Abstract
Background
The purpose of this study was to investigate the effects of sevoflurane on cancer immunosurveillance and metastasis in non-small-cell lung cancer (NSCLC).
Material/Methods
NCI-H23 cells, a human NSCLC cell line, were incubated with or without sevoflurane at the concentrations of 0, 12.5, 25, 50, 100, and 200 μM for 6 h. Cell viability, the expression of natural killer group 2, member D ligands (NKG2D ligands: UL16-binding proteins 1–3 [ULBP1–3] and major histocompatibility complex class I chain-related molecules A/B [MICA/B]), the expression of matrix metalloproteinases (MMPs), NK cell-mediated cytotoxicity, and cancer cell migration were measured.
Results
At 12.5, 25, 50, and 100 μM, sevoflurane increased the expression of NKG2D ligands (ULBP2–3 and MICA, ULBP1–3, ULBP1–3, and ULBP1, respectively). Sevoflurane decreased the expression of NKG2D ligands at 200 μM (MICA/B). NK cell-mediated lysis of NCI-H23 cells at 200 μM sevoflurane was significantly reduced compared with the control (P=0.025; target cell: effect cell=1: 10). Sevoflurane increased the expression of MMP-1, -2, and -9 and increased cell migration in NCI-H23 cells at 50, 100, and 200 μM (P=0.001, 0.035, and 0.039, respectively, compared with the control after 18 h of wound formation).
Conclusions
Sevoflurane could suppress NKG2D-mediated NK cell cytotoxicity and increased expression of MMPs and migration in NCI-H23 cells. Further research is needed to determine the effects of sevoflurane on cancer immunosurveillance and metastasis in NSCLC.
MeSH Keywords: Anesthesia, Inhalation; Killer Cells, Natural; Matrix Metalloproteinases; Neoplasm Metastasis
Background
Natural killer (NK) cells, which originate from the lymphatic system of white blood cells, are components of innate immunity [1–3]. NK cells play a pivotal role in the defense against oncologic disease and viral infections [1]. NK cells interact with target cells through a balance of inhibitory and activating signals, and these signals are transmitted from the engagement of NK cell surface receptors and their ligands on target cells [4,5]. NK cell inhibitory receptors, natural killer receptor group 2, member A (NKG2A) and killer immunoglobulin-like receptors, bind to major histocompatibility complex (MHC) class I molecules and deliver inhibitory signals [4,5]. Natural killer receptor group 2, member D (NKG2D) binds to its ligands and delivers activating signals [4,5]. NKG2D serves as a master activating NK cell receptor in this process because it is present in all NK cells and its activation signal can override other inhibitory signals [6,7]. NKG2D has multiple ligands, including MHC class I chain-related A/B (MICA/B) and UL16-binding proteins (ULBPs) [8,9]. In contrast to normal cells, cancer cells express NKG2D ligands on their surface, which makes them more susceptible to detection and elimination by NK cells [7].
To evade NKG2D-mediated immune surveillance, cancer cells secrete matrix metalloproteinases (MMPs) that remove NKG2D ligands expressed on the cancer cell surface [6,8,10]. MMPs also degrade the extracellular matrix and promote cancer invasion, migration, and metastasis [11,12]. Therefore, MMPs play a fundamental role in cancer recurrence, which is strongly associated with advanced cancer stage and poor prognosis [10,12].
As surgery and general anesthesia are frequently inevitable in cancer treatment, extensive research has been conducted to determine the effect of volatile anesthetics on cancer recurrence and treatment outcomes. The results of recent meta-analyses suggest that volatile anesthetics could be associated with cancer recurrence and poor outcomes in comparison with total intravenous anesthesia [13,14]. However, the results concerning the effect of volatile anesthetics on the prognosis of non-small-cell lung cancer (NSCLC) remain inconclusive [15].
Accordingly, the present study aimed to investigate the effect of sevoflurane on NK cell-mediated immunosurveillance and metastatic potential in NSCLC in a dose-response manner.
Material and Methods
Cell lines and reagents
NCI-H23 (Korean Cell Line Bank, Seoul, Korea), a human NSCLC cell line, was maintained in Roswell-Park-Memorial-Institute (RPMI) 1640 medium (WELGENE, Gyeongsan, Korea) with fetal bovine serum (10%; WELGENE) and penicillin (1%; WELGENE). The NK92 cell line (American Type Culture Collection, Rockville, MD), a human NK cell line, was maintained in the α-minimum essential medium with fetal bovine serum (12.5%), horse serum (12.5%), recombinant human interleukin-2 (200 U/mL), 2-mercaptoethanol (0.1 mM), and l-glutamine (2 mM). The cells were incubated at 37°C humidified air containing 5% CO2.
Sevoflurane exposure
Sevoflurane solutions were prepared following a previously reported method [16,17]. Briefly, sevoflurane solution (Sevoprane; Ilsung Pharmaceuticals, Seoul, Korea) was diluted in RPMI 1640 medium and stirred for 30 min to make a 1 mM sevoflurane solution. This 1 mM sevoflurane solution was then serially diluted to make 200, 100, 50, 25, and 12.5 μM sevoflurane solutions immediately before the experiments. Sevoflurane exposure was performed on NCI-H23 for 6 h and, to compensate for concentration reduction due to evaporation, each sevoflurane solution was replaced on an hourly basis. Previous study reported that sevoflurane solutions remain relatively stable at below 10% additional loss for 1 h [16]. Corresponding concentrations of distilled water in RPMI 1640 medium were used as controls (0 μM). NCI-H23 cells were harvested after 18 h of sevoflurane exposure to measure the expression levels of mRNA, while NCI-H23 cells were harvested after 24 h of sevoflurane exposure to measure the expression levels of protein using the flow cytometry assay, immunofluorescence, and western blot analysis.
