Abstract
Wastewater-based epidemiology (WBE) is successful in the detection of the spread of SARS-CoV-2. This review examines the methods used and results of recent studies on the quantification of SARS-CoV-2 in wastewater. WBE becomes essential, especially with virus transmission path uncertainty, limitations on the number of clinical tests that could be conducted, and a relatively long period for infected people to show symptoms. Wastewater surveillance was used to show the effect of lockdown on the virus spread. A WBE framework tailored for SARS-CoV-2 that incorporates lessons learnt from the reviewed studies was developed. Results of the review helped outline challenges facing the detection of SARS-CoV-2 in wastewater samples. A comparison between the various studies with regards to sample concentration and virus quantification was conducted. Five different primers sets were used for qPCR quantification; however, due to limited data availability, there is no consensus on the most sensitive primer. Correlating the slope of the relationship between the number of gene copies vs. the cumulative number of infections normalized to the total population served with the average new cases, suggests that qPCR results could help estimating the number of new infections. The correlation is improved when a lag period was introduced to account for asymptomatic infections. Based on lessons learnt from recent studies, it is recommended that future applications should consider the following: 1) ensuring occupational safety in managing sewage collection and processing, 2) evaluating the effectiveness of greywater disinfection, 3) measuring viral RNA decay due to biological and chemical activities during collection and treatment, 4) assessing the effectiveness of digital PCR, and 5) conducting large scale international studies that follow standardized protocols.
Keywords: SARS-CoV-2, COVID-19, Virus concentration, Wastewater-based epidemiology, Wastewater virus surveillance
Graphical abstract
Highlights
-
•
Wastewater concentration methods & SARS-CoV-2 detection were reviewed.
-
•
Issues related to SARS-CoV-2 detection assays were discussed.
-
•
Tailored framework for SARS-CoV-2 WBE was proposed.
-
•
Based on lessons learnt, insights into future directions were highlighted.
1. Introduction
The world is currently battling the coronavirus disease 2019 (COVID-19) pandemic spanning 190 countries as of September 26, 2020 (“COVID-19 Map,” 2020). As of November 9, 2020, COVID-19 has led to over 50 million infections and >1.25 million deaths, with an average case fatality rate (CFR) of 2.49% since its emergence in Wuhan, China in December 2019 (“COVID-19 Map,” 2020; Gorbalenya et al., 2020; Uddin et al., 2020). Coronaviruses are a group of RNA viruses that cause respiratory tract infections in mammals and birds. Since the 1960's, six strains of coronaviruses have entered the human species besides SARS-CoV-2 of which the first four caused only mild upper respiratory tract infections. However, the latest two, SARS-CoV and MERS-CoV were observed to be highly pathogenic, involving lower respiratory tract infections in addition to the upper airways (Uddin et al., 2020). Discovery of yet another strain of coronavirus in China that is genetically-related to SARS-CoV and responsible for COVID-19 led to it being named SARS coronavirus 2 (SARS-CoV-2), causing both upper and lower respiratory tract infections (Gorbalenya et al., 2020). COVID-19 was declared a public health emergency of international concern by the World Health Organization (WHO) early in the pandemic and continues to remain that way (Uddin et al., 2020). Till date, there are no WHO-approved medications to treat the disease or any effective vaccines, while affected individuals are managed with supportive care and mechanical ventilation. Like SARS-CoV, SARS-CoV-2 has its origins in bats but is highly infectious in humans (Gorbalenya et al., 2020; Uddin et al., 2020; Zhou et al., 2020). The primary mode of SARS-CoV-2 transmission is via respiratory droplets from infected individuals (Cevik et al., 2020). Clinical presentation of COVID-19 has shown that most people (~80%) are either asymptomatic or develop only mild symptoms that include, cough, fever, fatigue, myalgia, and temporary loss of the senses of smell and taste (Cevik et al., 2020).
SARS-CoV-2 is not the only viral outbreak that the world has seen recently. Other important viral outbreaks in the last twenty years globally include: Ebola in 2000 (Okware et al., 2002), SARS-CoV in 2003 (Drosten et al., 2003), swine flu (H1N1 influenza virus) in 2009 (CDC, 2009) - the second H1N1 outbreak after the Spanish flu of 1918 that infected nearly 700 to 1.2 billion people and killed anywhere from 150,000 to 575,000 (CDC, 2012; Roos, 2011), MERS-CoV in 2012 (Assiri et al., 2013), Zika in 2015 (Sikka et al., 2016), and Nipah in 2018 (Yadav et al., 2019). The One Health approach stipulated by the WHO recommends collecting information on viruses circulating in animals and the environment as a crucial step for the detection of future pandemics and selection of potential human vaccines. Thus, due to the increased frequency of animal virus outbreaks into humans, there is an urgent need to develop rapid surveillance systems to follow pre-existing viral infections and discover/track newly emerging viral infections in the human populations (new zoonosis) in an efficient and effective manner such as wastewater surveillance and wastewater-based epidemiology (WBE).
