Abstract
Background
Liver fibrosis is a consequence of chronic inflammation and is associated with protein changes within the hepatocytes structure. In this study, we aimed to investigate if this is reflected by the urinary proteome and can be explored to diagnose liver fibrosis in patients with chronic liver disease.
Methods
In a multicentre combined cross-sectional and prospective diagnostic test validation study, 129 patients with varying degrees of liver fibrosis and 223 controls without liver fibrosis were recruited. Additionally, 41 patients with no liver, but kidney fibrosis were included to evaluate interference with expressions of kidney fibrosis. Urinary low molecular weight proteome was analysed by capillary electrophoresis coupled to mass spectrometry (CE-MS) and a support vector machine marker model was established by integration of peptide markers for liver fibrosis.
Findings
CE-MS enabled identification of 50 urinary peptides associated with liver fibrosis. When combined into a classifier, LivFib-50, it separated patients with liver fibrosis (N = 31) from non-liver disease controls (N = 123) in cross-sectional diagnostic phase II evaluation with an area under the curve (AUC) of 0.94 (95% confidence intervals (CI): 0.89–0.97, p<0.0001). When adjusted for age, LivFib-50 demonstrated an AUC of 0.94 (95% CI: 0.89–0.97, p<0.0001) in chronic liver disease patients with (N = 19) or without (N = 17) liver fibrosis progression. In this prospective diagnostic phase III validation set, age-adjusted LivFib-50 showed 84.2% sensitivity (95% CI: 60.4–96.6) and 82.4% specificity (95% CI: 56.6–96.2) for detection of liver fibrosis. The sequence-identified peptides are mainly fragments of collagen chains, uromodulin and Na/K-transporting ATPase subunit γ. We also identified ten putative proteolytic cleavage sites, eight were specific for matrix metallopeptidases and two for cathepsins.
Interpretation
In liver fibrosis, urinary peptides profiling offers potential diagnostic markers and leads to discovery of proteolytic sites that could be targets for developing anti-fibrotic therapy.
Keywords: Liver fibrosis, Capillary electrophoresis mass spectrometry, Urinary peptide marker, Diagnosis
1. Introduction
Liver fibrosis is the replacementof healthy liver tissue with a scarred and fibrotic tissue. It is caused by a variety of hepatic insults leading to hepatocellular death, followed by hepatic tissue degeneration and regeneration [1]. In developed countries, the common causes of liver fibrosis include alcohol abuse, hepatitis C virus (HCV) infection and non-alcoholic steatohepatitis (NASH).
Research in context.
Evidence before this study
Liver fibrosis develops slowly as a consequence of progressive chronic inflammation, it has clinical and life-threatening implications to the patients affected. It is usually associated with changes of proteins in the liver. Liver fibrosis can be silent in patients and is often diagnosed late. Noninvasive diagnosis of liver fibrosis in usual practice is still lacking, With the current definitive tool being an invasive histological assessment via a liver biopsy.
Added value of this study
In this study, the application of capillary electrophoresis mass spectrometry enabled identification of 50 urinary peptides significantly associated with liver fibrosis. Urinary peptides in liver fibrosis were mainly fragments of collagen chains. Other discovered urinary peptides included the homeostasis markers such as uromodulin and Na/K-transporting ATPase subunit γ. These discovered peptides offer potential for diagnosis of liver fibrosis using urine samples and enhance our understanding about the pathogenesis of liver fibrosis and possible relevant treatment options.
Implications of all the available evidence
Noninvasive urinary detection of liver fibrosis is an attractive tool for diagnosing and/or screening patients at risk for liver fibrosis in the primary care. Additionally, this study identified eight proteolytic cleavage sites specific for matrix metallopeptidases and two cathepsins which are responsible for degrading extracellular matrix proteins. These sites could be potential therapeutic targets for antifibrotic therapy.
Alt-text: Unlabelled box
NASH is the severe form of non-alcoholic fatty liver disease (NAFLD), for which patients with obesity, type 2 diabetes mellitus, hyperlipidaemia and hypertension are predisposed [2]. Advanced liver fibrosis may progressively result in liver cirrhosis (LC) and chronic liver failure, and often requires liver transplantation as the last curative treatment option.
Diagnosis and staging of liver fibrosis rely on histological assessment of liver biopsies as the gold standard [3], [4], [5]. However, liver biopsy is associated with discomfort for the patient and risk of complications requiring post-biopsy care (e.g. intraperitoneal bleeding, visceral perforation, and infections) or even death in rare cases [3]. Histological grading has further limitations, including variability due to sampling (sampling error) and the experience of the investigator [3], [4], [5].
Other diagnostic modalities used with increasing frequency in clinical practice are vibration-controlled transient ultrasound elastography (Fibroscan® device; Echosens, Paris, France) which measures liver stiffness [5], [6], [7]. This approach has limitations since it does not provide information about current fibrotic activity, whilst there are also financial barriers to its use, as it carries a substantial initial cost (about €100,000) and recurrent (for consumables and specialists time) costs[7,8]. Additional tools used to diagnose advanced liver fibrosis include surrogate markers related to liver function like AST/ALT ratio and NAFLD fibrosis score. These markers appear to be only useful in advanced liver fibrosis with poor performance at early stages [9]. Recent research has focused not only on imaging tools, but also on non-invasive biomarkers to determine the stage of liver injury/fibrosis [[4], [5], [6],10]. These efforts have led to the identification of a plethora of possible serum biomarker candidates [[4], [5], [6],[11], [12], [13]]. Shortcomings unfortunately are common in these studies; several of these involved a small number of patients and samples, thus potentially suffering from clinical and therapeutic confounders, while they also have not been decisively confirmed in independent and representative prospective patient cohorts.
Morphologically liver fibrosis is characterized by an excessive deposition of collagen-rich extracellular matrix (ECM) components in the liver, which is mainly caused by the trans-differentiation of hepatic stellate cells into collagen-producing cells and reduced degradation of collagen fibrils [1,14]. Recent studies in the context of chronic kidney disease and heart failure, we could demonstrate the significant association of specific urinary collagen peptides with disease, and with fibrosis [15,16]. Based on these findings, we hypothesized that specific urinary collagen-derived peptides may be valuable in the detection of liver fibrosis. The hypothesis was tested in this study, aimed at finding urinary peptides associated with liver fibrosis, and in a second step combining these in a classifier that enables non-invasive identification of patients with advanced fibrosis, which can be silent with patients presenting late with complications. Urine is easily obtained and can be applied in primary care to discover unidentified cases.
Capillary electrophoresis coupled to mass spectrometry (CE-MS) has emerged in recent years as a hybrid technology using capillary electrophoresis (CE) instead of liquid chromatography for sensitive (1 fmol) and high-resolution low molecular weight protein and peptide separation before mass spectrometry (MS) [17]. Notably, this method enables profiling of the urinary proteomic content in a mass range of 0.8 to 20 kilodalton (kDa) [17]. In addition, a database containing currently more than 70,000 CE-MS datasets is available for further in silico validation of findings [18].
In this study, we used CE-MS to search in the low molecular weight proteome of urine from patients with NAFLD, NASH, LC and hepatocellular carcinoma (HCC) for peptide markers sensitive for liver fibrosis, and tested them for their use in staging and risk stratification of liver fibrosis. The aim of this study was to explore value diagnostic utility of urinary peptides in patients with liver fibrosis. To reach this aim, our study design followed not a simple discovery and validation approach, instead different steps of biomarker selection were applied to ensure accurate diagnosis of liver fibrosis in patients with liver disease. Also, it was based on two independent validation cohorts, a cross-sectional cohort of patients with a long history of liver fibrosis and a prospective cohort of liver disease patients under continuous surveillance subdivided into groups with and without clinical signs of liver fibrosis. The cross-sectional cohort was used for performance evaluation and exploratory confounder analysis, whereas the prospective cohort was used to evaluate clinical applicability of the proteomic model and to determine important test characteristics in an unbiased fashion in advance to the planning of further prospective clinical studies.
2. Methods
2.1. Ethics
Participants in the study were recruited between 2014 and 2019 from both University Hospital Coventry and Warwickshire, UK and Hannover Medical School, Germany. In the UK, the study was approved by both the Coventry and Warwickshire and North East-York Research National Health Service Ethics Committees (Reference numbers 09/H1211/38 and 19/NE/0213). In Germany, the study was approved by the Ethics Committee of the Medical School Hannover (Reference number: 901). Study conformed to the World Medical Association Declaration of Helsinki, with all study participants providing written informed consent.
2.2. Study design
In the discovery phase, we prospectively recruited: (i) 19 patients with NAFLD; (ii) 9 patients with NASH, but without LC manifestations; (iii) 13 patients with NASH and well established LC; (iv) 19 patients with histologically confirmed cirrhosis not related to NASH, mainly of viral or alcoholic origin; and (v) 19 patients with HCC and LC.
Differential diagnosis of these patients was established by a combination of liver ultrasound, Fibroscan and laboratory markers (e.g. AST/PLT-ratio). Liver histology assessment was performed when there was uncertainty of liver fibrosis. In order to reduce confounding factors, we only included stable cirrhotic patients from the liver outpatient clinics; none of whom had recently or were currently receiving a course of antibiotics. Patients with type 2 diabetes or other gastrointestinal conditions (e.g. inflammatory bowel disease or coeliac disease) were excluded.