Viability test
NCI-H23 cells were plated in 96-well plates at a density of 1×104 cells per well. The cells were treated with 100 μL of 0, 100, and 200 μM sevoflurane solution for 6 h. After 24 and 48 h of the treatment, the cells were incubated with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (Sigma-Aldrich, St. Louis, MO, USA) solution for 4 h. Then, the supernatant solution was suctioned, and decrystallization was performed with dimethyl sulfoxide. The absorbance was detected using a microplate-spectrophotometer (μQuant; Bio-Tek Instruments Inc., Winooski, VT, USA).
Total RNA extraction, reverse transcriptase-polymerase chain reaction (RT-PCR), and multiplex-PCR
RNeasy® Mini Kit (Qiagen GmbH, Hilden, Germany) was used for the total RNA extraction. One microgram of extracted total RNA, 2.5 mM concentration of each deoxynucleotide triphosphate (Takara Shuzo, Shiga, Japan), and 100 pmol of random primers (Takara Shuzo) were incubated at 65°C for 5 min and chilled at 4°C for 4 min to synthesize cDNA. Then, 0.5 μL of M-MLV reverse transcriptase and 2 μL of the 5×reaction buffer (Promega Co., Fitchburg, WI, USA) were added and incubated at 35°C for 50 min. Multiplex-PCR was performed using the resulting cDNA and QIAGEN® Multiplex PCR Kit (Qiagen GmbH). The primer sets used to evaluate the expression of NKG2D ligand and MMP genes were as follows: ribosomal protein L19 (RPL19; degradation marker), MICA, MICB, ULBP1–3, and β-actin (ACTB; loading control) genes; and RPL19, MMP1–2, MMP9, and ACTB, respectively (Table 1). The MultiNA microchip-electrophoresis system (Shimadzu Biotech, Kyoto, Japan) was used for quantification. In order to normalize the gene expression level, the mRNA band intensity of each NKG2D ligand or MMP was divided by the band intensity of the ACTB. Subsequently, relative gene expression ratios of specific genes were computed by a comparison with samples and the control.
Table 1.
List of primer used in multiplex RT-PCR.
| Name | Polarity | Sequence (5′→3′) | Amplicon length (bp) |
|---|---|---|---|
| MICA | Sense | TTGAGCCGCTGAGAGGGTGGC | 460 |
| Anti-sense | GGGAGAGGAAGAGCTCCCCATC | ||
|
| |||
| MICB | Sense | GCCCCCTGACCCCTTGTTCC | 358 |
| Anti-sense | GGGCTGGTCAACTTGGCGAAA | ||
|
| |||
| ULBP1 | Sense | TGGCTGGTCCCGGGCAGGAT | 266 |
| Anti-sense | GAATGTCAAGCAGTTGCCCTTTAAGGAAA | ||
|
| |||
| ULBP2 | Sense | TCAAACTCGCCCTTCTGTCTGGC | 194 |
| Anti-sense | GCAGGAATTCCATCAGGTAGCACCA | ||
|
| |||
| ULBP3 | Sense | AGGTCTTATCTATGGGTCACCTAGAAG | 132 |
| Anti-sense | TGAAATCCTCCAGCTCAGTGTCAGC | ||
|
| |||
| MMP1 | Sense | AGACAAAGGCAAGTTGAAAAGCGGA | 195 |
| Anti-sense | TTGCTCCCAGCGAGGGTTCC | ||
|
| |||
| MMP2 | Sense | ACGGACTCCTGGCTCATGCC | 305 |
| Anti-sense | CTGTCCTTCAGCGTTGCCGC | ||
|
| |||
| MMP9 | Sense | CGACCCGAGCTGACTCGACG | 390 |
| Anti-sense | GCGGTGTGGTGGTGGTTGGA | ||
|
| |||
| RPL19 | Sense | ATGCTCAGGCTTCAGAAGAGGCTCG | 550 |
| Anti-sense | TGATGATCTCCTCCTTCTTGGCCTG | ||
|
| |||
| ACTB | Sense | TCCATCCTGGCCTCGCTGTC | 93 |
| Anti-sense | GCATTTGCGGTGGACGATGG | ||
RT-PCR – reverse transcriptase-polymerase chain reaction; MICA/B – MHC class I chain-related molecules A/B; ULBP – UL16-binding proteins; MMP – matrix metallopeptidase; RPL19 – ribosomal protein L19; ACTB – β-actin.
Analysis of NKG2D ligand expression by flow cytometry
To determine the surface expression of NKG2D ligands, the NCI-H23 cells were incubated with 10 μg/mL of mouse anti-MICA, anti-MICB, or anti-ULBP1–3 antibody or the corresponding isotype controls (anti-IgG2a/b; R&D Systems, Minneapolis, MN, USA). Then, the cells were incubated with goat anti-mouse phycoerythrin-conjugated antibody (BD Pharmingen Inc., San Diego, CA, USA). The mean fluorescence intensities were measured using a FACS Canto™ II flow cytometer (BD Biosciences, San Jose, CA, USA) and quantified using the FlowJo software (v10.6.1, TreeStar Inc., Ashland, OR, USA). Relative expression ratios were calculated by comparing the mean fluorescence intensity values between the treated samples and the control sample.
