Abstract
Extended blood group genotyping is an invaluable tool used for prevention of alloimmunization. Genotyping is particularly suitable when antigens are weak, specific antisera are unavailable, or accurate phenotyping is problematic because of a disease state or recent transfusions. In addition, genotyping facilitates establishment of mass-scale patient-matched donor databases. However, standardization of genotyping technologies has been hindered by the lack of reference panels. A well-characterized renewable reference panel for standardization of blood group genotyping was developed. The panel consists of genomic DNA lyophilized and stored in glass vials. Genomic DNA was extracted in bulk from immortalized lymphoblastoid cell lines, generated by Epstein-Barr virus transformation of peripheral blood lymphocytes harvested from volunteer blood donors. The panel was validated by an international collaborative study involving 28 laboratories that tested each DNA panel member for 41 polymorphisms associated with 17 blood group systems. Overall, analysis of genotyping results showed >98% agreement with the expected outcomes, demonstrating suitability of the material for use as reference. Highest levels of discordance were observed for the genes CR1, CD55, BSG, and RHD. Although limited, observed inconsistencies and procedural limitations reinforce the importance of reference reagents to standardize and harmonize results. Results of stability and accelerated degradation studies support the suitability of this panel for use as reference reagent for blood group genotyping assay development and standardization.
Nearly 350 antigens, organized in 36 blood group systems, are recognized by the International Society of Blood Transfusion, and the genetic background for most of them is known.1 As a result, blood group genotyping methods are becoming widely used to overcome serologic typing limitations encountered by immunohematology laboratories.2,3
Red blood cell (RBC) antigen matching between donors and recipients by genotyping is an effective strategy to identify antigen-negative donor units for prevention of alloimmunization and for transfusion into chronically transfused, alloimmunized patients, such as those with sickle cell disease and thalassemia.4–6 Molecular typing also predicts the correct RBC antigens of patients with a positive direct antiglobulin test result,7 identifies a fetus at risk of hemolytic disease by the prediction of fetal RhD phenotype and other RBC antigens (ie, Rhc/RhC, RhE, and K) in fetal DNA,8–13 allows typing when serologic reagents are unavailable, and aids identification of Rh variants.3,14 Large-scale RBC genotyping of blood donor populations allows identification of individuals with rare and multiple antigen-negative phenotypes,15,16 ensuring better transfusion outcomes for alloimmunized patients.14
Molecular methods used for RBC genotyping vary from conventional PCR-based methods to mass-scale genotyping. Commercially available RBC genotyping platforms differ in format, allele content, processing steps, result interpretation algorithms, cost, and regulatory status. The preferred method of a laboratory is influenced by cost, throughput capabilities, turnaround time, and availability of specific genotypes.
The clinical use of any genotyping assay requires validation of results, which is achieved using reference reagents. However, there is a paucity of validated reference reagents for RBC antigen genotyping. The World Health Organization Reference Reagents for RBC genotyping, distributed by the UK National Institute for Biological Standards and Control, consist of four reference samples with limited allele coverage.17,18 Consequently, kit manufacturers, genotyping laboratories, and proficiency schemes frequently use existing clinical materials as reference,19 which are often limited in volume availability and characterization, potentially compromising the quality of results and patient care. To address this issue, we have developed and validated a renewable panel designed to contain the least number of samples with the greatest number of genotypes for use as controls for prediction of certain RBC antigens, as recommended by the Consortium for Blood Group Genes.20 The panel is available to researchers, kit manufacturers (for development and validation of assay kits), and genotyping laboratories (for test evaluation and monitoring of performance).
Materials and Methods
Sample Selection and Preparation
The study was approved by the BloodCenter of Wisconsin (Milwaukee, WI) institutional review board and the US Food and Drug Administration Research Involving Human Subjects Committee, and informed consent was signed by the participants. A total of 53 donors were recruited,21 whose blood types were represented in the Consortium for Blood Group Genes list based on their historical RBC antigen profile, determined by serologic screening (Table 1) and by genotyping performed at the BloodCenter of Wisconsin.22 Their blood was collected and screened for bloodborne pathogens, according to current US regulations, by BloodCenter of Wisconsin; samples were anonymized and sent to the Office of Blood Research and Review/Center for Biologics Evaluation and Research/US Food and Drug Administration laboratory. Whole blood was separated into plasma, erythrocytes, and leukocytes by centrifugation; and peripheral blood mononuclear cells were isolated using Ficoll-Paque Plus (Fisher, Pittsburgh, PA). Aliquots of each component were cryopreserved, and genomic DNA (gDNA) was extracted from buffy coats using a FlexiGene DNA Kit (Qiagen, Germantown, MD).
Table 1.
Phenotypes of the 18 Panel Members, Determined by Established Immunohematological Techniques
| Panel member | ABO | MNS | RH | KEL | FY | JK | DI | YT* | DO* | CO* | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | N | S | s | U | D | C | c | E | e | Cw | K | k | kpa | kpb | Jsa | Jsb | Fya | Fyb | Jka | Jkb | Dia | Yta | Dob | Coa | Cob | ||
| 10006 | A | + | 0 | 0 | + | + | + | 0 | + | + | + | NA | 0 | + | NA | NA | NA | + | 0 | + | 0 | + | NA | NA | NA | 0 | NA |
| 10007 | A | + | 0 | 0 | + | NA | + | + | + | 0 | + | NA | 0 | + | + | 0 | NA | NA | + | + | 0 | NA | NA | NA | NA | NA | NA |
| 10012 | A | + | 0 | + | + | NA | + | + | + | + | + | NA | 0 | + | NA | NA | NA | + | 0 | + | 0 | + | NA | NA | NA | NA | NA |
| 10014 | O | + | + | 0 | + | NA | 0 | 0 | + | 0 | + | NA | + | 0 | 0 | NA | NA | NA | 0 | + | + | + | NA | NA | NA | NA | 0 |
| 10018 | O | + | + | 0 | + | NA | + | + | + | 0 | + | NA | + | 0 | 0 | NA | NA | + | + | + | + | + | NA | NA | NA | NA | 0 |
| 10020 | B | + | 0 | 0 | + | NA | + | + | 0 | 0 | + | NA | 0 | + | NA | NA | NA | NA | 0 | + | 0 | + | NA | 0 | NA | NA | NA |
| 10029 | O | + | + | + | 0 | NA | + | + | + | + | + | 0 | + | + | NA | + | NA | NA | + | + | 0 | + | + | + | NA | NA | NA |
| 10030 | A | + | + | 0 | + | NA | + | + | + | 0 | + | + | 0 | + | NA | NA | NA | NA | + | 0 | 0 | + | NA | NA | NA | NA | NA |
| 10035 | A | + | + | + | + | NA | + | 0 | + | + | + | NA | 0 | + | NA | NA | NA | NA | + | W | + | + | NA | NA | NA | NA | NA |
| 10038 | O | 0 | + | 0 | + | + | + | 0 | + | + | + | NA | 0 | NA | 0 | + | + | NA | 0 | 0 | + | 0 | NA | NA | NA | NA | NA |
| 10044 | O | 0 | + | 0 | + | + | + | 0 | + | 0 | + | NA | 0 | NA | NA | + | NA | 0 | 0 | 0 | + | 0 | NA | + | NA | + | NA |
| 10047 | O | + | + | + | + | + | 0 | 0 | + | 0 | + | NA | 0 | + | 0 | + | NA | + | 0 | 0 | + | 0 | NA | NA | + | NA | 0 |
| 10048 | A | 0 | + | 0 | + | + | + | 0 | NA | 0 | NA | NA | 0 | NA | NA | NA | 0 | + | 0 | 0 | + | 0 | NA | NA | NA | NA | NA |
| 10050 | B | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | NA | + | + | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 10052 | O | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | NA | NA | + | NA | NA | NA | NA | NA | NA | NA | + | NA | + | NA |
| 10053 | O | + | 0 | 0 | + | NA | + | 0 | NA | 0 | + | NA | 0 | NA | NA | + | NA | + | 0 | 0 | + | 0 | NA | NA | NA | NA | NA |
| 10056 | A | 0 | + | 0 | + | + | 0 | 0 | NA | NA | + | NA | 0 | NA | NA | + | NA | + | 0 | 0 | + | 0 | NA | NA | NA | NA | NA |
| 10058 | O | + | + | 0 | + | NA | + | 0 | NA | + | + | NA | 0 | NA | NA | + | NA | + | + | NA | NA | 0 | NA | + | NA | + | NA |
No serology data are available for the following blood group systems: CROM, IN, KN, LU, LW, OK, and SC.
No commercial reagents are available for these and the following antigens: Dib, Ytb, Doa, VS, and V.
presence of the corresponding antigen; 0, absence of the corresponding antigen; NA, not available; W, weak expression.