Wastewater surveillance is a much cheaper and an efficient means of tracking infectious agents in communities (Hart and Halden, 2020), since it overcomes the need to test a large proportion of the population individually, a task which can be tedious, expensive, and time consuming. The main requirement for this type of surveillance is that the pathogen to be monitored should survive the stomach acidity and be excreted in the feces; i.e., it should be an enteric virus that travels through the gastrointestinal tract. Other than the poliovirus, such waterborne viral pathogens include adenoviruses, hepatitis A & E viruses, coxsackie and rotaviruses (Edition, 2011). Such enteric viruses are excreted in feces at high levels (up to 1011 particles per gram of stool), and thus are easy to detect by wastewater surveillance (Fong and Lipp, 2005). Similarly, several studies have shown that the SARS-CoV-2 is excreted into the feces despite being a respiratory virus primarily, making its tracking amenable to wastewater surveillance (Y. Wu et al., 2020; Xiao et al., 2020; Xu et al., 2020).
In addition to surveillance, WBE has been utilized to relate certain viral strains in wastewater and those in clinical samples (Bisseux et al., 2018; Carducci et al., 2006; Weil et al., 2017). Thus, WBE is an essential tool to investigate the existence and spread of both older human pathogens, such as poliovirus, as well as newer ones such as SARS-CoV-2, to determine the spread of the pathogen, presence of their hotspots, and the effectiveness of control measures within a community (Barcelo, 2020). Identifying the rates of SARS-CoV-2 infection in communities has been difficult considering that about 80% of the infected people may remain either asymptomatic or develop mild symptoms (Heneghan et al., 2020). WBE has been applied in the past for non-enveloped viruses with a particular focus on polioviruses (Blomqvist et al., 2012; Brouwer et al., 2018; El Bassioni et al., 2003; Lago et al., 2003; Manor et al., 2014; Matsuura et al., 2000; Nakamura et al., 2015; O'Reilly et al., 2018; Van der Avoort et al., 1995; Yoshida et al., 2002) and to a lesser extent to enteroviruses (Apostol et al., 2012), echovirus (Tao et al., 2011), adenoviruses (Amdiouni et al., 2012), polyomavirus (Bofill-Mas et al., 2000), and Hepatitis E (Clemente-Casares et al., 2003).
This review presents an in-depth analysis of the current literature and data regarding the detection of SARS-CoV-2 in wastewater as a surveillance tool for tracking the transmission of SARS-CoV-2 and predicting its future breakouts in the communities. Recent reviews have examined the state-of-the-art in detecting coronaviruses and other enveloped viruses in wastewater (Carducci et al., 2020; Foladori et al., 2020; Kitajima et al., 2020; Lu et al., 2020). The studies reviewed were published early in the pandemic, thus the number of references focusing on SARS-CoV-2 in particular were limited. The sheer volume of studies published since March 2020 warrants an in-depth review that compares the techniques used and links their results. The study focuses on SARS-CoV-2 surveillance in relation to WBE. In particular, this review offers: 1) a comparative analysis of the methods of wastewater concentration and virus detection, 2) a deep data analysis of the studies already published on SARS-CoV-2 wastewater surveillance, 3) a discussion on issues related to virus detection assays such as sensitivity and specificity and the ability to predict future community outbreaks, 4) an overview of issues related to quality control and safety, 5) a tailored framework for SARS-CoV-2 wastewater-based epidemiology, and finally 6) an insight into future directions based on current lessons learnt including the implications of reusing greywater.
2. Wastewater surveillance for viral pathogens
Global urbanization mixed with rapid population growth and frequent emergence of pathogenic viruses in the human population calls for the development of rapid surveillance systems to monitor the spread of viruses as well as the infection status of the population (Mao et al., 2020). For pandemics, finding a cure takes time and other means of intervention are required to reduce the impact on the society. One of these is the need to determine the distribution and magnitude of infected cases (Daughton, 2020). This becomes critical especially if there is uncertainty about the transmission path of the virus, limitations on the number of clinical tests that could be conducted, and a relatively long period for infected people to show symptoms.
Using WBE to determine the spread of SARS-CoV-2
Recently, application of WBE has been proposed to track the magnitude and distribution of infection by SARS-CoV-2 (a non-enveloped virus) in a community through testing viral biomarker levels in samples collected from the sewage network (Daughton, 2020; Sims and Kasprzyk-Hordern, 2020). In this context, Xagoraraki and O'Brien (2020) provided a description of the requirements for the development and realization of WBE to be utilized as a tool for identifying and predicting viral outbreaks. In addition to its use as an early warning for an outbreak, it could be used to generate spatial and temporal heat maps regarding the distribution of the viral biomarker in a community. This could be utilized to carry out quantitative microbial risk assessment to identify the number of infected people in different zones. Kitajima et al. (2020) reviewed the potential use of wastewater surveillance to assess the epidemiology of SARS-CoV-2 and stressed on the need for further research to establish the methodology. Using computational analysis and modeling, Hart and Halden (2020) have demonstrated that WBE is feasible to apply and is significantly cheaper and faster than clinical screening. Nonetheless, these authors suggested a two-step process for management of a viral pandemic in which WBE is used to map and enumerate infected cases followed by clinical testing for identification of infected people in hotspots of the WBE map.
WBE to assess effectiveness of viral control measures
To evaluate the effectiveness of control measures, wastewater surveillance of poliovirus has been used as the “proof-of-concept” to demonstrate the success of vaccination against poliovirus (Hovi et al., 2012). Other studies have applied WBE to evaluate the immunization efficacy of poliovirus (Blomqvist et al., 2012; Lago et al., 2003; Nakamura et al., 2015). For SARS-CoV-2, Nemudryi et al. (2020) monitored the virus in pre-treated wastewater over a period of 17 days. They observed that the viral RNA had steadily decreased during the last week of monitoring and suggested that this could be attributed to the mandated social isolation. Wurtzer et al. (2020a) confirmed that the lockdown led to lower levels of SARS-CoV-2 viral RNA.