Immunofluorescence assay
The cells were incubated on fibronectin-coated coverslips. After 24 h of treatment, the cells were fixed with 4% paraformaldehyde for 20 min and washed with phosphate-buffered saline (PBS) and 0.5% bovine serum albumin. Blocking was performed with 5% bovine serum albumin for 1 h. After having been washed with PBS, the cells were incubated overnight at 4°C with the following primary antibodies: goat anti-ULBP2 polyclonal antibody (1: 100; R&D Systems) and mouse anti-MICA/B monoclonal antibody (1: 50; Santa Cruz Biotechnology, Santa Cruz, CA, USA). The cells were then washed with PBS and incubated with fluorescein isothiocyanate-conjugated secondary antibody (anti-goat or anti-mouse; Sigma-Aldrich) for 1 h in the dark. The cells were washed with PBS and stained with 4′,6-diamidino-2-phenylindole for 10 min. After the cells were washed with PBS, the coverslips were mounted onto glass slides with a fluorescence mounting medium (Dako, Carpinteria, CA, USA). Snapshot images were obtained using Leica TCS SP8 confocal microscope (×400 magnification; Leica Microsystems AG, Wetzlar, Germany). The intensity was quantified by measuring the mean gray values of regions of interest using the Leica LAS AF software (Leica Microsystems AG, Wetzlar, Germany) under identical exposure settings. Relative immunofluorescence intensity ratios were calculated by comparing the mean gray values between the treated samples and the control sample.
NK cell-mediated cytotoxicity assay
The target cells, 1×105 cells of NCI-H23, were labeled with Vybrant® carboxyfluorescein succinimidyl ester Cell Tracer Kit (Invitrogen, Eugene, OR, USA). The labeled target cells were co-cultured with NK92 cells, the effector cells, for 4 h at 1: 1 or 1: 10 ratio (i.e., target cells vs. effector cells: 1×105 cells: 1×105 or 1×105 cells: 1×106). These co-cultured cells were then stained with 1 μ/mL of propidium iodide (Sigma-Aldrich). The assay was performed using a FACS Canto™ II flow cytometer. The specific lysis (%) was calculated using Equation (1):
Western blot analysis
To evaluate the expression of MMPs, western blot analysis was performed. The cells were lysed with PRO-PREP™ solution (Intron Biotechnology, Sungnam, Korea), and equal amounts of cell extracts were analyzed using 4% to 20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins were transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA) and blocked with 4% nonfat milk in Tris-buffered saline and 0.1% Tween 20 at room temperature. Primary antibodies (MMP 1–2: Cell Signaling, Beverly, MA, USA; MMP9: Santa Cruz Biotechnology), secondary antibodies (Enzo Life Sciences, Minneapolis, MN, USA), and Amersham ECL Select Western blot detection reagent (GE Healthcare Life Sciences, Buckinghamshire, UK) were used. Anti-ACTB antibody (Sigma-Aldrich) was used for each probing. ImageJ software (version 1.52a; National Institutes of Health, Bethesda, MD, USA) was used for band intensity quantification. In order to calculate relative protein expression ratios, the protein expressions in the treated cells were divided by those of the control cells.
Cell migration: wound healing assay
NCI-H23 cells were plated on 6-well plates at a density of 1.5×105 cells per well and incubated at 37°C, with 5% CO2, in humidified air until a confluent monolayer was formed. After 6 h of treatment, a 20-μL pipette tip was used to form a straight scratch in each well of the monolayer of the cell. The monolayer was then washed with RPMI 1640 medium to remove cell debris, and the cells were returned to 37°C, 5% CO2, and humidified atmosphere. Snapshot images were obtained at 0, 18, 24, and 38 h after scratch formation using a Nikon TE-300 inverted microscope (×40 magnification; Nikon, Tokyo, Japan) and NIS-Elements F 3.0 (Nikon). The migration rate was measured through the wound closure (%) calculated using Equation (2):
where WA0h is the area of the wound measured immediately after scratch (0 h), and WAΔh is the area of the wound measured at Δh hours after the scratch was performed. ImageJ software was used to calculate the wound area.
Statistical analysis
All analyses were performed using IBM SPSS Statistics (version 22; IBM Corporation, Armonk, NY, USA). The variables were presented as mean±standard error of the mean (SEM). After a normality test, independent t test or Mann-Whitney U test was performed. Two-sided P-values below 0.05 were considered statistically significant.
Results
Viability
Sevoflurane was not found to affect the viability of NCI-H23 cells. No significant differences in cell viability of the control and treatment groups were observed. After 24 h of the treatment, the mean absorbances (SEM) at 450 nm were as follows: control, 0.560 (0.032); 100 μM, 0.487 (0.018); and 200 μM, 0.548 (0.033); (P=0.076 and 0.804; sevoflurane 100 and 200 μM vs. the control, respectively; n=6 per each group). After 48 h of the treatment, the mean absorbances (SEM) at 450 nm were as follows: control, 0.770 (0.062); 100 μM, 0.739 (0.022); and 200 μM, 0.689 (0.015); (P=0.651 and 0.250; sevoflurane 100 and 200 μM vs. the control, respectively; n=6 per each group).
