Abstract
Background
We examined the current biomarker landscape in breast cancer when programmed death‐ligand 1 (PD‐L1) testing is integrated with comprehensive genomic profiling (CGP).
Material and Methods
We analyzed data from samples of 312 consecutive patients with breast carcinoma tested with both CGP and PD‐L1 (SP142) immunohistochemistry (IHC) during routine clinical care. These samples were stratified into hormone receptor positive (HR+)/human epidermal growth factor receptor negative (HER2−; n = 159), HER2‐positive (n = 32), and triple‐negative breast cancer (TNBC) cohorts (n = 121).
Results
We found that in the TNBC cohort, 43% (52/121) were immunocyte PD‐L1–positive, and in the HR+/HER2− cohort, 30% (48/159) had PIK3CA companion diagnostics mutations, and hence were potentially eligible for atezolizumab plus nab‐paclitaxel or alpelisib plus fulvestrant, respectively. Of the remaining 212 patients, 10.4% (22/212) had a BRCA1/2 mutation, which, if confirmed by germline testing, would allow olaparib plus talazoparib therapy. Of the remaining 190 patients, 169 (88.9%) were positive for another therapy‐associated marker or a marker that would potentially qualify the patient for a clinical trial. In addition, we examined the relationship between immunocyte PD‐L1 positivity and different tumor mutation burden (TMB) cutoffs and found that when a TMB cutoff of ≥9 mutations per Mb was applied (cutoff determined based on prior publication), 11.6% (14/121) patients were TMB ≥9 mutations/Mb and of these, TMB ≥9 mutations per Mb, 71.4% (10/14) were also positive for PD‐L1 IHC.
Conclusion
Our integrated PD‐L1 and CGP methodology identified 32% of the tested patients as potentially eligible for at least one of the two new Food and Drug Administration approved therapies, atezolizumab or alpelisib, and an additional 61.2% (191/312) had other biomarker‐guided potential therapeutic options.
Implications for Practice
This integrated programmed death‐ligand 1 immunohistochemistry and comprehensive genomic profiling methodology identified 32% of the tested patients as eligible for at least one of the two new Food and Drug Administration‐approved therapies, atezolizumab or alpelisib, and an additional 61.2% (191/312) had other biomarker‐guided potential therapeutic options. These findings suggest new research opportunities to evaluate the predictive utility of other commonly seen PIK3CA mutations in hormone receptor‐positive breast cancers and to standardize tumor mutation burden cutoffs to evaluate its potentially predictive role in triple‐negative breast cancer.
Keywords: Comprehensive genomic profiling, PD‐L1 immunohistochemistry, Biomarkers, Breast carcinoma
Short abstract
This article reports the current landscape of biomarkers in breast cancer, focusing on patients using comprehensive genomic profiling and PD‐L1 immunohistochemistry in addition to the previous standard of care diagnostics for hormone receptor and human epidermal receptor 2 identification. The relationship between different immunotherapy biomarkers in patients with triple‐negative breast cancer is examined.
Introduction
Breast cancer remains one of the leading causes of cancer death, with an estimated 268,600 newly diagnosed patients in the U.S. in 2019 [1]. Targeted therapy has been used in patients with breast carcinoma since 1978, with the approval of tamoxifen use on hormone receptor (HR)‐positive patients, and later in 1998, with the approval of trastuzumab by the U.S. Food and Drug Administration (FDA) for human epidermal receptor 2 (HER2) (also known as ERBB2) immunohistochemistry (IHC)‐based positive patients [2]. Because of the necessity of identifying patients with breast carcinoma for these targeted therapies, HR and HER2 testing is standard of care for patients with breast cancer, and patients are often stratified into HR‐positive (+) and/or HER2+, or triple‐negative (Estrogen Receptor−, Progesterone Receptor−, HER2−) breast cancer (TNBC) [3].
The past few years saw a rapid expansion of clinically validated targeted therapies in breast cancer. Pembrolizumab is approved by the FDA for microsatellite unstable cancers, including breast cancer, and larotrectinib for cancers with NTRK fusion genes. Phase II clinical trial data also demonstrated activity of neratinib in patients with breast cancer with activating HER2 mutations. However, these genomic alterations are rare in breast cancer. Recently, two new therapies were approved by the FDA for patients with breast cancer, both with accompanying companion diagnostics (CDx), and a much broader target population [4]. Atezolizumab plus nab‐paclitaxel is now available as a first‐line treatment for patients with locally advanced or metastatic TNBC with positive programmed death‐ligand 1 (PD‐L1) immune cell expression, as identified by the VENTANA SP142 CDx IHC assay, either in the primary tumor or in a metastatic lesion. In the IMpassion130 clinical trial, the difference in median overall survival for patients with PD‐L1 positivity was more than 10 months between the patients treated with atezolizumab plus nab‐paclitaxel versus placebo plus nab‐paclitaxel [5]. No benefit from atezolizumab in PD‐L1–negative patients was observed, which highlights the importance of patient selection based on PD‐L1 immune cell expression.
The second therapy with an accompanying CDx approved by the FDA in 2019 for patients with breast carcinoma was alpelisib plus fulvestrant, which is now available to patients with HR+/HER2‐negative (−) breast carcinoma if their tumors harbor certain PIK3CA mutations. In the SOLAR‐1 clinical trial, the overall response among patients with PIK3CA mutations treated with alpelisib plus fulvestrant was 26.6%, compared with the 12.8% treated with placebo plus fulvestrant, and progression free survival also improved significantly by 5 months (11 vs. 5.7, p < .001) [6]. The first companion diagnostic approved for this therapy was from QIAGEN in May 2019 and consisted of a polymerase chain reaction kit that detects 11 mutations in the PIK3CA gene (Exon 7: C420R; Exon 9: E542K, E545A, E545D [1635G > T only], E545G, E545K, Q546E, Q546R; and Exon 20: H1047L, H1047R, H1047Y) [4]. In December of 2019, Foundation Medicine's FoundationOne CDx (F1CDx) comprehensive genomic profiling (CGP) assay also received FDA approval as a CDx for alpelisib in breast cancer. The same 11 mutations as the QIAGEN kit were approved as a CDx in the F1CDx approval.
