Abstract
Background and Aims
Dysbiosis of the gut microbiota is a well-known correlate of the pathogenesis of inflammatory bowel disease [IBD]. However, few studies have examined the microbiome in very early-onset [VEO] IBD, which is defined as onset of IBD before 6 years of age. Here we focus on the viral portion of the microbiome—the virome—to assess possible viral associations with disease processes, reasoning that any viruses potentially associated with IBD might grow more robustly in younger subjects, and so be more detectable.
Methods
Virus-like particles [VLPs] were purified from stool samples collected from patients with VEO-IBD [n = 54] and healthy controls [n = 23], and characterized by DNA and RNA sequencing and VLP particle counts.
Results
The total number of VLPs was not significantly different between VEO-IBD and healthy controls. For bacterial viruses, the VEO-IBD subjects were found to have a higher ratio of Caudovirales vs to Microviridae compared to healthy controls. An increase in Caudovirales was also associated with immunosuppressive therapy. For viruses infecting human cells, Anelloviridae showed higher prevalence in VEO-IBD compared to healthy controls. Within the VEO-IBD group, higher levels of Anelloviridae DNA were also positively associated with immunosuppressive treatment. To search for new viruses, short sequences enriched in VEO-IBD samples were identified, and some could be validated in an independent cohort, although none was clearly viral; this provides sequence tags to interrogate in future studies.
Conclusions
These data thus document perturbations to normal viral populations associated with VEO-IBD, and provide a biomarker—Anelloviridae DNA levels—potentially useful for reporting the effectiveness of immunosuppression.
Keywords: Very early-onset inflammatory bowel disease, VEO-IBD, virome, microbiome, metagenome
1. Introduction
Inflammatory bowel diseases [IBD], including Crohn disease [CD], ulcerative colitis [UC] and IBD-undetermined [IBD-U], are complex chronic intestinal inflammatory disorders which are hypothesized to be driven by pathological interactions among the host, environmental factors and gut microbes.1–9 Children with very early-onset IBD [VEO-IBD], defined as those diagnosed at <6 years of age, often have distinctive phenotypes and some have disease courses that are more severe and refractory to conventional medications than in older patients.10 A subset of patients with VEO-IBD have causative monogenic or digenic drivers of disease, but the rapid increase in incidence, particularly in this young age group, suggests that environmental factors, including the intestinal microbiota, play a major role as well.10–13
The human gut is colonized by multiple types of organisms, including bacteria, archaea, fungi, microeukaryotes and viruses.5,14,15 Some members of the gut virome replicate in bacterial cells [bacteriophages] while others replicate in eukaryotic cells [eukaryotic viruses], including gut tissue.16–22 The dysbiotic gut microbiota has been well characterized for IBD, including reductions of Bacteroidetes and Firmicutes and the expansion of Proteobacteria, as well as changes in bacterial microbiome function.2,5,8,23–25 However, fewer studies have investigated gut viral communities in IBD.26–28
Some recent studies have suggested outgrowth of specific lineages of bacteriophages and eukaryotic viruses in IBD. Among phages, Caudovirales and Microviridae are the predominant families in the healthy human gut. Caudovirales are tailed phages including the Siphoviridae, Myoviridae and Podoviridae; Caudovirales usually have double stranded DNA genomes. The Microviridae form icosahedral particles and have smaller single-stranded DNA genomes.20,29 An expansion of Caudovirales, the tailed bacteriophage, was observed in both paediatric and adult-onset IBD, although the results varied in different disease types [CD or UC]23 as well as by age and anatomical location of disease.26,28
Viruses that replicate in human cells have also been reported to be altered in abundance in gut samples from subjects with IBD.28Anelloviridae, a family of ubiquitous small, single-stranded DNA viruses present in most humans, have been reported to be more prevalent in IBD patients compared to healthy controls.28 A greater abundance of Anelloviridae has also been found among immunocompromised patients, such as the recipients of lung transplants and human immunodeficicncy virus [HIV[-positive patients.24,30–35 The prevalence of Anelloviridae in IBD may be particularly pertinent given that IBD patients are commonly treated with immunosuppressive agents. Several further eukaryotic viruses have been explored as possible causative agents of IBD, including herpesviruses,36,37 rotavirus,38 norovirus,39,40 influenza41 and measles virus,42 but no direct link has been found.
A complication is that sequence databases contain only a small fraction of the global virome, so that the majority of sequence reads in a typical virome sample are unattributed.18,28 Thus, further characterization of bacteriophages and eukaryotic viral populations are desirable to understand the dynamics of the microbiota in IBD and explore for possible contributors to its pathogenesis.
Here we performed a cross-sectional analysis of the gut virome using stool samples from a VEO-IBD cohort. We hypothesized that the influence of viruses might be more pronounced in the disease process of children with VEO-IBD, possibly including a mechanistic role in disease development. We purified virus-like particles [VLPs] and [1] evaluated the total particle density by epifluorescence staining, [2] used metagenomic sequencing of VLP DNA and RNA to analyse viral population structure, and [3] explored for potential novel viruses associated with VEO-IBD. Our primary aim was to characterize the virome in subjects with VEO-IBD relative to healthy paediatric controls. Our secondary aim was to associate the virome composition with clinical characteristics and disease activity in the VEO-IBD cohort.
2. Methods
2.1. Human subjects
This study was approved by the Institutional Review Board of The Children’s Hospital of Philadelphia [Protocols 14-010826 and 15-011817]. Subjects were identified from the Children’s Hospital of Philadelphia outpatient clinics and Emergency Department. Patients and families gave informed consent to participate in the study. Inclusion criteria for the VEO-IBD cohort included children under age 18 years with a confirmed diagnosis of IBD before age 6 years by clinical history, oesophagogastroduodenoscopy and colonoscopy, laboratory studies, and radiological examinations. Exclusion criteria for VEO-IBD subjects included concurrent intestinal comorbidity including positive coeliac serology and histology, Hirschsprung disease, eosinophilic oesophagitis, immunodeficiency, short bowel syndrome and infection. For healthy controls, the exclusion criteria included any chronic health condition including IBD, infections including Clostridium difficile, recent antibiotic use in the past 3 months and siblings with a history of IBD. Fresh stool specimens were collected and aliquoted into faeces collection tubes [Sarstedt]. All samples were stored at −80°C.
2.2. Metadata
To associate with the virome analyses, clinical characteristics were collected from the electronic medical records, including IBD diagnosis, age of diagnosis, disease location/extent according to the Paris Classification,43 phenotype as applicable, current IBD medication use, current antibiotic use and antibiotic use within 30 days prior to enrollment in the study. Additionally, disease activity was characterized by faecal calprotectin, when an additional stool sample was available for measuring. Active disease was defined as faecal calprotectin ≥250 µg per gram faeces and inactive disease was defined as <250 µg per gram faeces.44
2.3. Calprotectin
Calprotectin levels were measured from faecal samples using the QUANTA Lite Calprotectin Extended Range ELISA [enzyme-linked immunosorbent assay] kit [Inova Diagnostics], strictly in accordance with the manufacturer’s protocol. To extract samples, 100 mg faeces was mixed 1:50 [w/v] with extraction buffer, vortexed for 30 s and then homogenized for 25 min on a shaker. One millilitre of the homogenate was centrifuged for 20 min at 10 000 g and the cleared supernatant was stored at −20°C until the ELISAs were performed. The ELISAs were conducted on cleared supernatants diluted 1:400 in the provided dilution buffer. The supplied calibrators and controls were run alongside the samples on each plate. Absorbance measurements at 450 nm were taken on the EnSpire Multimodal Plate Reader [Perkin Elmer] and calprotectin levels were calculated based on a standard curve generated using the 4-parameter logistic regression model.