Expression of NKG2D Ligands
mRNA expression of NKG2D ligands
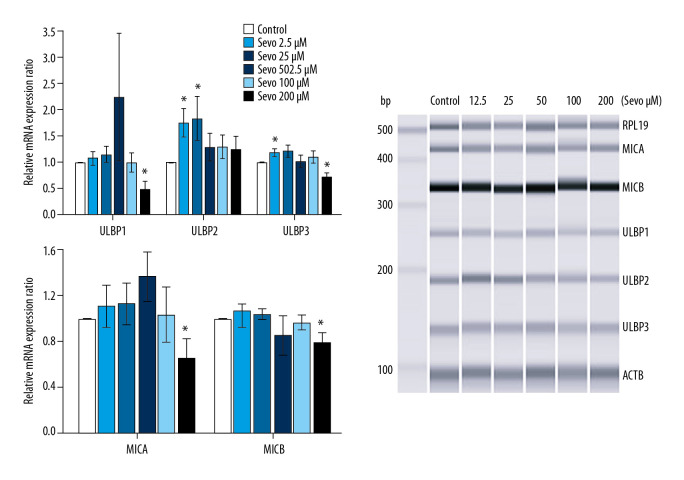
Sevoflurane 12.5 and 25 μM upregulated mRNA expression of NKG2D ligands, while 200 μM sevoflurane downregulated mRNA expression of NKG2D ligands (Figure 1; n=6 per each group). The relative mRNA expression ratio of ULBP1 at sevoflurane 200 μM was downregulated compared with the control (P=0.048). The relative mRNA expression ratio of ULBP2 with sevoflurane at 12.5 and 25 μM was upregulated compared with the control (P=0.048 and 0.002, respectively). Relative mRNA expression ratio of ULBP3 was upregulated compared with the control at sevoflurane 12.5 μM (P=0.048) and downregulated compared with the control at sevoflurane 200 μM (P=0.048). The relative mRNA expression ratio of MICA and MICB at sevoflurane 200 μM was downregulated compared with the control (P=0.048 and 0.048, respectively).
Figure 1.
Gene expression of natural killer group 2, member D (NKG2D) ligands. The variables are presented as mean±SEM (n=6 per each group). Sevo – sevoflurane; ULBP – UL16-binding proteins; MICA/B – MHC class I chain-related molecules A/B; RPL19 – ribosomal protein L19; ACTB – β-actin. * P<0.05 compared with the control.
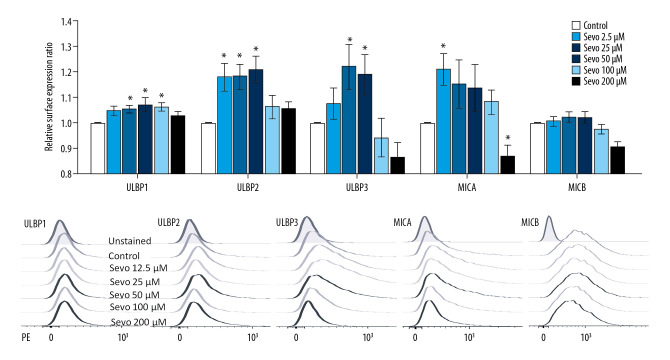
Surface expressions of NKG2D ligands
In line with mRNA expression, sevoflurane 12.5 to 100 μM upregulated the expression of NKG2D ligands, while 200 μM sevoflurane downregulated mRNA expression of NKG2D ligands (Figure 2; n=9 per each group). The relative surface expression ratio of ULBP1 was upregulated at sevoflurane 25, 50, and 100 μM compared with the control (P<0.001, P=0.007, and P=0.007, respectively). The relative surface expression ratio of ULBP2 and 3 at sevoflurane 12.5, 25, and 50 μM was upregulated compared with the control (ULBP2: P=0.002, P<0.001, and P<0.001; ULBP3: P=0.035, P=0.035, and P<0.001, respectively). The relative surface expression ratio of MICA was upregulated compared with the control at sevoflurane 12.5 μM (P<0.001) and downregulated compared with the control at sevoflurane 200 μM (P=0.002). The relative surface expression ratio of MICB at sevoflurane 200 μM was downregulated compared with the control (P<0.001).
Figure 2.
Surface expression of natural killer group 2, member D (NKG2D) ligands by flow cytometry. The variables are presented as mean±SEM (n=9 per each group). Sevo – sevoflurane; ULBP – UL16-binding proteins; MICA/B – MHC class I chain-related molecules A/B; PE – phycoerythrin. * P<0.05 compared with the control.
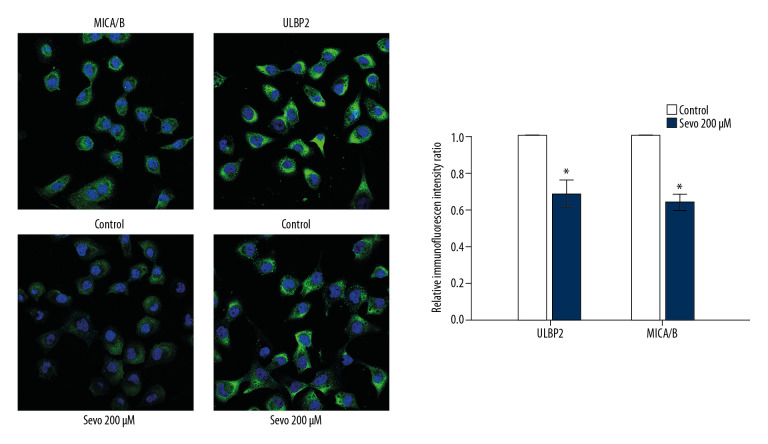
Immunofluorescence assay
Relative immunofluorescence intensity ratio of ULBP2 and MICA/B at sevoflurane 200 μM also decreased compared with the control (Figure 3; n=3 per each group, P=0.037 and P=0.037, respectively).
Figure 3.
Immunofluorescence for surface expressions of natural killer group 2, member D (NKG2D) ligands. ULBP2 and MICA/B are green and nuclei are blue. The variables are presented as mean ± SEM (n=3 per each group). Sevo – sevoflurane; ULBP – UL16-binding proteins; MICA/B – MHC class I chain-related molecules A/B. * P<0.05 compared with the control.