Olaparib and talazoparib were also recently approved as single‐agent therapies for metastatic breast cancers that harbor germline BRCA1/2 mutations [7, 8]. In addition, presence of a germline and/or somatic BRCA1/2 mutation could potentially allow a patient to be eligible for one of the 10 ongoing clinical trials (ranges from phase I to phase III trials) in which this biomarker is an eligibility criterion (available on https://clinicaltrials.gov/). According to the 2019 National Comprehensive Cancer Network (NCCN) guidelines, BRCA1/2 germline mutation testing in HER2− breast carcinoma is now recommended as a predictive test (in addition to its well‐established use as a diagnostic test for inherited susceptibility to breast cancer) [9]. Somatic tumor mutation profiling cannot definitely distinguish between germline versus acquired somatic BRCA1/2 mutations. However, detection of BRCA1/2 mutation in cancer tissues warrant subsequent germline testing and is one of the indications for germline screening according to NCCN guidelines.
With these two new therapeutic options, which both require a CDx, and the recent expansion of BRCA1/2 mutation testing, we sought to examine the current landscape of biomarkers in patients with breast cancer using CGP and PD‐L1 IHC in addition to the previous standard of care diagnostics of HR and HER2 identification. In addition, we examine the relationship between the different immunotherapy biomarkers in patients with TNBC.
Material and Methods
Breast Carcinoma Cohort
We performed a retrospective analysis of 312 consecutive patients with breast carcinoma who were tested with both CGP (F1CDx) and PD‐L1 IHC (SP142 CDx IHC) between March 2019 and June 2019. Approval for this study was obtained from the Western Institutional Review Board Protocol No. 20152817. All cases were submitted to Foundation Medicine for CGP and PD‐L1 IHC during routine clinical care. HR and HER2 status were determined from a combination of accompanying pathology reports from the outside institution and HER2 amplification status based on our F1CDx assay. Age, sex, and site of specimen of patient were extracted from accompanying pathology reports.
PD‐L1 SP142 CDx Immunohistochemistry Testing
All PD‐L1 IHC testing was performed using the VENTANA SP142 CDx assay per manufacturer's instructions in a Clinical Laboratory Improvement Amendments (CLIA)‐certified and College of American Pathologists (CAP)‐accredited reference laboratory (Foundation Medicine, Morrisville, NC). VENTANA SP142 CDx assay consists of the rabbit monoclonal anti‐PD‐L1 SP142 clone, the OptiView DAB IHC detection kit, the Opti‐View Amplification Kit stained on the VENTANA BenchMark ULTRA instrument using the staining protocol provided by the package insert and interpreted with the guidelines of the VENTANA interpretation guide [10, 11]. All cases have an accompanying H&E‐stained patient slide, negative regent control‐stained patient slide with an on‐slide tonsil control, and a VENTANA PD‐L1 SP142‐stained patient slide with an on‐slide tonsil control. PD‐L1 IHC slides were interpreted by board‐certified pathologists using the tumor‐infiltrating immune cell (IC) percentage scoring method, where IC% = proportion of tumor area that is occupied by PD‐L1 staining IC of any intensity. Each case was interpreted by one of six pathologists who were trained specifically in the SP142 PD‐L1 TNBC CDx assay interpretation, and borderline cases (close to 1% IC positive) were reviewed by at least two pathologists to arrive at a consensus score. Tumor‐infiltrating immune cells consist of lymphocytes, macrophages, dendritic cells, and granulocytes. In general, IC stains with a dark, granular punctate pattern; however, different staining patterns such as membranous staining can also be present as explained in the VENTANA interpretation guide. Tumor area for the purposes of this assay was defined as tumor cells and associated peritumoral and intratumoral stroma. The CDx cutoff for atezolizumab plus nab‐paclitaxel for TNBC is an IC score of ≥1%. Tumor cells usually stain with a linear pattern, but tumor cell staining percentage is not considered in the TNBC CDx cutoff for atezolizumab plus nab‐paclitaxel.
Comprehensive Genomic Profiling of Breast Carcinoma Samples
CGP was performed using the FDA‐approved FoundationOne CDx assay in a CLIA‐certified and CAP‐accredited laboratory (Foundation Medicine, Cambridge, MA) using previously described methods [12]. F1CDx uses a next generation sequencing platform and a hybrid capture methodology that detects base substitutions, insertions and deletions, and copy number alterations in 324 genes and select gene rearrangements, as well as tumor mutation burden (TMB) and microsatellite instability (MSI). Each sample had an accompanying H&E slide and was reviewed by a board‐certified pathologist under light microscopy for presence of adequate tumor (≥ 20% of nucleated cells are tumor cells) before sequencing and for review of genomic findings after sequencing for final report approval. With the F1CDx assay, the CDx claim for alpelisib in patients with breast cancer is specific to certain PIK3CA gene mutations (Exon 7: C420R; Exon 9: E542K, E545A, E545D [1635G > T only], E545G, E545K, Q546E, Q546R; and Exon 20: H1047L, H1047R, H1047Y). However, the F1CDx assay also identifies additional pathogenic PIK3CA gene mutations not currently FDA approved as targetable mutations by alpelisib. Tumor mutational burden was determined on 0.8 Mb of sequenced DNA, and assessment of microsatellite instability was performed from DNA sequencing across 114 loci as previously described [13].
Integrative Analysis of CGP and PD‐L1 Biomarkers in Breast Carcinoma
To examine the potential impact of testing with both CGP and PD‐L1 IHC, we examined the rates of PD‐L1 IC positivity in the TNBC disease subset (n = 121), PIK3CA CDx mutation in the HR+/HER2− disease subsets (n = 159), and BRCA1/2 mutations for the overall breast carcinoma cohort (n = 312). We also examined additional biomarkers (ABIOs) with a biomarker‐associated therapy or an active clinical trial based on their biomarker status (actionability associated with short variants, copy number alterations, and/or rearrangements). Examples of ABIOs include PTEN loss or mutations for potential eligibility of everolimus and 10 clinical trials; and CCND1 amplifications for potential eligibility of abemaciclib, palbociclib, and/or ribociclib and 10 clinical trials.
Next, we examined the genomic biomarker landscape of our patient cohort by extracting the top 20 genes with mutations in each disease subset (HR+/HER2−, HER2+, TNBC) and representing them in comutation plots. We next analyzed the top 20 genes in the total cohort of 312 patient cases and compared these 20 genes between the three disease subsets (HR+/HER2−, HER2+, TNBC) using the Fischer's exact test to compare HR+/HER2− versus HER2+, HR+/HER2− versus TNBC, and HER2+ versus TNBC. The p value was adjusted for multiple comparisons using the Bonferroni method, and p < .05 was considered significant [14]. We also examined the top 20 genes of the TNBC disease subset and compared the TNBC PD‐L1+ and TNBC PD‐L1− disease subset using the Fisher's exact test. In addition, we examined TMB in TNBC PD‐L1+ and TNBC PD‐L1− disease subsets. For the purposes of these analyses, HER2 alterations were not included because they were previously extracted from a combination of accompanying pathology reports and HER2 amplification status and would be redundant.