2.4. VLP purification
VLPs were purified as in previous work.45 Briefly, 150 mg of stool was homogenized in 25 mL of SM buffer [50 mM Tris-HCl pH 7.5, 100 mM NaCl, 8 mM MgSO4]. The homogenate was then spun down and filtered through a 0.2-µm pore-size filter [Thermo Fisher Scientific]. The filtrate was concentrated using a 100-kDa molecular-mass Amicon Ultra-15 Centrifugal filter [Millipore] and resuspended in 25 mL SM buffer. A second round of concentration was performed, yielding a final volume of ~500 µL concentrate. The concentrate containing VLPs was treated with DNase I and RNase [Roche] at 37°C for 30 min to eliminate non-encapsulated nucleic acids. A 200-µL VLP preparation was used for viral nucleic acid extraction immediately after DNase I and RNase treatment; the rest of the VLP preparation was stored at 4°C for up to 3 months. To detect enveloped viruses, no chloroform was used during VLP purification.
2.5. Epifluorescence staining
A total of 50 µL of isolated VLPs was suspended in 5–10 mL SM buffer and filtered onto a 0.02-μm Anodisc polycarbonate filter [Whatman]. Filters were stained with 2 × SYBR Gold [Thermo Fisher Scientific] for 15 min, then washed with H2O for several seconds. After air-drying, the filter was mounted on a glass slide with 15 µL of mountant buffer [100 µL 10% ascorbic acid + 4.9 mL pH 7.4 PBS + 5 mL 100% glycerol; filtered at 0.02 µm]. Viruses were counted in five to ten fields of view selected randomly on each filter, and mean values were calculated. The filter was visualized using a motorized inverted IX81 microscope [Shinjuku] for fluorescence. VLPs were counted using ImageJ software [Particle counting function]. Stained particles <0.5 µm in diameter were considered as VLPs [larger particles were excluded]. A serial dilution of VLPs was sometimes used to obtain more accurate quantification. Lambda phage cultures with known plaque-forming unit [PFU] counts were used as a positive control for software adjustment, such as image colour, saturation, level and contrast. VLPs mock isolated from only SM buffer were used as negative controls.
2.6. Viral DNA and RNA preparation
Viral DNA and RNA were extracted from VLPs using the AllPrep DNA/RNA Mini kit [Qiagen] following the manufacturer’s instructions. DNA was stored at −20°C and RNA at −80°C until used. Viral DNA was subjected to DNA whole genome amplification using the GenomiPhi V2 Amplification kit [GE Healthcare]. RNA was treated with DNAse [Roche] for 20 min at 37°C, followed by reverse transcription. A SuperScript III First Strand Synthesis kit [Thermo Fisher Scientific] and Primer A [5′-GTTTCCCAGTCACGATCNNNNNNNNN-3′] were used for the first-strand cDNA synthesis.46 DNA Polymerase I, Large [Klenow] Fragment [New England BioLab] was then used for second-strand cDNA synthesis. The cDNA product was then amplified by adding Primer B [5′-GTTTCCCAGTCACGATC-3′] and AccuPrime Taq High Fidelity DNA polymerase [Thermo Fisher Scientific] with the following reaction conditions: 75.5 µL of molecular-grade H2O, 10 µL of 10× PCR Buffer I, 4 µL of 50 mM MgCl2, 2.5 µL of 10 mM dNTPs, 1 µL of 100 µM Primer B, 1 µL Taq and 6 µL cDNA product. The PCR programme was 40 cycles 94°C for 2 min, 94°C for 30 s, 40°C for 30 s, 50°C for 30 s and 72°C for 1 min. Amplified DNA and cDNA were stored at −20°C.
2.7. Virome library construction and sequencing
DNA concentration was evaluated by a Quant-iT PicoGreen dsDNA Assay kit [Thermo Fisher Scientific], and fluorescence was measured by an EnVision Multilabel Plate Reader. An illumina Nextera XT Samples Prep kit [Illumina] was used for library construction. Quantification of libraries was performed by both a Quant-iT PicoGreen dsDNA Assay kit and a KAPA Library Quantification kit [Kapa Biosystems]. The size distribution of the libraries was checked using a 5300 Fragment Analyzer [Agilent]. Libraries were pooled for sequencing. The concentration of the pooled libraries was assessed using Qubit [Invitrogen], and the size distributions of the pooled libraries were measured by an Agilent Technology 2100 Bioanalyzer. Sequences were acquired using both the Illumina MiSeq [250-bp paired-end reads, Illumina] and HiSeq [125-bp paired-end reads, Illumina].
2.8. Virome sequence read quality control
The Sunbeam pipeline47 with a custom Sunbeam extension [https://github.com/guanxiangliang/sbx_dedup] was used for read quality control. Low-quality reads and adapter sequences were removed and trimmed by Trimmomatic48 with Sunbeam default options [leading: 3, trailing: 3, slidingwindow: [4, 15], minlen: 36], and duplicate identical sequences [inferred PCR replicates] were filtered out by BBmap [https://jgi.doe.gov/data-and-tools/bbtools/]. Host reads were identified by BWA-MEM [Sunbeam default options: pct_id >0.5; frac >0.6] using the human genome [GRCh38]. Reads from phix174 were also removed.
2.9. Read assembly and taxonomic assignment
The quality-controlled reads from samples were pooled for assembly. DNA and RNA libraries were assembled separately. Reads were assembled into contigs using megahit with default options [--min-count: 2, --k-list: [21, 29, 39, 59, 79, 99, 119, 141]].49 To exclude contigs resulting from contamination, any contigs with read numbers greater than 25 in more than one negative control sample were removed from the analysis.
Contigs with lengths greater than 500 bp were selected to predict open reading frames [ORFs] using Prodigal in ‘meta’ mode.50 The predicted ORFs were mapped to the UniProt viral protein database [TrEMBL and Swiss-Prot database] using BLASTP with an E value < 1e-5.51
The taxonomy of each contig was assigned based on a voting system described previously,29 to compile attributions over multiple reading frames to assign each contig to the nearest database viral species. The ORFs in a contig were given taxonomic ranking based on the best-hit viral protein in the UniProt reference database. If more than 50% of the predicted ORFs in a contig were viral ORFs, the contig was classified as viral. All taxonomic assignments of each ORF within the same contig were then compared, and the contig was annotated by the majority vote of ORF taxonomy assignments. Some contigs shared the same taxonomic assignments; the contig table was collapsed by taxonomic identity to yield pooled values for each taxon.
We analysed the differentially present taxa, assigned at the species level, and used edgeR52 to assess significance. EdgeR was designed to analyse data with a negative binomial distribution, as in our virome data. The significantly differentially present taxa were determined by false discovery rate [FDR] < 0.05 based on Benjamini and Hochberg multiple testing correction as well as fold change >1.5.
2.10. Profiling eukaryotic viruses
The eukaryotic viral genomes and segments in the RefSeq and Viral Neighbor databases were retrieved from the Reference Viral Database [RVDB].53 The virome sequences were mapped to these genomes to estimate genome coverage using Bowtie2 with the ‘global’ alignment option.54 The output sam files were processed by Samtools,55 Bedtools56 and custom code [https://github.com/guanxiangliang/liang2019VEO] to quantify the fraction of the genome covered. We used percentage coverage for genome detection,34 because amplification during library construction can produce many copies of short genome regions, yielding many sequences but with low genome coverage.
2.11. K-mer-based analysis
The quality-controlled reads were merged from both ends. Jellyfish [version 2.2.3] was used to count k-mers with k = 31 [Maximum option in the software]. Only 31-mers with at least four counts were reported and used in the analysis. A presence/absence data matrix was used, and Fisher’s Exact test was performed to determine significant differences between groups, followed by multiple comparison p-value correction. Virome data from the integrative Human Microbiome Project [iHMP] were downloaded by grabseqs.57 The chi-squared test was used for a similar analysis of the iHMP dataset.