Sample Characterization
Allelic discrimination assays were performed for each of the 38 polymorphisms associated with 17 blood group systems in all gDNA samples tested. Fifteen predesigned and 23 custom-designed TaqMan genotyping assays (Applied Biosystems, Foster City, CA) were run for each sample on the StepOnePlus Real-Time PCR System (Applied Biosystems). Sanger sequencing was also performed to characterize 41 polymorphisms, including single-nucleotide polymorphisms (SNPs) and insertion/deletion polymorphisms23,24 (Table 2). A PCR with sequence-specific primers (PCR-SSP) assay was performed to identify the presence/absence of a 109-bp insertion in intron 2 of the RHCE gene. The RHD deletion and its zygosity were determined by a PCR-SSP assay and a PCR-restriction fragment length polymorphism assay, as previously described.25
Table 2.
Primers Used for Amplification and Sequencing
| Primer name | Sequence |
|---|---|
| CO-PCR-F | 5′-TCAGAGGGAATTGAGCACCC-3′ |
| CO-PCR-R | 5′-GACACCCCACTCACGTCAT-3′ |
| CO-Seq-F | 5′-AAGAAGCTCTTCTGGAGGGCA-3′ |
| CO-Seq-R | 5′-AAGCGAGTTCCCAGTCAGGGA-3′ |
| CR-PCR-F | 5′-TGCATTCAAAGCTCCCACAT-3′ |
| CR-PCR-R | 5′-TCATTGAATGACTGCAACCC-3′ |
| CR-Seq-F1 | 5′-GGCATTTATAAGCATCTCTTGTTGGT-3′ |
| CR-Seq-R1 | 5′-CATATAGACCTGAGGGTAAGAAT-3′ |
| DI-PCR-F1 | 5′-AGATGTGCCCTACGTCAAGC-3′ |
| DI-PCR-R1 | 5′-TTAGGGGTCCAGCTCACTCA-3′ |
| DI-Seq-F1 | 5′-GGTACAGGACCCTTTTCTGGCT-3′ |
| DI-Seq-R1 | 5′-TGTCTGCCTCTTGACGCTCT-3′ |
| DO-PCR-F1 | 5′-CTGCAACCACATTCACCATCTG-3′ |
| DO-PCR-R1 | 5′-TCTGTGATCCTGAGTGGCCT-3′ |
| DO-Seq-F1 | 5′-AGTACCAAGGCTGTAGCAAACAG-3′ |
| DO-Seq-R1 | 5′-AATACCTTTTAGCAGCTGACAGTT-3′ |
| Duffy-PCR-F | 5′-CTGAGTGTAGTCCCAACCAGCCAA-3′ |
| Duffy-PCR-R | 5′-AGAGCTGCGAGTGCTACCTAGC-3′ |
| Duffy Seq-F1 | 5′-CAAGGCCAGTGACCCCCATA-3′ |
| Duffy Seq-R1 | 5′-GGCAAACAGCACGGGAAATGAG-3′ |
| Duffy Seq-F2 | 5′-GATGGCCTCCTCTGGGTATGTCCT-3′ |
| Duffy Seq-R2 | 5′-GCACCACAATGCTGAAGAGG-3′ |
| Kpab-PCR-F1 | 5′-GCCCCTCAACCCTTAATCCC-3′ |
| Kpab-PCR-R1 | 5′-TAGCAGCATCTCCTCCACCA-3′ |
| Kpab-Seq-F1 | 5′-TTGTGTCCATCTCCCAAGGC-3′ |
| Kpab-Seq-R1 | 5′-AGGGACTGAGAAGGGCTCAG-3′ |
| KEL578PCR-F | 5′-TTTAGTCCTCACTCCCATGCTTCC-3′ |
| KEL578PCR-R | 5′-TATCACACAGGTGTCCTCTCTTCC-3′ |
| KEL578SeqF | 5′-CCTAGAGGAATCGAAGGGCG-3′ |
| KEL578SeqR | 5′-CCAACTGTGTCTTCGCCAGT-3′ |
| JS-PCR-F2 | 5′-TTCCACCCTGGCTATCCCAG-3′ |
| JS-PCR-R1 | 5′-AGAAGGGCCATCAGGCTCTA-3′ |
| JS-Seq-F1 | 5′-TGCTGAGAGATTCTGGGGGT-3′ |
| JS-Seq-R1 | 5′-ATGGCCCTTGCTCACTGGTT-3′ |
| JK-PCR-F2 | 5′-CTATACCAGTGGGAGTTGGTCAGA-3′ |
| JK-PCR-R1 | 5′-GGGGAGCTTTGAGAAGGCAT-3′ |
| JK-Seq-F1 | 5′-ATTTTCCTGGGAGCCATCCT-3′ |
| JK-Seq-R1 | 5′-ACAGCAAGTGGGCTCAAGCC-3′ |
| KN-PCR-F1 | 5′-ACCATACTCTTCCTTCTCTCAGTCA-3′ |
| KN-PCR-R1 | 5′-CCCTCACACCCAGCAAAGTC-3′ |
| KN-Seq-F1 | 5′-GCAACAATAGAATAGAACATCTTTTCAC-3′ |
| KN-Seq-R1 | 5′-CACTCACCCCTGGAGCAGTGT-3′ |
| LU-PCR-F3 | 5′-AAAGAGGCAGAGCCAGGAGCTGCA-3′ |
| LU-PCR-R3 | 5′-TCACCACGCACACGTAGTCTCG-3′ |
| LU-Seq-F3 | 5′-GGGACACCCGGAGCTGAGAG-3′ |
| LU-Seq-R3 | 5′-TCACCACGCACACGTAGTCTCG-3′ |
| LW-PCR-F2 | 5′-CTTATCTCTAGAGCCGGCCCT-3′ |
| LW-PCR-R2 | 5′-AAGGGGGAGCCTCTGAGTGA-3′ |
| LW-Seq-F1 | 5′-TCTGTTCCCTCTGTCGCTGCT-3′ |
| LW-Seq-R1 | 5′-GCTCCCTATAGAGCGACTGTCA-3′ |
| IN-PCR-F1 | 5′-AGGTTCATGCCATTCTCCTG-3′ |
| IN-PCR-R1 | 5′-CCAGGAGAGCTCTGTGGAAG-3′ |
| IN-Seq-F1 | 5′-GGAGTCTGTCCTAAACTGAACTT-3′ |
| IN-Seq-R1 | 5′-GTCGGGTGCTGGTCTCTTACC-3′ |
| IN-Seq-F2 | 5′-TGTTAACCAGGCTGGTCTTGAG-3′ |
| IN-Seq-R2 | 5′-GGAAAGGAGCCTTCCAGTTC-3′ |
| OK-PCR-2–4F | 5′-GAGGTGTGGGGTTCATCAGT-3′ |
| OK-PCR-2–4R | 5′-TCCCCCTCGTTGATGTGTTC-3′ |
| OK-Seq-F | 5′-CCAGAGGTGTGGGGTTCATC-3′ |
| OK-Seq-R | 5′-TCAGGCCAACAACTTCTCCCGACA-3′ |
| SC-PCR-F1 | 5′-GGCGTCCCCAGAATAAGGAA-3′ |
| SC-PCR-R1 | 5′-GACACTTCCCTCTTGGGCAT-3′ |
| SC-Seq-F1 | 5′-TGGCAATCCTTTCCAGAGTCCTT-3′ |
| SC-Seq-R1 | 5′-ATTCCGGCATCAGAGATCTTCA-3′ |
| YT-PCR-F2 | 5′-CAGGAGAACGTGGCAGCCTT-3′ |
| YT-PCR-R | 5′-GGAGGACTTCTGGGACTTCTG-3′ |
| YT-Seq-F | 5′-ACCAGCGCAGGTCCTGGTGAA-3′ |
| YT-Seq-R | 5′-AATGGGCCTGGAGAAGCCCTCAT-3′ |
| CwCx-PCR-F4 | 5′-CCTCTTCTGAGCTTCAGTTTCCTTATTT-3′ |
| CwCx-PCR-R4 | 5′-GCTTCTAAAGGAAAGCTTACATTGTTGA-3′ |
| CwCx-Seq-F2 | 5′-CTCCATAGACAGGCCAGCACAG-3′ |
| CwCx-Seq-R2 | 5′-GAAGATGGGGGAATCTTTTCCTC-3′ |
| VVS-EePCR-F5 | 5′-CTGGGCAACAGAGCAAGAGTCC-3′ |
| VVS-EePCR-R5 | 5′-AGAAACGGGGTTCAAACTC-3′ |
| VVS-EeSeq-F | 5′-CCATCACAGAGCAGGTTCAGG-3′ |
| VVS-EeSeq-R | 5′-GGGGTGGGGAGGGGCATAAA-3′ |
| VVS-Fprimer 6 | 5′-AACAAACTCCCCATTGATGTGAGTA-3′ |
| VVS-Rprimer 6 | 5′-GCTCTGTGTTTGTGGGGTCACAG-3′ |
| VVS-Fprimer 3 | 5′-CCTTAGTGCCCATCCCCATTT-3′ |
| VVS-SeqR2 | 5′-ATCCAAGGTAGGGGCTGGACAT-3′ |
| RHCl09-PCRF3 | 5′-GTGCCACTTGACTTGGGACT-3′ |
| RHCl09-PCRR3 | 5′-GTGGACCCAATGCCTCTG-3′ |
| l09ins-Seq-3F | 5′-GGTACAATCATAGCTCATTGCTATAGC-3′ |
| l09ins-Seq-3R | 5′-CTATGATTGTACCACTGGGAAGTGAC-3′ |
| RHc307PCR-F3 | 5′-GCTTCCCCCTCCTCCTTCTCAC-3′ |
| RHc307PCR-R1 | 5′-TGAGAGGCCTTGAGAGGTCC-3′ |
| RHc307SeqF1-S | 5′-TGGGCTTCCTCACCTCAAAT-3′ |
| RHc307PCR-F4 | 5′-GTGCGAAAACAGTTGGTGATTATT-GATAAG-3′ |
| RHc307PCR-R4 | 5′-GGCAATATCCCAGATCTTCTGGAACC-3′ |
| RHc307SeqF4 | 5′-CAGTTGAGAACATTGAGGCTCA-3′ |
| RHc307SeqR4 | 5′-TTTCGGGGTCCATTCCCTCT-3′ |
| MN-PCR-F2 | 5′-GCTTTATCTGTAAACCTCTGCTATGC-3′ |
| MN-PCR-R3 | 5′-TGAGGTGACTGCGTGGACATAG-3′ |
| MN-Seq-F1 | 5′-GAGGGAATTTGTCTTTTGCAAT-3′ |
| MN-Seq-R2 | 5′-GGGTCTGAGCTGAACTCAGTTT-3′ |
| Ss-PCR-F3 | 5′-TAAGGCAACCATACTATCAATTGCTA-3′ |
| Ss-PCR-R3 | 5′-AGGCTTGGCCTCCCAAAATTATA-3′ |
| Ss-Seq-F2 | 5′-TGGCATCTCTGTGGAGTAATGGC-3′ |
| Ss-Seq-R2 | 5′-GTTAACAACATATGCTCTTCTG-TTTTAAG-3 |
| SSI-PCR-F5 | 5′-TGGGTCTGGAATCAGAAGCC-3′ |
| SSI-PCR-R5 | 5′-GACTTCTATGTGTCCAGTTGAAAAAG-3′ |
| SSI-Seq-F2 | 5′-CACATAATAGTATGTTAACTGTACTTTG-3′ |
| SSI-Seq-R4 | 5′-GAATTTTATGCAGTTCTGTTTCTCTTC-3′ |
| ABO-PCR-F6 | 5′-GGGCTGGGAATGATTTG-3′ |