Finally, we examined immunotherapy biomarkers with PD‐L1 IHC, TMB, and MSI. For TMB, we examined the cutoffs of 5, 9, 10, and 20 mutations per Mb in correlation to PD‐L1 status. Using 5 mutations per Mb (mean TMB) as a baseline, we examined 10 mutations/Mb (2× baseline) and 20 mutations per Mb (4× baseline) as exploratory cutoffs for TMB. In addition, we specifically examined a TMB ≥9 mutations per Mb cutoff as previously described by Alva et al. that showed a disease control rate of 37% in patients that had TMB ≥9 mutations per Mb using the F1CDx assay and treated with pembrolizumab [15].
Results
Patient Cohort
A total of 312 patients with breast carcinoma had combined CGP and PD‐L1 IHC testing at Foundation Medicine between March 2019 and June 2019. The median age was 57 years, with a range between 28 and over 80 years. A total of 26.6% (83/312) of the patients were at least 65 years, and 74.4% (229/312) of the patients were younger than 65 years. All patients in this cohort were female, except for two male patients. A majority of the specimens received were from a metastatic site, 61.9% (193/312). Of the 312 samples tested, 51% (159/312) were HR+/HER2−, 10% (32/312) were HER2+ (of the 32 HER2+ patients, 53.1% [17/32] were also HR+), and 39% (121/312) were TNBC (Fig. 1). Detailed patient characteristics are in Table 1.
Figure 1.
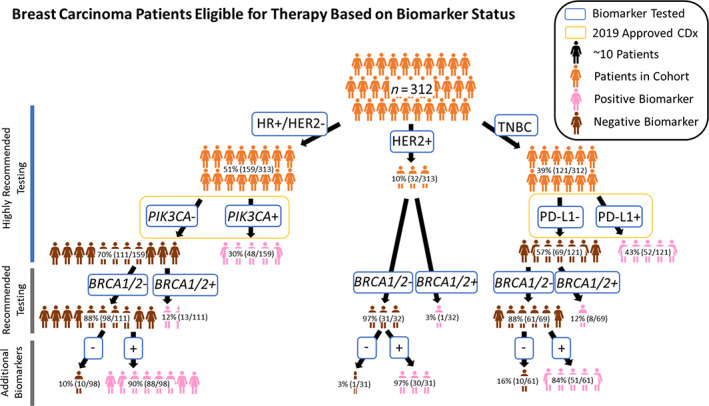
Represents patients with breast carcinoma eligible for therapy based on biomarker status. Patients with breast carcinoma were stratified into HR+/HER2−, HER2+, and triple‐negative breast cancer (TNBC) groups and tested with standard of care diagnostics. Next, in the HR+/HER2− cohort, patients were stratified into PIK3CA+ versus PIK3CA− groups, and patients with TNBC were stratified into programmed death‐ligand 1 (PD‐L1) + versus PD‐L1− groups. The remaining patients that were not positive for PIK3CA and/or PD‐L1 were stratified into whether they had a BRCA1/2 mutation. Last, the patients without positivity in one of the above‐mentioned biomarkers were examined for positivity in a biomarker that was clinically actionable or potentially clinically actionable.
Abbreviations: −, negative; +, positive; CDx, companion diagnostics; HR, hormone receptor.
Table 1.
Demographic and histological characteristics of 312 patients with breast carcinoma
| Characteristics | Overall (n = 312), n (%) | HR+/HER2− (n = 159), n (%) | HER2+ (n = 32), n (%) | TNBC (n = 121), n (%) |
|---|---|---|---|---|
| Median age, yr | 57 | 58 | 57 | 56 |
| Gender, female | 310 (99.4) | 157 (98.7) | 32 (100) | 121 (100) |
| Specimen from metastatic sites | 193 (61.9) | 114 (71.7) | 20 (62.5) | 59 (48.8) |
| Breast carcinoma histologic subtype | ||||
| Carcinoma (NOS) | 159 (51.0) | 93 (58.5) | 17 (53.1) | 49 (40.5) |
| Invasive ductal carcinoma | 132 (42.3) | 53 (33.3) | 14 (43.8) | 65 (53.7) |
| Invasive lobular carcinoma | 17 (5.4) | 12 (7.5) | 1 (3.1) | 4 (3.3) |
| Metaplastic carcinoma | 3 (1.0) | 1 (0.6) | 0 (0) | 2 (1.7) |
| Myoepithelial carcinoma | 1 (0.3) | 0 (0) | 0 (0) | 1 (0.8) |
Abbreviations: −, negative; +, positive; HR, hormone receptor; NOS, not otherwise specified; TNBC, triple‐negative breast cancer.
Clinically Actionable Biomarkers Detected by CGP and PD‐L1 Immunohistochemistry
As expected, detection of potentially actionable MSI‐high (MSI‐H) status, NTRK fusions, and somatic mutations in HER2 were low. In the overall cohort of 312 patients, 0.3% (1/312) were MSI‐H, 0.3% (1/312) had a NTRK3 fusion (partner gene CDK12), and 2.6% (8/312) had a somatic HER2 mutation (not amplification [2 cases: S310F; 2 cases: L755S; 1 case: L755_T759del; 1 case: P780_Y781insGSP; 1 case: V777L; and 1 case: L755S and S310F]).
In the HR+/HER2− cohort, 38% (60/159) were positive for a PIK3CA mutation. The top two PIK3CA mutations were E542K (33.3%, 20/60) and H1047R (26.7%, 16/60), and of the 60 HR+/HER2− PIK3CA positive cases, 80% (48/60) had 1 of the 11 CDx PIK3CA mutations and 18.3% (11/60) of samples had more than one PIK3CA mutation (Table 2). An example of a E545K PIK3CA mutation is shown in supplemental online Figure 1A. Of the remaining 159 HR‐positive patients who were PIK3CA CDx negative, 12% (13/111) were positive for a BRCA1/2 mutation, and of the 98 patients that were negative for a biomarker previously mentioned, 90.0% (88/98) were positive for an ABIO examined (e.g., PIK3CA [non‐CDx mutation] 12.2% [12/98], PTEN loss/mutations 13.3% [13/98], and CCND1 amplifications 29.6% [29/98]; Fig. 1).
Table 2.