Relationship between viral strains available in wastewater and clinical samples
One of the applications of WBE has been to provide spatial and temporal viral surveillance. For example, Bofill-Mas et al. (2000) found through studying the epidemiologic patterns of certain polyomavirus strains that they are endemic to certain regions in Europe and Africa. Meanwhile, Clemente-Casares et al. (2003) detected Hepatitis E virus in the USA and some European countries although these countries were not previously considered endemic for the virus. Furthermore, WBE was found to play a fundamental role in detecting and investigating the silent introduction of wild poliovirus type 1 in Israel (Manor et al., 2014).
2.1. Factors affecting viral stability in wastewater systems
Several factors could influence the fate of viruses in wastewater such as inactivation, decay, dispersion, or retardation. Factors affecting viral survival should be useful from an occupational safety perspective, while those related to the fate of the viral biomarkers are useful for identification of infected areas within a community. Some information is currently available about certain types of viruses, but limited information is available about human coronaviruses and none is available about SARS-CoV-2. Therefore, a review of the available data may serve as a guidance for the time being till actual data becomes available.
Effect of temperature
Temperature is the most significant factor affecting the survival of viruses. Several studies have investigated the effect of temperature on the survival of viruses in water using different water types. Differences included sterilized/unsterilized reagent water (Casanova et al., 2009; Duan et al., 2003), filtered/unfiltered tap water (Gundy et al., 2008), filtered/unfiltered sewage (Gundy et al., 2008; Wang et al., 2005), pasteurized/unpasteurized sewage (Casanova et al., 2009; Casanova and Weaver, 2015), and surface water (Adcock et al., 2009; Casanova et al., 2009; Nazir et al., 2010). We compared results obtained from the studies reported earlier, with emphasis on those that related the effect of temperature on the survival of viruses in sewage.
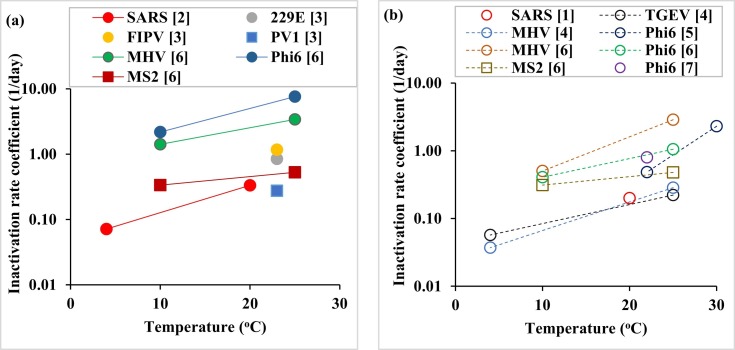
As can be seen from Fig. 1a, as the temperature increases, the inactivation rate coefficient increases, resulting in a shorter lifetime of the virus in sewage. The reduction in viral survival with the increase in temperature has been mainly attributed to denaturation of proteins and increased activity of extracellular enzymes (John and Rose, 2005). The behavior, however, was observed to be dependent on the virus type; as expected, the non-enveloped viruses like PV1 and MS2 survived longer than enveloped ones, including some coronaviruses like 229E (Fig. 1a). However, the enveloped SARS-CoV had a much lower inactivation rate coefficient than other enveloped viruses, such as MHV and ɸ6, and close to that of non-enveloped viruses (PV1 and MS2), suggesting that among these viruses, SARS-CoV was nearly the most stable with temperature in sewage water.
Effect of availability of other microorganisms
Fig. 1.
Inactivation rate coefficient of viruses and virus surrogates in sewage (a) and pasteurized sewage (b) based on reported log inactivation values. When multiple log inactivation values were reported, an average inactivation rate coefficient value was determined. SARS: Severe acute respiratory syndrome coronavirus (SARS-CoV), 229E: Human coronavirus 229E, FIPV: Feline infectious peritonitis virus, PV1: Poliovirus 1, MHV: Mouse hepatitis virus, TGEV: transmissible gastroenteritis virus, Phi6: Pseudomonas phage Phi6, MS2: Enterobacteria phage MS2. Rectangles refer to non-enveloped viruses (PV1 and MS2), while the rest are for enveloped viruses or enveloped virus surrogates. Note that the value for SARS in (b) is for sterilized water. [1] (Duan et al., 2003), [2] (Wang et al., 2005), [3] (Gundy et al., 2008), [4] (Casanova et al., 2009), [5] (Casanova and Weaver, 2015), [6] (Ye et al., 2016), [7] (Aquino de Carvalho et al., 2017).
Another factor that affects the survival of viruses in wastewater is the presence of other microorganisms. Fig. 1b shows how the inactivation rate coefficient of viruses or virus surrogates changes with temperature in the absence of other microorganisms in wastewater. The survival trend of viruses and virus surrogates with temperature (Fig. 1b) was similar to the case with unpasteurized wastewater (Fig. 1a), but the inactivation rate coefficient values had lower magnitudes. This indicates that the presence of other microorganisms speeded up the inactivation of viruses in wastewater.