| ABO-PCR-R6 | 5′-GGTGTCCCCCTCCTGCTATC-3′ |
| ABO-PCR-F1 | 5′-GGCAGAAGCTGAGTGGAGTT-3′ |
| ABO-O-SeqR2 | 5′-GCTCAGTAAGATGCTGC-3′ |
| ABO-PCR-F7 | 5′-CCCCGTCCGCCTGCCTTGCAG-3′ |
| ABO-PCR-R7 = R2 | 5′-GGGCCTAGGCTTCAGTTACTC-3′ |
| ABO-Bl-SeqF2 | 5′-CTACTATGTCTTCACCGAC-3′ |
| ABO-PCR-R1 | 5′-ACTCACAACAGGACGGACAA-3′ |
| DS4-S23 | 5′-GCCGACACTCACTGCTCTTAC-3′ |
| DS4-AS23 | 5′-TGAACCTGCTCTGTGAAGTGC-3′ |
| DS4-Seq23 | 5′-GGGAGATTTTTTCAGCCAG-3′ |
| RhD-Intron4-F24 | 5′-TAAGCACTTCACAGAGCAGG-3′ |
| Rh-I5R24 | 5′-TATGTGTGCTAGTCCTGTTAGAC-3′ |
| DS6-S23 | 5′-CAGGGTTGCCTTGTTCCCA-3′ |
| DS6-AS23 | 5′-CTTCAGCCAAAGCAGAGGAGG-3′ |
Production of Immortalized Cell Lines and Bulk DNA Preparation
Immortalized cell lines were generated by Epstein-Barr virus transformation, according to standard protocols, under contract with KamTek, Inc. (Birmingham, AL). Fresh peripheral blood mononuclear cells were transformed, expanded, and cryopreserved. The lymphoblastoid cell lines (LCLs) were further expanded and used to obtain bulk gDNA. Cell line expansion and large-scale DNA extraction/purification were performed under contracts with KamTek, Inc., and Rockland Immunochemicals, Inc. (Limerick, PA). The suitability of the resulting DNA was evaluated for genotyping using selected TaqMan and PCR assays—the same assays that we used for the sample characterization.
Reference DNA Panel Design and Formulation
The complete genotyping profiles were determined for each cell line, covering 41 polymorphisms of interest (Table 3), which served as the basis for selection of a subset of 18 lines that encompassed all genetic variants present in the original group of 53 blood donors (Table 4). Although every panel member has been tested for the presence of a 37-bp duplication in the RHD gene, known as the RHD pseudogene (RHDΨ), none of them has been validated as representative standard for this allele. The bulk DNA isolated from the 18 cell lines was diluted to the final concentration of 1 μg/mL in 0.40 mmol/L tris and 40 μmol/L EDTA containing 3 mg/mL trehalose, and 1-mL aliquots were dispensed into 3-mL glass vials, followed by assessment of the coefficients of variation of the fill. After lyophilization, vials were sealed with rubber stoppers and aluminum caps at the ISO 17025-certified testing facility of the Office of Compliance and Biologics Quality/Center for Biologics Evaluation and Research/US Food and Drug Administration using a VirTis Benchmark Lyophilizer (SP Scientific, Gardiner, NY). Residual moisture was determined by a methanol extraction Karl Fischer coulometric method using a nonpyridine reagent and a coulometric titrator (Mettler-Toledo, Columbus, OH). After freeze drying, materials were stored at −20°C. To determine if freeze drying affected DNA integrity, four DNA panel members were reconstituted and their performance was compared with that of the same materials prelyophilization.
Table 3.
Coverage of RBC Genotypes by the Entire Panel
| Blood group system | Predicted antigen | Gene* | Reference sequence† | Polymorphism | Variants covered by panel‡ | ||
|---|---|---|---|---|---|---|---|
| ABO | A2§ | ABO | NM_020469.2 | c.1061delC | C/C | C/delC | |
| NM_020469.2 | c.526C>G | C/C | G/C | ||||
| B§ | NM_020469.2 | c.703G>A | G/G | G/A | |||
| NM_020469.2 | c.796C>A | C/C | C/A | ||||
| NM_020469.2 | c.803G>C | G/G | G/C | ||||
| O§ | NM_020469.2 | c.261delG | G/G | G/delG | delG/delG | ||
| MNS | M/N | GYPA | NM_002099.5 | c.59C>T | C/C | C/T | T/T |
| S/second | GYPB | NM_002100.5 | c.143C>T | C/C | C/T | T/T | |
| U+var | NM_002100.5 | c.230C>T | C/C | ||||
| NM_002100.5 | c.270+5G>T | G/G | |||||
| RH | D+/D− | RHD | NM_016124.4 | RHDdel | RHD/RHD | RHD/RHDdel | RHDdel/RHDdel |
| NM_016124.4 | 37-bp Duplicate exon 4 | Not present | |||||
| C/c | RHCE | NG_009208 | 109-bp Intron 2 insertion | Ins +/+ | Ins +/− | Ins −/− | |
| NM_020485.4 | c.307C>T | C/C | C/T | T/T | |||
| E/e | NM_020485.4 | c.676G>C | G/G | C/G | |||
| Cw−/Cw+ | NM_020485.4 | c.122A>G | A/A | A/G | |||
| Cx−/Cx+ | NM_020485.4 | c.106G>A | G/G | ||||
| V−/VS− | NM_020485.4 | c.733C>G | C/C | C/G | G/G | ||
| V+/VS+ | |||||||
| V+/V− (in the presence of VS) | NM_020485.4 | c.1006G>T | G/G | ||||
| LU | Lua/Lub | BCAM | NM_005581.4 | c.230G>A | G/G | G/A | A/A |
| KEL | K/k | KEL | NM_000420.2 | c.578C>T | C/C | C/T | T/T |
| Kpa/kpb | NM_000420.2 | c.841C>T | C/C | C/T | T/T | ||
| Jsa/Jsb | NM_000420.2 | c.1790T>C | T/T | C/T | C/C | ||
| FY | Fya/Fyb | ACKR1 | NM_002036.3 | c.125G>A | G/G | A/G | A/A |
| Fy(bw) | NM_002036.3 | c.265C>T | C/C | C/T | |||
| Fy silenced in RBC | NM_002036.3 | c.−67T>C | T/T | T/C | C/C | ||
| JK | Jka/Jkb | SLC14A1 | NM_015865.6 | c.838G>A | G/G | G/A | A/A |
| DI | Dia/Dib | SLC4A1 | NM_000342.3 | c.2561C>T | C/C | C/T | |
| YT | Yta/Ytb | ACHE | NM_001302621.1 | c.1057C>A | C/C | C/A | A/A |
| SC | Sc1/Sc2 | ERMAP | NM_001017922.1 | c.169G>A | G/G | G/A | |
| DO | Doa/Dob | ART4 | NM_021071.2 | c.793A>G | A/A | A/G | G/G |
| Hy+/Hy− | NM_021071.2 | c.323G>T | G/G | G/T | |||
| Jo(a+)/Jo(a−) | NM_021071.2 | c.350C>T | C/C | C/T | T/T | ||
| CO | Coa/Cob | AQP1 | NM_198098.3 | c.134C>T | C/C | C/T | T/T |
| LW | Lwa/Lwb | ICAM4 | NM_001544.4 | c.299A>G | A/A | A/G | |
| CROM | Cr(a+)/Cr(a−) | CD55 | NM_000574.3 | c.679G>C | G/G | G/C | |
| KN | Kna/Knb | CR1 | NM_000573.3 | c.4681G>A | G/G | G/A | |
| McCa/McCb | NM_000573.3 | c.4768A>G | A/A | A/G | |||
| SIa/Vil | NM_000573.3 | c.4801A>G | A/A | A/G | G/G | ||
| IN | Ina/Inb | CD44 | NM_001001391.1 | c.137G>C | G/G | ||
| OK | Ok(a)/Ok(a−) | BSG | NM_198589.2 | c.274G>A | G/G | ||
Empty cells denote genotypes that are not represented in the panel.