2.12. Quantification and statistical analysis
Statistical tests were conducted using R. Non-parametric tests were used for comparing two independent groups [Wilcoxon rank-sum test]. Non-parametric correlation was performed using Spearman’s rank-order correlation. Fisher’s Exact test was used to test the difference between two categorical variables. A chi-squared test was used to test the difference between two categorical variables in the iHMP dataset, which has a larger sample size. The p-values for multiple comparisons were corrected using the Benjamini–Hochberg FDR method; p < 0.05 or FDR < 0.05 was considered significant. All acquired data were included in the analyses.
2.13. Data and software availability
Sample information and raw sequences are available in the National Center for Biotechnology Information [NCBI] Sequence Read Archive under BioProject ID PRJNA 564995 [Supplementary Table 1]. All bioinformatic scripts are available on Github [https://github.com/guanxiangliang/liang2019VEO].
3. Results
3.1. Human subjects and virome sequencing
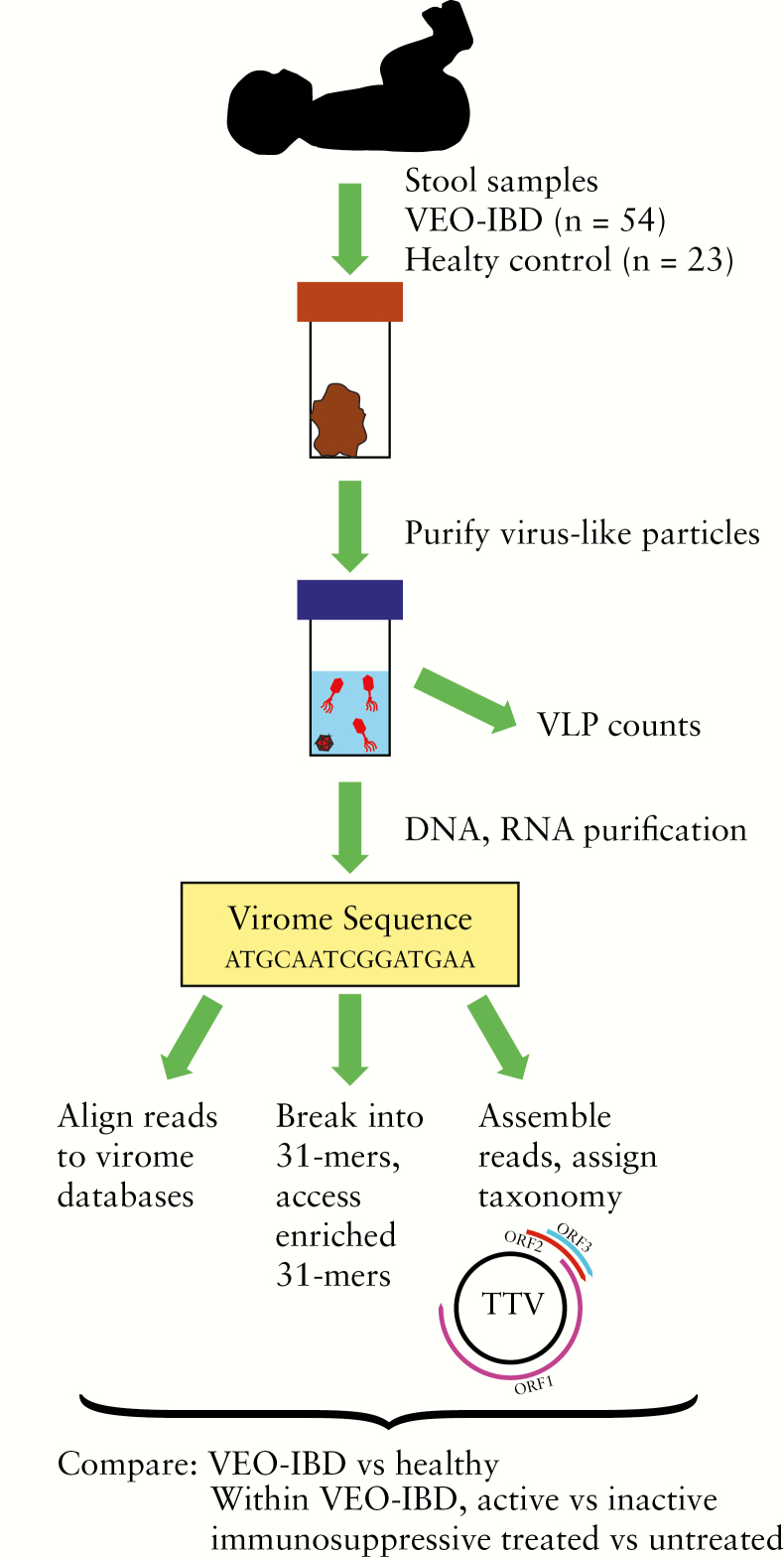
Samples were collected from VEO-IBD subjects [n = 54] and healthy controls [n = 23, Figure 1]. Of the VEO-IBD subjects, 95% [51/54] had primarily colonic disease and 59% [32/54] were diagnosed with CD, 39% [21/54] with IBD-U and 2% [1/54] with UC based on endoscopic, histological and radiological findings. Among the 54 subjects with VEO-IBD, 56% [30/54] were naïve to any immunosuppression at the time of sampling and 44% [24/54] were on immunosuppressive therapies including corticosteroids, anti-tumour necrosis factor α [anti-TNFα] medications, or immunomodulators; ten [19%] patients were treated with antibiotics, such as metronidazole, vancomycin or azithromycin [Supplementary Table 2]. Disease activity, measured by faecal calprotectin levels, was available for 36/54 [67%] of the VEO-IBD subjects. Active disease [calprotectin ≥ 250 µg per gram faeces] was found in 20 subjects, and inactive [<250 µg per gram faeces] in 16 subjects. Detailed subject information is given in Supplementary Table 2. Calprotectin level was not associated with age, sex or treatment [Supplementary Table 3]. All patients with VEO-IBD were diagnosed before age 6 years, although some samples were collected at older ages [up to age 16 years]. Sampling ages or the duration time between diagnosis and sampling were not associated with sex, treatment or calprotectin levels [Supplementary Table 3].
Figure 1.
Overview of cohort characteristics and data analysis.
VLPs were isolated from stool samples, followed by DNA and RNA extraction [Figure 1]. Nucleic acids were subjected to shotgun metagenome sequencing [Figure 1]. An average of 1.81 ± 0.65 million [mean ± SEM] dereplicated high-quality non-human reads were obtained per sequence library.
3.2. Total VLP density was not affected in VEO-IBD
To test the total VLP density in VEO-IBD, 25 VLP samples [13 from VEO-IBD samples and 12 from healthy controls] were randomly selected for epifluorescence staining analysis. Total VLP density was not significantly different between VEO-IBD samples [8.3 × 108 ± 3.8 × 108 counts per gram faeces, mean ± SEM] and healthy controls [4.7 × 108 ± 1.7 × 108 counts per gram faeces, p = 0.98, Wilcoxon rank-sum test]. In addition, VLP numbers were not detectably related to immunosuppressive treatment [p = 0.71, Wilcoxon rank-sum test, eight with immunosuppressive treatment and five without immunosuppressive treatment] or clinical disease activity [p = 1.0, Wilcoxon rank-sum test, four from active disease and four from inactive disease; p = 0.98, Spearman’s correlation].