Eighty-one normal individuals adjusted for age, gender and renal function and without clinical or biochemical evidence of liver disease served as controls. These were selected from epidemiological study populations ensuring absence of diabetes, heart disease, hypertension, hyperlipidaemia and obesity. Composition of the liver disease case and normal control groups together with the applied exclusion criteria and adjusted demographic variables are presented in Table 1.
Table 1.
Selected clinical and demographic characteristics of recruited study participants.
| Patient group | Discovery |
Validation Cross-sectional |
Validation Prospective |
Interference Testing | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Study phase | Normal controls (Age/Gender-adj.) |
Liver disease w/ fibrosis | P* | Normal controls | Liver disease w/ fibrosis |
P* | Liver disease w/o fibrosis |
Liver disease w/ fibrosis |
P* | Kidney fibrosis |
| No patients, n | 81 | 79 | — | 123 | 31 | — | 17 | 19 | — | 41 |
|
Age mean (range), y |
54 (21–76) |
56 (18–79) |
0.17 | 39 (20–77) |
56 (33–82) |
<0.0001 | 42 (23–84) |
64 (31–86) |
0.001 | 64 (18–85) |
|
Gender female/male, n |
30/51 | 34/45 | 0.52 | 62/61 | 5/26 | 0.0005 | 3/14 | 5/14 | 0.70 | 20/21 |
|
No peptides mean (range), 1 × 103 |
1.6 (0.9–3.2) |
2.3 (0.6–4.0) |
<0.0001 | 1.5 (0.8–2.7) |
2.1 (0.8–4.1) |
<0.0001 | 3.1 (1.5–4.5) |
2.9 (1.1–4.6) |
0.43 | 2.5 (1.2–4.5) |
|
WBC mean (range), 1 × 103/µL |
— | 5 (1–13) |
— | — | 7 (2–18) |
— | 6 (4–9) |
7 (2–11) |
0.49 | — |
|
Hb mean (range), g/dL |
— | 13 (8–18) |
— | — | 12 (10–16) |
— | 14 (11–16) |
13 (9–16) |
0.06 | — |
|
Plt mean (range), 1 × 109/L |
236 (126–326) |
145 (25–402) |
<0.0001 | 238 (130–490) |
187 (51–384) |
0.017 | 261 (152–470) |
177 (69–568) |
0.05 | — |
|
INR mean (range) |
— | 1.2 (0.9–2.0) |
— | — | 1.2 (1.0–1.7) |
— | 1.0 (0.9–1.2) |
1.1 (1.0–1.4) |
0.002 | — |
|
TC mean (range), mg/dL |
207 (122–312) |
329 (93–690) |
0.0007 | 197 (119–305) |
185 (77–321) |
0.61 | 197 (155–232) |
378 (197–540) |
0.0003 | 272 (96–561) |
|
HDL mean (range), mg/dL |
56 (27–105) |
121 (9–340) |
<0.0001 | 57 (34–97) |
36 (27–54) |
0.003 | 49 (31–70) |
141 (70–350) |
0.0002 | — |
|
TG mean (range), mg/dL |
— | 130 (46–400) |
— | — | 122 (48–294) |
— | 266 (80–549) |
149 (55–610) |
0.12 | — |
|
AST mean (range), U/L |
21 (13–65) |
50 (13–284) |
<0.0001 | 21 (12–46) |
77 (20–180) |
<0.0001 | 44 (16–90) |
59 (20–152) |
0.40 | — |
|
ALT mean (range), U/L |
— | 48 (11–288) |
— | — | 115 (22–1038) |
— | 45 (5–246) |
36 (15–72) |
0.33 | — |
|
ALP mean (range), U/L |
— | 128 (44–797) |
— | — | 278 (71–693) |
— | 88 (45–358) |
179 (44–516) |
0.0006 | 68 (19–174) |
|
GGT mean (range), U/L |
— | 142 (15–719) |
— | — | 335 (49–1269) |
— | 133 (17–374) |
279 (22–682) |
0.11 | — |
|
Bilirubin mean (range), µmol/L |
— | 25 (4–163) |
— | — | 48 (10–210) |
— | 8 (3–14) |
22 (4–65) |
0.003 | — |
|
Albumin mean (range), g/L |
— | 40 (26–49) |
— | — | 32 (16–57) |
— | 46 (40–51) |
38 (23–52) |
0.001 | — |
|
AFP mean (range), µg/L |
— | 429 (1–22,826) |
— | — | 171 (1–1493) |
— | 17,330 (3–103,965) |
1414 (1–24,400) |
0.36 | — |
Difference between liver fibrosis cases and non-liver fibrosis controls included in the discovery, cross-sectional and prospective validation cohorts by two-tailed probability for continuous data and significance level by Fisher exact test for categorical data.
Diagnosis was established by a combination of liver ultrasound, Fibroscan, laboratory markers, e.g. AST/PLT-ratio, and histology. Patients with type 2 diabetes or other gastrointestinal conditions (e.g. inflammatory bowel disease or coeliac disease) were excluded. Controls were adjusted for age, gender and renal function and without clinical or biochemical evidence of liver disease served as controls. We ensured absence of diabetes, heart disease, hypertension, hyperlipidemia and obesity.
Abbreviations: AFP, alpha-fetoprotein; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma-glutamyltransferase; Hb, hemoglobin; HDL, high density lipoprotein; INR, International Normalized Ratio; Plt, platelets; TC, total cholesterol; TG, triglycerides; WBC, white blood cells.
In the validation phase, we recruited 123 normal control subjects without evidence of liver disease and 19 patients with non-fibrotic liver diseases of various aetiologies including two with NAFLD, three with NASH and three with HCC as non-LC control groups. Fifty patients with biopsy-proven LC of whom 11 had concomitant HCC, 13 benign biliary disorders, 9 chronic pancreatitis, 4 cholangiocarcinoma and 4 pancreatic cancer served as LC case group. Thirty-one of these patients were selected cross-sectionally based on their long-term clinical course and were used as an initial patient set for performance evaluation of the LivFib-50 classification model followed by an additional confounder analysis. The other 19 patients of this liver fibrosis case group with much more diversified LC aetiologies and LC disease expressions were recently enrolled and used for final prospective and unbiased (by correction of confounders) evaluation of LivFib-50 classification values. Finally, a set of 41 patients with no liver, but kidney fibrosis was included to evaluate potential interference of the liver fibrosis-specific urinary peptide profile with expressions of kidney fibrosis.
2.3. Sample preparation
Urine was collected from all study participants in standard universal specimen containers (Newport, UK) and frozen to −80 °C, after collection, for subsequent batch analysis. For proteomic analysis, samples were prepared as previously described [19]. In brief, a 0.7 mL aliquot was thawed immediately before use and diluted with 0.7 mL 2 M urea, 10 mM NH4OH containing 0.02% Sodium Dodecyl Sulfate. To remove proteins of higher molecular mass (e.g. albumin and immunoglobulin G) the sample was filtered using a Centrisart ultracentrifugation filter device (20 kDa molecular weight cut-off; Sartorius, Goettingen, Germany) at 3000 rcf until 1.1 ml filtrate was obtained.
Subsequently, the filtrate was loaded onto a PD-10 desalting column (GE Healthcare, München, Germany), and equilibrated in 0.01% NH4OH in HPLC-grade H2O (Roth, Karlsruhe, Germany) in order to decrease matrix effects by removing urea, electrolytes, and salts, and also to enrich polypeptides. Finally, all samples were lyophilized, stored at 4 °C, and resuspended in HPLC-grade H2O shortly before CE-MS analysis.
2.4. CE-MS analysis and data processing
CE-MS analysis was performed using a P/ACE MDQ capillary electrophoresis system (Beckman Coulter, Fullerton, USA) on-line coupled to a Micro Time-of-Flight MS (Bruker Daltonic, Bremen, Germany) [19]. The ESI sprayer (Agilent Technologies, Palo Alto, USA) was grounded, and the ion spray interface potential was set between −4.0 and −4.5 kV. Data acquisition and MS acquisition methods were automatically controlled by the CE via contact-close-relays. Spectra were accumulated every 3 s over a range of m/z 350 to 3000. Details on accuracy, precision, selectivity, sensitivity, stability, and reproducibility of the CE-MS method have been established [19].
Mass spectral ion peaks, representing identical molecules at different charge states, were deconvoluted into single masses using MosaiquesVisu software [19]. For noise filtering, signals with z > 1 observed in a minimum of 3 consecutive spectra with a signal-to-noise ratio of at least 4 were considered. MosaiquesVisu employs a probabilistic clustering algorithm and uses both isotopic distribution (for z ≤ 6) and conjugated masses for charge-state determination of peptides/proteins.
The resulting peak list characterizes each polypeptide by its mass and its migration time. Time-of-flight-MS data were calibrated utilizing 150 reference mass data points and 452 reference migration time data points by applying global and local linear regression, respectively. Ion signal intensity (amplitude) showed variability, mostly due to different amounts of salt and peptides in the sample and were normalized. Reference signals of 29 highly abundant peptides were used as “internal standard” peptides for calibration using local linear regression. This procedure was shown to be an easy and reliable method to address both analytical and dilution variances in a single calibration step [20]. The obtained peak list characterizes each polypeptide by its calibrated molecular mass [Da], calibrated CE migration time [min] and normalized signal intensity.