As defined by the Human Genome Organization Gene Nomenclature Committee (http://www.genenames.org, last accessed December 8, 2018).
Publicly available nucleotide sequences, as reported in GenBank (https://www.ncbi.nlm.nih.gov/genbank, last accessed February 7, 2019).26
Details about the blood group allele terminology available at the International Society of Blood Transfusion website (http://www.isbtweb.org/working-parties/red-cell-immunogenetics-and-blood-group-terminology, last accessed December 8, 2018).
Analysis of multiple single-nucleotide polymorphisms and insertion/deletion polymorphisms is needed to accurately predict the phenotype. RBC, red blood cell.
Table 4.
Genotypes of the 18 DNA Panel Members, Determined as Described in Materials and Methods
| DNA panel member† | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Polymorphism* | 10006 | 10007 | 10012 | 10014 | 10018 | 10020 | 10029 | 10030 | 10035 | 10038 | 10044 | 10047 | 10048 | 10050 | 10052 | 10053 | 10056 | 10058 |
| ABO | c.1061delC | C/C | C/C | C/delC | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/delC | C/C | C/C | C/C | C/C | C/C |
| c.526C>G | C/C | C/C | C/C | C/C | C/C | C/G | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/G | C/C | C/C | C/C | C/C | |
| c.703G>A | G/G | G/G | G/G | G/G | G/G | G/A | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/A | G/G | G/G | G/G | G/G | |
| c.796C>A | C/C | C/C | C/C | C/C | C/C | C/A | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/A | C/C | C/C | C/C | C/C | |
| c.803G>C | G/G | G/G | G/G | G/G | G/G | G/C | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/C | G/G | G/G | G/G | G/G | |
| c.261delG | G/delG | G/delG | G/G | delG/delG | delG/delG | G/delG | delG/delG | G/delG | G/delG | delG/delG | delG/delG | delG/delG | G/G | G/delG | delG/delG | delG/delG | G/G | delG/delG | |
| GYPA | c.59C>T | C/C | C/C | C/C | C/C | C/T | C/C | C/T | C/T | C/T | T/T | T/T | C/T | T/T | T/T | T/T | C/C | T/T | C/T |
| GYPB | c.143C>T | C/C | C/C | T/T | C/C | C/C | C/C | T/T | C/C | C/T | C/C | C/C | C/T | C/C | C/T | C/C | C/C | C/C | C/C |
| c.230C>T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | |
| c.270+5G>T | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | |
| RHD | RHDdel‡ | +/− | +/− | +/+ | −/− | +/+ | +/+ | +/+ | +/+ | +/− | +/+ | +/+ | −/−§ | +/+§ | +/+ | +/+ | +/+ | −/−§ | +/+§ |
| RHCE | 109-bp Intron 2 insertion¶ | −/− | +/− | +/− | −/− | +/− | +/+ | +/− | +/− | −/− | −/− | −/− | −/− | −/− | +/− | +/− | −/− | −/− | −/− |
| c.307C>T | C/C | T/C | T/C | C/C | T/C | T/T | T/C | T/C | C/C | C/C | C/C | C/C | C/C | T/C | T/C | C/C | C/C | C/C | |
| c.676G>C | C/G | G/G | C/G | G/G | G/G | G/G | C/G | G/G | C/G | C/G | G/G | G/G | G/G | C/G | C/G | G/G | G/G | C/G | |
| c.122A>G | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/G | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | |
| c.106G>A | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | |
| c.733C>G | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | G/G | G/C | G/C | |
| c.1006G>T | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | |
| BCAM | c.230G>A | G/G | G/G | A/A | G/A | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G |
| KEL | c.578C>T | C/C | C/C | C/C | T/T | T/T | C/C | C/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C |
| c.841C>T | C/C | T/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/T | C/C | C/C | C/C | C/C | |
| c.1790T>C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/C | C/C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | |
| ACKR1 | c.125G>A | A/A | A/G | A/A | A/A | A/G | A/A | A/G | G/G | A/G | A/A | A/A | A/A | A/A | A/G | A/G | A/A | A/A | A/A |
| c.265C>T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | |
| c.−67T>C | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | T/T | C/C | C/C | C/C | C/C | T/T | T/T | C/C | C/C | T/C | |
| SLC14A1 | c.838G>A | A/A | A/A | A/A | A/G | A/G | A/A | A/A | A/A | A/G | G/G | G/G | G/G | G/G | A/G | A/G | G/G | G/G | G/G |
| SLC4A1 | c.2561C>T | C/C | C/C | C/C | C/C | C/C | T/C | T/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C |
| ACHE | c.1057C>A | C/C | C/C | C/C | C/C | C/C | A/A | C/C | C/C | C/A | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C |
| ERMAP | c.169G>A | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/A | G/G | G/G | G/G |
| ART4 | c.793A>G | A/G | G/G | A/G | A/G | A/G | A/G | A/G | A/A | G/G | A/G | G/G | A/G | A/A | A/G | A/G | G/G | A/G | A/A |
| c.323G>T | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/T | G/G | G/G | G/G | G/G | G/G | G/G | |
| c.350C>T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/T | T/T | C/C | C/C | C/C | C/C | C/T | |
| AQP1 | c.134C>T | T/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/T | C/C | C/C | C/C |
| ICAM4 | c.299A>G | A/A | A/A | A/A | A/A | A/G | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A |
| CD55 | c.679G>C | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/C | G/G |
| CR1 | c.4681G>A | G/G | G/G | G/G | G/A | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/A | G/G | G/G | G/G |
| c.4768A>G | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/G | A/G | A/G | A/G | A/A | A/A | A/A | A/A | A/A | |
| c.4801A>G | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/A | A/G | A/G | G/G | G/G | A/A | A/A | A/G | A/G | G/G | |
| CD44 | c.137G>C | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G |
| BSG | c.274G>A | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G |
Nucleotide position refers to the position within the cDNA sequence (see reference sequence in Table 3).
The DNA reference reagents are maintained at the Center for Biologics Evaluation and Research/US Food and Drug Administration.
+/+ Indicates no RHD deletion; +/−, deletion of one RHD allele; and −/−, deletion of both RHD alleles.
Caution in the interpretation of results should be exercised if an assay targeting upstream and downstream Rhesus boxes is used for determination of this genotype.
+/+ Indicates presence of a 109-bp intron 2 insertion in both RHCE alleles; +/−, presence of a 109-bp intron 2 insertion in one RHCE allele; and −/−, absence of a 109-bp intron 2 insertion.