3.3. An altered ratio between Caudovirales and Microviridae correlated with immunosuppression
To assess virome composition, a cross-assembly of the metagenomic sequences was performed and analysed. A total of 106 898 DNA contigs and 1398 RNA contigs of length >500 bp were built from the pool of all sequence reads. Taxonomy assignments of the contigs yielded 24 474 DNA viral contigs representing 2326 apparent species and 92 RNA viral contigs representing 30 apparent species. All the RNA contigs annotated as viral were attributed to eukaryotic viruses, reflecting the known scarcity of RNA phages in available databases.58
When comparing VEO-IBD to healthy controls, there were no differences in global ecological parameters observed for DNA viruses, including viral richness [p = 0.31, Wilcoxon rank-sum test] and Shannon diversity [p = 0.29, Wilcoxon rank-sum test]. The Bray–Curtis distances between communities were calculated based on the relative abundance table of viral species, and no significant differences in viral population structure were detected [p = 0.06, PERMANOVA].
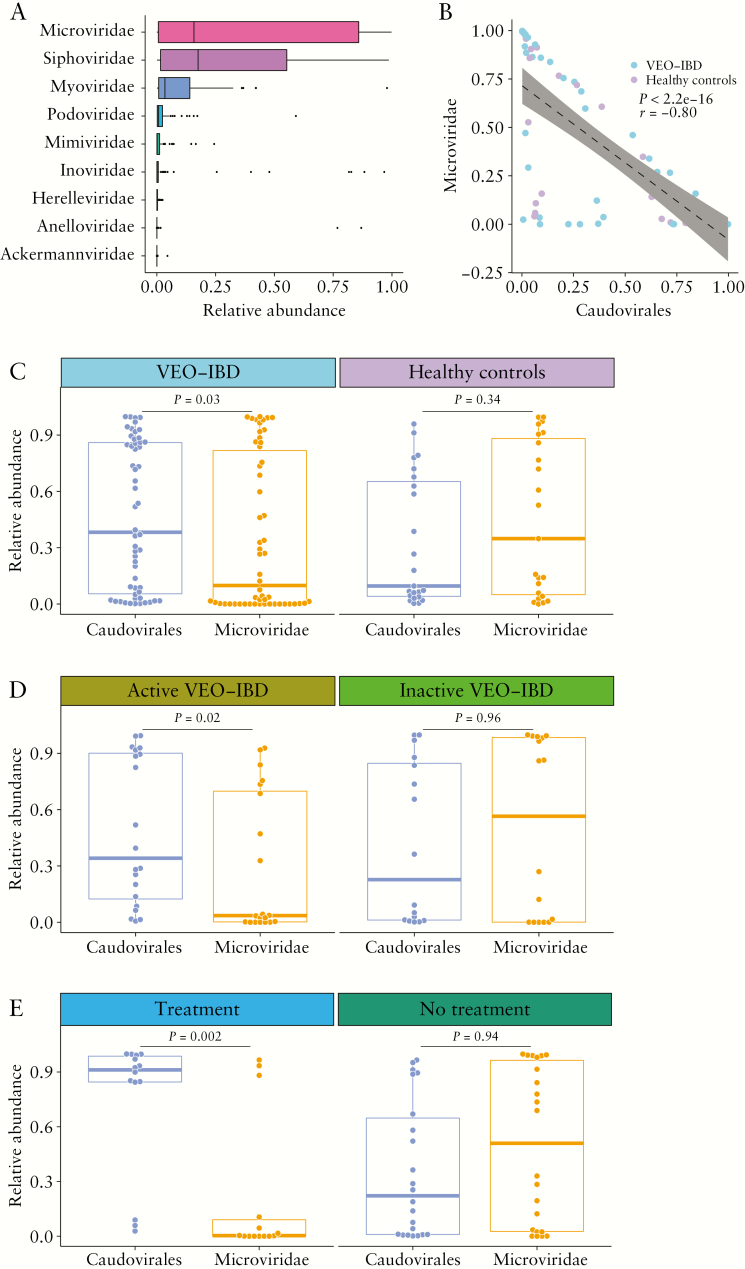
Caudovirales [e.g. Siphoviridae, Myoviridae, Podoviridae, Herelleviridae and Ackermannviridae], Microvirirdae, Mimiviridae, Inoviridae and Anelloviridae were the most abundant DNA viral taxa identified [Figure 2A]. Caudovirales or Microviridae were the most abundant taxa in 70 [91%] subjects, and Anelloviridae or Inoviridae were dominant in seven [9%] subjects. While previous studies have reported an increased richness of Caudovirales28 and increased abundance of Caudovirales in paediatric subjects and adults with UC,59 we found no significant difference in the abundance [p = 0.09, Wilcoxon rank-sum test] or richness [p = 0.38, Wilcoxon rank-sum test] of Caudovirales between healthy controls and VEO-IBD samples.
Figure 2.
Comparing bacteriophage populations in samples from VEO-IBD patients vs healthy controls. [A] Boxplot of the relative abundance of different viral taxa at the family level. [B] Correlation between the abundance of Caudovirales and Microviridae. The p- and r-values were calculated by Spearman’s correlation test. Linear regression with 95% confidence interval is shown. [C] Comparison between the abundance of Caudovirales and Microviridae in VEO-IBD patients [n = 54] and healthy controls [n = 23]. [D] Comparison between the abundance of Caudovirales and Microviridae in active VEO-IBD patients [n = 20] and inactive VEO-IBD patients [n = 16]. [E] Comparison between the abundance of Caudovirales and Microviridae in VEO-IBD patients with immunosuppressive treatment [n = 24] and without immunosuppressive treatment [n = 30]. The Wilcoxon rank-sum test was used throughout to test for differences.
However, comparing the ratio of the two most abundant phage groups did yield a significant difference, consistent with a previous report.28 A strong negative correlation between Caudovirales and Microviridae was observed in the VEO-IBD cohort [p < 2.2e-16, r = −0.8, Spearman’s rank-order correlation, Figure 2B]. In healthy controls, the abundance of Caudovirales was similar to that of Microviridae [p = 0.34, Wilcoxon rank-sum test, Figure 2C], while in the samples from the VEO-IBD subjects, the abundance of Caudovirales was significantly higher than Microviridae [p = 0.03, Wilcoxon rank-sum test, Figure 2C].
To test whether the altered ratio between Caudovirales and Microviridae was associated with disease activity, we compared the viral abundances in the samples from subjects with active VEO-IBD separately from those with inactive VEO-IBD. A significant difference was found within the active VEO-IBD group [p = 0.02, Wilcoxon rank-sum test, Figure 2D]. No difference was found in the inactive VEO-IBD group [p = 0.96, Wilcoxon rank-sum test, Figure 2D].
We next investigated the effects of immunosuppressive treatments on the ratio of Caudovirales and Microviridae. A significantly higher level of Caudovirales compared to Microviridae was detected in samples from immunosuppressed subjects [p = 0.002, Wilcoxon rank-sum test, Figure 2E] but not untreated subjects [p = 0.94, Wilcoxon rank-sum test, Figure 2E]. We then tested whether the altered ratio between Caudovirales and Microviridae was associated with disease activity within the immunosuppressive untreated group [n = 22, with calprotectin data]. The abundance of Caudovirales was similar to Microviridae in both active [p = 0.84, Wilcoxon rank-sum test] and inactive VEO-IBD samples [p = 0.25, Wilcoxon rank-sum test]. Thus, a difference could be detected in phage populations between VEO-IBD patients and controls associated with immunosuppressive treatment.