All detected peptides were deposited, matched, and annotated in a Microsoft SQL database allowing further statistical analysis. For clustering, peptides in different samples were considered identical, if mass deviation was <50 parts per million for small (<4000 Da) or 75 parts per million for larger peptides. Due to analyte diffusion effects, CE peak widths increase with CE migration time. For data clustering, this effect was considered by linearly increasing cluster widths over the entire electropherogram (19 min to 45 min) from 2 to 5%.
2.5. Peptide sequencing
Peptide sequencing was carried out both on a Dionex Ultimate 3000 RSLS nanoflow system (Dionex, Camberly, UK) and a Beckman CE/Orbitrap Q Exactive plus combination (Thermo Scientific, Waltham, MA) [21]. Spectra files were analyzed with Proteome Discoverer 2.4 allowing a precursor mass tolerance of 5 ppm and a fragment mass tolerance of 0.05 Da. This was followed by a search against the UniProt human non-redundant database without any protease specificity or fixed modification. Oxidation of methionine and proline were considered as variable modifications. Only sequences with high confidence (Xcorr ≥ 1.9) and without unmodified cysteine were accepted (due to the application of non-reducing conditions). A strong correlation between peptide charge at the CE operating pH of 2 deduced from the number of basic amino acids in the annotated peptide sequence and the migration time was used as another criterion to prevent false sequence assignments [22].
2.6. Protease assessment
In silico protease assessment was performed using Proteasix (www.proteasix.org), a web-based tool for investigation of proteolytic events involved in naturally occurring peptide generation [23]. Observed specific proteases responsible for cleavage of N- or C- terminus of a peptide were retrieved from CutDB proteolytic event database available at www.cutdb.burnham.org [24]. Protease activity was assessed in the patient's CE-MS peptide profiles to gain fold changes between LC cases versus normal controls based on the average of associated peptide intensities. This method is described in detail by Voigtländer et al. [25].
2.7. Tissue transcriptomic data analysis
Tissue transcriptomics data were retrieved from the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and from the EMBL-EBI Expression Atlas (https://www.ebi.ac.uk/gxa/home). This includes expression profiling data by array from 1) 22 patients with cirrhosis (LC) and 14 histologically normal livers (GEO accession: GSE77627), 2) four biological replicates of histologically normal kidney and three biological replicates of histologically normal liver of human origin (E-MTAB-2836, [26]), as well as 3) four biological replicates of histologically normal kidney and four biological replicates of histologically normal liver of human origin (E-MTAB-5782, [27]). The GEO data sets were analysed with GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). Analysis was conducted using default settings, with false discovery rate adjustment of p-values based on the method by Benjamini & Hochberg [28]. For the presentation of E-MTAB data, values are expressed in transcripts per million (TPM) units.
2.8. Statistical analysis
Clinical and demographic data, as well as peptide distributions in the samples of the patients included in the different patient subgroups are presented either as mean ± standard deviation or as percentage. For intergroup comparison, continuous data were compared by Student's t-test or Mann Whitney rank-sum test, and categorical data by Chi square or Fisher's exact test after testing for normal distribution. A two-sided p value < 0.05 was considered to be statistically significant.
For peptide marker identification in the discovery phase of the study, the distribution differences of the urinary peptides between the patients with liver disease and normal controls were based on natural logarithm transformed intensities and a non-parametric Wilcoxon rank sum test. For correction of p-values due to multiple testing the method by Benjamini and Hochberg was used [28].
Receiver-operating-characteristic (ROC) curves were generated for the classification of the patient samples with the LivFib-50 classifier. The ROC curve was obtained by plotting all sensitivity values (true positive fraction) on the y axis against their equivalent (1-specificity) values (false positive fraction) on the x axis for all available thresholds. The area under the ROC curve (AUC) was evaluated as it provides a single measure of overall accuracy independent of any threshold. Calculation of 95% confidence intervals (Cl) was based on exact binomial calculations and the optimal balance of sensitivity and specificity was determined based on the Youden index.
All statistical analyses were carried out using MedCalc version 12.7.5.0 (MedCalc Software; Mariakerke, Belgium).
2.9. Role of funding source
This manuscript was supported by research grant from the medical and life sciences research fund (MLSRF) which was awarded to AB to help with research consumables. It is charity that supports research and education to enhance human health and had no role in study design, data collection, data analyses, interpretation, or writing of report.
3. Results
3.1. Liver fibrosis peptide biomarker identification and support vector machine model establishment
For the identification of urinary peptide markers associated with fibrosis, we investigated the low molecular weight urine proteome from 79 patients with chronic liver diseases and 81 age and gender-matched normal subjects by CE-MS. Age- and gender-adjustment of controls was implemented in the study design of the discovery phase to ensure that only those peptides that are unaffected by age or gender were selected as marker for liver fibrosis.
For age and gender adjustment, 81 normal controls were selected from a pool of 204 normal controls by using the R-script “MatchIt” [29], which conducts nearest neighbour matching with logistic regression used to estimate the propensity score. The age of the 81 selected normal controls was subsequently compared with that of the 79 liver fibrosis case patients by a Mann-Whitney rank sum test and resulted in a p-value of 0.18. The difference in gender was proven to be statistically insignificant (p = 0.63) by a Chi-squared test. For renal function it was verified that all patients have no renal dysfunction at the date of urine collection as indicated by an eGFR-value above 60 mL/min/1.73m2.
We specifically searched for those fibrosis markers showing a gradual increase or decrease in their CE-MS-detected amplitude signals from normal over non-fibrotic NAFLD and NASH to LC. For this purpose, we followed a three-step strategy which is schematically presented in Fig. 1.
Fig. 1.
Study flow chart. A total of 391 patients was included in the discovery and validation phases of the LivFib-50 peptide marker model for the detection of progressive liver fibrosis. Biomarker selection was carried out in three sequential steps resulting in 50 urinary peptides with differential and graded expression ranging from disease-free normal individuals. over NAFLD and NASH patients without LC to patients with well-established LC. Combining the peptide markers to the LivFib-50 model was followed by a first evaluation of the model's classification performance and confounder analysis in patients with LC and normal controls. Since the classification factors significantly correlate with the age of the patients, the LivFib-50 classification model was adjusted for age on these patient groups by logistic regression. In a final validation phase the age-adjusted LivFib-50 classification model was validated in an independent group of patients with liver disease, with or without LC and interference of classification was tested in a set of patients with renal fibrosis, but no liver fibrosis.
In the first selection step, we statistically compared the CE-MS peptide profiles of the patients with liver diseases to those of the normal control subjects by a group-wise non-parametric Wilcoxon rank sum test. From a total of 21,559 annotated peptides in the mass range of 0.8 to 20 kDa, 854 peptides were identified with a frequency threshold of 30% and p-values below 0.05. Out of these, 732 remained significant after adjustment of the false discovery rate for multiple testing by the Benjamini and Hochberg procedure [28]. The list of peptides displaying differences in abundance between liver disease and healthy control as generated in this first selection step is presented in Supplementary Table 1 together with their CE-MS properties and statistical characteristics. The amino acid sequence for those of these peptides for which sequences could be assigned with high confidence is also presented in Supplementary Table 1.
In the second selection step, we correlated the abundance of the 732 preselected peptides with a severity score of liver disease, assigning 0 for normal controls, 1 for patients with NAFLD, 2 for patients with NASH (both without signs of liver fibrosis), and 3 for patients with LC to select peptides that appear associated with progression of liver fibrosis. By selecting a threshold above 0.2 or below −0.2 for Spearman rho correlation (FDR-adjusted p-values < 0.05), 500 out of the 732 peptides selected in step 1 fulfilled also the selection criteria from step 2. Out of the selected peptides, 198 showed negative correlation and 302 were positively correlated to the severity scores of the liver insult. The Spearman rho correlation factors of the 500 peptides passing selection step 2 together with their FDR-adjusted p-values are also presented in Supplementary Table 1. In this way, selection step 2 was used as a first step to identify those liver disease-associated peptides that show a progression in their abundance depending on the presence of liver fibrosis.
In the final biomarker selection step, we searched the list of the preselected 500 peptides for those fibrosis associated markers showing a strictly linear decrease or increase from normal controls over the NAFLD and NASH patients to the LC-positive patients when applying a Kruskal-Wallis rank sum test. This last, very stringent selection step reduced the number of fibrosis grading markers to 50 peptides. The final list of fibrosis markers together with their statistical characteristics of all three selection steps is presented in Supplementary Table 2.