Stability and Accelerated Degradation Studies
Three of the 18 DNA panel members were arbitrarily chosen, and on the day of lyophilization, they were placed at −80°C, −20°C, 4°C, 22°C, 37°C, and 45°C. One vial of each was used to reconstitute the materials and perform tests on day 0. Reconstituted day 0 vials were kept at 4°C and tested along with other samples at each time point to establish open-vial stability. The accelerated degradation study included the following temperatures and time points: 25°C, 37°C, and 45°C at 1, 2, 4, and 8 weeks. The long-term stability study included vials stored at 20°C, 4°C, and 25°C and tested at 3 months, 6 months, and 1 year using vials stored at −80°C at each time point for baseline values. For each temperature/time point combination, DNA panel members were resuspended in 40 μL of water, resulting in gDNA at 25 ng/μL, and the following tests were performed: DNA quantification; agarose gel electrophoresis using 1% agarose gel to detect visible degradation of gDNA; and real-time quantitative PCR assay of a 432-bp fragment of human growth hormone gene to quantify the percentage loss in the amount of DNA.27
Collaborative Validation Study
Forty-nine laboratories from 19 countries were invited to participate in the study; 31 laboratories from 13 countries accepted the invitation and declared which of the 41 SNPs and insertion/deletion polymorphisms they would evaluate. Each collaborator received three vials of each lyophilized panel member and was asked to test the panel members in three independent runs, preferably on different days, using their routine blood genotyping assays. The participants were asked to report the results using a specifically designed worksheet with predefined values and to provide the original reports produced by their genotyping platform(s). On study completion, all participating laboratories received a study report with preliminary results, which allowed every collaborator to investigate possible reasons for discrepant results and correct previously overlooked discrepancies.
Data Analysis
Data reported back by the participants were analyzed and compared with the expected results. We classified discordant results as follows: i) no calls, when the result was not determined; ii) reporting errors, when mistakes and typos were made when transcribing original reports to the electronic worksheet or when there was an inadvertent error in the workflow not caused by genotyping platform; and iii) other discrepancies. In addition, there were cases in which the correct genotypes had not been determined due to inherent limitations of the assay used. Agreement with expected results was calculated by polymorphism/phenotype, test method/platform, and testing laboratory/site.
Results
Genotypes for the polymorphisms associated with 17 blood group systems were determined for all 53 donors, from whom successful transformation produced 51 LCLs. Eighteen of the 51 LCLs were selected for expansion, bulk gDNA extraction, and inclusion in the panel formulation. The blood group genotypes of the 18 DNA panel members are shown in Table 2, and the historical phenotypes are presented in Table 1. No discrepancies were observed between historical erythrocyte phenotypes and the predicted phenotypes in any of the panel members.
DNA profiles obtained for peripheral blood mononuclear cells were compared with those of their corresponding immortalized cell lines. Results of allelic discrimination assays indicated that the transformation process had no effect on the target sequences. Freeze drying under the used conditions lowered the purity of the DNA, as estimated by ratios of absorbance at 260 and 280 nm (1.5 to 1.6 versus 1.8 to 1.9 for original DNA stocks), but did not affect the accuracy of genotyping.
Stability and Accelerated Degradation Studies
Storage of the vials containing the DNA at different temperatures for up to 1 year did not lead to degradation of the DNA at any of the tested time points, as verified by agarose gel electrophoreses and real-time quantitative PCR assay. The concentration of DNA remained stable for both lyophilized materials and open vials stored at 4°C, never decreasing to <24.9 ng/μL. Moreover, Ct values obtained by real-time quantitative PCR did not increase with time in DNA panel members stored at elevated temperatures (−20°C, 4°C, and 25°C) when compared with those stored at −80°C as baseline, indicating stability of the DNA.
Collaborative Studies for Panel Validation
Twenty-eight of the 31 participants from 13 countries reported their results in the panel validation study (Supplemental Appendix S1). Two laboratories provided two sets of results, each produced by different methods, and these sets were considered separately. Raw data sets, in addition to standardized results, were provided by 18 collaborators; and the data were used to analyze performance.
Participants performed their tests in triplicate (42.9%), duplicate (14.3%), and single (42.9%) runs, with the number of polymorphisms tested per laboratory ranging from 6 to 39. There were no intralaboratory discrepancies when more than one run was performed on the same panel member, with the exception of RHDΨ and NM_000420.2:c.578C>T, for which one collaborator reported inconsistent results between replicates. Twenty-eight data sets contained genotyping results for FY, KEL, and RH blood group systems, whereas only three collaborators tested for OK (Figure 1).
Figure 1.
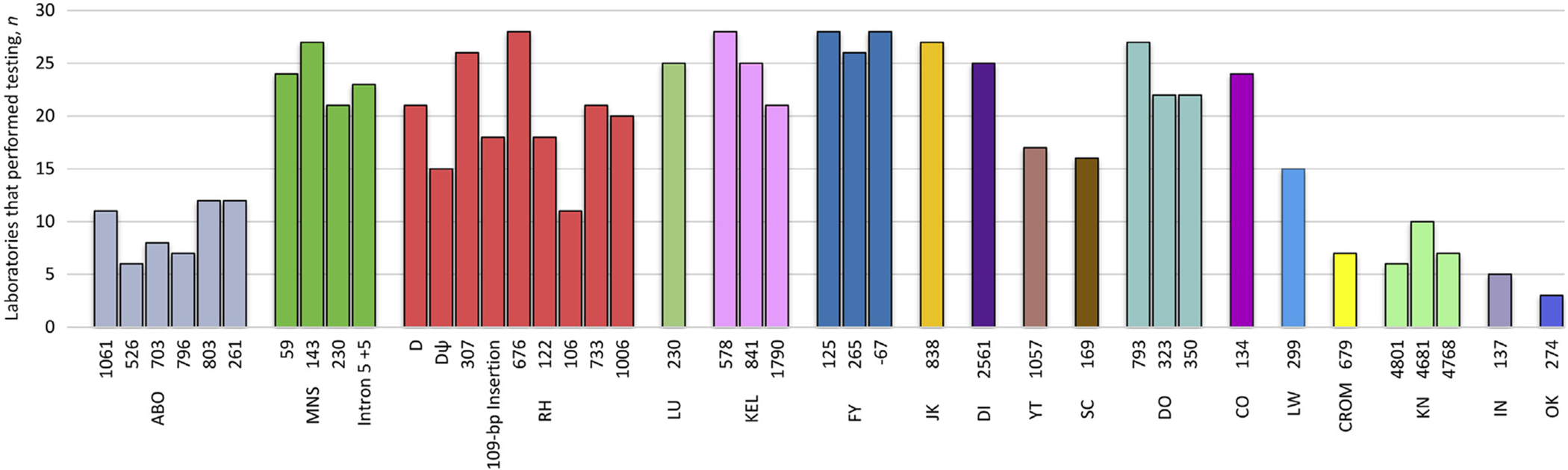
Blood group polymorphisms analyzed by collaborators. Polymorphisms are grouped by blood group system, which is indicated under each cluster using International Society of Blood Transfusion nomenclature.
Collaborators used a wide variety of genotyping techniques, including droplet digital PCR, the human erythrocyte antigen, RHD, and RHCE BeadChip arrays from Immucor (Peachtree Corners, GA); HI-FI Blood by AXO Science (Villeurbanne, France); high-resolution melting analysis, ID-CORE XT, and BLOODchip REFERENCE by Progenika (Derio, Spain); matrix-assisted laser desorption/ionization-based assays, such as Hemo ID, from Agena Bioscience (San Diego, CA); next-generation sequencing, PCR-restriction fragment length polymorphism, PCR-SSP, either single or multiplex, RBC-Ready Gene, and RBC-FluoGene by inno-train Diagnostik GmbH (Kronberg im Taunus, Germany); real-time PCR-based assays; Sanger sequencing; and SNaPshot (Applied Biosystems). Twelve collaborators had a single genotyping method established in their laboratories, and 16 participants had two to four methods that were generally used in a complementary manner.
Participants collectively performed 743 genotyping tests, producing 13,374 results. Of these genotyping results, 35 (0.26%) were no call/undetermined, 71 (0.53%) were errors in reporting, 97 (0.73%) were discrepant, and 70 (0.52%) were produced by the assays limited in their ability to distinguish homozygous from heterozygous variants for some targets (Table 5). To predict the phenotype of RhD, RhCc, and Fy antigens, more than one polymorphism is often evaluated. Consequently, the agreement with expected results in these three cases was analyzed on the basis of predicted phenotypes rather than individual genotypes, which decreased the number of discordant results to 34 for no call, 64 for reporting errors, and 85 for discrepant. In addition, 72 results (0.54%) were not reported by a collaborator in the predesigned worksheet but were present and correct in their genotyping platform’s original report.
Table 5.
Assays Designed to Determine the Presence or Absence of a Target, and Not Zygosity
| Gene | Polymorphism* | Method | Allele detected (antigen associated) |
|---|---|---|---|
| ABO | c.803G>C | I | 803C [B, cisAB, B(A) and O]† |
| c.703G>A | 703A [B, cisAB, B(A), O, and A2]† | ||
| c.1061delC | 1061delC (A2 and O)† | ||
| GYPB | c.230C>T | I | 230T (S silenced) |
| c.270+5G>T | 270+5T (S silenced) | ||
| RHD | RHDdel | A, D, F, I, and J | RHD (RhD) |
| RHCE | c.676G>C | J | 676G (Rhe) |
| c.122A>G | I and J | 122G (CW) | |
| c.106G>A | J | 106A (CX) | |
| c.733C>G | I and J | 733G (VS) | |
| c.1006G>T | I and J | 1006T (V) | |
| ART4 | c.323G>T | I | 323T (Hy) |
| c.350C>T | 350T (Joa) |
Nucleotide position refers to the position within the cDNA sequence (see reference sequence in Table 3).