Effect of pH
Survival of viruses in wastewater is also influenced by pH. A majority of studies on the effect of pH on viral survival have been conducted in vitro. The human coronavirus 229E was found to be most stable at pH 6 and remained so within the pH range of 5–8 at 33 °C (Lamarre and Talbot, 1989). The transmissible gastroenteritis virus (TGEV) was found to be most stable at pH 6.5 and remained stable within the pH range of 6.5–8 (Pocock and Garwes, 1975). Similarly, the stability of murine hepatitis virus (MHV) was optimal at pH 6 (Daniel and Talbot, 1987), and the canine coronavirus was found to be more stable at pH 6–6.5, but it lost stability at extreme acidic conditions (Pratelli, 2008). A recent study has shown that SARS-CoV-2 survives over a wide range of pH (3−10) without any noticeable loss for a testing period of 1 h, but it is still not clear how its stability changes over a longer duration (Chin et al., 2020).
Effect of suspended solids and biofilm
Suspended solids in wastewater could also influence the survival of coronaviruses by increasing the chances for the viruses to adsorb on these particles and thus become more protected (Gundy et al., 2008). Sorption of viruses to solid material in a wastewater system (suspended solids in solution or biofilm around the pipes) depends on the virus type, the properties of the sorbent, and the characteristics of the solution. The general consensus is that sorption is enhanced by the presence of clay particles and cations in solution, but decreases with the increase in competing compounds like dissolved organic matter (Jin and Flury, 2002). Sorption of viruses has also been found to depend on the pH of the solution in relation to the isoelectric point (IEP) of the virus. If the pH of the solution is the same as the IEP of the virus, the virus has a net neutral charge on its outer surface. For solutions with a pH less than the virus IEP, the virus will have a net positive charge and will tend to adsorb to negatively charged surfaces, while at a pH higher than the IEP, the virus particles will have a net negative charge and will tend to adsorb to positively charged surfaces (Xagoraraki et al., 2014). Sorption may also be affected by the hydrophobicity of the virus, with some studies considering this as a key factor that affects sorption of viruses during transport through soil material (Powelson et al., 1990). Thus, the virus interaction with the adsorbent may be electrostatic in nature (at pH values that sufficiently deviate from the IEP of the virus) or hydrophobic (at pH values close to the IEP) (Dika et al., 2013; Zerda et al., 1985). The role of these two mechanisms for sorption of four bacteriophages to model surface has been linked to the surface properties of the viral capsid (Armanious et al., 2016). The importance of virus retention due to sorption in the sewer system (possibly on biofilm) is illustrated through a study in which a onetime virus pulse injected at a certain location in the system remained detected down-flow over a 4-day period (Ranta et al., 2001).
Effect of disinfecting agents
Another factor that could influence viral stability in wastewater is the presence and levels of disinfecting agents like residual chlorine. Free chlorine was found to inactivate SARS-CoV better than chlorine dioxide (Wang et al., 2005). Free residue chlorine over 0.5 mg/L for chlorine or 2.19 mg/L for chlorine dioxide in wastewater ensures complete inactivation of SARS-CoV, but it does not inactivate completely Escherichia coli (E. coli) or f2 phage (Wang et al., 2005).
2.2. Factors affecting SARS-CoV-2 RNA fate in wastewater systems
Current WBE approaches for SARS-CoV-2 depend on the detection of the viral RNA. The fate of SARS-CoV-2 RNA in wastewater systems will be influenced by several transfer and transform mechanisms. Although the decay of SARS-CoV-2 RNA in wastewater is expected to be mainly due to biological activity (Balboa et al., 2020), chemical decay should not be excluded (Núñez-Delgado, 2020). Boehm et al. (2019) investigated the decay of viruses in surface waters. The decay rate was reported to range from 0.07 to 0.9 per day. The lowest decay rate was associated with noroviruses and the highest was with Enteroviruses. Nevertheless, the decay rate of viruses in wastewater could be amplified due to biological and possibly chemical reactions that may be not be encountered in surface waters. On the other hand, sorption on the suspended solids present in the wastewater (Balboa et al., 2020; Núñez-Delgado, 2020) and on the biofilm layer on the pipe's internal surface could also influence the movement of the SARS-CoV-2 RNA in the sewer network. Parameters related to transport mechanisms of the RNA in the wastewater network could be derived from laboratory experiments as they are not available in literature. These parameters are expected to be dependent on the physical, chemical, and biological characteristics of the wastewater.
2.3. WBE framework for SARS-CoV-2
SARS-CoV-2 is not truly an enteric virus; however, it can infect cells of the gastrointestinal track (Xiao et al., 2020) and long-term survival of two surrogate coronaviruses has already been shown in 2009 over several days in both water and pasteurized sewage (Casanova et al., 2009). Furthermore, the pH stability (Chin et al., 2020) and fecal excretion of SARS-CoV-2 has now been well-established (Heller et al., 2020; Pan et al., 2020; Y. Wu et al., 2020; Xiao et al., 2020; Xu et al., 2020), suggesting the use of wastewater as an important tool for SARS-CoV-2 surveillance (Venugopal et al., 2020).