3.4. Viral species assignments differing between VEO-IBD and healthy controls
Seventy-four taxa were differentially present in VEO-IBD samples compared to healthy controls [FDR < 0.05, edgeR, Supplementary Table 4]. Among the 74 taxa, 18 were enriched in healthy controls, and 56 were enriched in VEO-IBD [Supplementary Table 4]. Among viruses that infect human cells, there was enrichment in VEO-IBD samples of an unnamed Anelloviridae species and Torque teno virus 8; all others belonged to phages including Xanthomonas citri phage CP2 and Escherichia coli O157 typing phage 15. These together with the unnamed Anelloviridae species were enriched in VEO-IBD with >30-fold change. Arthrobacter phage Correa and Indivirus ILV1 were enriched in healthy controls with >10 -fold change.
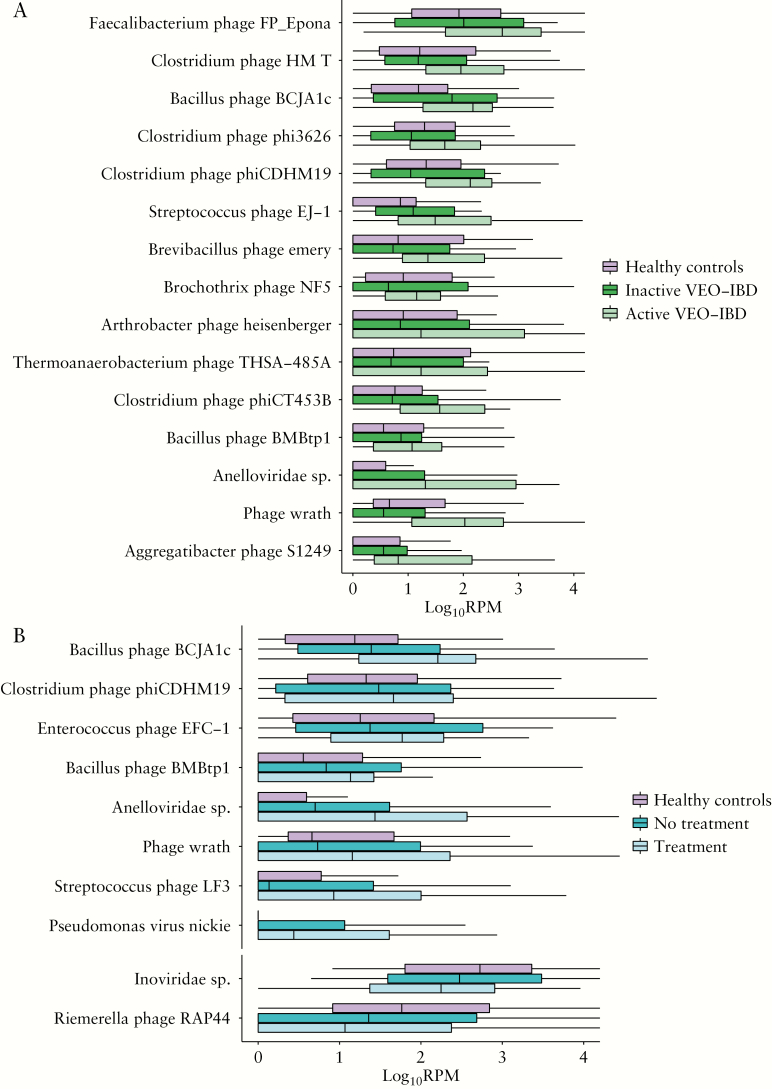
To identify viral taxa that may be correlated with VEO-IBD disease activity, taxa were quantified that differed for active vs inactive disease. Forty-five viral taxa were enriched in either active or inactive VEO-IBD [FDR < 0.05, edgeR, Supplementary Table 5]. We found 15 out of 45 taxa were enriched in active VEO-IBD and also enriched in the VEO-IBD cohort compared to healthy controls [Figure 3A]. The 15 taxa enriched in active VEO-IBD included an unnamed Anelloviridae species, and 14 phages belonging to Caudovirales [Figure 3A].
Figure 3.
Viral taxa that are differentially present in VEO-IBD vs healthy controls. The y-axis at the top presents reads per million total mapped reads [RPM] of each taxon with log10 transformation. The x-axis at the left presents each different taxon. [A] Viral taxa differed among healthy controls, and inactive and active patients. [B] Viral taxa differed among healthy controls, and immunosuppressive untreated and treated patients.
Taxa associated with immunosuppressive treatments were next identified. Ninety-six viral taxa were enriched in either immunosuppressive treated or untreated patients [FDR < 0.05, edgeR, Supplementary Table 6]. We found eight out of 96 taxa, including an unnamed Anelloviridae species, and seven phages belonging to Caudovirales, were enriched in immunosuppressive treated patients and also enriched in the VEO-IBD cohort compared to healthy controls [Figure 3B]. Two phages, Inoviridae species and Riemerella phage RAP44, were enriched in untreated patients and were also enriched in healthy controls when compared to VEO-IBD [Figure 3B].
3.5. Eukaryotic virus colonization in VEO-IBD
To investigate the colonization of eukaryotic viruses in VEO-IBD, both DNA and RNA metagenomic sequences were mapped to a custom database with eukaryotic viral genomes, which were collected from the RefSeq and Neighbor assembly databases.60 In this analysis, to call a virus as present, we required that 33% of the genome sequence be covered by our metagenomic virome reads.34 We favour use of percentage coverage as a metric and not counts of reads because empirical experience shows that artefacts can often involve large numbers of sequence reads aligning to short regions of viral genomes, so that simply counting reads aligning can be misleading. This value of 33% represents a robust but not excessively strict threshold.
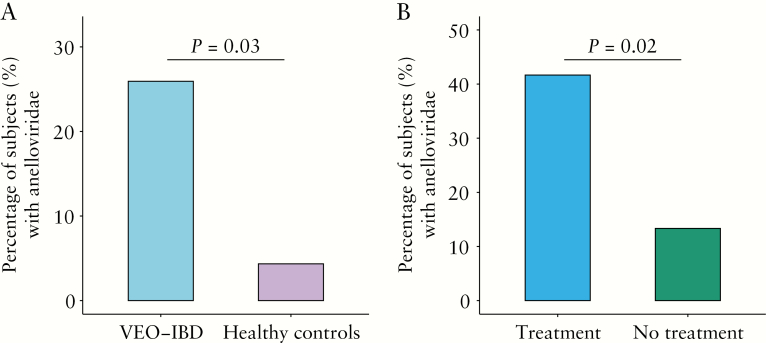
For DNA viruses, Anelloviridae, Geminiviridae, Genomoviridae and Polyomaviridae were all detected in at least one sample. Only Anelloviridae were enriched in VEO-IBD in this analysis [p = 0.03, Fisher’s Exact test, Figure 4A]. We note that our methods are particularly sensitive at recovering small circular DNA viruses, which includes all of the above, because we used a pre-amplification step that can lead to rolling circle replication of small circular genomes. Colonization with Anelloviridae, analysed over the entire VEO-IBD cohort, was not correlated with disease activity [p = 1, Fisher’s Exact test]. In another analytical approach, comparison using the abundance of Anelloviridae sequence reads also yielded a significant difference between VEO-IBD and healthy controls [p = 0.02; Wilcoxon rank-sum test] and showed a trend toward a correlation with disease activity [p = 0.06; Wilcoxon rank-sum test].
Figure 4.
Anelloviridae prevalence. [A] Comparison between the prevalence of Anelloviridae in VEO-IBD patients [n = 54] and healthy controls [n = 23]. The y-axis indicates the percentage of positive subjects with Anelloviridae. Positive calls required that at least one Anelloviridae genome was more than 33% covered by sequence reads. Samples were compared using Fisher’s exact test. [B] Comparison between the prevalence of Anelloviridae in VEO-IBD patients with immunosuppressive treatment [n = 24] and VEO-IBD patients without immunosuppressive treatment [n = 30]. The y-axis is as in A. Samples were compared using Fisher’s exact test.