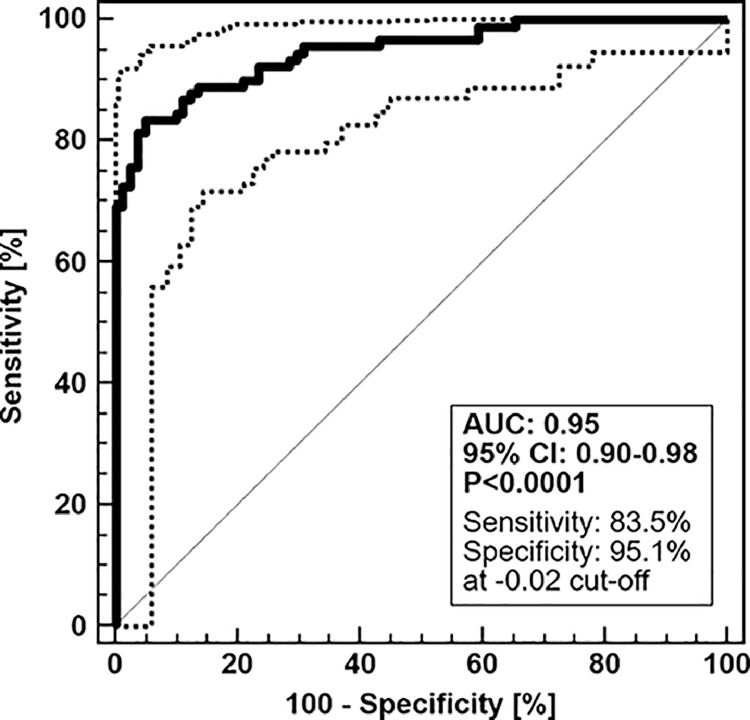
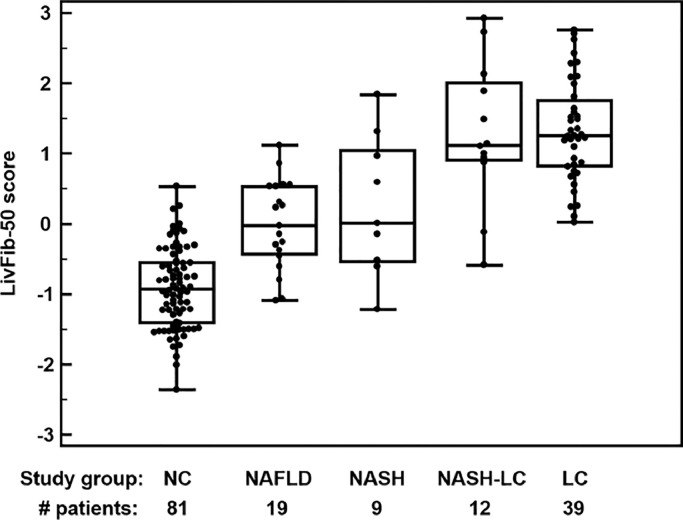
These 50 liver fibrosis peptide markers were combined into a support vector machine (SVM)-based peptide multi-marker panel for non-invasive detection of liver fibrosis. The peptide marker model was trained by SVM using a Radial Basis Function (RBF) kernel using a cost C value of 9.7 and a gamma value of 0.00073 as penalty settings. In combination, these two parameters control the trade-off between allowing training errors and forcing rigid margins. After optimization of the SVM parameters, the peptide marker pattern designated as LivFib-50 including the 50 peptides resulted in an AUC of 0.95 and 95% (CI) in the range of 0.90 to 0.98 (p<0.0001) on the 19 NAFLD, 9 NASH and 51 LC (out of which 12 are NASH-LC) patients and 81 normal control subjects in receiver operating characteristics analysis (ROC) after take-one-out total cross-validation. The ROC curve and a table of ROC characteristics are presented in Fig. 2A. As presented in the group-specific Box-and-Whisker distribution plot of Fig. 2B, the established peptide marker pattern not only allowed differentiation of the normal controls from NAFLD, NASH and LC, but also the classification scores for NAFLD and NASH-LC are significantly different, with higher values in the case of LC.
Fig. 2A.
ROC curve and ROC characteristics for the discovery set. Patients with NAFLD, NASH, LC or HCC were treated as case group (N = 79) and were compared to non-diseased age- and gender-matched normal controls (N = 81). Bootstrapping of the classification results was performed by leave one out total cross-validation. Dotted lines represent the upper and lower bounds of the confidence interval.
Fig. 2B.
Box and Whisker distribution plots for classification of the different patient groups of the discovery set with the LivFib-50 model. A Kruskal-Wallis test was performed for rank sum differences in the LivFib-50 classification scores and revealed significant differences in post-hoc analysis between normal controls to all liver diseased patient groups (p < 0.0001) as well as between patients with combined NASH and LC (NASH-LC) compared to NASH without concomitant LC (p = 0.04).
3.2. Adjustment of the LivFib-50 peptide marker model for patient age
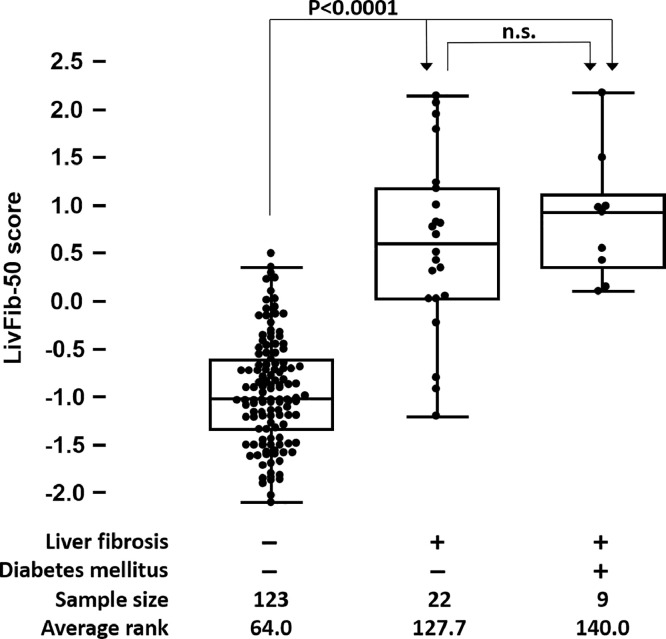
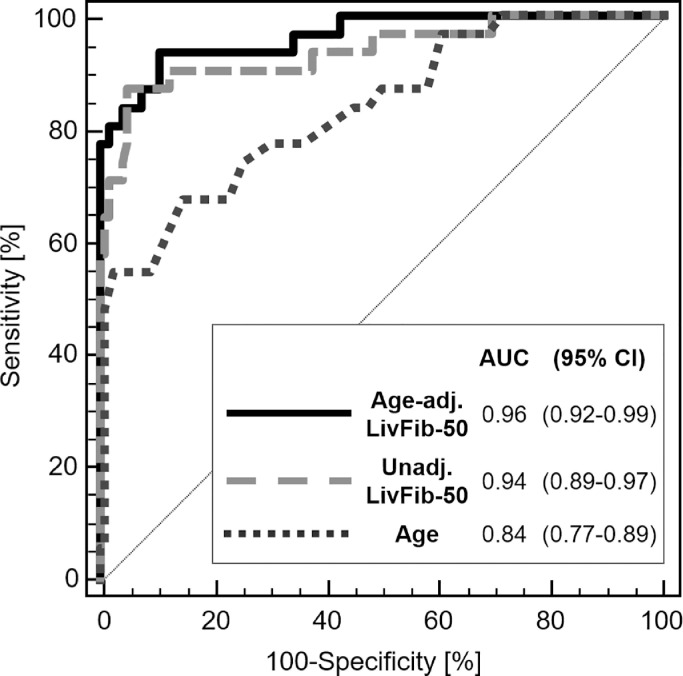
In a first validation phase, LivFib-50 was tested for generalizability on independent cross-sectional datasets of 31 patients with LC and 123 normal controls. ROC analysis revealed an AUC of 0.94 (95% CI: 0.89–0.97, p<0.0001) in this diagnostic phase II testing. Besides performance evaluation, correlation of LivFib-50 classification values to relevant clinical and demographic patient characteristics was done in this first validation cohort and not in the training cohort since the former represents an independent patient group free of any overfitting bias and thus without the need for any bootstrapping. Confounder analysis revealed correlation of LivFib-50 classification to age (r = 0.43, p < 0.0001) as the only confounder with impact on the LivFib-50 classification result. Most importantly and with the results shown in Fig. 3A, a potential impact of diabetes mellitus (DM) was excluded by dividing the 31 patients with LC into those with (N = 9) and without (N = 22) DM and by performing a global Kruskal Wallis analysis also including the 123 patients without signs of liver disease. As a result of this exploratory confounder analysis on this first validation set, an age-adjusted LivFib-50 model was constructed by logistic regression of the LivFib-50 classification values and age on the basis of their odds ratios of 27.54 (95% CI: 6.87–110.33) and 1.16 (95% CI: 1.06–1.27) resulting in a logit function for an age-adjusted LivFib-50 score of −7.37 + 3.32 ⨯ LivFib-50 classification factor + 0.15 ⨯ patient age. As presented in Fig. 3B, the age-adjusted LivFib-50 classification model significantly improved classification performance relative to the LivFib-50 peptide marker model alone by an AUC increase of 0.027 (AUC = 0.96, p = 0.044) on the 31 LC cases in comparison to the 123 normal controls.
Fig. 3A.
Distribution of classification scores of the LivFib-50 marker pattern in normal liver and liver fibrosis groups of the first validation set of patients. The liver fibrosis group (N = 31) was further divided into those with (N = 9) or without (N = 22) concomitant diabetes mellitus in order to evaluate the impact of diabetes mellitus on the LivFib-50 classification results. A post hoc test was performed for average rank differences between the three different subgroups (each with p< 0.05) after a significant result in the global Kruskal-Wallis test. The abbreviation n.s. indicates a non-significant result.