Prevalence of specific CDx and non‐CDx PIK3CA alterations in 159 HR+/HER2– patients
| CDx PIK3CA mutations | Percentage of HR+/HER2− cases (n = 159), n (%) | Non‐CDx PIK3CA mutations, n (%) | Percentage of HR+/HER2− cases (n = 159), n (%) |
|---|---|---|---|
| E542K | 20 (12.6) | N345K | 5 (3.1) |
| H1047R | 16 (10.1) | H450_L455del | 1 (0.6) |
| C420R | 1 (0.6) | H450_P458del | 1 (0.6) |
| E545A | 1 (0.6) | R88Q | 1 (0.6) |
| E545D [1635G > T only] | 1 (0.6) | Mixed a | 4 (2.5) |
| E545K | 1 (0.6) | ||
| H1047L | 1 (0.6) | ||
| E545G | 0 (0) | ||
| Q546E | 0 (0) | ||
| Q546R | 0 (0) | ||
| H1047Y | 0 (0) | ||
| Mixed b | 7 (4.4) |
Case: 1. E545Q, N345K; 2. D1017H, G1049R; 3. E545Q, N345K; 4. Q546K, T1025A
Case: 1. E542K, E453K, C420R, Q546K; 2. E542K, M1004I; 3. E545K, M1043I; 4. E545K, E726K; 5. H1047R, Q969K; 6. H1047R, E453K; 7. H1047R, D350G.
Abbreviations: −, negative; +, positive; CDx, companion diagnostics; HR, hormone receptor.
For the HER2+ samples, 3.1% (1/32) patients were positive for a BRCA1/2 mutation, and for the remaining 31 patients, 30 (96.8%) patients were positive for an ABIO examined (PTEN loss or mutations 3.2% [1/31], and CCND1 amplifications 12.9% [4/31]).
Finally, in the TNBC cohort, 43% (52/121) were positive for PD‐L1 IHC and of the remaining 69 patients, 11.6% (8/69) were positive for a BRCA1/2 mutation. Supplemental online Figure 1B (H&E) and C (corresponding PD‐L1 SP142 IHC) represent an example of a patient with TNBC with greater than 1% of immune cells staining in the tumor area and hence positive for the PD‐L1 SP142 IHC CDx. Of the 61 patients without positivity in one of the biomarkers previously mentioned, 83.6% (51/61) were positive for an ABIO examined (PTEN loss/mutations 16.4% [10/61], and CCND1 amplifications 6.6% [4/61]). Although not a companion diagnostic in HER2+ and HR+/HER2− patients, the prevalence of PD‐L1 SP142 positivity (based on an IC ≥1% cutoff) was 43.8% (14/32) in the HER2+ cohort and 22.0% (35/124) in the HR+/HER2− cohort.
Genomic Landscape of Patients with Breast Carcinoma
The genomic profiles of each of the 312 patients we examined are shown in the comutation plots (Figs. 2, 3). Here, we observed that although there was some overlap in the top 20 genes in each disease subset (HR+/HER2−, HER2+, and TNBC) there were some differences. For the HR+/HER2− disease subset, the top 5 genes in descending order were PIK3CA, TP53, RAD21, NBN, and CCND1; for the HER2+ disease subset, they were TP53, CDK12, PIK3CA, MYC, and RAD21; and for the TNBC disease subset they were TP53, RAD21, MYC, PIK3CA, and NBN. This observation was also true for the TNBC PD‐L1+ and PD‐L1− disease subset in which the top five genes in the PD‐L1+ cohort in descending order were TP53, RAD21, MYC, DDR2, and FH; for the PD‐L1− cohort, they were TP53, RAD21, MYC, NBN, and RB1, although no significant difference was found between the two disease subsets, as explained below.
Figure 2.
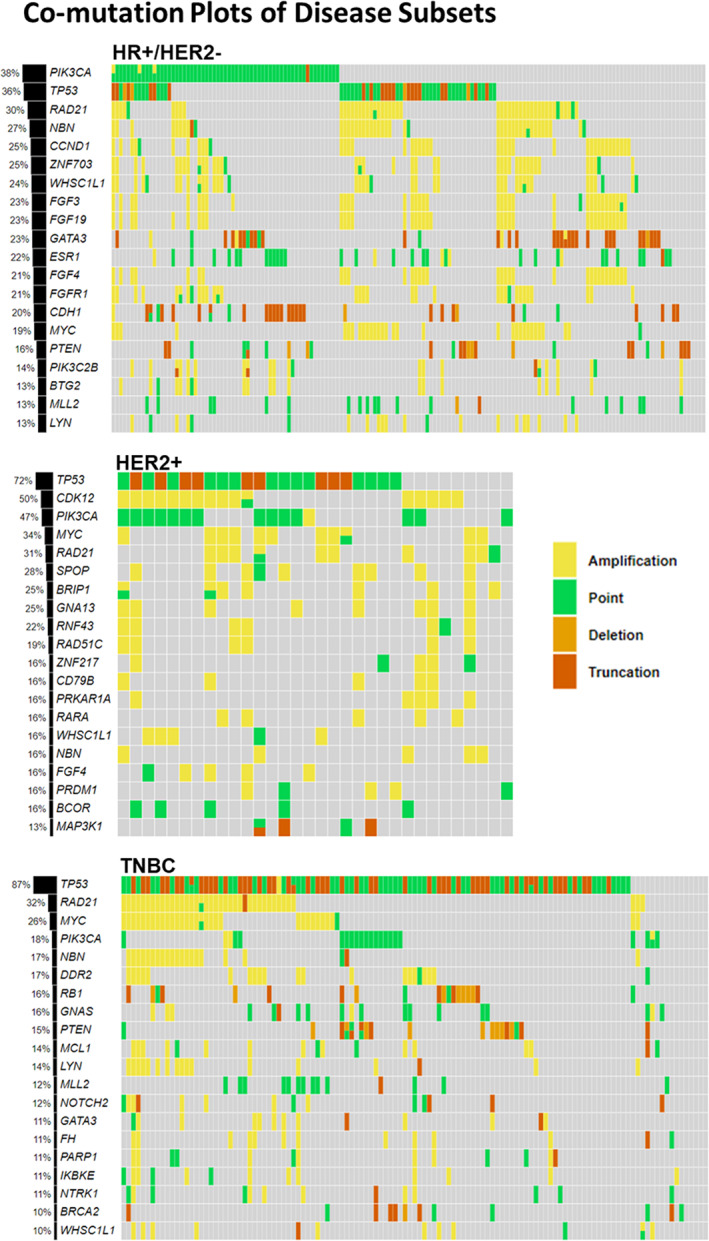
Comutation plots of the top 20 genes for each breast carcinoma cohort stratified based on biomarker status of HR and HER2. Here, we see that although there was some overlap in the top 20 genes in each cohort (HR+/HER2−, HER2+, and TNBC) there were some differences. For the HR+/HER2− cohort, the top 5 genes in descending order were PIK3CA, TP53, RAD21, NBN, and CCND1; for the HER2+ cohort, they were TP53, CDK12, PIK3CA, MYC, and RAD21; and for the TNBC cohort, they were TP53, RAD21, MYC, PIK3CA, and NBN. Abbreviations: −, negative; +, positive; HR, hormone receptor; TNBC, triple‐negative breast cancer.