Analysis of multiple single-nucleotide polymorphisms and insertion/deletion polymorphisms is needed to accurately predict the phenotype.
Agreement with expected results was calculated for each allele (Table 6), method used (Table 7), and testing laboratory (Table 8). The number of no calls, reporting errors, and discrepant results was subtracted from the total number of panel members analyzed for polymorphism/phenotype by all participants, and the resulting value was divided by the total and presented as agreement rate in Table 6. The agreement was 100% for 10 SNPs: NM_002100.5:c.143C>T, NM_020485.4:c.106G>A, NM_020485.4:c.122A>G, NM_000420.2:c.841C>T, NM_000420.2:c.1790T>C, NM_001017922.1:c.169G>A, NM_001544.4:c.299A>G, NM_000573.3:c.4768A>G, NM_000573.3:c.4801A>G, and NM_001001391.1:c.137G>C (Table 6). For 24 polymorphisms or predicted phenotypes, the agreement ranged from 95% to 99.8%; for the polymorphisms NM_016124.4 RHD del, NM_000573.3:c.4681G>A, NM_000574.3:c.679G>C, and NM_198589.2:c.274G>A, the agreement rates were 94.2%, 91.1%, 85.7%, and 66.7%, respectively. When only no call and discrepant results were considered in the analysis, 100% agreement was identified for 15 polymorphisms and 97% to 99.9% agreement was identified for 19 polymorphisms or predicted phenotypes. The agreement for NM_000573.3:c.4681G>A remained at 91%, and the agreement for NM_000574.3:c.679G>C remained at 85.7%; interestingly, for the NM_198589.2:c.274G>A polymorphism, the agreement rate increased to 98%.
Table 6.
Agreement Rate of Allele Identification by Study Participants, Calculated for Each Blood Group Allele
| Blood group system | Polymorphism* | Laboratories that performed testing, n | Results reported | Reporting errors | No call | Discrepant results | Agreement rate, % |
|---|---|---|---|---|---|---|---|
| ABO | c.1061delC | 11 | 198 | 1 | 1 | 98.99 | |
| ABO | c.526C>G | 6 | 108 | 2 | 98.15 | ||
| ABO | c.703G>A | 8 | 144 | 1 | 2 | 97.92 | |
| ABO | c.796C>A | 7 | 126 | 2 | 98.41 | ||
| ABO | c.803G>C | 12 | 216 | 2 | 99.07 | ||
| ABO | c.261delG | 12 | 216 | 1 | 2 | 98.61 | |
| MNS | c.59C>T | 24 | 432 | 3 | 1 | 99.07 | |
| MNS | c.143C>T | 27 | 486 | 100 | |||
| MNS | c.230C>T | 21 | 378 | 16 | 95.77 | ||
| MNS | c.270+5G>T | 23 | 414 | 16 | 96.14 | ||
| RH | RHD+/RHD−† | 21 | 378 | 1 | 4 | 17 | 94.18 |
| RH | 37-bp Duplicate exon 4 | 15 | 270 | 10 | 96.3 | ||
| RH | RhC/Rhc‡ | 26 | 468 | 1 | 6 | 98.5 | |
| RH | c.676G>C | 28 | 504 | 2 | 1 | 99.4 | |
| RH | c.122A>G | 18 | 324 | 100 | |||
| RH | c.106G>A | 11 | 198 | 100 | |||
| RH | c.733C>G | 21 | 378 | 1 | 2 | 99.21 | |
| RH | c.1006G>T | 20 | 360 | 4 | 98.89 | ||
| LU | c.230G>A | 25 | 450 | 1 | 3 | 1 | 98.89 |
| KEL | c.578C>T | 28 | 504 | 4 | 99.21 | ||
| KEL | c.841C>T | 25 | 450 | 100 | |||
| KEL | c.1790T>C | 21 | 378 | 100 | |||
| FY | § | 28 | 504 | 6 | 98.81 | ||
| JK | c.838G>A | 27 | 486 | 1 | 3 | 99.18 | |
| DI | c.2561C>T | 25 | 450 | 1 | 99.78 | ||
| YT | c.1057C>A | 17 | 306 | 2 | 99.35 | ||
| SC | c.169G>A | 16 | 288 | 100 | |||
| DO | c.793A>G | 27 | 486 | 3 | 99.38 | ||
| DO | c.323G>T | 22 | 396 | 1 | 99.75 | ||
| DO | c.350C>T | 22 | 396 | 2 | 2 | 98.99 | |
| CO | c.134C>T | 24 | 432 | 1 | 1 | 99.54 | |
| LW | c.299A>G | 15 | 270 | 100 | |||
| CROM | c.679G>C | 7 | 126 | 18 | 85.71 | ||
| KN | c.4681G>A | 10 | 180 | 16 | 91.11 | ||
| KN | c.4768A>G | 7 | 126 | 100 | |||
| KN | c.4801A>G | 6 | 108 | 100 | |||
| IN | c.137G>C | 5 | 90 | 100 | |||
| OK | c.274G>A | 3 | 54 | 17 | 1 | 66.67 | |
| Total | 12,078 | 64 | 34 | 85 | 98.48 |
Blank cells denote 0 events observed.
Nucleotide position refers to the position within the cDNA sequence (see reference sequence in Table 3).
Predicted by determining RHD deletion or RHD zygosity.
Predicted by testing single-nucleotide polymorphism c.307T>C and 109-bp insertion within intron 2 of RHCE (NM_020485.4).
Predicted by testing single-nucleotide polymorphisms c.125G>A, c.265C>T, and c.−67T>C of the ACKR1 gene (NM_002036.3). The total number of results reflects reported results for RhD, RhCc, and Fy predicted phenotypes.
Table 7.
Agreement Rates Calculated for Each Method
| Method | Laboratories using the method, n | Results reported | No call | Discrepant results | Agreement rate, % |
|---|---|---|---|---|---|
| A | 1 | 468 | 0 | 0 | 100.00 |
| B | 1 | 18 | 0 | 0 | 100.00 |
| C | 1 | 18 | 0 | 0 | 100.00 |
| D | 8 | 3060 | 1 | 0 | 99.97 |
| E | 3 | 1296 | 0 | 1 | 99.92 |
| F | 3 | 1386 | 0 | 3 | 99.78 |
| G | 2 | 1350 | 2 | 1 | 99.78 |
| H | 1 | 306 | 0 | 1 | 99.67 |
| I | 6 | 2038 | 2 | 21 | 98.87 |
| J | 8 | 2434 | 4 | 32 | 98.52 |
| K | 5 | 666 | 1 | 9 | 98.50 |
| L | 6 | 958 | 20 | 10 | 96.87 |
| M | 1 | 270 | 8 | 13 | 92.22 |
Table 8.
Agreement Rates Calculated for Each Laboratory
| Tests performed, n | Genotyping results | ||
|---|---|---|---|
| Concordant | Reported | Agreement rate, % | |
| 37 | 663 | 666 | 99.55 |
| 36 | 630 | 648 | 97.22 |
| 35 | 607 | 630 | 96.35 |
| 34 | 612 | 612 | 100.00 |
| 31 | 548 | 558 | 98.21 |
| 30 | 540 | 540 | 100.00 |
| 28 | 502 | 504 | 99.60 |
| 28 | 500 | 504 | 99.21 |
| 28 | 500 | 504 | 99.21 |
| 26 | 460 | 468 | 98.29 |
| 25 | 450 | 450 | 100.00 |
| 23 | 414 | 414 | 100.00 |
| 22 | 382 | 396 | 96.46 |
| 22 | 395 | 396 | 99.75 |
| 22 | 396 | 396 | 100.00 |
| 21 | 377 | 378 | 99.74 |
| 21 | 374 | 378 | 98.94 |
| 21 | 378 | 378 | 100.00 |
| 21 | 378 | 378 | 100.00 |
| 21 | 378 | 378 | 100.00 |
| 21 | 377 | 378 | 99.74 |
| 21 | 378 | 378 | 100.00 |
| 21 | 378 | 378 | 100.00 |
| 18 | 324 | 324 | 100.00 |
| 18 | 324 | 324 | 100.00 |
| 13 | 213 | 234 | 91.03 |
| 10 | 180 | 180 | 100.00 |
| 9 | 162 | 162 | 100.00 |
| 4 | 72 | 72 | 100.00 |
| 4 | 67 | 72 | 93.06 |
| 671 | 11,959 | 12,078 | 98.88 |
The agreement rates by method were calculated as a percentage of tests performed on the DNA panel members by a particular method that produced expected results. Two of the three methods that had 100% agreement of results were used to analyze 18 DNA panel members for only one polymorphism, which precludes reaching meaningful conclusions regarding these methods’ performance. For the remainder of the methods used, agreement ranged from 92.22% to 99.97% (Table 7).