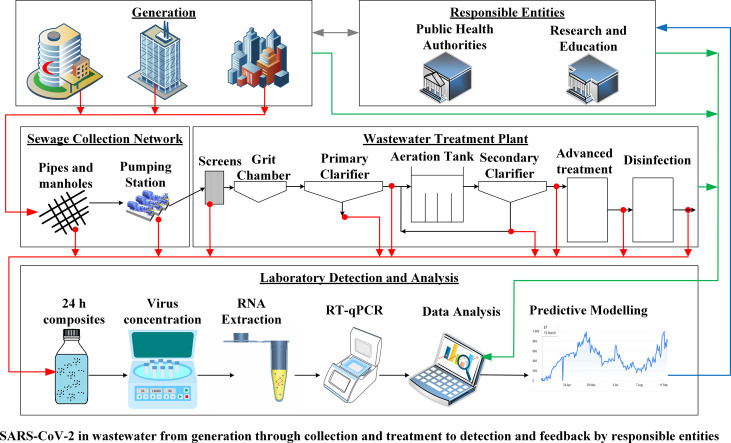
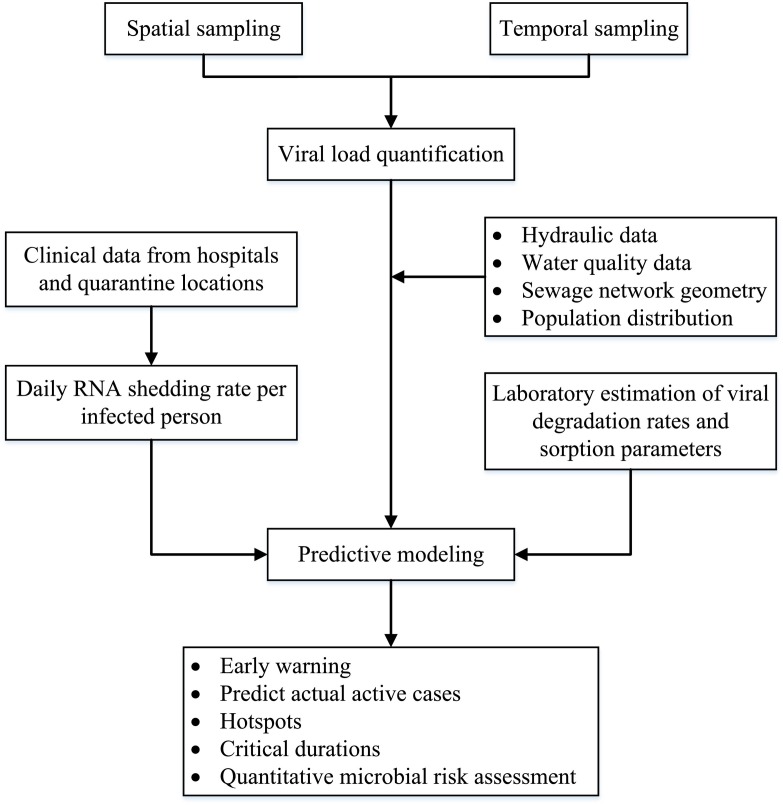
A framework for a WBE system is shown in Fig. 2 . Spatial and temporal wastewater samples are collected from the sewage network and the wastewater treatment plant serving a particular region. The samples should be 24-hour flow-weighted-composite ones and not grab samples as the latter could cause inaccurate determination of viral load (Curtis et al., 2020). Samples are further analyzed for SARS-CoV-2 RNA concentration using one of the established analytical methods. The concentration at the inlet of the wastewater treatment plant reflects the level of infection across the served region and thus could be utilized as an early alert of an outbreak. Alternatively, primary sludge samples at wastewater treatment plants could be used for this purpose as it has been demonstrated that the use of primary sludge provides better results when the concentration in the influent water is low (Balboa et al., 2020).
Fig. 2.
Framework for a wastewater-based epidemiology system.
In order to relate the viral RNA concentration to the number of infected people, several other steps should be considered. One of these steps is to know the daily RNA shedding rate per infected person, which was estimated at 5 thousand to 40 million copies/mL (Foladori et al., 2020). This could be either obtained from previous studies, if available, or otherwise by analyzing sewage samples collected from the closest network point receiving sewage from a hospital or a quarantine location hosting a known number of infected cases. One of the challenges here is related to variations of the shedding rates during the phase of infection. Both asymptomatic and symptomatic patients contribute to the viral load, but for the latter group the viral loads will be higher (Bai et al., 2020; Zou et al., 2020). Another point that needs to be considered is the contribution of different excretion routes (fecal, urine, and other bodily fluids) to the RNA load in the wastewater (Daughton, 2020).
A complementary step in the WBE framework is to use hydraulic modeling to predict the number of infected people contributing to the measured RNA load at the different sampling points based on incoming flowrates. One of the challenges is the stability of SARS-CoV-2 RNA. To address this issue, the used model should incorporate fate and transport mechanisms, including decay that could affect RNA load in the sewage network.
Other challenges that face the use of WBE for SARS-CoV-2 detection are the uncertainty regarding the population served and wastewater flows as well as the time-lag between sample collection and analysis (Sims and Kasprzyk-Hordern, 2020). Uncertainty related to contributing population could be high for internal and external tourism. Therefore, data about the land-use (residential, commercial, industrial, etc.) in the study region could be used to come up with better predictions of the number of infections. As for the uncertainty in wastewater flow, consideration should be given to wet versus dry conditions. This becomes important for combined sewers or domestic sewers that receive an appreciable amount of stormwater. The issue of the delay in obtaining real time data is unavoidable with the current SARS-CoV-2 RNA quantification protocols, which requires one day for sample collection and two days for sample analysis. This time-lag is further amplified with the lack of insufficient resources to process a required large number of samples.