Figure 3.
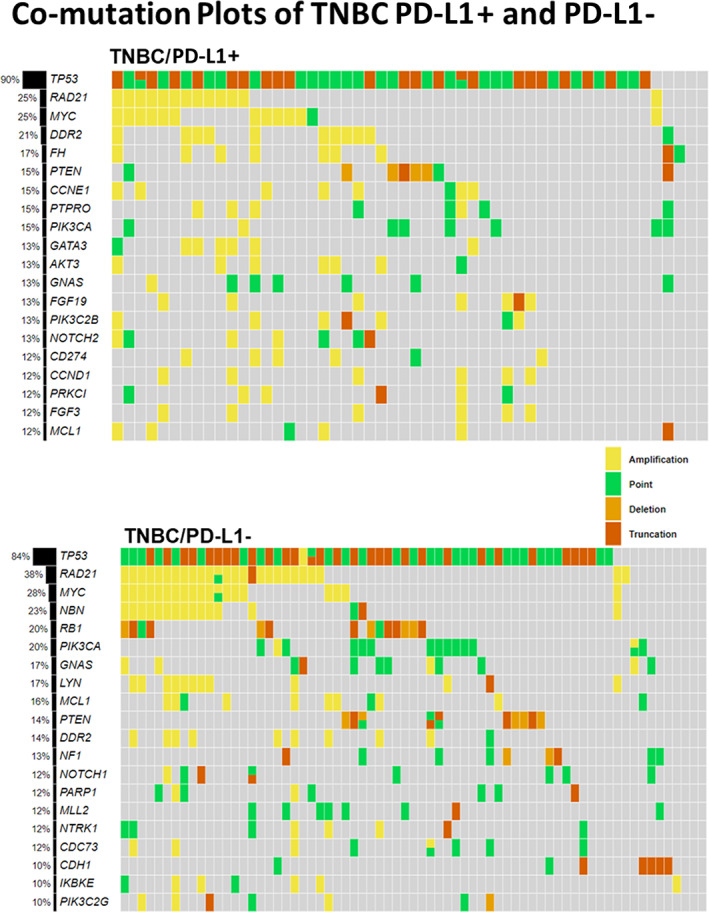
Comutation plots of the top 20 genes for the TNBC PD‐L1+ and TNBC PD‐L1− breast carcinoma cohort. Here, we see that although there was some overlap in the top 20 genes, in each cohort there were some differences. In the TNBC breast carcinoma cases, the top five genes in the PD‐L1+ cohort in descending order were TP53, RAD21, MYC, DDR2, and FH; and for the PD‐L1− cohort, they were TP53, RAD21, MYC, NBN, and RB1. Abbreviations: −, negative; +, positive; PD‐L1, programmed death‐ligand 1; TNBC, triple‐negative breast cancer.
Mutation frequencies were determined for each disease subset, and Fisher's exact test was performed to compare gene mutation frequencies for the HR+/HER2− versus HER2+, HR+/HER2− versus TNBC, and HER2+ versus TNBC disease subset (Fig. 4; supplemental online Table 1). The p value was adjusted for multiple comparisons using the Bonferroni method, and p < .05 was considered significant. It was found that there was a significantly higher TP53 (p < .001) rate of mutation in the TNBC disease subset versus the HR+/HER2− disease subset and that there were significantly higher PIK3CA (p = .020), CCND1 (p = .015), ZNF703 (p = .003), and ESR1 (p < .001) rates of mutation in the HR+/HER2− disease subset when compared with the TNBC disease subset. In the TNBC disease subset, further comparison between TNBC PD‐L1+ and TNBC PD‐L1− samples revealed no significant difference in gene mutation frequencies at individual gene level. (supplemental online Table 2). Median TMB was the same (4 mutations per Mb) between the TNBC PD‐L1+ and TNBC PD‐L1− disease subset, with a higher mean (5.5 mutations per Mb) in the TNBC PD‐L1− disease subset when compared with the mean (4.4 mutations per Mb) in the TNBC PD‐L1+ disease subset.
Figure 4.
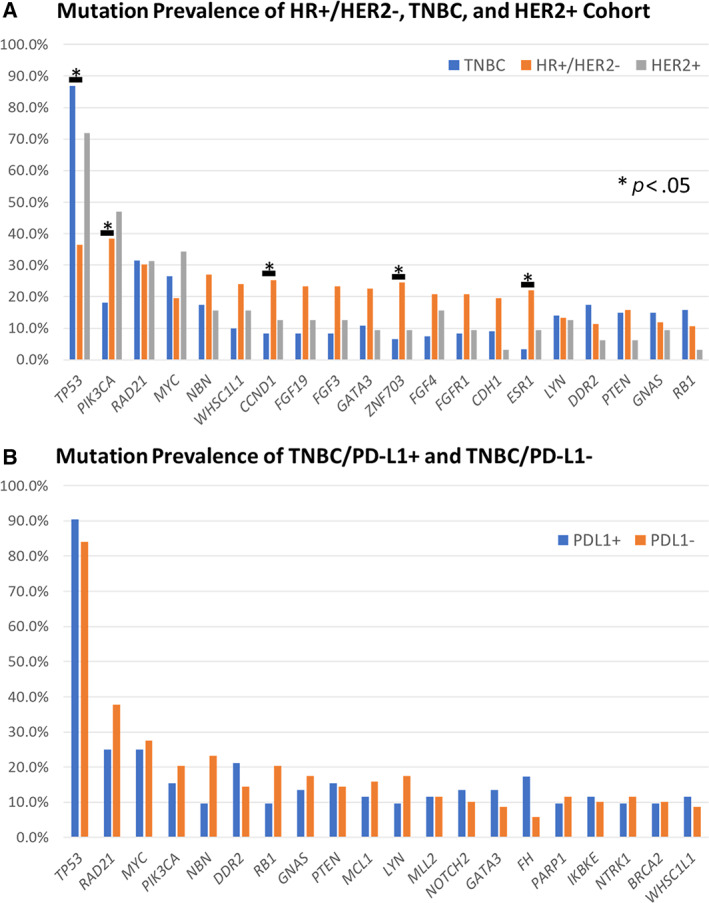
Mutational prevalence of different breast carcinoma patient cohorts based on HR, HER2, and PD‐L1 status. (A): Mutation prevalence of top 20 genes in HR+/HER2−, TNBC, and HER2+ cohorts. Mutation prevalence was determined for each cohort, and Fisher's exact test was performed to compare HR+/HER2− versus HER2+, HR+/HER2− versus TNBC, and HER2+ versus TNBC cohorts (supplemental online Table 1). The p value was adjusted for multiple comparisons using the Bonferroni method, and p < .05 was considered significant. It was found that there was a significantly higher TP53 (p < .001) rate of mutation in the TNBC disease subset vs the HR+/HER2− disease subset and that there were significantly higher PIK3CA (p = .020), CCND1(p = .015), ZNF703 (p = .003), and ESR1 (p < .001) rates of mutation in the HR+/HER2− disease subset when compared with the TNBC disease subset. (B): The top 20 genes in the TNBC cohort were also extracted and no significant difference was discovered using the Fisher's exact test when comparing the programmed death‐ligand 1 (PD‐L1) + and PD‐L1− cohort (supplemental online Table 2). Bars represent genes with significant differences in rate of mutation. Abbreviations: −, negative; +, positive; HR, hormone receptor; TNBC, triple‐negative breast cancer.