The agreement rates with the expected results by laboratory were determined as a percentage of tests performed on DNA panel members by a laboratory that were concordant; no calls and discrepant results were treated as errors (ie, they were subtracted from the total number of tests performed, and the resulting value was divided by the total to determine percentage). Of 30 laboratories, 15 had a 100% agreement rate, 7 had 99.0% to 99.9% agreement, 4 had 97.1% to 98.9% agreement, and 4 had <97.0% agreement (Table 8).
Discussion
We developed and evaluated a panel consisting of 18 DNA panel members, covering 41 polymorphisms in 17 blood group systems. LCLs were produced from well-characterized blood donors to generate a continuous supply of stable and reliable gDNA material for RBC genotyping. This panel is well suited as a tool for development and validation of new genotyping assays, calibration of established tests, and proficiency testing in genotyping laboratories.
A total of 28 laboratories participated in this study, returning 30 sets of molecular genotyping data. A total of 97 (0.73%) of 13,374 were discordant from the expected results. Overall, the agreement with the expected results was high and depended on the testing laboratory, method used, and polymorphism evaluated. In general, polymorphisms that were tested less frequently were associated with the lowest and highest result agreement rates. Agreement rates by method varied from 92.2% to 100%. Both the lowest and the highest agreement rates were observed with methods used for a relatively low number of tests performed in the panel members.
The raw data provided by 18 laboratories were helpful in the analysis because, of 12,078 genotypes/predicted phenotypes, 64 results from various blood group systems (OK, MNS, RH, DO, DI, ABO, and LU) showed transcribing errors when compared with the raw data from the original platform output. In addition, it is possible that some reporting inaccuracies were overlooked because some collaborators did not provide feedback to the preliminary study report. Therefore, lower agreement rates observed for some laboratories that did not provide original platform output or did not respond to the call for feedback (average agreement rate for those was 96.84% compared with 99.39% for the laboratories that responded to the feedback request or provided the original results) might be explained by uncovered reporting errors.
Because some blood group system phenotypes are predicted on the basis of more than a single polymorphism, RBC genotyping methods and platforms are often designed to produce accurate phenotype predictions and not genotype readings. Taken separately, genotyping results were, in many cases, discrepant or inconclusive; however, when analyzed together, they predicted correct phenotypes. For instance, the NM_002036.3:c.125G>A genotype was reported as undetermined for some samples by one of the collaborators. This collaborator’s method is specific for the wild-type GATA box, so that the FY*B-specific sequence is not amplified if the GATA box mutation is present (NM_002036.3:c.−67T>C). The combined test results of c.−67T>C and c.125G>A allowed for the correct determination of FY genotype and the predicted phenotype associated with the DNA panel members. Similarly, three of the methods used by participants to analyze the nucleotide polymorphism NM_020485.4:c.307C>T associated with RhC and Rhc phenotypes generated discrepant or indeterminate genotypes because of the high sequence homology between the RHD and RHCE genes. However, the assays also identified the presence of the RHCE*C-specific 109-bp insertion into intron 2, which allowed correct prediction of the C/c phenotype and inference of the NM_020485.4:c.307C>T genotype. Consequently, errors produced by these assays were not considered as such for the purposes of the analysis.
Discrimination between RhD-positive and RhD-negative phenotypes was performed by 21 laboratories; of these laboratories, 14 performed an RHD zygosity test by assays that target the upstream and downstream Rhesus boxes surrounding the RHD gene, and six collaborators had different outcomes when analyzing four of the DNA panel members. It is well known that some RH variant alleles carry genetic alterations in Rhesus box regions.28–30 Our hypothesis is that these genetic variations might have affected the outcomes of the assays28,29,31,32 and caused discrepant RHD zygosity results for those DNA panel members. Therefore, end users are advised to exercise caution when using these panel members as standards in RHD zygosity assays targeting upstream and downstream Rhesus boxes.
Interference of an RHD variant with an assay outcome was observed in the determination of the NM_020485.4:c.733C>G SNP in panel member 10044 by a real-time allelic discrimination PCR assay. To interrogate this SNP, a collaborator used RHCE nt.697C to specifically amplify the RHCE allele. However, the panel member 10044 also has nt.697C in the RHD gene because of the presence of the RHD*DAU-5 allele33 encoded by the polymorphisms NM_016124.4:c.667T>G, c.697G>C, and c.1136C>T. Presumably, the assay was not correctly detecting the polymorphism NM_020485.4:c.733C>G because the RHD*DAU-5 sequence was masquerading as RHCE.
RHDΨ was tested by 16 laboratories by a variety of methods; erratic results for 10 (10007, 10012, 10020, 10029, 10030, 10044, 10048, 10050, 10052, and 10058) of 18 DNA panel members were observed by one collaborator. Because these samples are not representative of the RHDΨ allele, they should not be used as reference reagents for RHDΨ.
Some polymorphisms of the KN, OK, and CROM blood group systems were associated with a lower agreement rate when compared with most of the polymorphisms included in this panel. The polymorphism NM_000573.3:c.4681G>A showed the lowest agreement rate (91.11%). Interestingly, results provided for this SNP by one collaborator were discrepant for 16 of 18 panel members. Because discrepant results were found only for samples with a homozygous genotype and raw data were not provided by this laboratory, we hypothesize that interpretation of wild-type and mutated alleles was erroneous. The agreement rate for polymorphism NM_198589.2:c.274G>A increased from 66.7% to 98% when reporting errors that were not considered in the analysis. The results for this SNP from laboratory 17 were initially considered incorrect in our analysis and later reclassified as reporting errors for 17 samples and as a no call for 1 sample after the collaborator reanalyzed the data using the correct chromosome location of the SNP as a reference. In addition, the agreement rate of 85.7% for NM_000574.3:c.679G>C resulted from no call results reported by one collaborator. In contrast, the genotypes were correctly determined by other laboratories using both similar and different methods. Therefore, the low agreement rate obtained for these polymorphisms may not be caused by the materials’ intrinsic qualities, but rather by limitations in assay performance.
Common issues experienced and reported by collaborators were the difficulty of opening the glass vials sealed with aluminum caps and resuspending lyophilized DNA in only 40 μL of water. One of the participants noted that purity of the provided DNA is lower than recommended for standard molecular typing techniques. Nevertheless, although there were also two reports of low DNA concentration, there were no instances of failure to perform genotyping assays because of quantity and/or quality of DNA.
As is evident from this study, existing red cell genotyping methods and platforms provide a high level of agreement/accuracy, which can be further improved by standardization and use of appropriate controls. With the advent of personalized medicine and genotyping becoming increasingly common, availability of well-characterized standards will aid development of new assays and allow more thorough proficiency testing. The high degree of agreement between genotypes and predicted phenotypes of the DNA panel members between the study participants, along with results of stability and accelerated degradation testing, supports the suitability of this panel for use as reference material for blood group genotyping. Extensive evaluation by several genotyping methods makes this DNA panel a unique tool to assist pretransfusion crossmatching, leading to improved clinical outcomes.
Supplementary Material
Acknowledgments
We thank all members of the Collaborative Study Group, who contributed greatly to this study by performing evaluation of reagents and providing feedback: Carine P. Arnoni, Tatiane A. de Paula Vendrame, Gregor Bein, Ulrich J. Sachs, Maria G. Aravechia, Carolina B. Bub, Mike Bunce, David Pye, Lilian Castilho, Mayra D. de Macedo, Jessica Constanzo, Marie-Claire Chevrier, Nathalie Desjardins, Benjamin Corgier, Nelly da Silva, Agnès Mailloux, Meghan Delaney, Gayle Teramura, Samantha Harris, Sarah Heidl, Gregory A. Denomme, Kathleen Bensing, Andrea Doescher, Tadeja D. Drnovsek, Anja Lukan, Willy A. Flegel, Kshitij Srivastava, Rainer Frank, Sabrina König, Christoph Gassner, Stefan Meyer, Nadine Trost, Catherine Hyland, Yew-Wah Liew, Naomi Roots, Jill Johnsen, Debbie Nickerson, Marsha Wheeler, Margaret Keller, Trina Horn, Jessica Keller, Sofia L. Crottet, Christine Henny, Shirley Modan, Gorka Ochoa, Roser Hoffman, Åsa Hellberg, Lis Nertsberg, Martin L. Olsson, Cédric Vrignaud, Thierry Peyrard, Maryse St-Louis, Josée Lavoie, Geneviève Laflamme, Yoshihiko Tani, Mitsunobu Tanaka, Anthony Trinkle, Stephanie Goe, Connie Westhoff, Sunitha Vege, Michael Wittig, Andre Franke, and Ping Chun Wu; and Simleen Kaur and members of the Laboratory of Microbiology, In-Vivo Testing and Standards (US Food and Drug Administration), for assistance with filling and lyophilization of the DNA.
