located at chromosome 22q12.2, taurine up-regulated gene 1 (TUG1) plays a pivotal role in the development and progression of many cancers.8 For instance, it promotes the development and progression of pancreatic cancer through the targeted regulation of miR-29c.9 Its epigenetic characteristics for osteosarcoma make it possible to become a target for personalized drug therapy.10 This gene may also play a carcinogenic role in cholangiocarcinoma, and its high expression is possibly related to the prognosis of the disease, but the mechanism has not been thoroughly explored.11
Therefore, in this study, the mechanism of action of TUG1 in cholangiocarcinoma was further investigated, so as to provide a new target direction for diagnosing and treating the disease.
Materials and Methods
General Information
Eighty-two patients who underwent cholangiocarcinoma resection at our hospital from January 2015 to 2017 were enrolled. With their consent, their cholangiocarcinoma tissues and adjacent tissues (n=82 each) were taken out during the operation for detection. Inclusion criteria: patients who were confirmed with cholangiocarcinoma by pathology; patients who were diagnosed with cholangiocarcinoma for the first time. Exclusion criteria: patients who had been treated with radiotherapy and chemotherapy; patients complicated with other malignant tumors; patients with severe renal dysfunction or serious infectious diseases; patients who had refused to provide experimental specimens.
The research was conducted according to the principles of the World Medical Association Declaration of Helsinki. This study was approved by the Ethical Medical Committee of the Cancer Hospital Affiliated to University of Chinese Academy of Sciences. All patients and their family members had consented to participation in the experiment and had signed the informed consent form.
Experimental Materials and Reagents
A human normal biliary epithelial cell line (HIBE) and human cholangiocarcinoma cell lines (RBE, QBC939, HuH28) were purchased from ATCC. qRT-PCR and reverse transcription kits were purchased from TransGen Biotech, Beijing, China. CCK-8 assay kits were purchased from Promega, USA. Transwell kits were purchased from Shanghai Yantuo Biotechnology Co., Ltd. Phosphate buffer solution (PBS) and fetal bovine serum (FBS) were purchased from Gibco, Rockville, MD, USA. Trizol reagents were purchased from Beijing Biolab Science and Technology Co., Ltd. Dual-luciferase reporter gene assay (DLRGA) kits were purchased from Beijing Biolab Science and Technology Co., Ltd. Bax, Bcl-2, Caspase-3, and β-Actin antibodies were purchased from Cell Signaling Technology, Boston, Massachusetts, USA. Goat anti-rabbit IgG secondary antibody was purchased from Thermo Fisher Scientific, Shanghai. RIPA and BCA protein assay kits were purchased from Thermo Fisher Scientific, Waltham, MA, USA. An ECL developing solution was purchased from Thermo Fisher Scientific, Waltham, MA, USA. All primers were designed and synthesized by Sangon Biotech (Shanghai) Co., Ltd.
Cell Culture and Transfection
RBE, QBC939, HuH28, CCLP1, and HIBE cells were placed in a DMEM (10% PBS) for culture (37°C, 5% CO2). After adherently growing and fused to 85%, they were digested with 0.25% pancreatin. Next, they were continuously cultured in the medium for passage. TUG1 and miR-29a expression in the cells was detected. After that, QBC939 and HuH28 cells were selected and then respectively transfected with miR-29a-inhibitor (suppression sequence), miR-29a-mimics (overexpression sequence), miR negative control (miR-NC), targeted inhibition TUG1 RNA plasmids (si-TUG1), and targeted overexpression TUG1 RNA plasmids (pcDNA3.1 TUG1) using Lipofectamine™ 2000 kits,use Si-NC and pcDNA3.1 empty vectors as controls. Operating steps were strictly carried out according to the kit instruction.