NK cell-mediated cytotoxicity
No significant differences in NK cell-mediated lysis between the control and treatment groups in the target and effect cells at the 1: 1 ratio were observed. NK cell-mediated lysis of NCI-H23 at sevoflurane 200 μM was significantly reduced compared with the control in target cells and effect cells at the 1: 10 ratio (Figure 4; P=0.025, n=6).
Figure 4.
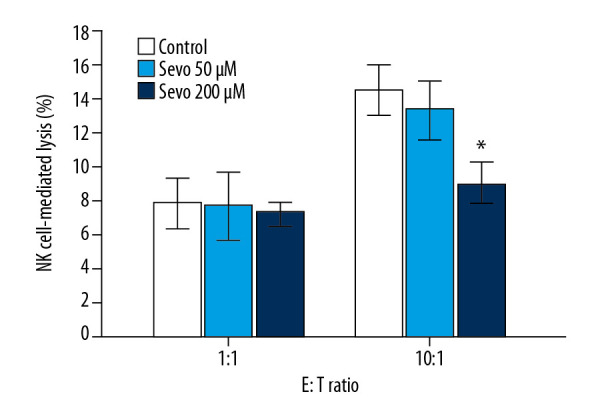
Natural killer (NK) cell-mediated cytotoxicity assay by flow cytometry. The variables are presented as mean±SEM (n=6 per each group). Sevo – sevoflurane; E – effect cells (NK-92); T – target cells (NCI-H23). * P<0.05 compared with the control.
Expression of MMPs
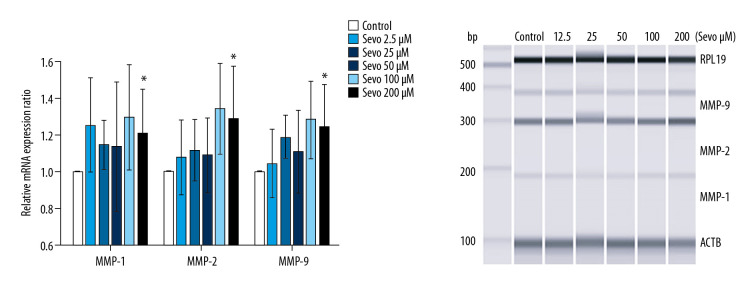
mRNA expression of MMPs
Sevoflurane upregulated mRNA expression of MMP-1, -2, and -9 in NCI-H23 cells (Figure 5; n=6 per each group). At sevoflurane 200 μM, all relative mRNA expression ratios of MMP-1, -2, and -9 were upregulated compared with the control (P=0.048, P=0.048, and P=0.048, respectively).
Figure 5.
Gene expression of MMP. The variables are presented as mean±SEM (n=6 per each group). Sevo – sevoflurane; MMP – matrix metalloproteinase; RPL19 – ribosomal protein L19; ACTB – β-actin. * P<0.05 compared with the control.
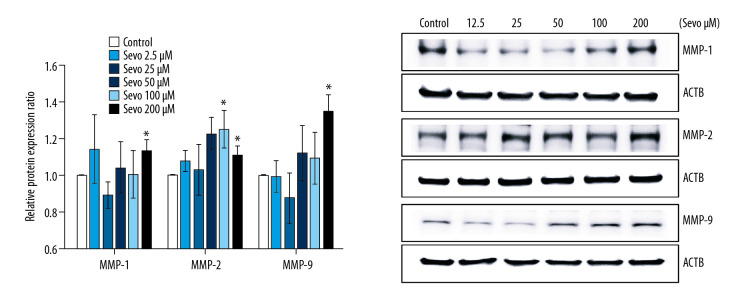
Protein expression of MMPs
Sevoflurane increased protein expression of MMP-1, -2, and -9 in NCI-H23 cells (Figure 6; n=6 per each group). Relative protein expression ratio of MMP-1 and -9 was upregulated compared with the control at sevoflurane 200 μM (P=0.048 and P=0.002). The relative protein expression ratio of MMP-2 was upregulated compared with the control at sevoflurane 100 and 200 μM (P=0.048 and P=0.048, respectively).
Figure 6.
Protein expression of MMP. The variables are presented as mean±SEM (n=6 per each group). Sevo – sevoflurane; MMP – matrix metalloproteinase; ACTB – β-actin. * P<0.05 compared with the control.
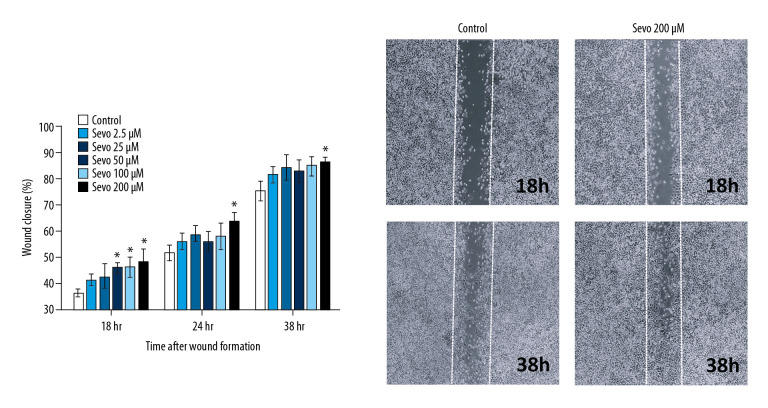
Cell migration
Sevoflurane increased cell migration in NCI-H23 cells (Figure 7; n=8 per each group). In sevoflurane 50, 100, and 200 μM, the mean wound closure was increased compared with the control after 18 h of wound formation (P=0.001, P=0.035, and P=0.039, respectively). After 24 and 38 h of wound formation, an increase in the mean wound closure was observed at sevoflurane 200 μM compared with the control (P=0.017 and P=0.018, respectively).