Among RNA viruses, Astroviridae, Caliciviridae, Closteroviridae, Picobirnaviridae, Picornaviridae, Pospiviroidae, Tombusviridae and Virgaviridae were found in at least one subject. None was enriched in VEO-IBD or associated with disease activity.
3.6. Anelloviridae and immunosuppression
We next investigated whether there was an association between abundance of Anelloviridae and immunosuppression in VEO-IBD, as previously reported in other conditions.24,30–34 We pooled all VEO-IBD subjects on immunosuppressive therapy, including anti-TNFα therapies, immunomodulators and corticosteroids, and compared them to subjects who were treatment-naive [Supplementary Table 2]. We found that Anelloviridae prevalence was indeed increased in subjects treated with immunosuppressive agents [p = 0.02, Fisher’s Exact test, Figure 4B].
An alternative hypothesis would be that higher levels of Anelloviridae were associated with disease activity, which was higher in the subjects treated with immunosuppressive therapy. We thus tested within the untreated group [n = 22, with calprotectin data] to see whether there was any increase in Anelloviridae abundance associated with higher disease activity. Of the 13 with active disease, one showed Anelloviridae representation that passed our threshold of 33% coverage, and in the inactive group [n = 9], two showed detectable Anelloviridae colonization. Thus, there was no evidence for a positive association of Anelloviridae colonization with disease activity [p = 0.54, Wilcoxon rank-sum test], although our power to detect differences was low. Thus, we favour the idea that Anelloviridae colonization is promoted by immunosuppression.
3.7. Assessing possible association of previously unknown viruses with VEO-IBD
We next investigated the hypothesis that previously unknown viruses are associated with IBD, and so carried out analyses that did not depend on alignment to known viral genomes in existing databases. In one approach, we interrogated all of the virome contigs [including contigs annotated as non-viral and contigs shorter than 500 bp] to determine whether any sequences were selectively enriched in the VEO-IBD specimens. A total of 203 480 DNA contigs and 6606 RNA contigs were assessed in the enrichment analysis. A presence/absence matrix was constructed for each sample using the percentage coverage data. Similar to the eukaryotic virus analysis, to call a contig as present, we required that 33% of the contig sequence be covered by the metagenomic virome reads. Binary presence–absence data were evaluated using Fisher’s Exact test. We found that no DNA contig was over-represented in VEO-IBD patients, while only one RNA contig was enriched in VEO-IBD samples [FDR < 0.05]. However, this RNA contig could be mapped to the rRNA-23S rRNA of a bacterial genome [family Enterobacteriaceae], and therefore was not a viral contig, but rather appeared to be a contaminant. We conclude that there was no strong evidence for enrichment of any virome contigs in the VEO-IBD samples.
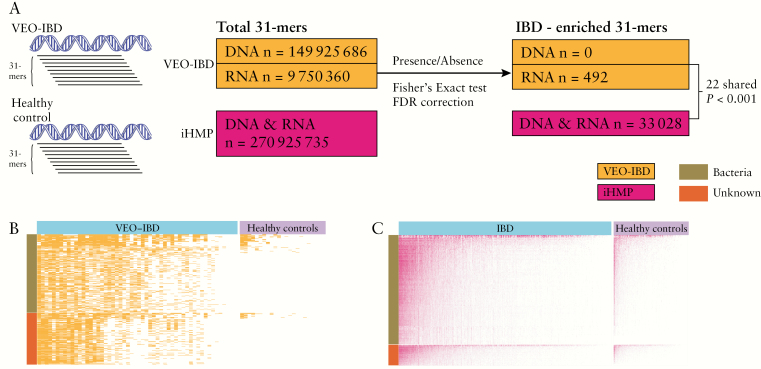
Next, a k-mer-based analysis was performed to look for short sequences associated with VEO-IBD, again using an approach that does not rely on alignment to databases of known viral sequences or on contig assembly [Figure 5A]. We selected the maximum k-mer size [31-mers] available in the Jellyfish package,61 reasoning that this would provide the greatest specificity while minimizing k-mer numbers to enhance computational feasibility.
Figure 5.
K-mer-based analysis. [A] Diagram of the analytical approach. [B] Presence/absence heatmap of differentially present RNA 31-mers between VEO-IBD and healthy controls. Red-coloured cells indicate presence while white-coloured cells indicate absence. The y-axis indicates different 31-mers [n = 492], and the y-axis coloir bar represents the nucleotide database annotation of 31-mers. [C] As in B, but showing differentially present 31-mers between IBD and healthy controls from the iHMP dataset.
A total of 149 925 686 31-mer sequences were identified in the DNA VLP dataset, and 9 750 360 31-mer sequences were identified in RNA VLP sequences [Figure 5A]. For the DNA 31-mers, pairwise comparison between VEO-IBD and healthy control samples, followed by p-value correction for multiple comparisons, revealed that none was significantly enriched in VEO-IBD compared to healthy controls [Figure 5A]. The NCBI nt database annotation of the top 500 VEO-IBD enriched DNA 31-mer sequences [according to ascending uncorrected p-values] showed that 225 of the nominally enriched DNA 31-mers could be annotated [Supplementary Table 7]. Seventy-one of the annotated 31-mers were mapped to Anelloviridae [Supplementary Table 7], consistent with the finding above that Anelloviridae were enriched in VEO-IBD. Of the remainder, 137 31-mers mapped to bacterial genomes and 17 could not be mapped [Supplementary Table 7].
For RNA 31-mers, 492 were found to be enriched in VEO-IBD after correction for multiple comparisons [FDR < 0.05, Fisher’s Exact test, Figure 5A and B]. Of these, 298 could be mapped to the NCBI nt database,62 and were annotated as matching bacterial or fungal genomes [Figure 5B and Supplementary Table 8]. The remaining 194 31-mers could not be annotated [Figure 5A]. Attempts to assemble these sequences did not yield contigs.
To interrogate enriched k-mers further, the analysis was repeated using the IBD virome dataset from the iHMP.5 In this work, acellular extracts were prepared from faeces, DNA and RNA were isolated, and the two together were reverse transcribed and worked up for virome sequence analysis—thus, for these data RNA and DNA sequences are pooled. The study assessed 523 IBD subjects [332 CD and 191 UC] and 180 healthy controls.5 A total of 270 925 735 31-mer sequences were identified in the iHMP dataset [Figure 5A]. Statistical analysis revealed that 33 028 31-mers were enriched in samples from IBD subjects [FDR < 0.05, chi-square test, Figure 5A and C, Supplementary Table 9]. Of these, 22 were in common between the iHMP virome IBD 31-mers and our VEO-IBD 31-mers [Figure 5A]. This represents more overlap than expected by chance—1000 simulations were carried out drawing 492 31-mers from the VEO-IBD data and 33 028 31-mers from the iHMP data, and in no case was there more than one 31-mer in common. Of the 22 31-mers found to be in common, 20 annotated to bacterial rRNA genes of the genus Haemophilus, which is a proteobacterium previously implicated in IBD.5 The remaining two 31-mers could not be annotated by alignment to databases and were not included in any of our contigs. Thus, these two sequences are of unknown importance, but may be of interest as probes to query future virome samples from IBD patients.
4. Discussion
Recent studies of the human gut virome have demonstrated alterations in viral populations in subjects with chronic diseases, including hypertension,63 diabetes,64,65 colorectal cancer66 and IBD.26,28 Here, we focus for the first time on VEO-IBD, following the idea that possible viral involvement might be particularly evident in early-onset disease. We analysed both bacteriophages and eukaryotic viruses, and both DNA and RNA viruses, revealing several differences between patients with VEO-IBD and healthy controls, and also among different subgroups within the VEO-IBD cohort.