Real-Time Quantitative PCR (RT-qPCR)
The Trizol reagents were first used to extract total RNA from the tissues and cells, and then the RNA (5 μg each) was taken out for the reverse transcription into cDNA based on the kit instruction. The synthesized cDNA (1 μL) was taken out for PCR amplification. Conditions for the reaction were pre-denaturation at 95°C for 10 s and then cycling (denaturation at 94°C for 10 s, and annealing and extension at 60°C for 30 s) for 40 times. Each sample was provided with 3 same wells. The experiment was repeatedly performed for 3 times. U6 and β-Actin were the internal references of miR-29a and TUG1, respectively. 2−ΔΔct was used to analyze the data. (See Table 1)
Table 1.
Primer Sequences
| Forward Primers | Reverse Primers | |
|---|---|---|
| TUG1 | 5ʹ-TAGCAGTTCCCCAATCCTTG-3’ | 5ʹ-CACAAATTCCCATCATTCCC-3’ |
| miR-29a | 5ʹ-CGCGGATCCTGGATTTAGTAAGATTTGGGC-3’ | 5ʹ–CCGGAATTCACATGCAATTCAGGTCAGTG-3’ |
| U6 | 5ʹ-CTCGCTTCGGCAGCACATATACT-3’ | 5ʹ-ACGCTTCACGAATTTGCGTGTC-3’ |
| β-Actin | 5ʹ-GACCTCTATGCCAACACAGT-3’ | 5ʹ-AGTACTTGCGCTCAGGAGGA-3’ |
Cell Proliferation
The proliferation of QBC939 and HuH28 cells was assessed by the CCK-8 assay kits. After the cells (transfected for 48 hours) were collected, they were inoculated into a 96-well plate at 1×104 cells/well, and then cultured at 37°C and with 5% CO2. After the cells adherently grew, each well was added with CCK-8 solution (10 μL) at 0, 24, 48 and 72 hours, respectively. Then, the cells were continuously cultured in an incubator (37°C, 5% CO2) for 2 hours. Finally, optical value (OD) values at 450 nm were determined by a microplate reader to detect cell proliferation and plot the growth curve. The experiment was repeatedly conducted for 3 times.
Cell Invasion
The Transwell kits were used to determine cell invasion. The upper chamber was added with DMEM culture solution (200 μL; containing 3x104 cells), while the lower one was added with DMEM (500 mL; containing 20% FBS), both of which were cultured at 37°C for 48 hours. The matrix and cells that did not penetrate the membrane surface in the upper chamber were wiped off. After the cells were washed with PBS for 3 times, they were fixed with paraformaldehyde for 10 min, cleaned with double distilled water for 3 times, and finally stained with 0.1% crystal violet after they were dried. The cell invasion was observed by a microscope.
Cell Apoptosis
After digested with 0.25% trypsin, the cells were rinsed with PBS for twice, and then added with binding buffer (100 μL) to prepare a 1*106 cells/mL suspension. Next, the suspension was sequentially added with AnnexinV-FITC and PI, and then incubated in dark for 5 min at room temperature. The FACSVerse flow cytometer system was used for detection, and the experiment was conducted for 3 times to obtain the average value.
Western Blotting (WB)
Total protein was first extracted from the cells using RIPA lysis buffer, and then the BCA method was used to detect its concentration. The protein was separated with 12% SDS-PAGE and then transferred to the PVDF membrane, after its concentration was adjusted to 4 μg/μL. Next, the membrane was sealed with 5% skimmed milk powder for 2 hours, and then added with primary antibodies [Bax (1: 500), Bcl-2 (1: 500), Caspase-3 (1: 500),β-actin (1: 1000)] for sealing at 4°C all night. After washed to remove the antibodies, it was added with a horseradish peroxidase-labeled secondary antibody [goat anti-rabbit (1: 1000)], incubated at 37°C for 1 hour, and then rinsed with PBS over 5 min for 3 times. After developed in a dark room, the membrane luminesced with the ECL developing solution and was developed, after excess liquid on it was absorbed dry with filter papers.