Despite the above challenges, a well-designed WBE system should provide information that could be utilized for early warning, for spatial distribution of infected cases, for identification of critical duration, and for qualitative risk assessment. Both the detection limit and the sensitivity of the used analytical method for viral quantification are important. The lower the method detection limit, the earlier a warning could be issued. The higher the sensitivity of the method, the better the estimate of the infected cases.
Daughton (2020) classified the approaches to WBE into three types based on the level of information required. These include (1) a qualitative approach to assess if infection is present or not, (2) a semi-quantitative approach to assess the relative levels of infection in different zones within a community, and (3) a quantitative approach to assess the absolute levels of infection in different zones within a community and could be utilized for comparison of level of infection across communities. The qualitative approach could be based on samples collected at the inlet of the wastewater treatment plant (WWTP). However, for the semi-quantitative and the quantitative approaches, wastewater samples need to be collected and analyzed at different locations within the sewer network. The spatial distribution of the sampling points and the frequency of sampling should be related to the objectives of the surveillance program. A higher number of sampling locations should lead to better resolution, while more frequent sampling should detect temporal fluctuations. If the number of samples are more than the capacity of a certain laboratory in the region, then sample analysis could be distributed among different laboratories. These laboratories should use a similar standardized analytical protocol to avoid improper interpretation of the results across different laboratories.
3. Methods of detection and quantification of SARS-CoV-2 in water and wastewater
3.1. SARS-CoV-2 genetics
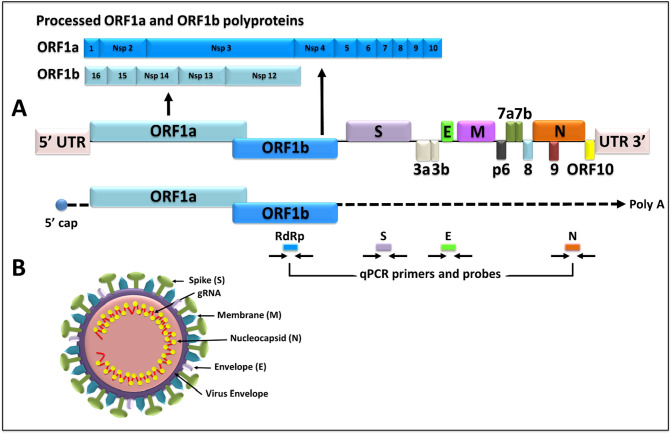
SARS-CoV-2 is an enveloped, positive sense, single-stranded betacoronavirus that has a genome of ~30 kilobases (kb) (Fig. 3 ; summarized in Kim et al., 2020; Y. Wu et al., 2020; A. Wu et al., 2020; F. Wu et al., 2020). The first two-thirds of the genome (~20 kb) codes for the replicase complex consisting of two large genes, ORF1a and ORF1b which code for 16 non-structural proteins (named Nsps 1–16) important for virus replication (Fig. 3A). The last one-third of the genome (~10 kb) codes for the structural and accessory genes of the virus, including four structural proteins that create the virus particle, and accessory genes that are not needed for virus replication but seem to have roles in viral pathogenesis (Fig. 3B). The four structural genes of the virus include: the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The spike is a ~150 kD trimeric, heavily glycosylated protein that allows virus entry into permissive pulmonary epithelial and a number of other cell types expressing the angiotensin converting enzyme 2 (ACE2) which acts as the viral receptor (Li et al., 2020). The S protein is cleaved by a host protease into a surface (S1) domain that interacts with ACE2 and a transmembrane domain (S2) that allows virus fusion with the host cell membrane. S is also the viral protein against which a lot of current vaccine development efforts are being targeted since antibodies against S can be neutralizing, inhibiting the first step of virus lifecycle which is entry into target cells. E is a small (~8–12 kD) protein that is highly divergent between viral strains, but with a common structure which is not well-studied. Similar to the HIV-1 Vpu protein, E is important for virus assembly and release of virus particles. It also has an ion channel activity dispensable for virus replication, but important for viral pathogenesis. The M protein (~25–30 kDa in size) is the most abundant protein of the virus important for virion shape. Finally, the N protein is the largest structural protein of the virus which is the only one present in the nucleocapsid. It interacts with the genomic RNA, giving it its classical “helical” structure that is characteristic of coronaviruses (Fig. 3).
Fig. 3.
Schematic representation of SARS-CoV-2 genome and virus particle. (A) SARS-CoV-2 full-length genome, location of all the genes and proteolytic processing of the polyproteins encoded by the ORF1a and ORF1b genes. The genome is followed by schematic of the full-length genomic RNA (capped and polyadenylated, poly A), and location of the primers and probes being currently used for real time qPCR assays for virus detection. (B) Graphic representation of the virus particle along with its major structural proteins.