Fig. 3B.
Classification of normal controls without clinical signs of liver disease (NC, N = 123) and those with clinical manifestations of liver cirrhosis (LC, N = 31) with the age-adjusted LivFib-50 peptide marker model in comparison to the proteomic model without age adjustment and age alone. Age adjustment of the LivFib-50 peptide marker model was performed using logistic regression. Significance P values for each ROC curve were determined to be <0.0001.
3.3. Validation of the age-adjusted LivFib-50 classification model
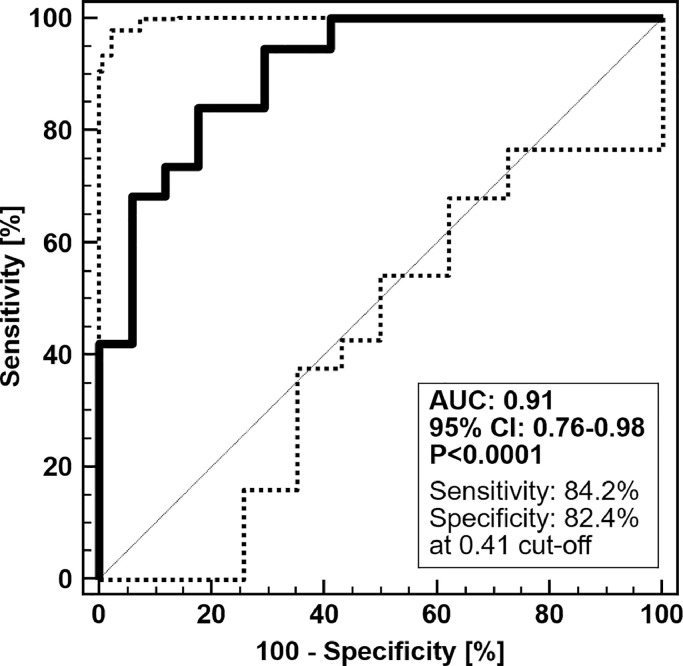
For validation in a clinical setting of intended use, the age-adjusted LivFib-50 model was tested in an independent set of 36 prospectively collected chronic liver disease patients, 19 with and 17 without LC. This second validation set is essential for the diagnostic test validation process in order to compensate for potential classification bias by the fitted logistic regression model algorithm. As demonstrated by an AUC of 0.91 and a 95% CI range of 0.76 to 0.98 at a significance level of p<0.0001 (Fig. 4A), patients with LC are clearly distinguishable in their LivFib-50 model scores from patients without LC. Without age-adjustment the LivFib-50 model resulted in an AUC of 0.87 (95% CI: 0.71–0.96, p<0.0001) on this second validation set (data not shown). The positive (PPV) and negative (NPV) predictive values were determined to be 67% (95% CI: 42–85%) and 92% (95% CI: 81–97%), respectively. Of note, we corrected the PPV and NPV for a 30% prevalence of liver fibrosis in chronic liver disease patients, since our prospective validation cohort does not fully reflect the reported value for this patient collective [30].
Fig. 4A.
ROC curve and ROC characteristics of the age-adjusted LivFib-50 peptide marker model for patients with LC in the absence or presence of HCC (N = 19) compared to non-fibrotic control patients (N = 17) in independent validation. Of note, the three HCC patients without LC manifestations were treated as controls.
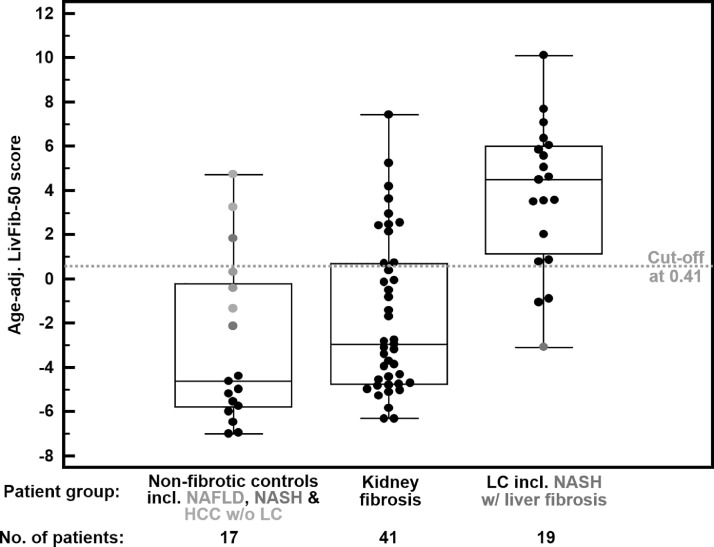
Applying the optimal classification threshold based on the Youden index at 0.41, classification of this validation cohort resulted in a sensitivity of 84.2% (95% CI: 60.4–96.6) and a specificity of 82.4% (95% CI: 56.6–96.2). As revealed by the Box-and-Whisker distribution plot of LibFib-50 classification values in Fig. 4B, there is a clear separation between the fibrotic and non-fibrotic patient groups irrespective of concomitant HCC. A clear distinction from the case group is also observed for patients with no liver, but kidney fibrosis as clearly demonstrated by a specificity of 73% for LivFib-50 positivity in this interference risk group.
Fig. 4B.
Box and Whisker distribution plots for classification of the different patient groups of the validation set with the age-adjusted LivFib-50 classification model. The validation set consists of patients without clinical signs of liver fibrosis (N = 17), patients with kidney fibrosis (N = 41) and patients with LC or fibrosis (N = 19).
3.4. Sequencing of liver fibrosis peptide markers reveal differential urinary excretion of collagen chain fragments
We were able to obtain high confidence amino acid sequence information for 22 of these 50 peptides. All liver fibrosis peptide markers that could be successfully resolved for their amino acid sequence are presented in Table 2. These peptides are mainly fragments of collagen α−1(I) (n = 14) and five other collagen fragments. Of these, 10 showed increased and nine showed reduced excretion during fibrosis. Other peptides are derived from the renal homeostasis proteins such as uromodulin (n = 2) and Na/K-transporting ATPase subunit γ (n = 1), with decreased urinary levels in the former and increased urinary levels in the latter case.
Table 2.
Amino acid alignment of all sequence-identified naturally occurring urinary peptides included in the LivFib-50 peptide marker model due to their graded association with progressive liver fibrosis. Peptide markers are for the most part overlapping fragments derived from the triple helical region of the collagen α−1(I) chain. Opposite regulation of overlapping fragments might be attributed to changes in the activity of extracellular matrix degrading proteases during fibrosis progression.
| Peptide ID† | Sequence‡ | AA§ | Regulation in liver fibrosisǁ | Protein name |
|---|---|---|---|---|
| 6546 | PpGPpGKNGDDGEAGKP | 222–238 | ↓ | Collagen α−1(I) chain (COL1A1) |
| 9627 | PpGPpGKNGDDGEAGKpGRp | 222–241 | ↓ | |
| 13,021 | LDGAKGDAGpAGpKGEpGSpGENGApG | 273–299 | ↑ | |
| 5810 | PpGEAGKpGEQGVpGD | 651–666 | ↑ | |
| 3793 | GEAGKPGEQGVPGD | 653–666 | ↓ | |
| 8462 | GANGApGNDGAKGDAGApGApG | 698–719 | ↓ | |
| 4419 | ApGDRGEpGPpGPAG | 798–812 | ↓ | |
| 2136 | GDRGEpGPpGPA | 800–811 | ↓ | |
| 14,801 | GPpGADGQPGAKGEpGDAGAKGDAGpPGPAGP | 815–846 | ↓ | |
| 11,753 | GADGQpGAKGEPGDAGAKGDAGPpGP | 818–843 | ↑ | |
| 13,342 | GADGQpGAKGEpGDAGAKGDAGpPGPAGP | 818–846 | ↑ | |
| 10,953 | DGQpGAKGEpGDAGAKGDAGPpGP | 820–843 | ↑ | |
| 3079 | EKGSpGADGpAGAP | 933–946 | ↑ | |
| 13,730 | AGPpGAPGApGAPGPVGPAGKSGDRGETGP | 1042–1071 | ↑ | |
| 11,744 | LQGLpGTGGPpGENGKpGEpGPKG | 640–663 | ↓ | Collagen α−1(III) chain (COL3A1) |
| 9061 | GApGApGGKGDAGApGERGPpG | 666–687 | ↑ | |
| 16,811 | GERGSpGGpGAAGFpGARGLpGpPGSNGNPGPpGp | 861–895 | ↑ | |
| 13,779 | DDILASPPRLPEPQPYPGAPHHSS | 1534–1557 | ↑ | Collagen α−1(XVIII) chain (COL18A1) |
| 16,419 | EAGRDGNpGNDGPpGRDGQpGHkGERGYPG | 923–952 | ↓ | Collagen α−2(I) chain (COL1A2) |
| 2740 | YLGGSPKGDVDP | 5–16 | ↑ | Na/K-transporting ATPase subunit γ (FXYD2) |
| 5833 | SGSVIDQSRVLNLGP | 589–603 | ↓ | Uromodulin (UMOD) |
| 5241 | DQSRVLNLGPITR | 594–606 | ↓ |
Peptide identification numbers.
Lower case p and k indicates hydroxyproline and hydroxylysine.