DLRGA
The starBase v2.0 bioinformatics database (http://starbase.sysu.edu.cn/index.php) was applied to search for candidate miRNAs that could bind to TUG1. Subsequently, oligonucleotides that contained target sequences for TUG1 were first amplified and then cloned into pmirGLO plasmids (WT). pmirGLO-TUG1-3ʹUTR wild type (Wt) and pmirGLO-TUG1-3ʹUTR mutant (Mut) were respectively established and then transferred into the downstream of luciferase reporter genes, so as to sequence and identify the constructed plasmids. The Lipofectamine 2000 was used to co-transfect luciferase reporter plasmids and miR-29a-mimics or miR-NC into HuH28 cells. After the cells were cultured for 24 hours, they were collected and their luciferase activities were tested by the luciferase reporter gene assay kits. The results were statistically analyzed.
Statistical Methods
In this study, SPSS19.0 was used to statistically analyze the collected data. GraphPad 7 was used to plot the required figures. The comparison between groups was conducted by an independent t test, and the comparison between multiple groups was conducted by one-way analysis of variance (ANOVA), with post hoc pairwise comparison conducted by LSD-t test. The comparison of expression between multiple time points was conducted by repeated measures ANOVA, and Bonferroni was used for post hoc test. Pearson Correlation Coefficient was used for correlation analysis. When P<0.05, the difference was statistically significant.
Results
TUG1 Overexpression in Cholangiocarcinoma Tissues and Cells
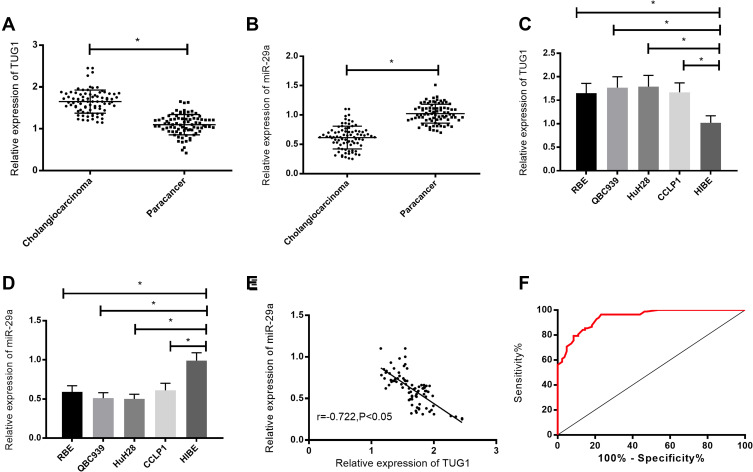
According to the qPCR, compared with adjacent tissues, TUG1 was highly expressed (P<0.05), while miR-29a was lowly expressed in cholangiocarcinoma tissues and cells (P<0.05). Moreover, TUG1 expression in QBC939 and HuH28 cells was higher than that in other cholangiocarcinoma cells, so the two cells were selected for follow-up experiments. According to the receiver operating curve (ROC), the area under the curve (AUC) of TUG1 for diagnosing cholangiocarcinoma was 0.942. Results of the correlation analysis showed that TUG1 expression was negatively correlated with miR-29a expression (r=−0.722, P<0.05). See Figure 1.
Figure 1.
Expression and significance of TUG1 in cholangiocarcinoma tissues and cells. (A) TUG1 expression in cholangiocarcinoma tissues. (B) miR-29a expression in cholangiocarcinoma tissues. (C) TUG1 expression in cholangiocarcinoma cells. (D) miR-29a expression in cholangiocarcinoma cells. (E) The correlation analysis of TUG1 with miR-29a expression in cholangiocarcinoma tissues. (F) The ROC analysis of TUG1 for diagnosing cholangiocarcinoma. * indicates P<0.05.