3.2. SARS-CoV-2 replication
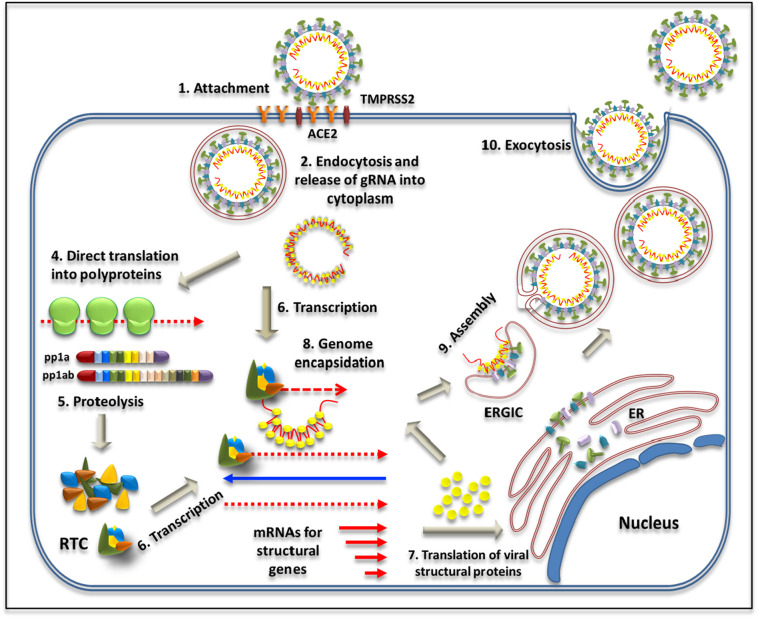
SARS-CoV-2 life cycle initiates with infection of susceptible cells using the ACE2 receptor and the viral spike protein that protrudes out from the virus particle (Fig. 4 : Step 1; reviewed in Fehr and Perlman, 2015; Y. Wu et al., 2020; A. Wu et al., 2020; F. Wu et al., 2020). The virus is internalized via endocytosis and the genomic RNA released into the cytoplasm following cleavage of the spike protein into S1 and S2 subunits by TMPRRS2, a host protease that exposes the fusion peptide present within the S2 domain (Steps 2 and 3). Since the viral gRNA is capped and polyadenylated like cellular mRNAs, ORF1a and ORF1b are translated immediately into polyproteins 1a and 1b that produce the replication-transcription complex (RTC) responsible for genome replication and transcription of other genes via a (−) strand RNA intermediate shown in blue (Steps 4–6). Translation of the N protein in the cytoplasm initiates the process of virion assembly by capturing the newly-synthesized viral genome (Steps 7 & 8). The M, E, and S structural proteins are expressed within the endoplasmic reticulum, followed by their transport via the secretory pathway consisting of the endoplasmic reticulum–Golgi intermediate compartment (ERGIC) (Steps 8 & 9). These structural proteins complete the assembly process by interacting with the gRNA-N complex while still being part of the ERGIC (Step 9), finally forming the progeny virions within vesicles that are released as infectious virions from the cells via exocytosis (Step 10).
Fig. 4.
Schematic representation of SARS-CoV-2 life cycle. See text for details. ACE2, angiotensin-converting enzyme-2; TMPRSS2, transmembrane serine protease 2; RTC, replication-transcription complex; ER, endoplasmic reticulum; ERGIC, endoplasmic reticulum–Golgi intermediate compartment. The blue arrow denotes (−) strand RNA transcribed as an intermediate in the genome replication process. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Molecular detection of viruses in wastewater
Presence of viruses can be detected based on assays that can either estimate infectious viral titers or the amount of viral genetic material. Since viruses in wastewater are found in extremely low concentrations, over the last three decades, amplification techniques based on PCR have replaced infectious cell culture assays that are laborious, time consuming, expensive, and not amenable to high throughput (Kim et al., 2020). Among the PCR techniques, many different types are available the use of which depends upon the needs of the investigators (reviewed in Kim et al., 2020; Y. Wu et al., 2020; A. Wu et al., 2020; F. Wu et al., 2020). For example, the classical PCR, real time quantitative PCR (qPCR), and reverse transcriptase quantitative PCR (RT-qPCR) are the “gold standard” for detection of low-level genetic material in a sensitive and specific manner, but requires a standard curve of known DNA for absolute quantification. Digital PCR is an improvement on this technique and can achieve the same results or even better without a standard curve (Fehr and Perlman, 2015); however, it is still a more “specialist-driven” technique that is expensive, not amenable to high throughput, requires instruments & chemistries not commonly available commercially or in routine clinical & research laboratories, and has many other technical and practical caveats that makes it a more impractical approach. But if it can be adapted into a more user-friendly methodology, it can become valuable in the years to come. Other permutations of PCR have been developed to enable testing of the infectious nature of the virus by cleverly integrating cell culturing of the virus inoculum from the environment followed by PCR (ICC PCR) or concentrating virus particles in an environmental sample via immunomagnetic separation prior to PCR (IMS PCR) and each of these types have been used successfully (A. Wu et al., 2020).