Figure 7.
0Cell migration by wound healing assay. The variables are presented as mean±SEM (n=8 per each group). Sevo – sevoflurane. * P<0.05 compared with the control.
Discussion
In the present study focused on NSCLC, sevoflurane increased the expression of NKG2D ligand at 12.5 to 100 μM, but decreased the expression of NKG2D at 200 μM. NK cell-mediated cell death was inhibited as the sevoflurane dose increased. Moreover, sevoflurane upregulated the expression of MMP-1, -2, and -9, and increased cell migration in NSCLC.
Lung cancer is one of the most common causes of death worldwide, with about 85% of cases being diagnosed as NSCLC [18,19]. However, despite significant advances in understanding the pathogenesis and progression mechanisms of NSCLC, the cure rate and survival rate remain low, especially in the case of metastasis [20]. In the early stages of NSCLC, surgical treatment is considered to be the most efficient treatment. However, the effects of surgery-related factors on NSCLC remain poorly understood [21,22].
Previous research demonstrated that sevoflurane directly inhibits NK cell activity [23–25]. Tazawa et al. [23] found that sevoflurane attenuated NK cell-mediated conjugation, polarization, and cytotoxicity by inhibiting leukocyte function-associated antigen-1 in NK cells. Furthermore, in a randomized controlled trial to study the effect of anesthetic techniques on human NK cell function and cytotoxicity, Buckley et al. [24] found that, in contrast to the serum from patients with propofol-paravertebral block, the serum from the patients with sevoflurane-opioid anesthesia downregulated the expression of NK cell-activating receptor CD16, reduced NK cell cytokine interleukin (IL)-1β and IL-10, and suppressed NK cell-mediated cell death against breast cancer cells. In addition, Cho et al. [25] demonstrated that the cytotoxicity of NK cells decreased postoperatively in patients who received sevoflurane-remifentanil. In the present study, the suppression of NK cell-mediated cytotoxicity by sevoflurane is consistent with the results of previous studies [23–25]; however, the research on the effects of sevoflurane on the expression of NK cell ligands remains scarce, which necessitates further evaluation of this field.
NKG2D ligands, known as “stress-induced ligands,” are expressed in damaged and transformed cells [9]. Cancer cells also express NKG2D ligands on their surface, which plays an important role in cancer immunosurveillance by sensitizing cancer cells to NK cell-mediated elimination [7]. While MICA, MICB, and ULBP1-6 have been identified as human NKG2D ligands, the roles of ULBP4–6 remain poorly understood [8,9].
The 50 μM sevoflurane increased the expression of NKG2D ligands in NSCLC, but no significant change was observed in NK cell-mediated cytotoxicity in co-culture assays with NK cells. Previous research demonstrated that there is a threshold for NK cell activation [7,26], and that the relationship between the expression levels of activating and inhibitory ligands for NK cells on target cells and susceptibility to NK cell-mediated cytotoxicity is nonlinear [26,27]. This finding suggests that the increased expression of the ligands may not have been sufficient to reach the threshold level for the NK cell activation in the present study. Other possible causes of the unexpected results may be the increased expression of some other genes encoding anti-apoptotic molecules or some molecules that transduce NK cell inhibitory signals such as MHC class 1 molecules [28].
MMPs, a family of zinc-dependent proteases, degrade and remodel the extracellular matrix, thereby creating a favorable microenvironment for cancer cell angiogenesis, migration, and invasion [10,29]. Proteolytic activities of MMPs also remove NKG2D ligands from the cancer cell surface, which helps cancer cells escape from NK cell-mediated surveillance [6,8,10]. Ultimately, this series of actions by MMPs might lead to cancer recurrence and metastasis [10,29]. Specifically, MMP-1, -2, and -9 were reported to play key roles in NSCLC progression and metastasis and are strongly associated with a poor survival rate [10].
In the present study, sevoflurane not only increased the transcription and translation of MMP-1, -2, and -9, but also increased NSCLC cell migration ability depending on the exposed dose. Consistent with our results, Wang et al. [30] found that sevoflurane anesthesia during lung cancer surgery increased the serum MMP-9 level compared with propofol anesthesia. In contrast, in a study using A549 cells, another NSCLC cell line, Liang et al. [31] demonstrated that sevoflurane inhibited MMP-2 and -9 expression and cancer cell invasion and migration in a dose- and time-dependent manner. These inconsistent findings could be due to the differences in study subjects and the protocol of sevoflurane exposure, which highlights the need for further in-depth research on the effects of sevoflurane on the expression of MMPs.
Previous studies found that sevoflurane could cause the accumulation of intracellular reactive oxygen species (ROS) and the loss of ROS homeostasis [32–34]. ROS act as inducing molecules of C-X-C chemokine receptor type 4 (CXCR4), resulting in an increased expression of CXCR4 [35,36]. The signal delivered through CXCR4 activates phosphoinositol-3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) signaling pathways, and upregulates the expression of MMPs [35–38]. Therefore, an increase in ROS triggered by sevoflurane could underlie the upregulated expression of MMP-1, -2, and -9. With regard to the downregulation of NKG2D ligands at a high concentration of sevoflurane, the result could have been partially caused by the increased removal of cell surface NKG2D ligands by MMP-1, -2, and -9. However, further research is needed to explain how a high dose of sevoflurane inhibits the transcription of NKG2D ligands.