Effects of TUG1 on Proliferation, Invasion, and Apoptosis of Cholangiocarcinoma Cells
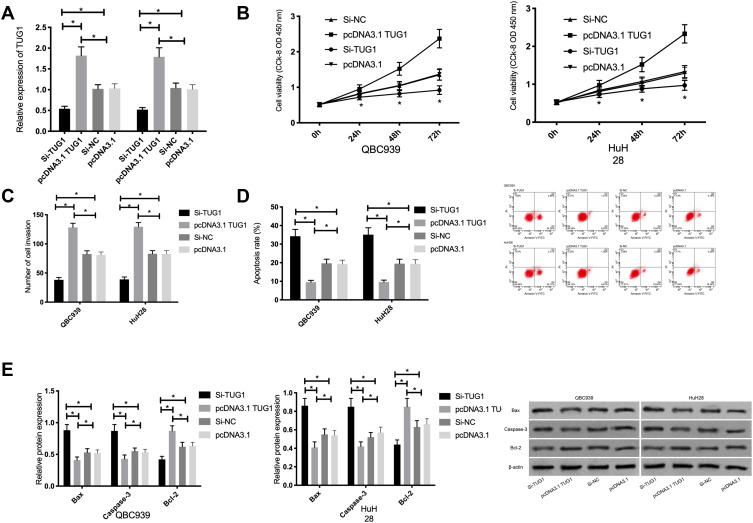
QBC939 and HuH28 cells were transfected with Si-TUG1 and pcDNA3.1 TUG1. Compared with the Si-NC group, TUG1 expression was remarkably lower in the Si-TUG1 group.Compared with the pcDNA3.1 group,TUG1 expression was remarkably higher in the pcDNA3.1 TUG1 group. According to the detection of cell biological behaviors, compared with those in the Si-NC group, cell proliferation and invasion remarkably reduced, the apoptotic rate remarkably increased, and Bax and Caspase-3 expression remarkably increased, as well as Bcl-2 expression remarkably decreased in the Si-TUG1 group (P<0.05);.Compared with the pcDNA3.1 group, the proliferation and invasion remarkably increased, the apoptotic rate remarkably decreased, and Bax and Caspase-3 expression remarkably decreased, as well as Bcl-2 expression remarkably increased in the pcDNA3.1 TUG1 group (P<0.05). See Figure 2.
Figure 2.
Effects of TUG1 on proliferation, invasion, and apoptosis of cholangiocarcinoma cells. (A) TUG1 expression in cholangiocarcinoma cells after transfection. (B) Effects of TUG1 on the proliferation of cholangiocarcinoma cells. (C) Effects of TUG1 on the invasion of cholangiocarcinoma cells. (D) Effects of TUG1 on the apoptotic rate of cholangiocarcinoma cells. (E) Effect of TUG1 on the expression of apoptosis-related proteins in cholangiocarcinoma cells. * indicates P<0.05.
Effects of miR-29a on Proliferation, Invasion, and Apoptosis of Cholangiocarcinoma Cells
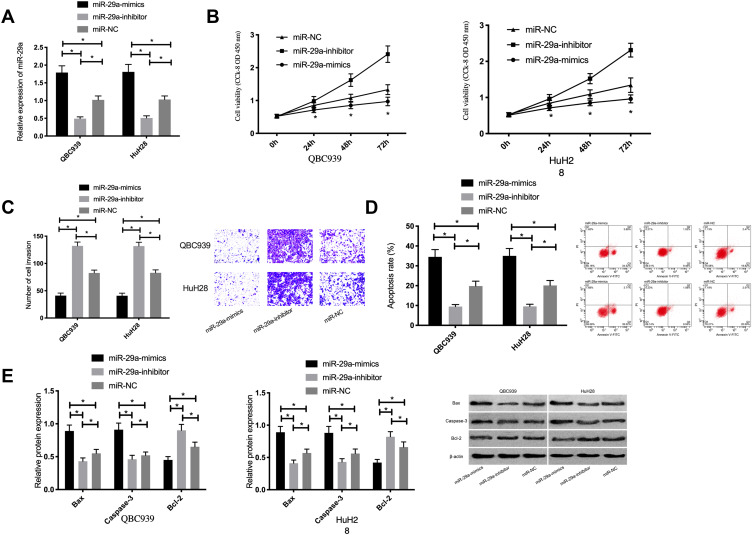
Compared with those in the miR-NC group, miR-29a expression was remarkably lower in the miR-29a-inhibitor group, but remarkably higher in the miR-29a-mimics group. According to the detection of cell biological behaviors, compared with those in the miR-NC group, cell proliferation and invasion remarkably reduced, the apoptotic rate remarkably increased, and Bax and Caspase-3 expression remarkably increased, as well as Bcl-2 expression remarkably decreased in the miR-29a-mimics group (P<0.05); the proliferation and invasion remarkably increased, the apoptotic rate remarkably decreased, and Bax and Caspase-3 expression remarkably decreased, as well as Bcl-2 expression remarkably increased in the miR-29a-inhibitor group (P<0.05). See Figure 3.