We did not find a significant difference in the total number of stool VLPs when comparing patients with VEO-IBD to healthy controls. There are approximately 108–1010 VLPs per gram faeces in the human gut detected in our epifluorescence assays.19,67,68 The total density of VLPs has been reported to be elevated in the gut mucosal tissue of CD patients compared to controls.69 Here we saw a trend toward higher levels in VEO-IBD [about two-fold higher; only a subgroup of the cohort was analysed], but sample-to-sample variation was high enough that we did not detect a significant difference.
A decrease in stool bacterial diversity in IBD has been reported in many studies,28,70,71 but the association of IBD with intestinal virome diversity has been less consistent, with disease- and cohort-specific results.26,28,72 Similar to other studies of paediatric IBD,26 here there were no detectable differences in total viral diversity and richness in VEO-IBD samples from similarly aged healthy children.
We found a strong negative correlation between Caudovirales and Microviridae, with higher Caudovirales in the VEO-IBD cohort. Within the VEO-IBD group, we detected an elevated ratio of Caudovirales associated with disease activity. Several Caudovirales species were associated with VEO-IBD. Expansion of Caudovirales in the gut virome of subjects with IBD was first reported by Norman et al.28 In another study, the same data were pooled with another and re-analysed using database-independent methods, showing that virulent bacteriophages, Microviridae and crAss-like phages, were replaced by temperate bacteriophages, Siphoviridae and Myoviridae, in IBD.73 In another recent virome analysis of a paediatric IBD cohort, a higher abundance of Caudovirales vs Microviridae was found in comparisons of subjects with IBD to healthy controls.26 The small and circular genomes of Microviridae are likely to be amplified more efficiently by our methods compared to the genomes of Caudovirales—thus, the ratio of Caudovirales to Microviridae in VEO-IBD may in fact be underestimated. The reason for the higher ratio of Caudovirales to Microviridae in IBD is unclear. One possibility is that Caudovirales more commonly target the Proteobacteria-dominated communities common in IBD, while Microviridae grow preferentially within commensal bacterial communities associated with health. A definitive analysis of this will require development of much more efficient methods to determine the hosts of phages identified in metagenomic data.
The prevalence of Anelloviridae was greater in VEO-IBD, which is consistent with a previous study of adult subjects.28 Within VEO-IBD patients, we detected a pronounced increase in those undergoing immunosuppressive treatment, including anti-TNFα therapies, immunomodulators and corticosteroids. The data thus suggest that Anelloviridae abundance is related to reduced immune surveillance rather than the pathogenesis of VEO-IBD, paralleling studies of organ transplantation and HIV-positive patients.24,30
Known viruses can be identified by aligning sequence reads to database genomes, but detection of previously unknown viruses requires specialized methods. Two methods were used in this study to detect the possible presence of viruses lacking sequence resemblance to known lineages: alignment to contigs and k-mer-based analysis. For the contig-based analysis, we assembled contigs from pooled DNA or RNA data, then aligned reads to them to query possible enrichment in the VEO-IBD or control samples. No contigs scored as enriched in the DNA data. One was enriched in RNA data, but alignment showed that the contig encoded an rRNA of Enterobacteriaceae. This is consistent with the expansion of Proteobacteria in IBD patients, and the presence of the sequence in our RNA data as a contaminant.2,5,8,23–25
Our second approach was based on breaking all of the reads into short 31-mer sequences and looking for 31-mers enriched in VEO-IBD vs controls. No DNA 31-mer was differentially present, but 492 RNA 31-mers were enriched in VEO-IBD. However, these 31-mers were not found in all VEO-IBD patients, and they could not be assembled into contigs. Further steps were taken to test the enrichment in an iHMP IBD virome dataset. We found 33 028 31-mers were enriched in the iHMP data. However, none of the 31-mers can be mapped to virus-like sequences. For both the VEO-IBD data and the iHMP data, 31-mers could be found that did not map to any known sequence, and two of these were in common between datasets. Further analysis of the reads containing the two 31-mers showed that some aligned to bacterial database sequences detectably, while other reads remained unattributed. Thus, we did not find any sequences that are strong candidates for markers of novel viruses associated with IBD, but it will nevertheless be of interest to retest our unknown sequences [Supplementary Tables 8 and 9] against future IBD-related virome cohorts and suitable controls.
We tested the association between virome data and metadata such as age, sex, race, immunosuppressive treatment, antibiotic use, calprotectin level, and duration of time between diagnosis and sampling [Supplementary Table 3]. The majority of comparisons were non-significant, but they do still contribute to our understanding of the gut virome in VEO-IBD patients.
This study has several limitations. [1] The cross-sectional design would have been enhanced by longitudinal follow up. [2] We analysed stool samples only—comparison of tissue samples might have allowed more sensitive detection of some viral lineages. [3] Although our subjects were diagnosed with VEO-IBD prior to age 6 years, there was a wide range of time between diagnosis and sample collection, so that any initiating environmental triggers may have ebbed in abundance or been obscured by subsequent treatments. [4] Unavoidably, the VEO-IBD patient population was heterogeneous, with a variety of medication exposures. Lastly, [5] the inability to assign phage sequences to bacterial hosts efficiently limits the interpretation of phage data.
In conclusion, a moderate alteration in stool virome was observed associated with VEO-IBD. We found altered ratios between Caudovirales and Microviridae, with an increased abundance of Caudovirales in VEO-IBD, which was also related to immunosuppressive treatment. Anelloviridae were enriched in VEO-IBD, particularly in subjects undergoing immunosuppressive treatment. No clearcut markers of unknown viruses enriched in IBD were identified, although some short IBD-enriched sequence motifs were found that warrant investigation in future cohorts.
Supplementary Material
Acknowledgments
We thank Jian You and co-workers for help with imaging. We are grateful to members of the Bushman laboratory for help and suggestions, and Laurie Zimmerman for artwork.
Funding
This work was supported by National Institutes of Health grants R61-HL137063 [F.D.B.], R01-HL113252 [F.D.B.] and K23DK100461-01A1 [J.R.K.]. The project described was also supported by the Penn Center for AIDS Research [P30 AI 045008] [F.D.B.], the PennCHOP Microbiome Program [F.D.B., R.N.B.], and a Tobacco Formula grant under the Commonwealth Universal Research Enhancement [C.U.R.E] programme [grant number SAP 4100068710] [F.D.B., R.N.B.].
Conflict of Interest
The authors declare no competing interests.
Author Contributions
G.L. carried out most of the experimentation, sequencing and analysis; L.R.K., H.Z. and L.M. assisted with manipulations; K.B. assisted with analysis; M.A.C., J.K., S.M., A.G. and N.D. carried out sample and metadata collection; G.L., M.A.C., J.R.K., L.A., J.B., R.N.B. and F.D.B. conceived the project and prepared the manuscript. All authors contributed to reviewing and editing of the manuscript.