According to the results of a previous study, during balanced anesthesia, the arterial sevoflurane concentration is about 100 to 300 μM [39]. The average age for lung cancer diagnosis is about 70 years old, and this type of cancer is very rarely diagnosed in younger patients [40]. Patients become significantly more sensitive to anesthetic agents with aging [41]. In elderly patients, a lower concentration of sevoflurane is usually required to achieve an adequate depth of anesthesia, and the effect of sevoflurane is frequently prolonged [41,42]. In addition, the deeper level of anesthesia is closely associated with an increase in postoperative morbidity and mortality in elderly patients [43,44]. In consideration of these points, anesthetic management using a relatively low concentration of an anesthetic agent is recommended in elderly patients. These clinical characteristics were considered in the determination of the range of sevoflurane concentrations used in the present study.
The present study has several limitations. First, considering that this is an in vitro study, our results are not directly applicable to animals or humans. Second, since the present study did not elucidate the molecular mechanism of NKG2D ligands and expression of MMPs, further research is needed to better understand the impact of sevoflurane on NKG2D ligands and expression of MMPs.
Conclusions
Sevoflurane could suppress NKG2D-mediated NK cell cytotoxicity and increase the expression of MMPs and migration in NCI-H23 cells. Further research is needed to determine the effects of sevoflurane on cancer immunosurveillance and metastasis in NSCLC.
Footnotes
Conflict of interests
None.
Source of support: This study was supported by Biomedical Research Institute Grant (2019B018), Pusan National University Hospital
References
- 1.Stollings LM, Jia L, Tang P, et al. Immune modulation by volatile anesthetics. Anesthesiology. 2016;125(2):399–411. doi: 10.1097/ALN.0000000000001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivier E, Raulet DH, Moretta A, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vivier E, Ugolini S, Blaise D, et al. Targeting natural killer cells and natural killer T cells in cancer. Nat Rev Immunol. 2012;12(4):239–52. doi: 10.1038/nri3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moretta L, Locatelli F, Pende D, et al. Killer Ig-like receptor-mediated control of natural killer cell alloreactivity in haploidentical hematopoietic stem cell transplantation. Blood. 2011;117(3):764–71. doi: 10.1182/blood-2010-08-264085. [DOI] [PubMed] [Google Scholar]
- 5.Backström E, Kristensson K, Ljunggren HG. Activation of natural killer cells: Underlying molecular mechanisms revealed. Scand J Immunol. 2004;60(1–2):14–22. doi: 10.1111/j.0300-9475.2004.01475.x. [DOI] [PubMed] [Google Scholar]
- 6.López-Soto A, Huergo-Zapico L, et al. NKG2D signaling in cancer immunosurveillance. Int J Cancer. 2015;136(8):1741–50. doi: 10.1002/ijc.28775. [DOI] [PubMed] [Google Scholar]
- 7.Dhar P, Wu JD. NKG2D and its ligands in cancer. Curr Opin Immunol. 2018;51:55–61. doi: 10.1016/j.coi.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheppard S, Ferry A, Guedes J, Guerra N. The paradoxical role of NKG2D in cancer immunity. Front Immunol. 2018;9:1808. doi: 10.3389/fimmu.2018.01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmiedel D, Mandelboim O. NKG2D ligands – critical targets for cancer immune escape and therapy. Front Immunol. 2018;9:2040. doi: 10.3389/fimmu.2018.02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merchant N, Nagaraju GP, Rajitha B, et al. Matrix metalloproteinases: Their functional role in lung cancer. Carcinogenesis. 2017;38(8):766–80. doi: 10.1093/carcin/bgx063. [DOI] [PubMed] [Google Scholar]
- 11.Zingoni A, Vulpis E, Nardone I, et al. Targeting NKG2D and NKp30 ligands shedding to improve NK cell-based immunotherapy. Crit Rev Immunol. 2016;36(6):445–60. doi: 10.1615/CritRevImmunol.2017020166. [DOI] [PubMed] [Google Scholar]
- 12.Rundhaug JE. Matrix metalloproteinases, angiogenesis, and cancer. Clin Cancer Res. 2003;9(2):551–54. [PubMed] [Google Scholar]
- 13.Yap A, Lopez-Olivo MA, Dubowitz J, et al. Global Onco-Anesthesia Research Collaboration Group. Anesthetic technique and cancer outcomes: A meta-analysis of total intravenous versus volatile anesthesia. Can J Anaesth. 2019;66(5):546–61. doi: 10.1007/s12630-019-01330-x. [DOI] [PubMed] [Google Scholar]
- 14.Soltanizadeh S, Degett TH, Gögenur I. Outcomes of cancer surgery after inhalational and intravenous anesthesia: A systematic review. J Clin Anesth. 2017;42:19–25. doi: 10.1016/j.jclinane.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Oh TK, Kim K, Jheon S, et al. Long-term oncologic outcomes for patients undergoing volatile versus intravenous anesthesia for non-small cell lung cancer surgery: A retrospective propensity matching analysis. Cancer Control. 2018;25(1) doi: 10.1177/1073274818775360. 1073274818775360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama T, Penheiter AR, Penheiter SG, et al. Differential effects of volatile anesthetics on M3 muscarinic receptor coupling to the Galphaq heterotrimeric G protein. Anesthesiology. 2006;105(2):313–24. doi: 10.1097/00000542-200608000-00014. [DOI] [PubMed] [Google Scholar]
- 17.Ecimovic P, McHugh B, Murray D, et al. Effects of sevoflurane on breast cancer cell function in vitro. Anticancer Res. 2013;33(10):4255–60. [PubMed] [Google Scholar]