Figure 3.
Effects of miR-29a on proliferation, invasion, and apoptosis of cholangiocarcinoma cells. (A) miR-29a expression in cholangiocarcinoma cells after transfection. (B) Effects of miR-29a on the proliferation of cholangiocarcinoma cells. (C) Effects of miR-29a on the invasion of cholangiocarcinoma cells. (D) Effects of miR-29a on the apoptotic rate of cholangiocarcinoma cells. (E) Effect of miR-29a on the expression of apoptosis-related proteins in cholangiocarcinoma cells. * indicates P<0.05.
Regulatory Relationship Between TUG1 and miR-29a
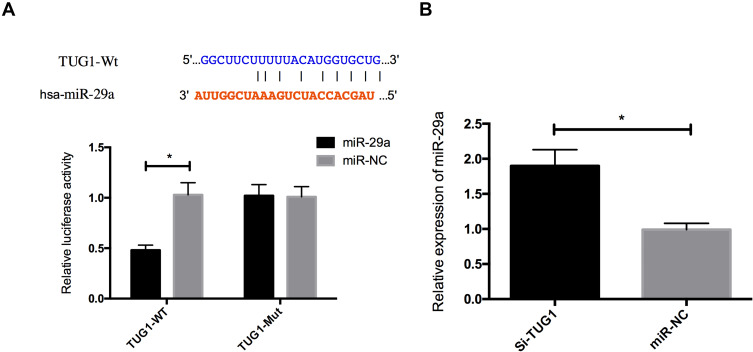
According to the bioinformatics prediction, TUG1 may have a direct effect on miR-29a, and their binding sequences had a similar binding site. For verifying whether TUG1 could bind to miR-29a 3ʹUTR, the two genes were co-transfected into HuH28 cells. According to the DLRGA, the luciferase activity of miR-29a remarkably reduced after the co-transfection, which suggested that TUG1 could specifically bind to miR-29a 3ʹUTR and regulate its expression activity and levels. Besides, we compared the expression of endogenous miR-29a between the miR-NC and si-TUG1 groups, and found that miR-29a expression increased in the QBC939 and HuH28 cells with TUG1 inhibition. See Figure 4.
Figure 4.
Regulatory relationship between TUG1 and miR-29a. (A) Dual luciferase reporter genes and binding sites. (B) Effects of TUG1 on miR-29a expression. * indicates P<0.05.