Amino acid positions according to UniProt Knowledge Base numbering.
Regulation determined sequentially from normal controls over NAFLD and NASH (all without clinical signs of liver cirrhosis) to clinically well-documented liver cirrhosis by the Kruskal-Wallis rank sum test.
Abbreviations: AA, amino acid sequence; Da, Dalton; MS, mass spectrometry; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis.
Collagen fragments were reported as biomarkers for a variety of other pathologies, especially for association with fibrosis in kidney disease and possibly heart failure. To estimate a potential interference, we have investigated the degree of overlap with the chronic kidney disease-specific classifier CKD273 [31] and the heart failure-specific classifier HF1 [32]. As a result, an overlap of six peptides (12%) was detected for CKD273. Three (6%) thereof had the same direction of regulation compared to controls. In the case of HF1, there was an overlap of five peptides (10%) of which four (8%) showed the same trend. Based on these results, interference of LivFib-50 with CKD273 and HF1 is estimated as low, which will however be further investigated in subsequent studies.
Following the hypothesis that naturally occurring peptides for the most part arise from proteolytical degradation of their parent proteins, we searched for observed protease cleavage sites in the CutDB database on the basis of the octamer amino acid sequence motifs around the N- and C-terminal ends of the 22 sequence-identified fibrosis peptide markers. As presented in Supplementary Table 3, we identified ten putative proteolytic cleavage sites, eight with specificity for matrix metallopeptidases and two for cathepsins. Protease activity was subsequently assessed in the patient's CE-MS peptide profiles based on the average of associated peptide intensities in the 31 LC cases compared to the 123 normal controls of the cross-sectional validation set. This analysis revealed a fold change (FC) in the activity between LC cases and normal controls of 0.86 (p = 0.0005) for MMP2, 0.89 (p = 0.0016) for MMP3, 0.56 (p<0.0001) for MMP13, 0.90 (p = 0.0034) for MMP14 and 0.74 (p<0.0001) for CTSB. All other proteases listed in Supplementary Table 3 were not identified as being significant by this kind of analysis.
Next, we analyzed tissue transcriptomics data of 22 cirrhotic and 14 normal human livers retrieved from the NCBI GEO database (GSE77627) to compare it with our proteomic results. As presented in Supplementary Table 4, we identified significant differences in gene expression levels between cirrhotic and normal tissue for the collagen chains COL1A2 (FC: 0.66, p = 0.04) and COL3A1 (FC: 0.28, p = 5.26E-06), both with a decreased fold change ratio in LC, whereas COL18A1 (FC: 1.49, p = 0.026) showed increased expression in LC. For the protease CTSB a significant decrease in its expression levels was found, with the fold change in LC being 0.45 (p = 4.31E-04). The relevant MMP's demonstrate an increase in expression, as in the case of MMP2 (FC: 3.08, p = 0.002), or no significant expression differences between cirrhotic and normal livers, as in the case of the MMP's 3, 13 and 14. Finally, as shown for two independent transcriptomic data sets in Supplementary Figure 1, expression levels of COL1A1, COL3A1 and COL18A1 are higher, whereas those of COL1A2 are lower in histologically normal livers than histologically normal kidneys of humans.
4. Discussion
This is the first study using CE-MS to identify urinary peptides as biomarkers for liver fibrosis. The findings reported here demonstrate feasibility to identify specific peptide markers significantly associated with liver fibrosis, which, can be combined into a multi-marker panel for accurate detection of liver fibrosis, irrespective of other comorbidities. Our initial hypothesis that the transition from a healthy liver state to NAFLD and further to NASH on the molecular level is mainly characterized by fibrosis which displays in urinary collagen fragments could be verified in this study [1,2,33]. To select specific fibrosis marker(s), our search strategy for urinary biomarkers was not restricted to a statistical group comparison of peptide distributions in patients with NAFLD and NASH compared to normal individuals, but also extended to patients with LC (both alcohol and non-alcohol aetiology). Aside from this group-wise comparison, our fibrosis marker search strategy was further adapted to peptides that showed gradual differences in their excretion profiles between normal liver states, NAFLD, NASH and LC, representing groups with none, mild, moderate to advanced stages of fibrosis. Validation of the findings was performed in a different set of samples, further strengthening the validity of our findings.
Even after applying highly stringent criteria, CE-MS-based urinary proteome analysis, enabled the definition of a set of 50 naturally occurring peptides in the molecular mass range of 0.8 to 20 kDa that showed significant differences in their excretion profiles with high confidence (due to adjustment of p-values by the Benjamini and Hochberg procedure [28]) between normal subjects and patients with NAFLD, NASH or LC. Moreover, these peptides are correlated with the severity of liver injury from normal liver state over NAFLD and NASH to LC, as demonstrated by Spearman Rho correlation factors above 0.2 or below −0.2.
The amino acid sequence could be resolved from 22 of these 50 peptides. Reasons for not retrieving the amino acid sequence for all peptides include the inability to identify all possible post-translational modifications, e.g. due to high glycan heterogeneity [34], and resistance of the peptide to its total fragmentation even at high collision energies in tandem mass spectrometry. This is of no relevance for the diagnostic use of the peptide marker model, as indicated for example by clinical application of a combined bile and urine proteome test for cholangiocarcinoma diagnosis based on similar peptide marker models [35], [36], [37], and is based on the fact that the peptide markers are clearly defined in the CE-MS as the device for operating the diagnostic test by their molecular mass and by their capillary electrophoresis migration time.
In liver fibrosis, normal hepatic tissue is replaced by connective tissue. Morphologically liver fibrosis is characterized by an excessive deposition of collagen-rich ECM components in the liver, which is thought to be mainly caused by the trans-differentiation of hepatic stellate cells into collagen-producing cells and reduced degradation of collagen fibrils [1,14]. Since urine in its low molecular weight protein composition consists to a large degree of collagen fragments, partially arising from glomerular filtration and endosomal-exosomal passage of blood circulating collagen fragments, we hypothesize that progressive fibrosis can be detected by the profiling of the urinary low molecular weight proteome.
A further advantage of the approach chosen is the ability to adjust for confounders based on additional data available from the CE-MS database. As such, we could also test the specificity of the age-adjusted LivFib-50 peptide model for liver fibrosis. When investigating data from patients with chronic kidney disease and kidney fibrosis, the later defined as ≥ 2.5% of fibrosis per total tissue area, 30 out of 41 (73%) of these patients scored negative for liver fibrosis. This is in line with the finding, that no impact of DM on LivFib-50 classification was observed in a global Kruskal-Wallis rank sum test when subdividing the LC patients of our cross-sectional study cohort into those with or without DM.
Not unexpectedly, due to the impact of dysregulated ECM remodeling on the progression of liver fibrosis, sequencing of the peptide markers included in the LivFib-50 classification model revealed a predominant abundance of peptides derived from fibrillar collagen chains, and particularly from collagen α−1(I) (COL1A1) and collagen α−1(III) (COL3A1). Amino acid alignment of the 14 COL1A1 peptides to the linear protein sequence revealed that some of the peptides are overlapping fragments of certain areas within the triple helical COL1A1 region. In respect to the differences in peptide distributions between patients with liver disease and normal controls, even overlapping peptides demonstrate opposite regulation. The restriction of the peptide markers to certain protein regions and the non-directional type of regulation of the COL1A1 peptide markers might be explained by sites of increased proteolytical accessibility and changes in the activity of ECM degrading proteases during fibrosis progression. In this respect, the comparison of the octamer amino acid motifs surrounding the N- and C-terminal ends of the 50 fibrosis peptide markers with already reported proteolytic cleavage sites reported in the CutDB, proteolytic event database[24] was performed and resulted in the identification of ten putative proteolytic cleavage sites, eight specific for matrix metallopeptidases and two for cathepsins [38,39].
Our results on proteases predicted to be responsible for cleavage of our collagen markers are in agreement with the role of matrix metallopeptidases and cathepsins in liver fibrotic disease processes [40,41]. Both are essential together with their tissue inhibitors of metallopeptidases (TIMP) counterparts in maintaining the structure of the hepatic extracellular matrix. Their normal function becomes impaired in the presence of chronic liver inflammation leading to fibrogenesis.
To provide further evidence, we have compared our findings with tissue transcriptomics data of human cirrhotic and normal livers and found evidence for differential regulation of the targeted collagen chains and predicted proteases also in liver tissue. It must be stated, however, that the regulation of protease activity and the degradation of the ECM leading to the release of endogenous collagen peptides into circulation and then into urine is much more complex than simply explained by changes in tissue gene expression levels. To determine the origin of the collagen peptide markers, we compared gene expression levels of their collagen precursors in normal livers versus normal kidneys [26,27] and found higher expression levels of COL1A1, COL3A1 and COL18A1 in the liver than in the kidney. Together with the observation that a significant part of the urinary collagen peptides is derived from extrarenal origin [42], this provides evidence that the identified collagen peptide markers are derived from fibrotic processes in the liver.
Finally, when the activity of the predicted proteases was assessed based on the associated peptide marker expressions in the patient's CE-MS peptide profiles [25], significance could be reached for the MMP's 2, 3, 13 and 14, as well as CTSB. Since for all of these proteases a decrease in activity was predicted in the case of LC, this leads us to the hypothesis based on literature findings [43,44] that mainly TIMP-2 might be the inducer and/or hepatorenal connector leading to the observed changes associated with liver fibrosis in the urinary peptidome.