- 18.Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–94. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 20.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–54. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 21.Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol. 2016;11(1):39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 23.Tazawa K, Koutsogiannaki S, Chamberlain M, Yuki K. The effect of different anesthetics on tumor cytotoxicity by natural killer cells. Toxicol Lett. 2017;266:23–31. doi: 10.1016/j.toxlet.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckley A, McQuaid S, Johnson P, Buggy DJ. Effect of anaesthetic technique on the natural killer cell anti-tumour activity of serum from women undergoing breast cancer surgery: A pilot study. Br J Anaesth. 2014;113(Suppl 1):i56–62. doi: 10.1093/bja/aeu200. [DOI] [PubMed] [Google Scholar]
- 25.Cho JS, Lee MH, Kim SI, et al. The effects of perioperative anesthesia and analgesia on immune function in patients undergoing breast cancer resection: A prospective randomized study. Int J Med Sci. 2017;14(10):970–76. doi: 10.7150/ijms.20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes TD, El-Sherbiny YM, Davison A, et al. A human NK cell activation/inhibition threshold allows small changes in the target cell surface phenotype to dramatically alter susceptibility to NK cells. J Immunol. 2011;186(3):1538–45. doi: 10.4049/jimmunol.1000951. [DOI] [PubMed] [Google Scholar]
- 27.Kim J, Son Y, Park S, et al. Increase of NKG2D ligands and sensitivity to NK cell-mediated cytotoxicity of tumor cells by heat shock and ionizing radiation. Exp Mol Med. 2006;38(5):474–84. doi: 10.1038/emm.2006.56. [DOI] [PubMed] [Google Scholar]
- 28.Paul S, Lal G. The molecular mechanism of natural killer cells function and its importance in cancer immunotherapy. Front Immunol. 2017;8:1124. doi: 10.3389/fimmu.2017.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278(1):16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Liu J, Gao J, Zheng X. Comparison of the effects of sevoflurane and propofol anesthesia on pulmonary function, MMP-9 and postoperative cognition in patients receiving lung cancer resection. Oncol Lett. 2019;17(3):3399–405. doi: 10.3892/ol.2019.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang H, Gu M, Yang C, et al. Sevoflurane inhibits invasion and migration of lung cancer cells by inactivating the p38 MAPK signaling pathway. J Anesth. 2012;26(3):381–92. doi: 10.1007/s00540-011-1317-y. [DOI] [PubMed] [Google Scholar]
- 32.Rocha TL, Dias-Junior CA, Possomato-Vieira JS, et al. Sevoflurane induces DNA damage whereas isoflurane leads to higher antioxidative status in anesthetized rats. Biomed Res Int. 2015;2015 doi: 10.1155/2015/264971. 264971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cukurova Z, Cetingok H, Ozturk S, et al. DNA damage effects of inhalation anesthetics in human bronchoalveolar cells. Medicine. 2019;98(32):e16518. doi: 10.1097/MD.0000000000016518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong CH, Liu TZ, Chye SM, et al. Sevoflurane-induced oxidative stress and cellular injury in human peripheral polymorphonuclear neutrophils. Food Chem Toxicol. 2006;44(8):1399–1407. doi: 10.1016/j.fct.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Chetram MA, Hinton CV. ROS-mediated regulation of CXCR4 in cancer. Front Biol. 2013;8(3):273–78. doi: 10.1007/s11515-012-1204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Deng Y, Feng J, Ye W. Oxidative preconditioning promotes bone marrow mesenchymal stem cells migration and prevents apoptosis. Cell Biol Int. 2009;33(3):411–18. doi: 10.1016/j.cellbi.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Otsuka S, Bebb G. The CXCR4/SDF-1 chemokine receptor axis: A new target therapeutic for non-small cell lung cancer. J Thorac Oncol. 2008;3(12):1379–83. doi: 10.1097/JTO.0b013e31818dda9d. [DOI] [PubMed] [Google Scholar]
- 38.Chu CY, Cha ST, Chang CC, et al. Involvement of matrix metalloproteinase-13 in stromal-cell-derived factor 1 alpha-directed invasion of human basal cell carcinoma cells. Oncogene. 2007;26(17):2491–501. doi: 10.1038/sj.onc.1210040. [DOI] [PubMed] [Google Scholar]
- 39.Frink EJ, Jr, Malan TP, Jr, Brown EA, et al. Plasma inorganic fluoride levels with sevoflurane anesthesia in morbidly obese and nonobese patients. Anesth Analg. 1993;76(6):1333–37. doi: 10.1213/00000539-199376060-00026. [DOI] [PubMed] [Google Scholar]
- 40.Brown JS, Eraut D, Trask C, Davison AG. Age and the treatment of lung cancer. Thorax. 1996;51(6):564–68. doi: 10.1136/thx.51.6.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanonidou Z, Karystianou G. Anesthesia for the elderly. Hippokratia. 2007;11(4):175–77. [PMC free article] [PubMed] [Google Scholar]
- 42.Mapleson WW. Effect of age on MAC in humans: A meta-analysis. Br J Anaesth. 1996;76(2):179–85. doi: 10.1093/bja/76.2.179. [DOI] [PubMed] [Google Scholar]
- 43.Monk TG, Saini V, Weldon BC, Sigl JC. Anesthetic management and one-year mortality after noncardiac surgery. Anesth Analg. 2005;100(1):4–10. doi: 10.1213/01.ANE.0000147519.82841.5E. [DOI] [PubMed] [Google Scholar]
- 44.Chan MTC, Cheng BCP, Lee TMC, Gin T CODA Trial Group. BIS-guided anesthesia decreases postoperative delirium and cognitive decline. J Neurosurg Anesthesiol. 2013;25(1):33–42. doi: 10.1097/ANA.0b013e3182712fba. [DOI] [PubMed] [Google Scholar]