Rescue Experiment
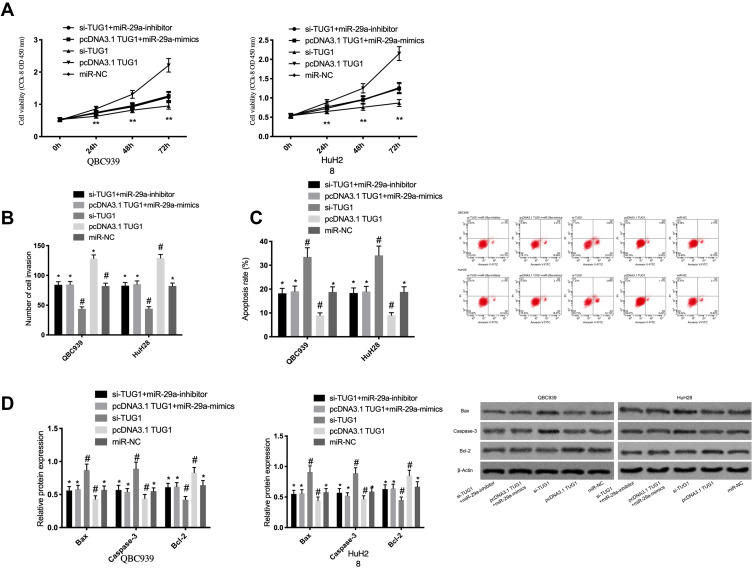
For further confirming that TUG1 affects cholangiocarcinoma cells by regulating miR-29a, we co-transfected pcDNA3.1 TUG1 and miR-29a-mimics or si-TUG1 and miR-29a-inhibitor into QBC939 and HuH28 cells. According to the results, pcDNA3.1 TUG1 reversed the effects of miR-29a-mimics on inhibiting the cells’ proliferation and invasion and on promoting their apoptosis, while si-TUG1 could offset the effects of miR-29a-inhibitor on the promotion. See Figure 5.
Figure 5.
Rescue experiment. (A) Effects of TUG1 and miR-29a co-transfection on the proliferation of cholangiocarcinoma cells. (B) Effects of TUG1 and miR-29a co-transfection on the invasion of cholangiocarcinoma cells. (C) Effects of TUG1 and miR-29a co-transfection on the apoptotic rate of cholangiocarcinoma cells. (D) Effects of TUG1 and miR-29a co-transfection on apoptosis-related proteins in cholangiocarcinoma cells. * was compared with #, P<0.05; ** indicates P<0.05.
Discussion
With a very high mortality rate, cholangiocarcinoma is a malignant solid tumor of the digestive tract and is one of the most fatal cancers.12 Although the effects of surgical treatment have been effective in recent years, the easy metastasis and strong invasion of the disease possibly cause many treated patients to have a poor prognosis due to tumor metastasis.13,14 Therefore, it is of great significance to explore the mechanisms of development and progression of cholangiocarcinoma.
With the development of molecular biology in recent years, LncRNAs have been gradually valued by the medical community, so the role and mechanism of LncRNA TUG1 in cholangiocarcinoma were explored. In our study, TUG1 was abnormally overexpressed in cholangiocarcinoma tissues and cells, and the further analysis found that it had a high diagnostic value for the disease. It has been reported that TUG1 is a LncRNA that is abnormally expressed in various tumor diseases. For example, it is highly expressed in acute myeloid leukemia and its up-regulation is related to the poor prognosis of patients.15 Additionally, it is highly expressed in pancreatic cancer according to gene sequencing.16 These findings are consistent with our conclusions. For further exploring the effects of TUG1 on cholangiocarcinoma cells, we intervened its expression in them. Their proliferation and invasion were remarkably inhibited after inhibiting TUG1 expression, and their apoptotic rate remarkably increased, but reverse cell phenotypes were obtained after overexpressing TUG1. These findings show that TUG1 plays a role as an oncogene in cholangiocarcinoma. TUG1 has been found to play the role in many studies. For instance, it promotes the proliferation and inhibits the apoptosis of ovarian cancer cells through regulating AURKA,17 and promotes the proliferation and migration of renal carcinoma cells through regulating YAP.18 These findings are similar to the roles of TUG1 in our study. However, the specific mechanism of action of this gene in cholangiocarcinoma remains unclear.