Supported in part by an appointment to the Research Participation Program at the Center for Biologics Evaluation and Research (CBER), administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration (FDA) (E.S.); and CBER/Office of Blood Research and Review and Office of Minority Health US FDA intramural funds FDA Critical PathFY2014 and FDA Modernizing Science FY2015-2017.
Footnotes
Disclosures: None declared.
Supplemental Data
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2019.02.003.
References
- 1.Storry JR, Castilho L, Chen Q, Daniels G, Denomme G, Flegel WA, Gassner C, de Haas M, Hyland C, Keller M, Lomas-Francis C, Moulds JM, Nogues N, Olsson ML, Peyrard T, van der Schoot CE, Tani Y, Thornton N, Wagner F, Wendel S, Westhoff C, Yahalom V: International society of blood transfusion working party on red cell immunogenetics and terminology: report of the Seoul and London meetings. ISBT Sci Ser 2016, 11:118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hillyer CD, Shaz BH, Winkler AM, Reid M: Integrating molecular technologies for red blood cell typing and compatibility testing into blood centers and transfusion services. Transfus Med Rev 2008, 22: 117–132 [DOI] [PubMed] [Google Scholar]
- 3.Denomme GA, Flegel WA: Applying molecular immunohematology discoveries to standards of practice in blood banks: now is the time. Transfusion 2008, 48:2461–2475 [DOI] [PubMed] [Google Scholar]
- 4.Casas J, Friedman DF, Jackson T, Vege S, Westhoff CM, Chou ST: Changing practice: red blood cell typing by molecular methods for patients with sickle cell disease. Transfusion 2015, 55:1388–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putzulu R, Piccirillo N, Orlando N, Massini G, Maresca M, Scavone F, Ricerca BM, Zini G: The role of molecular typing and perfect match transfusion in sickle cell disease and thalassaemia: an innovative transfusion strategy. Transfus Apher Sci 2017, 56:234–237 [DOI] [PubMed] [Google Scholar]
- 6.Rozman P, Dovc T, Gassner C: Differentiation of autologous ABO, RHD, RHCE, KEL, JK, and FY blood group genotypes by analysis of peripheral blood samples of patients who have recently received multiple transfusions. Transfusion 2000, 40:936–942 [DOI] [PubMed] [Google Scholar]
- 7.El Kenz H, Efira A, Le PQ, Thiry C, Valsamis J, Azerad MA, Corazza F: Transfusion support of autoimmune hemolytic anemia: how could the blood group genotyping help? Transl Res 2014, 163:36–42 [DOI] [PubMed] [Google Scholar]
- 8.Fasano RM: Hemolytic disease of the fetus and newborn in the molecular era. Semin Fetal Neonatal Med 2016, 21:28–34 [DOI] [PubMed] [Google Scholar]
- 9.Cro F, Lapucci C, Vicari E, Salsi G, Rizzo N, Farina A: An innovative test for non-invasive Kell genotyping on circulating fetal DNA by means of the allelic discrimination of K1 and K2 antigens. Am J Reprod Immunol 2016, 76:499–503 [DOI] [PubMed] [Google Scholar]
- 10.Finning KM, Martin PG, Soothill PW, Avent ND: Prediction of fetal D status from maternal plasma: introduction of a new noninvasive fetal RHD genotyping service. Transfusion 2002, 42:1079–1085 [DOI] [PubMed] [Google Scholar]
- 11.Van der Schoot CE, Soussan AA, Koelewijn J, Bonsel G, Paget-Christiaens LG, de Haas M: Non-invasive antenatal RHD typing. Transfus Clin Biol 2006, 13:53–57 [DOI] [PubMed] [Google Scholar]
- 12.Clausen FB, Christiansen M, Steffensen R, Jorgensen S, Nielsen C, Jakobsen MA, Madsen RD, Jensen K, Krog GR, Rieneck K, Sprogoe U, Homburg KM, Grunnet N, Dziegiel MH: Report of the first nationally implemented clinical routine screening for fetal RHD in D-pregnant women to ascertain the requirement for antenatal RhD prophylaxis. Transfusion 2012, 52:752–758 [DOI] [PubMed] [Google Scholar]
- 13.Daniels G, Finning K, Martin P, Massey E: Noninvasive prenatal diagnosis of fetal blood group phenotypes: current practice and future prospects. Prenat Diagn 2009, 29:101–107 [DOI] [PubMed] [Google Scholar]
- 14.Svensson AM, Delaney M: Considerations of red blood cell molecular testing in transfusion medicine. Expert Rev Mol Diagn 2015, 15: 1455–1464 [DOI] [PubMed] [Google Scholar]
- 15.Denomme GA: Prospects for the provision of genotyped blood for transfusion. Br J Haematol 2013, 163:3–9 [DOI] [PubMed] [Google Scholar]
- 16.Fasano RM, Chou ST: Red blood cell antigen genotyping for sickle cell disease, thalassemia, and other transfusion complications. Transfus Med Rev 2016, 30:197–201 [DOI] [PubMed] [Google Scholar]
- 17.Boyle J, Thorpe SJ, Hawkins JR, Lockie C, Fox B, Matejtschuk P, Halls C, Metcalfe P, Rigsby P, Armstrong-Fisher S, Varzi AM, Urbaniak S, Daniels G: International reference reagents to standardise blood group genotyping: evaluation of candidate preparations in an international collaborative study. Vox Sang 2013, 104: 144–152 [DOI] [PubMed] [Google Scholar]
- 18.Kroll H, Carl B, Santoso S, Bux J, Bein G: Workshop report on the genotyping of blood cell alloantigens. Transfus Med 2001, 11: 211–219 [DOI] [PubMed] [Google Scholar]
- 19.Flegel WA, Chiosea I, Sachs UJ, Bein G: External quality assessment in molecular immunohematology: the INSTAND proficiency test program. Transfusion 2013, 53:2850–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denomme GA, Westhoff CM, Castilho L, Reid ME: Consortium for Blood Group Genes (CBGG): 2008 report. Immunohematology 2009, 25:75–80 [PubMed] [Google Scholar]
- 21.Denomme GA, Westhoff CM, Castilho LM, St-Louis M, Castro V, Reid ME: Consortium for Blood Group Genes (CBGG): 2009 report. Immunohematology 2010, 26:47–50 [PubMed] [Google Scholar]
- 22.Flegel WA, Gottschall JL, Denomme GA: Implementing mass-scale red cell genotyping at a blood center. Transfusion 2015, 55: 2610–2615. quiz 2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legler TJ, Maas JH, Kohler M, Wagner T, Daniels GL, Perco P, Panzer S: RHD sequencing: a new tool for decision making on transfusion therapy and provision of Rh prophylaxis. Transfus Med 2001, 11:383–388 [DOI] [PubMed] [Google Scholar]
- 24.Reid ME, Halter Hipsky C, Hue-Roye K, Hoppe C: Genomic analyses of RH alleles to improve transfusion therapy in patients with sickle cell disease. Blood Cells Mol Dis 2014, 52:195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner FF, Flegel WA: RHD gene deletion occurred in the Rhesus box. Blood 2000, 95:3662–3668 [PubMed] [Google Scholar]
- 26.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW: GenBank. Nucleic Acids Res 2016, 44(Database issue):D67–D72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer O, Hildebrandt M, Schulz B, Blasczyk R, Salama A: Simultaneous genotyping of human platelet antigens (HPA) 1 through 6 using new sequence-specific primers for HPA-5. Transfusion 1999, 39: 1256–1258 [DOI] [PubMed] [Google Scholar]
- 28.Wagner FF, Moulds JM, Flegel WA: Genetic mechanisms of Rhesus box variation. Transfusion 2005, 45:338–344 [DOI] [PubMed] [Google Scholar]
- 29.Westhoff CM, Vege S, Hipsky CH, Horn T, Hue-Roye K, Keller J, Velliquette R, Lomas-Francis C, Chou ST, Reid ME: RHCE*ceAG (254C>G, Ala85Gly) is prevalent in blacks, encodes a partial cephenotype, and is associated with discordant RHD zygosity. Transfusion 2015, 55:2624–2632 [DOI] [PubMed] [Google Scholar]
- 30.Perco P, Shao CP, Mayr WR, Panzer S, Legler TJ: Testing for the D zygosity with three different methods revealed altered Rhesus boxes and a new weak D type. Transfusion 2003, 43:335–339 [DOI] [PubMed] [Google Scholar]
- 31.Matheson KA, Denomme GA: Novel 3’Rhesus box sequences confound RHD zygosity assignment. Transfusion 2002, 42:645–650 [DOI] [PubMed] [Google Scholar]
- 32.Grootkerk-Tax MG, Maaskant-van Wijk PA, van Drunen J, van der Schoot CE: The highly variable RH locus in nonwhite persons hampers RHD zygosity determination but yields more insight into RH-related evolutionary events. Transfusion 2005, 45:327–337 [DOI] [PubMed] [Google Scholar]
- 33.Chen Q, Flegel WA: Random survey for RHD alleles among D+ European persons. Transfusion 2005, 45:1183–1191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