Immunotherapy Biomarkers Relationship
The mean and median (mutation per megabase) TMB were as follows: overall breast carcinoma (5.0, 4.0), HER2+ (5.1,4.0), HR+/HER2− (5.1, 3.0), and TNBC (4.9, 4.0). Currently, there is no established TMB cutoff for patients with breast carcinoma and so we explored several TMB cutoffs in this study. We explored the relationship between PD‐L1 immune cell positivity in relationship to a range of TMB cutoffs (5, 9, 10, and 20 mutations per Mb; Fig. 5). We found that at a lower TMB cutoff, such as 5 mutations per Mb, there was a higher percentage of patients that would be TMB+/PD‐L1− as opposed to a higher TMB cutoff, such as 20 mutations per Mb. In addition, we specifically examined a TMB cutoff of ≥9 mutations per Mb as previously described by Alva et al. and found that TMB ≥9 mutations per Mb was present for 11.5% (14/122) of cases, and of these cases, 71.4% (10/14) were PD‐L1+. This observation suggests that if TMB ≥9 mutations per Mb was used for treatment stratification, an additional 3.3% (4/121) of patients with TNBC could be potentially eligible for immunotherapy when compared with PD‐L1 IHC alone. In the overall breast carcinoma cohort, MSI was high in only one patient. That patient had TNBC with a TMB of greater than 20 mutations per Mb and was negative for PD‐L1.
Figure 5.
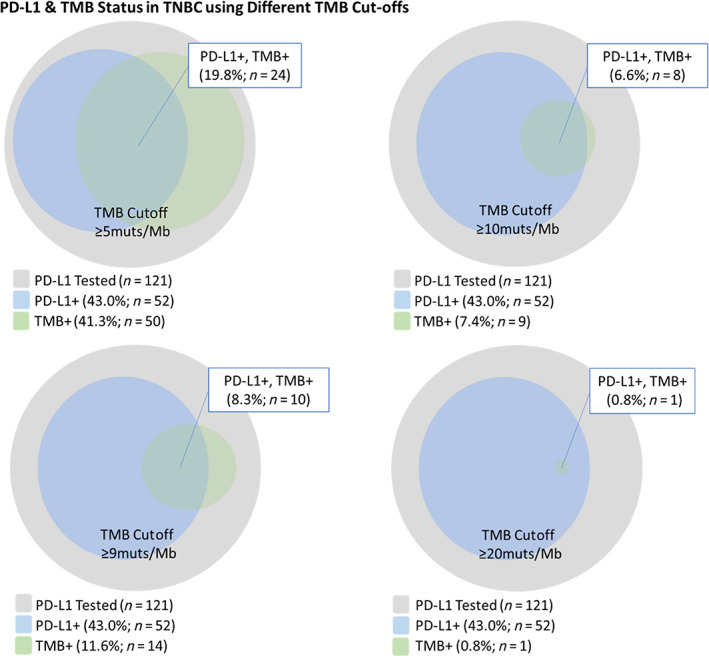
Figure of relationship between PD‐L1 status using the SP142 companion diagnostics immunohistochemistry assay and different exploratory TMB cutoffs (≥5, ≥9, ≥10, and ≥ 20 mutations per Mb). PD‐L1+ was defined as tumor‐infiltrating immune cell% ≥1% as per U.S. Food and Drug Administration companion diagnostic approval for atezolizumab plus nab‐pacitaxel. Abbreviations: +, positive; HR, hormone receptor; mut, mutation; TMB, tumor mutation burden; TNBC, triple‐negative breast cancer; PD‐L1, programmed death‐ligand 1.
Discussion
In this study, 32% (100/312) of patients were biomarker positive for an FDA‐approved companion diagnostic based on CGP and PD‐L1 IHC. Of the remaining 212 patients, 10.3% (22/212) had a BRCA1/2 mutation, and of the remaining 190 patients, 169 (88.9%) were biomarker positive for another biomarker‐associated therapy and/or clinical trial based on their genomic profile. These data provide real‐world evidence of the biomarker landscape when combining CGP with PD‐L1 IHC and highlight the importance of using these predictive biomarkers to provide the best treatment options for patients with breast cancer.
One specific example of the value of CGP is in patient A, a 59‐year‐old HR+/HER2− female patient with breast carcinoma, who had a PIK3CA E545K mutation (supplemental online Fig. 1A). In addition to the PIK3CA mutation that confers potential eligibility for alpelisib plus fulvestrant, the patient had a PTEN C124S mutation (potentially eligible with everolimus and 10 different clinical trials); a ARID1A Q944* mutation (potentially eligible for 6 different clinical trials), MAPK1 E322K (potentially eligible for 2 different clinical), a high TMB of 58 mutations per Mb (potential eligibility for multiple immunotherapies), and 8 other mutations that have no reportable therapeutic or clinical trial options. A specific example of the potential utility of CGP + PD‐L1 IHC is in patient B, a 49‐year‐old female patient with TNBC, who was positive for the PD‐L1 SP142 CDx assay and hence potentially eligible for atezolizumab plus nab‐pacitaxel based on the diagnostic test results (supplemental online Fig. 1B, C). This patient was microsatellite stable and had a TMB of 6 mutations per Mb. However, the patient also had an AKT2 amplification (potentially eligible for 10 different clinical trials), an MYC amplification (potentially eligible for 5 different clinical trials), and three other mutations that have no reportable therapeutic or clinical trial options. Therefore, in addition to this patient being potentially eligible for atezolizumab plus nab‐paclitaxel, the patient was also potentially eligible for 15 clinical trials based on the CGP findings.