Previous studies have found that LncRNAs exert their effects on cells through competitively binding to miRNAs.19,20 Therefore, for further analyzing the mechanism of action of TUG1 in cholangiocarcinoma, we predicted a binding site between LncRNA TUG1 and miR-29a through the online database Starbase. It has been previously confirmed that miR-29a plays the role of a tumor suppressor gene in many tumors. It inhibits glioblastoma cells from growth by regulating PDGF,21 and inhibits the proliferation, invasion, and migration of thyroid papillary cancer cells through targeting DPP4.22 Our study showed that miR-29a was lowly expressed in cholangiocarcinoma tissues and cells. After miR-29a was overexpressed in the cells, their proliferation and invasion were remarkably inhibited, but there were opposite cell phenotypes after inhibiting miR-29a expression. This indicates that miR-29a functions as a tumor suppressor gene in cholangiocarcinoma. Its role in the disease has been previously discussed. For example, it is poorly expressed in cholangiocarcinoma and affects its pathogenesis by targeting HDAC4,23 which is consistent with our conclusions. In a study on the mechanism of action of miR-29a in the disease, this miR inhibits the development and progression of tumors by inhibiting hedgehog signals and inflammatory pathways, but its upstream mechanism has not been discussed.24 After that, the DLRGA was performed to further confirm the correlation of TUG1 with miR-29a. The luciferase activity of miR-29a remarkably decreased after the co-transfection of TUG1 and miR-29a, and miR-29a expression up-regulated after inhibiting TUG1 expression. Subsequent co-transfection experiment showed that overexpressing miR-29a could reverse the effects of high TUG1 expression on cholangiocarcinoma cells.
In summary, TUG1 promotes the proliferation and invasion of cholangiocarcinoma cells and inhibits their apoptosis through the regulation of miR-29a expression, so it may be a new therapeutic target for cholangiocarcinoma. However, there are still deficiencies in this study. Firstly, experiments of tumor formation in nude mice were not conducted to explore the effects of TUG1 on tumor growth in vivo. Secondly, the downstream mechanism of miR-29a was not further verified. Therefore, we will conduct more basic experiments to further supplement our experiments.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Banales Jesus M, Marin Jose JG, Lamarca A, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esnaola NF, Meyer JE, Karachristos A, et al. Evaluation and management of intrahepatic and extrahepatic cholangiocarcinoma. Cancer. 2016;122(9):1349–1369. [DOI] [PubMed] [Google Scholar]
- 3.Li JD, Xu XF, Yu JJ, et al. Updated key points and clinical pathway for NCCN clinical practice guidelines in oncology: hepatobiliary cancers (Version 1. 2018). J Clin Hepatol. 2018;34(6):966–977. [Google Scholar]
- 4.Doussot A, Jarnagin WR, Azoulay D, et al. Improving actual survival after hepatectomy for intrahepatic cholangiocar- cinoma - still a long way to go. Hepatobiliary Surg Nutr. 2019;8(2):161–163. doi: 10.21037/hbsn.2018.11.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li CX, Zhang H, Wang K, et al. Preoperative bilirubin level predicts overall survival and tumor recurrence after resection for perihilar cholangiocarcinoma patients. Cancer Manag Res. 2019;11:10157–10165. doi: 10.2147/CMAR.S230620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanly DJ, Esteller M, Berdasco M. Interplay between long non- coding RNAs and epigenetic machinery: emerging targets in cancer? Philos Trans R Soc Lond B Biol Sci. 1748;2018:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Wang Z, Jiang X, et al. Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomed Pharmacother. 2017;92:17–23. doi: 10.1016/j.biopha.2017.05.061 [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Yang Y, Wang W, et al. Long noncoding RNA TUG1 promotes cell proliferation and migration of renal cell carcinoma via regulation of YAP. J Cell Biochem. 2018;119:9694–9706. [DOI] [PubMed] [Google Scholar]