Recently, diagnostic tests for cardiac fibrosis were proposed on the immunological detection of COL1A1 fragments in human serum. The tests are based on assessment of the carboxy-terminal propeptide (PICP) and the carboxy-terminal telopeptide (CITP) of COL1A1 [45]. Similarly, the LITMUS consortium reported the development of the two diagnostic panels FIBC3 and ABC3D both based on the detection of plasma levels of a PRO-C3 collagen neo-epitope for diagnosis and quantification of advanced liver fibrosis in patients with NAFLD [46]. These recent developments underpin the significance of circulating collagen fragments in non-invasive fibrosis testing. The LivFib-50 peptide marker model reported here is based on simultaneous detection of 50 urinary peptides by mass spectrometry. Such a multiplexed analysis has the advantage over single biomarker detection and it better compensates for biological and analytical variances making the diagnostic test more robust against interferences and variances caused by a complex biological matrix. Mass spectrometry in addition allows direct detection of the analyte in the sample and thus eliminates the need for antigen binding reagents with the potential to introduce bias by antigen cross reactivity.
In addition, the MS-based approach profits from the substantially higher selectivity: each peptide is detected based on its exact mass, while antibodies detect an epitope that may be present in multiple similar peptides, thereby severely compromising selectivity and accuracy. Due to its wide and adjustable measurement range and its independence from a sieving matrix and the requirement for continuous adaptation of electrospray conditions, CE-MS combined with stringent calibration and proteomic data normalization procedures is highly adapted for the use in routine clinical practice.
Over the last years, this was demonstrated in technical reports [19,20] and large-scale prospective and/or longitudinal clinical studies [47], [48], [49], [50], [51]. Recently, the value and applicability of CE-MS in the early detection of chronic kidney disease in a large multicentric randomized controlled trial has been demonstrated. In this respect, our results demonstrate that with further validation in larger prospective studies, urinary peptide detection can extend to screening in the primary care for liver fibrosis, which is lacking at present. This approach also offers insights into the pathophysiology of liver fibrosis and relevant peptidases involved in the hepatic ECM remodeling as well as potential antifibrotic therapeutic interventions in liver fibrosis.
Our study was limited with the variable degrees of liver fibrosis in patients recruited, hence our markers are indicative of presence of liver fibrosis rather than the stage of it. Development of classifiers for staging requires further investigation and validation against different histological grades of liver fibrosis. Nonetheless, these markers relate to changes within an individual patient rather than being a surrogate indication of liver function.
Another limitation of this study is that not all peptides included in the LivFib-50 model could be sequenced. In CE-MS, a peptide is identified simply by its physicochemical characteristics, namely number of positive charges at pH 2 and molecular mass. This is sufficient for its definite detection in a patient sample. Nevertheless, identification of endogenous peptide markers is desirable to understand the pathophysiological context behind their altered expression. Sequence identification is however challenging, most likely due to unknown post-translational modifications or resistance of larger peptides to fragmentation during the MS/MS. As such, sequence information can frequently not be assigned with the high confidence level. In the case where only the 21 sequenced peptides of the LivFib-50 model were used for classification of patients from the first validation set, an AUC of 0.84 (95% CI: 0.78–0.90, p<0.0001) was received under identical SVM settings, which is statistically inferior (p = 0.014 for the difference in areas) to that of 0.94 (95% CI: 0.89–0.97, p<0.0001) received with all LivFib-50 peptide markers. Thus, also the peptides for which the sequence could not be resolved until to date should be kept in the classification model.
Our findings indicate that specific biomarkers of liver fibrosis in the urine of patients with different liver diseases and different grades of liver injury/fibrosis exist. These urinary peptide markers can be combined into a multivariate model with high discriminatory potential. Further validation that qualifies the selected peptide markers for liver fibrosis and thorough evaluation of the established peptide multi-marker model from this initial study appear warranted to assess their exact value in predicting and staging liver fibrosis.
5. Contributors
All authors read and approved the final version of the manuscript. AB was involved in conceptualization, study design, patient recruitment, data curation, project administration and writing the original draft. JM was involved in investigation, methodology, visualization and writing the original draft. IK was involved in data curation and review & editing. TV was involved in conceptualization, data curation and review & editing. TB was involved in data curation and review & editing. JDM was involved in data curation, resources and review & editing. AL performed tissue transcriptomic analysis. MP was involved in investigation, methodology, software and review & editing. JS was involved in data curation, supervision and review & editing. HM was involved in project administration, supervision and writing original draft. MPM was involved in supervision and review & editing. RPA was involved in conceptualization, study design, project administration, resources, supervision and writing original draft.
Data sharing statement
All data are available in this manuscript and supplementary files
Declaration of Competing Interest
Harald Mischak is founder and co-owner of Mosaiques Diagnostics GmbH, which developed the CE-MS technology. Jochen Metzger, Agnieszka Latosinska and Martin Pejchinovski are employees of Mosaiques Diagnostics GmbH. All other authors declare no conflict of interest.
Acknowledgments
We would like to acknowledge the help of the consultant staff in the Hepatology departments in both Hannover Medical School and University Hospital Coventry and Warwickshire in identifying cases. We also thank Sister Subie Wurie for her help during recruitment of the cases. We also acknowledge Sean James (head of Arden tissue bank) and Parmjit Dahaley (tissue bank biomedical assistant) for their help in storage and transfer of samples. We also acknowledge the patients who consented to participate in this study and the support from the medical and life sciences research fund (MLSRF)
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103083.
Appendix. Supplementary materials
References
- 1.Seki E., Schwabe R.F. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066–1079. doi: 10.1002/hep.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalasani N., Younossi Z., Lavine J.E., Diehl A.M., Brunt E.M., Cusi K. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American association for the study of liver diseases, American college of gastroenterology, and the American gastroenterological association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 3.Nalbantoglu I.L., Brunt E.M. Role of liver biopsy in nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20:9026–9037. doi: 10.3748/wjg.v20.i27.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedossa P., Patel K. Biopsy and noninvasive methods to assess progression of nonalcoholic fatty liver disease. Gastroenterology. 2016;150 doi: 10.1053/j.gastro.2016.03.008. 1811-22.e4. [DOI] [PubMed] [Google Scholar]
- 5.Machado M.V., Cortez-Pinto H. Non-invasive diagnosis of non-alcoholic fatty liver disease. A critical appraisal. J Hepatol. 2013;58:1007–1019. doi: 10.1016/j.jhep.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 6.Castera L. Noninvasive evaluation of nonalcoholic fatty liver disease. Semin Liver Dis. 2015;35:291–303. doi: 10.1055/s-0035-1562948. [DOI] [PubMed] [Google Scholar]
- 7.Tapper E.B., Castera L., Afdhal N.H. FibroScan (vibration-controlled transient elastography): where does it stand in the United States practice. Clin Gastroenterol Hepatol. 2015;13:27–36. doi: 10.1016/j.cgh.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 8.Canavan C., Eisenburg J., Meng L., Corey K., Hur C. Ultrasound elastography for fibrosis surveillance is cost effective in patients with chronic hepatitis C virus in the UK. Dig Dis Sci. 2013;58:2691–2704. doi: 10.1007/s10620-013-2705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalasani N., Younossi Z., Lavine J.E., Charlton M., Cusi K., Rinella M. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American association for the study of liver diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 10.Grandison G.A., Angulo P. Can NASH be diagnosed, graded, and staged noninvasively? Clin Liver Dis. 2012;16:567–585. doi: 10.1016/j.cld.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ajmera V., Perito E.R., Bass N.M., Terrault N.A., Yates K.P., Gill R. Novel plasma biomarkers associated with liver disease severity in adults with nonalcoholic fatty liver disease. Hepatology. 2017;65:65–77. doi: 10.1002/hep.28776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barr J., Vázquez-Chantada M., Alonso C., Pérez-Cormenzana M., Mayo R., Galán A. Liquid chromatography-mass spectrometry-based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. J Proteome Res. 2010;9:4501–4512. doi: 10.1021/pr1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuman M.G., Cohen L.B., Nanau R.M. Biomarkers in nonalcoholic fatty liver disease. Can J Gastroenterol Hepatol. 2014;28:607–618. doi: 10.1155/2014/757929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bataller R., Brenner D.A. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magalhães P., Pejchinovski M., Markoska K., Banasik M., Klinger M., Švec-Billá D. Association of kidney fibrosis with urinary peptides: a path towards non-invasive liquid biopsies? Sci Rep. 2017;7:16915. doi: 10.1038/s41598-017-17083-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmakis D., Koeck T., Mullen W., Parissis J., Gogas B.D., Nikolaou M. Urine proteome analysis in heart failure with reduced ejection fraction complicated by chronic kidney disease: feasibility, and clinical and pathogenetic correlates. Eur J Heart Fail. 2016;18:822–829. doi: 10.1002/ejhf.544. [DOI] [PubMed] [Google Scholar]