After examining the potential treatment implications of combining CGP and PD‐L1 IHC, we examined the genomic landscape of patients with breast carcinoma and looked for differences between the different cohorts. Basal‐like is similar to TNBC in that they both lack the expression of ER, PR, and HER2, and Luminal A/B are similar to HR+/HER2− patients in that they are defined by being HR+, although Luminal B patients can be HER2+/−. Consistent with the literature, TP53 was mutated in a higher percentage of patients with TNBC (basal‐like; The Cancer Genome Atlas [TCGA] 84% vs. this data set 86.8%), PIK3CA was mutated in a moderate amount of HR+/HER2− (Luminal A) patients (TCGA 49% vs this data set 38.4%), and CCND1 was mutated in a higher amount of HR+/HER2− (Luminal A; TCGA 29% vs. our data set 25.2%) when compared with other subcohorts [16]. Also, ESR1 mutations occurred in a higher percentage in the HR+/HER2− cohort, which was consistent with a study by Niu et al., which found a 12.5% ESR1 mutation rate in patients with HR+ breast carcinoma. Although we do not have information on when the specimen was collected from the patient, the most likely reason for ESR1 mutation enrichment in patients with HR+ breast carcinoma is that the sample could have been taken after hormonal therapy, which induces the ESR1 mutation. Patients with TNBC do not receive treatment with hormonal therapy, so ESR1 mutations would not be expected. Similarly, ZNF703 amplifications have been shown to be higher in Luminal B breast cancer, which was reflected in our HR+/HER2− patients [17, 18]. Interestingly, all these mutations have either prognostic or therapeutic value in these specific cohorts of patients. Data suggest that TNBC patients with TP53 mutations have a poor prognosis [19]. PIK3CA mutations act in the PIK3CA/PTEN pathway and there are currently PIK3CA directed therapies on the market such as alpelisib for HR+/HER2− patients. CCND1 amplification has been shown to be a predictor of poor prognosis in ER+ breast carcinoma patients in multiple studies [20, 21, 22]. ESR1 mutations can potentially lead to hormonal therapy resistance for HR+ patients [23]. ZNF703 overexpression has been shown to have resistance to tamoxifen through activation of the Akt/mTOR signaling pathway in a study using luminal B‐type breast cancer cell lines [24].
Currently, the SP142 assay is the only FDA‐approved companion diagnostic for immunotherapy in patients with TNBC, although MSI‐high status qualifies patients for pembrolizumab therapy. TMB emerged recently as another promising marker that can only be performed using CGP or whole exome sequencing. In our TNBC cohort, MSI was only high in one patient, but depending on the four exploratory TMB cutoffs, the TMB high category ranged from 0.8% to 41.3%. Furthermore, the overlap between TMB and PD‐L1 positivity overlapped in more cases as the TMB cutoff increased. Although there are some data that support a TMB cutoff of 9 mutations per Mb for metastatic breast cancer, more clinical evidence is necessary to find the most optimal cutoff for TMB in patients with TNBC. Another question that arises from this study is if a single biomarker or a combination of biomarkers predicts the best response to immunotherapy for TNBC patients. For the PIK3CA CDx, there are currently only 11 mutations on the CDx label. In our cohort of patients, we were able to identify 12 additional cases (20%; 12/60) with PIK3CA mutations that had a pathogenic PIK3CA alteration. The ability of CGP to identify alterations outside of current FDA drug labels is one of the important advantages of CGP, as it allows for the real‐time practice of evidence‐based medicine and improved clinical trial enrollment in accordance with NCCN guidelines. In addition, we saw 18.3% of HR+/HER2− patients with more than one PIK3CA mutation. In a study by Vasan et al., the authors saw that patients with more than one PIK3CA mutation in cis were likely more sensitive to PIK3CA inhibitors [25].
The first limitation of this study is that we do not have strong clinical outcomes data to correlate with a TMB cutoff. However, we tried to mitigate this limitation by exploring multiple different exploratory cutoffs and also using the TMB cutoff of 9 mutations per Mb, which was observed by Alva et al. to have a disease control rate of 37% [15]. The second limitation in this study is that our center is a referral center and most of the patient samples we received are from advanced disease and were sent to us specifically for either CGP or PD‐L1 testing. This limitation is apparent in our subtype‐specific disease prevalence rates. For example, in this cohort of 312 patients, 39% (121/312) were patients with TNBC. In the general U.S. population, TNBC occurs in approximately 10%–20% of patients with breast carcinoma [26]. Our higher prevalence was most likely due to referral bias for patients with advanced disease and providers specifically ordering PD‐L1 CDx IHC testing for patients with TNBC.
Conclusion
These findings highlight the potential utility of combined CGP and PD‐L1 IHC for patients with breast carcinoma. Together, these techniques have the potential to help determine eligibility for two new therapies with accompanying companion diagnostics, as well as expand the biomarker‐guided therapeutic options available for patients with breast carcinoma. These findings also illustrate the need for studying the predictive value of common PIK3CA mutations not currently included in the CDx panel and establishing the optimal TMB cutoff for patients with TNBC that could potentially predict response to immunotherapy beyond PD‐L1 expression.
Author Contributions
Conception/Design: Richard S.P. Huang, Shakti H. Ramkissoon
Provision of study material or patients: Richard S.P. Huang, Eric Severson, Daniel L. Duncan, Amanda Hemmerich, Claire Edgerly, Erik Williams, Julia Elvin, Jo‐Anne Vergilio, Jonathan Killian, Douglas Lin, Matthew Hiemenz, Shakti H. Ramkissoon
Collection and/or assembly of data: Richard S.P. Huang, Xinyan Li, James Haberberger, Ethan Sokol, Jinpeng Xiao, Natalie Danziger
Data analysis and interpretation: All authors.
Manuscript writing: All authors.
Final approval of manuscript: All authors.