- 9.Yebin L, Ling T, Zhipeng Z, et al. Long noncoding RNA TUG1/miR-29c axis affects cell proliferation, invasion, and migration in human pancreatic cancer. Dis Markers. 2018;2018:6857042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Zhang T, Zhang Y, et al. Targeting the FOXM1-regulated long noncoding RNA TUG1 in osteosarcoma. Cancer Sci. 2018;109:3093–3104. doi: 10.1111/cas.13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, Kaiming L, Li Z, et al. The prognostic potential and carcinogenesis of long non-coding RNA TUG1 in human cholangiocarcinoma. Oncotarget. 2017;8:65823–65835. doi: 10.18632/oncotarget.19502 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Tatsuguchi T, Gotoh K, Kobayashi S, et al. Pathologic complete response after gemcitabine and S-1 chemotherapy for far advanced intrahepatic cholangiocarcinoma. Int Cancer Conf J. 2018;7:93–97. doi: 10.1007/s13691-018-0327-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunner Thomas B, Oliver B, Victor L, et al. Stereotactic body radiotherapy dose and its impact on local control and overall survival of patients for locally advanced intrahepatic and extrahepatic cholangiocarcinoma. Radiother Oncol. 2019;132:42–47. doi: 10.1016/j.radonc.2018.11.015 [DOI] [PubMed] [Google Scholar]
- 14.Seesai Y, Edward M, Shama V, et al. Trends in incidence of two major subtypes of liver and bile duct cancer: hepatocellular carcinoma and cholangiocarcinoma in Songkhla, Southern Thailand, 1989-2030. J Cancer Epidemiol. 2018;2018:8267059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, Bao H, Li H. Correlation of long non-coding RNA taurine-upregulated gene 1 with disease conditions and prognosis, as well as its effect on cell activities in acute myeloid leukemia. Cancer Biomark. 2018;23:569–577. doi: 10.3233/CBM-181834 [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Shuangni Y, Cuiping W, et al. Establishment of a non‑coding RNAomics screening platform for the regulation of KRAS in pancreatic cancer by RNA sequencing. Int J Oncol. 2018;53:2659–2670. [DOI] [PubMed] [Google Scholar]
- 17.Li T, Chen Y, Zhang J, et al. LncRNA TUG1 promotes cells proliferation and inhibits cells apoptosis through regulating AURKA in epithelial ovarian cancer cells. Medicine (Baltimore). 2018;97(36):e12131. doi: 10.1097/MD.0000000000012131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Yang Y, Wang W, et al. Long noncoding RNA TUG1 promotes cell proliferation and migration of renal cell carcinoma via regulation of YAP. J Cell Biochem. 2018;119:9694–9706. doi: 10.1002/jcb.27284 [DOI] [PubMed] [Google Scholar]
- 19.Shi F, Wang T, Liu Z, et al. LncRNA miR143HG up-regulates p53 in endometrial carcinoma by sponging miR-125a. Cancer Manag Res. 2019;11:10117–10123. doi: 10.2147/CMAR.S222215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian Y, Xia S, Ma M, et al. LINC00096 promotes the proliferation and invasion by sponging miR-383-5p and regulating RBM3 expression in triple-negative breast cancer. Onco Targets Ther. 2019;12:10569–10578. doi: 10.2147/OTT.S229659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Dodbele S, Park T, et al. MicroRNA-29a inhibits glioblastoma stem cells and tumor growth by regulating the PDGF pathway.. J Neurooncol. 2019;145:23–34. doi: 10.1007/s11060-019-03275-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Han J, Lv Y, et al. miR-29a inhibits proliferation, invasion, and migration of papillary thyroid cancer by targeting DPP4. Onco Targets Ther. 2019;12:4225–4233. doi: 10.2147/OTT.S201532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Li C, Jian Z, et al. TGF-β1 reduces miR-29a expression to promote tumorigenicity and metastasis of cholangiocarcinoma by targeting HDAC4. PLoS One. 2015;10(10):e0136703. doi: 10.1371/journal.pone.0136703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mott Justin L, Kurita S, Cazanave Sophie C, et al. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J Cell Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630 [DOI] [PMC free article] [PubMed] [Google Scholar]