- 17.Dakna M., He Z., Yu W.C., Mischak H., Kolch W. Technical, bioinformatical and statistical aspects of liquid chromatography-mass spectrometry (LC-MS) and capillary electrophoresis-mass spectrometry (CE-MS) based clinical proteomics: a critical assessment. J Chromatogr B Anal Technol Biomed Life Sci. 2009;877:1250–1258. doi: 10.1016/j.jchromb.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 18.Latosinska A., Siwy J., Mischak H., Frantzi M. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: the past, the present, and the future. Electrophoresis. 2019;40:2294–2308. doi: 10.1002/elps.201900091. [DOI] [PubMed] [Google Scholar]
- 19.Mischak H., Vlahou A., Ioannidis J.P. Technical aspects and inter-laboratory variability in native peptide profiling: the CE-MS experience. Clin Biochem. 2013;46:432–443. doi: 10.1016/j.clinbiochem.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 20.Jantos-Siwy J., Schiffer E., Brand K., Schumann G., Rossing K., Delles C. Quantitative urinary proteome analysis for biomarker evaluation in kidney disease. J Proteome Res. 2009;8:268–281. doi: 10.1021/pr800401m. [DOI] [PubMed] [Google Scholar]
- 21.Klein J., Papadopoulos T., Mischak H., Mullen W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis. 2014;35:1060–1064. doi: 10.1002/elps.201300327. [DOI] [PubMed] [Google Scholar]
- 22.Zürbig P., Renfrow M.B., Schiffer E., Novak J., Walden M., Wittke S. Biomarker discovery by CE-MS enables sequence analysis via MS/MS with platform-independent separation. Electrophoresis. 2006;27:2111–2125. doi: 10.1002/elps.200500827. [DOI] [PubMed] [Google Scholar]
- 23.Klein J., Eales J., Zürbig P., Vlahou A., Mischak H., Stevens R. Proteasix: a tool for automated and large-scale prediction of proteases involved in naturally occurring peptide generation. Proteomics. 2013;13:1077–1082. doi: 10.1002/pmic.201200493. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi Y., Eroshkin A., Gramatikova S., Gramatikoff K., Zhang Y., Smith J.W. CutDB: a proteolytic event database. Nucl Acids Res. 2007;35(Database issue):D546–D549. doi: 10.1093/nar/gkl813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voigtländer T., Metzger J., Husi H., Kirstein M.M., Pejchinovski M., Latosinska A. Bile and urine peptide marker profiles: access keys to molecular pathways and biological processes in cholangiocarcinoma. J Biomed Sci. 2020;27:13. doi: 10.1186/s12929-019-0599-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y., Orre L.M., Johansson H.J., Huss M., Boekel J., Vesterlund M. Discovery of coding regions in the human genome by integrated proteogenomics analysis workflow. Nat Commun. 2018;9:903. doi: 10.1038/s41467-018-03311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 29.Ho D.E., Imai K., King G., Stuart E.A. MatchIt: nonparametric preprocessing for parametric causal inference. J Stat Softw. 2011;42:1–28. [Google Scholar]
- 30.Pinzani M., Rombouts K., Colagrande S. Fibrosis in chronic liver diseases: diagnosis and management. J Hepatol. 2005;42(Suppl(1)):S22–S36. doi: 10.1016/j.jhep.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Argilés Á., Siwy J., Duranton F., Gayrard N., Dakna M., Lundin U. CKD273, a new proteomics classifier assessing CKD and its prognosis. PLoS ONE. 2013;8(5):e62837. doi: 10.1371/journal.pone.0062837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuznetsova T., Mischak H., Mullen W., Staessen J.A. Urinary proteome analysis in hypertensive patients with left ventricular diastolic dysfunction. Eur Heart J. 2012;33(18):2342–2350. doi: 10.1093/eurheartj/ehs185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuman M.G., Voiculescu M., Nanau R.M., Maor Y., Melzer E., Cohen L.B. Non-alcoholic steatohepatitis: clinical and translational research. J Pharm Pharm Sci. 2016;19:8–24. doi: 10.18433/J3F61F. [DOI] [PubMed] [Google Scholar]
- 34.Belczacka I., Pejchinovski M., Krochmal M., Magalhães P., Frantzi M., Mullen W. Urinary glycopeptide analysis for the investigation of novel biomarkers. Proteom Clin Appl. 2019;13 doi: 10.1002/prca.201800111. [DOI] [PubMed] [Google Scholar]
- 35.Lankisch T.O., Metzger J., Negm A.A., Vosskuhl K., Schiffer E., Siwy J. Bile proteomic profiles differentiate cholangiocarcinoma from primary sclerosing cholangitis and choledocholithiasis. Hepatology. 2011;53:875–884. doi: 10.1002/hep.24103. [DOI] [PubMed] [Google Scholar]
- 36.Metzger J., Negm A.A., Plentz R.R., Weismüller T.J., Wedemeyer J., Karlsen T.H. Urine proteomic analysis differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Gut. 2013;62:122–130. doi: 10.1136/gutjnl-2012-302047. [DOI] [PubMed] [Google Scholar]
- 37.Voigtländer T., Metzger J., Schönemeier B., Jäger M., Mischak H., Manns M.P., Lankisch T.O. A combined bile and urine proteomic test for cholangiocarcinoma diagnosis in patients with biliary strictures of unknown origin. United Eur Gastroenterol J. 2017;5:668–676. doi: 10.1177/2050640616687836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starr A.E., Bellac C.L., Dufour A., Goebeler V., Overall C.M. Biochemical characterization and Nterminomics analysis of leukolysin, the membrane-type 6 matrix metalloprotease (MMP25): chemokine and vimentin cleavages enhance cell migration and macrophage phagocytic activities. J Biol Chem. 2012;287:13382–13395. doi: 10.1074/jbc.M111.314179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreras M., Felbor U., Lenhard T., Olsen B.R., Delaissé J. Generation and degradation of human endostatin proteins by various proteinases. FEBS Lett; 486: 247–51. [DOI] [PubMed]
- 40.Hemmann S., Graf J., Roderfeld M., Roeb E. Expression of MMPs and TIMPs in liver fibrosis - a systematic review with special emphasis on anti-fibrotic strategies. J Hepatol. 2007;46:955–975. doi: 10.1016/j.jhep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Manchanda M., Das P., Gahlot G.P.S., Singh R., Roeb E., Roderfeld M. Cathepsin L and B as potential markers for liver fibrosis: insights from patients and experimental models. Clin Transl Gastroenterol. 2017;8:e99. doi: 10.1038/ctg.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magalhães P., Pontillo C., Pejchinovski M., Siwy J., Krochmal M., Makridakis M. Comparison of urine and plasma peptidome indicates selectivity in renal peptide handling. Proteomics Clin Appl. 2018;12(5) doi: 10.1002/prca.201700163. [DOI] [PubMed] [Google Scholar]
- 43.Arthur M.J., Iredale J.P., Mann D.A. Tissue inhibitors of metalloproteinases: role in liver fibrosis and alcoholic liver disease. Alcohol Clin Exp Res. 1999;23:940–943. doi: 10.1111/j.1530-0277.1999.tb04208.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang C.C., Hoffelt D.A.A., Merle U. Urinary cell cycle arrest biomarker [TIMP-2]•[IGFBP7] in patients with hepatorenal syndrome. Biomarkers. 2019;24:692–699. doi: 10.1080/1354750X.2019.1652347. [DOI] [PubMed] [Google Scholar]
- 45.Löfsjögård J., Kahan T., Díez J., López B., González A., Ravassa S. Usefulness of collagen carboxy-terminal propeptide and telopeptide to predict disturbances of long-term mortality in patients ≥60 years with heart failure and reduced ejection fraction. Am J Cardiol. 2017;119:2042–2048. doi: 10.1016/j.amjcard.2017.03.036. [DOI] [PubMed] [Google Scholar]
- 46.Boyle M., Tiniakos D., Schattenberg J.M., Ratziu V., Bugianessi E., Petta S. Performance of the PRO-C3 collagen neo-epitope biomarker in non-alcoholic fatty liver disease. JHEP Rep. 2019;1:188–198. doi: 10.1016/j.jhepr.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weissinger E.M., Metzger J., Dobbelstein C., Wolff D., Schleuning M., Kuzmina Z. Proteomic peptide profiling for preemptive diagnosis of acute graft-versus-host disease after allogeneic stem cell transplantation. Leukemia. 2014;28:842–852. doi: 10.1038/leu.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Good D.M., Zurbig P., Argiles A., Bauer H.W., Behrens G., Coon J.J. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteom. 2010;9:2424–2437. doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zurbig P., Jerums G., Hovind P., Macisaac R.J., Mischak H., Nielsen S.E. Urinary proteomics for early diagnosis in diabetic nephropathy. Diabetes. 2012;61:3304–3313. doi: 10.2337/db12-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein J., Lacroix C., Caubet C., Siwy J., Zurbig P., Dakna M. Fetal urinary peptides to predict postnatal outcome of renal disease in fetuses with posterior urethral valves (PUV) Sci Transl Med. 2013;5:198ra06. doi: 10.1126/scitranslmed.3005807. [DOI] [PubMed] [Google Scholar]
- 51.Tofte N., Lindhardt M., Adamova K., Bakker S.J.L., Beige J., Beulens J.W.J. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): a prospective observational study and embedded randomized placebo-controlled trial. Lancet Diabetes Endocrinol. 2020 doi: 10.1016/S2213-8587(20)30026-7. pii: S2213-8587(20)30026-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.