References
- 1. Chehoud C, Albenberg LG, Judge C, et al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis 2015;21:1948–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chu H, Khosravi A, Kusumawardhani IP, et al. Gene–microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 2016;352:1116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al.; IBDMDB Investigators Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang H, Fang M, Jostins L, et al.; International Inflammatory Bowel Disease Genetics Consortium Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- 8. Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015;18:489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 2017;152:327–339.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kelsen JR, Conrad MA, Dawany N, et al. The unique disease course of children with very early onset-inflammatory bowel disease. Inflamm Bowel Dis 2020;26:909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benchimol EI, Bernstein CN, Bitton A, et al. Trends in epidemiology of pediatric inflammatory bowel disease in canada: distributed network analysis of multiple population-based provincial health administrative databases. Am J Gastroenterol 2017;112:1120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uhlig HH, Schwerd T, Koletzko S, et al. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 2014;990–1007.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benchimol EI, Mack DR, Nguyen GC, et al. Incidence, outcomes, and health services burden of very early onset inflammatory bowel disease. Gastroenterology 2014;147:803–813.e7; quiz e14–5. [DOI] [PubMed] [Google Scholar]
- 14. Consortium THMP, Methé BA, Nelson KE, Pop M, et al. A framework for human microbiome research. Nature 2012;486:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Human Microbiome Consortium Project, Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arumugam M, Raes J, Pelletier E, et al.; MetaHIT Consortium Enterotypes of the human gut microbiome. Nature 2011;473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoffmann C, Dollive S, Grunberg S, et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 2013;8:e66019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aggarwala V, Liang G, Bushman FD. Viral communities of the human gut: metagenomic analysis of composition and dynamics. Mob DNA 2017;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reyes A, Haynes M, Hanson N, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010;466:334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 2011;21:1616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol 2012;10:607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barr JJ, Auro R, Furlan M, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A 2013;110:10771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Young JC, Chehoud C, Bittinger K, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am J Transplant 2015;15:200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity 2014;40:843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fernandes MA, Verstraete SG, Phan TG, et al. Enteric virome and bacterial microbiota in children with ulcerative colitis and Crohn disease. J Pediatr Gastroenterol Nutr 2019;68:30–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ungaro F, Massimino L, Furfaro F, et al. Metagenomic analysis of intestinal mucosa revealed a specific eukaryotic gut virome signature in early-diagnosed inflammatory bowel disease. Gut Microbes 2019;10:149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A 2013;110:12450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Monaco CL, Gootenberg DB, Zhao G, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 2016;19:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chris DM, Jesper E-O, Ole K, et al. TTV viral load as a marker for immune reconstitution after initiation of HAART in HIV-infected patients. HIV Clin Trials 2002;3:287–95. [DOI] [PubMed] [Google Scholar]
- 32. Devalle S, Rua F, Morgado MG, Niel C. Variations in the frequencies of torque teno virus subpopulations during HAART treatment in HIV-1-coinfected patients. Arch Virol 2009;154:1285–91. [DOI] [PubMed] [Google Scholar]
- 33. Abbas AA, Diamond JM, Chehoud C, et al. The perioperative lung transplant virome: torque teno viruses are elevated in donor lungs and show divergent dynamics in primary graft dysfunction. Am J Transplant 2017;17:1313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abbas AA, Young JC, Clarke EL, et al. Bidirectional transfer of Anelloviridae lineages between graft and host during lung transplantation. Am J Transplant 2019;19:1086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Vlaminck I, Khush KK, Strehl C, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 2013;155:1178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sipponen T, Turunen U, Lautenschlager I, Nieminen U, Arola J, Halme L. Human herpesvirus 6 and cytomegalovirus in ileocolonic mucosa in inflammatory bowel disease. Scand J Gastroenterol 2011;46:1324–33. [DOI] [PubMed] [Google Scholar]
- 37. Nelson DA, Petty CC, Bost KL. Infection with murine gammaherpesvirus 68 exacerbates inflammatory bowel disease in IL-10-deficient mice. Inflamm Res 2009;58:881–9. [DOI] [PubMed] [Google Scholar]
- 38. Kolho KL, Klemola P, Simonen-Tikka ML, Ollonen ML, Roivainen M. Enteric viral pathogens in children with inflammatory bowel disease. J Med Virol 2012;84:345–7. [DOI] [PubMed] [Google Scholar]
- 39. Khan RR, Lawson AD, Minnich LL, et al. Gastrointestinal norovirus infection associated with exacerbation of inflammatory bowel disease. J Pediatr Gastroenterol Nutr 2009;48:328–33. [DOI] [PubMed] [Google Scholar]
- 40. Basic M, Keubler LM, Buettner M, et al. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm Bowel Dis 2014;20:431–43. [DOI] [PubMed] [Google Scholar]
- 41. Tinsley A, Navabi S, Williams ED, et al. Increased risk of influenza and influenza-related complications among 140,480 patients with inflammatory bowel disease. Inflamm Bowel Dis 2019;25:369–76. [DOI] [PubMed] [Google Scholar]
- 42. Xia B, Crusius J, Meuwissen S, Pe?a A. Inflammatory bowel disease: definition, epidemiology, etiologic aspects, and immunogenetic studies. World J Gastroenterol 1998;4:446–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis 2011;17:1314–21. [DOI] [PubMed] [Google Scholar]
- 44. Naismith GD, Smith LA, Barry SJ, et al. A prospective evaluation of the predictive value of faecal calprotectin in quiescent Crohn’s disease. J Crohns Colitis 2014;8:1022–9. [DOI] [PubMed] [Google Scholar]
- 45. Chehoud C, Dryga A, Hwang Y, et al. Transfer of viral communities between human individuals during fecal microbiota transplantation. mBio 2016;7:e00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang D, Urisman A, Liu YT, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol 2003;1:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clarke E, Taylor LJ, Zhao C, Connell A, Bushman FD, Bittinger K. Sunbeam: a pipeline for next-generation metagenomic sequencing experiments. BioRxiv 2018;326363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015;31:1674–6. [DOI] [PubMed] [Google Scholar]
- 50. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010;11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pundir S, Magrane M, Martin MJ, O’Donovan C; UniProt Consortium Searching and navigating UniProt databases. Curr Protoc Bioinformatics 2015;50:1.27.1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goodacre N, Aljanahi A, Nandakumar S, Mikailov M, Khan AS. A reference viral database (RVDB) to enhance bioinformatics analysis of high-throughput sequencing for novel virus detection. MSphere 2018;3:e00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li H, Handsaker B, Wysoker A, et al.; 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Taylor LJ, Abbas A, Bushman FD. grabseqs: simple downloading of reads and metadata from multiple next-generation sequencing data repositories. Bioinformatics 2020;2017–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wolf YI, Kazlauskas D, Iranzo J, et al. Origins and evolution of the global RNA virome. MBio 2018;9:e02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zuo T, Lu X-J, Zhang Y, et al. Gut microbiota gut mucosal virome alterations in ulcerative colitis. Gut 2019;68:1169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016;44:D733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011;27:764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. NCBI Resource Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2016;44:D7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Han M, Yang P, Zhong C, Ning K. The human gut virome in hypertension. Front Microbiol 2018;9:3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao G, Vatanen T, Droit L, et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci U S A 2017;114:E6166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ma Y, You X, Mai G, Tokuyasu T, Liu C. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 2018;6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nakatsu G, Zhou H, Wu WKK, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 2018;155:529–541.e5. [DOI] [PubMed] [Google Scholar]
- 67. Kim MS, Park EJ, Roh SW, Bae JW. Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol 2011;77:8062–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shkoporov AN, Hill C. Bacteriophages of the human gut: the “Known Unknown” of the microbiome. Cell Host Microbe 2019;25:195–209. [DOI] [PubMed] [Google Scholar]
- 69. Lepage P, Colombet J, Marteau P, Sime-Ngando T, Doré J, Leclerc M. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 2008;57:424–5. [DOI] [PubMed] [Google Scholar]
- 70. Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006;55:205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ott SJ, Musfeldt M, Wenderoth DF, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004;53:685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pérez-Brocal V, García-López R, Nos P, Beltrán B, Moret I, Moya A. Metagenomic analysis of Crohn’s disease patients identifies changes in the virome and microbiome related to disease status and therapy, and detects potential interactions and biomarkers. Inflamm Bowel Dis 2015;21:2515–32. [DOI] [PubMed] [Google Scholar]
- 73. Clooney AG, Sutton TDS, Shkoporov AN, et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 2019;26:764–778.e5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.