Disclosures
Richard S. P. Huang: Foundation Medicine (E, OI); Xinyan Li: Foundation Medicine (E, OI); James Haberberger: Foundation Medicine (E, OI); Ethan Sokol: Foundation Medicine (E, OI); Eric Severson: Foundation Medicine (E, OI); Daniel L. Duncan: Foundation Medicine (E, OI); Amanda Hemmerich: Foundation Medicine (E, OI); Claire Edgerly: Foundation Medicine (E, OI); Erik Williams: Foundation Medicine (E, OI); Julia Elvin: Foundation Medicine (E, OI); Jo‐Anne Vergilio: Foundation Medicine (E, OI); Jonathan Keith Killian: Foundation Medicine (E, OI); Douglas Lin: Foundation Medicine (E, OI); Matthew Hiemenz: Foundation Medicine (E, OI); Jinpeng Xiao: Foundation Medicine (E, OI); Deborah McEwan: Foundation Medicine (E, OI); Oliver Holmes: Foundation Medicine (E, OI); Natalie Danziger: Foundation Medicine (E, OI); Rachel Erlich: Foundation Medicine (E, OI); Garrett Frampton: Foundation Medicine (E, OI); Kimberly McGregor: Foundation Medicine (E, OI); Prasanth Reddy: Foundation Medicine (E, OI); Dawn Cardeiro: Foundation Medicine (E, OI); Rachel Anhorn: Foundation Medicine (E, OI); Phar Jeffrey Venstrom: Foundation Medicine (E, OI); Brian Alexander: Foundation Medicine (E, OI); Charlotte Brown: Foundation Medicine (E, OI); Lajos Pusztai: AstraZeneca, Merck, Novartis, Bristol‐Myers Squibb, Genentech, Eisai, Pieris, Immunomedics, Seattle Genetics, Clovis, Syndax, H3Bio, Daiichi (C/A), Merck, AstraZeneca, Seattle Genetics (RF); Jeffrey S. Ross: Foundation Medicine (E, OI); Shakti H. Ramkissoon: Foundation Medicine (E, OI). Michael B. Cohen indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Supplemental Figure
Supplemental Tables
Acknowledgments
We thank Clarence Owens, Cierra Smith, Bethany Thompson, Natasha Oakley, and Panhia Vang for their contribution in processing all the PD‐L1 immunohistochemistry specimens.
Disclosures of potential conflicts of interest may be found at the end of this article.
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact Commercialreprints@wiley.com. For permission information contact permissions@wiley.com.
References
- 1. American Cancer Society . Facts & Figures 2019. Atlanta, GA: American Cancer Society; 2019. [Google Scholar]
- 2. Nounou M, ElAmrawy F, Ahmed N et al. Breast cancer: Conventional diagnosis and treatment modalities and recent patents and technologies. Breast Cancer (Auckl) 2015;27:17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Duffy M, Harbeck N, Nap M et al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur J Cancer 2017;75:284–298. [DOI] [PubMed] [Google Scholar]
- 4. Food and Drug Administration . List of cleared or approved companion diagnostic devices (in vitro and imaging tools). Available at https://www.fda.gov/medical-devices/vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-vitro-and-imaging-tools. Accessed September 27, 2019.
- 5. Schmid P, Adams S, Rugo H et al. Atezolizumab and nab‐paclitaxel in advanced triple‐negative breast cancer. N Engl J Med 2019;380:1929–1940. [DOI] [PubMed] [Google Scholar]
- 6. André F, Ciruelos E, Rubovszky G et al. Alpelisib for PIK3CA‐mutated, hormone receptor‐positive advanced breast cancer. N Engl J Med 2018;379:2108–2121. [DOI] [PubMed] [Google Scholar]
- 7. Robson M, Im SA, Senkus E et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017;377:523–533. [DOI] [PubMed] [Google Scholar]
- 8. Litton J, Rugo H, Ettl J et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med 2018;379:753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. NCCN Clinical Practice Guidelines in Oncology, Breast Carcinoma. Version 3.2019. National Comprehensive Cancer Network https://www.nccn.org/professionals/physician_gls/pdf/breast_blocks.pdf. Accessed November 20, 2019. [Google Scholar]
- 10. VENTANA. PD‐L1 (SP142) . Assay package insert. Available at https://www.accessdata.fda.gov/cdrh_docs/pdf16/p160002s009c.pdf. Accessed December 27, 2019.
- 11. VENTANA Medical Systems . VENTANA PD‐L1 (SP142). Assay interpretation guide for triple‐negative breast carcinoma (TNBC). Available at https://diagnostics.roche.com/content/dam/diagnostics/us/en/resource-center/VENTANA-PD-L1-(SP142)-Assay-Interpretation-Guide.pdf. Accessed December 27, 2019.
- 12. Frampton G, Fichtenholtz A, Otto G et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chalmers Z, Connelly C, Fabrizio D et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goeman J, Solari A. Multiple hypothesis testing in genomics. Stat Med 2014;20;33:1946–78. [DOI] [PubMed] [Google Scholar]
- 15. Alva A, Mangat P, Garrett‐Mayer E et al. Pembrolizumab (P) in patients (pts) with metastatic breast cancer (MBC) with high tumor mutational burden (HTMB): Results from the targeted agent and profiling utilization registry (TAPUR) study. J Clin Oncol 2019;37(suppl):1014a. [DOI] [PubMed] [Google Scholar]
- 16. Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumors. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Niu J, Andres G, Kramer K et al. Incidence and clinical significance of ESR1 mutations in heavily pretreated metastatic breast cancer patients. Onco Targets Ther 2015;8:3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holland D, Burleigh A, Git A et al. ZNF703 is a common Luminal B breast cancer oncogene that differentially regulates luminal and basal progenitors in human mammary epithelium. EMBO Mol Med 2011;3:167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Zhang X, Zhang Z et al. Association of p53 expression with poor prognosis in patients with triple‐negative breast invasive ductal carcinoma. Medicine (Baltimore). 2019;98:e15449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tobin NP, Bergh J. Analysis of cyclin D1 in breast cancer: A call to arms. Curr Breast Cancer Rep 2012;4:171–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aaltonen K, Amini R, Landberg G et al. Cyclin D1 expression is associated with poor prognostic features in estrogen receptor positive breast cancer. Breast Cancer Res Treat 2009;113:75–82. [DOI] [PubMed] [Google Scholar]
- 22. Roy P, Pratt N, Purdie C et al. High CCND1 amplification identifies a group of poor prognosis women with estrogen receptor positive breast cancer. Int J Cancer. 2010;127:355–360. [DOI] [PubMed] [Google Scholar]
- 23. Reinert T, Gonçalves R, Bines J. Implications of ESR1 mutations in hormone receptor‐positive breast cancer. Curr Treat Options Oncol 2018;19:24. [DOI] [PubMed] [Google Scholar]
- 24. Zhang X, Mu X, Huang O et al. Luminal breast cancer cell lines overexpressing ZNF703 are resistant to tamoxifen through activation of Akt/mTOR signaling. PLoS One 2013;8:e72053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasan N, Razavi P, Johnson J et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science. 2019;366:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Plasilova M, Hayse B, Killelea B et al. Features of triple‐negative breast cancer: Analysis of 38,813 cases from the national cancer database. Medicine (Baltimore) 2016;95:e4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Supplemental Figure
Supplemental Tables