3.4. Challenges of detection of viruses in wastewater
Unlike research samples and body fluids/tissues, detection of viruses in environmental samples, especially wastewater, presents its fair share of challenges. This is due to the presence of not only fecal and suspended solids in the sample, but also chemicals induced by domestic usage, urban and rural runoffs, industrial activities, etc., that create a complex state of the sample from which the viral genetic material must be isolated (Haramoto et al., 2018). This effort is further compounded by the fact that most viral pathogens exist at much diluted levels in wastewater, making concentration of the water sample a prerequisite for successful detection. However, depending upon the sample type (wastewater with or without biosolids or sludge), water concentration can lead to inadvertent concentration of other undesirable substances/chemicals that can inhibit downstream steps of virus detection. Hjelmsø et al. (2017) have addressed this issue by comparing different virus concentration and RNA isolation methods that are commonly used in research laboratories in a systematic manner. These included precipitation of the viral genetic material with polyethylene glycol (PEG; the most common and cheapest method of concentrating viral genetic material), but also others such as flocculation and filtration using positively charged membranes, monolithic adsorption, and glass wool filtration. For RNA extraction, they tested a number of commercially available kits. Their analyses revealed that PEG remained a very powerful means of concentrating different types of viruses from sewage and also identified two commercial kits (QIAamp Viral RNA Mini Kit or PowerViral® Environmental RNA/DNA Isolation Kit from Qiagen) with the ability to isolate viral genomes with the greatest diversity. The use of RNA extraction kits has the added advantage of facilitating high throughput analysis of sewage samples. In the absence of kits, other classical reagents such as TRIzol (a method that combines guanidinium thiocyanate withphenol/chloroform extraction) can also be used as well, providing high quality RNA with no loss of biodiversity of viruses (Chomczynski and Sacchi, 2006).
3.5. Molecular detection of SARS-CoV-2 in wastewater
Detection of SARS-CoV-2 RNA is currently the most sensitive and quantitative means of monitoring viral presence in the wastewater. Virus particles present in the wastewater protect the viral genome from decay by environmental factors (Fig. 3b). Thus, as long as there are intact virus particles in the wastewater, the RNA remains safe from decay and can be detected empirically. Given the infective nature of this virus, a first step in all viral RNA procedures should be inactivation of the virus for the safety of the technical staff. Other safety control measures reported by some studies include: wearing personal protective equipment throughout the investigation process, using UV to disinfect sample bottles before handling in the lab, and pasteurization of the sample at around 60 °C for a period of 30–90 min to destroy the virus's capsid before processing the sample (Ahmed et al., 2020a; Bar-Or et al., 2020; La Rosa et al., 2020; F. Wu et al., 2020). It is also becoming increasingly clear that for a virus like SARS-CoV-2 that is not a typical enteric virus, the type of sample being tested from the wastewater plant can influence the success of virus detection. For example, the latest study on wastewater detection of SARS-CoV-2 from Spain suggests that primary sludge rather than the influent wastewater may be a better source of virus isolation in WBE screening for SARS-CoV-2 (Balboa et al., 2020). This is most likely due to the presence of viral envelope which may offer better affinity of the virus particles towards biosolids present in the primary sludge (Balboa et al., 2020).
3.6. Real time qRT PCR
In terms of molecular techniques, real time qPCR combined with reverse transcription (qRT-PCR) is the most extensively-used technique employed for SARS-CoV-2 detection (Ahmed et al., 2020a; Balboa et al., 2020; Bar-Or et al., 2020; Haramoto et al., 2020; Kocamemi et al., 2020a, Kocamemi et al., 2020b; Kumar et al., 2020; La Rosa et al., 2020; Medema et al., 2020a, Medema et al., 2020b; Nemudryi et al., 2020; Peccia et al., 2020; Randazzo et al., 2020a, Randazzo et al., 2020b; Rimoldi et al., 2020; Sharif et al., 2020; Sherchan et al., 2020; F. Wu et al., 2020; Wurtzer et al., 2020a, Wurtzer et al., 2020b). It is a technique that uses antisense DNA probes and primers (small complementary pieces of DNA), to “amplify” viral RNA; however, since RNA itself cannot be directly amplified, it must first be converted into a DNA form (Bustin and Nolan, 2020; Hamza and Bibby, 2019; Jahne et al., 2020). This is accomplished by using the enzyme “reverse transcriptase (RT)”—hence the name RT qPCR. Briefly, the purified RNA from the wastewater sample is chemically converted into cDNA (complementary DNA) using the RT along with a poly dT primer that can bind to any RNA containing the polyadenylated tail and convert it into DNA (Fig. 5 ). Once converted into cDNA, investigators can use fluorescently-labelled probes and unlabeled primers to further amplify the DNA using commercially-available chemistries, depending upon the particular real time PCR instrument being used. This technique has shown great success as can be seen by the slew of studies that are emerging since the start of the pandemic (Ahmed et al., 2020a; Balboa et al., 2020; Bar-Or et al., 2020; Haramoto et al., 2020; Kocamemi et al., 2020a, Kocamemi et al., 2020b; Kumar et al., 2020; La Rosa et al., 2020; Medema et al., 2020a, Medema et al., 2020b; Nemudryi et al., 2020; Peccia et al., 2020; Randazzo et al., 2020a, Randazzo et al., 2020b; Rimoldi et al., 2020; Sharif et al., 2020; Sherchan et al., 2020; F. Wu et al., 2020; Wurtzer et al., 2020a, Wurtzer et al., 2020b). However, digital PCR has also been tested and found to be valuable, especially when the viral loads are lower which happens during early stages of the virus spread in the community or when the virus is waning (Suo et al., 2020).
Fig. 5.
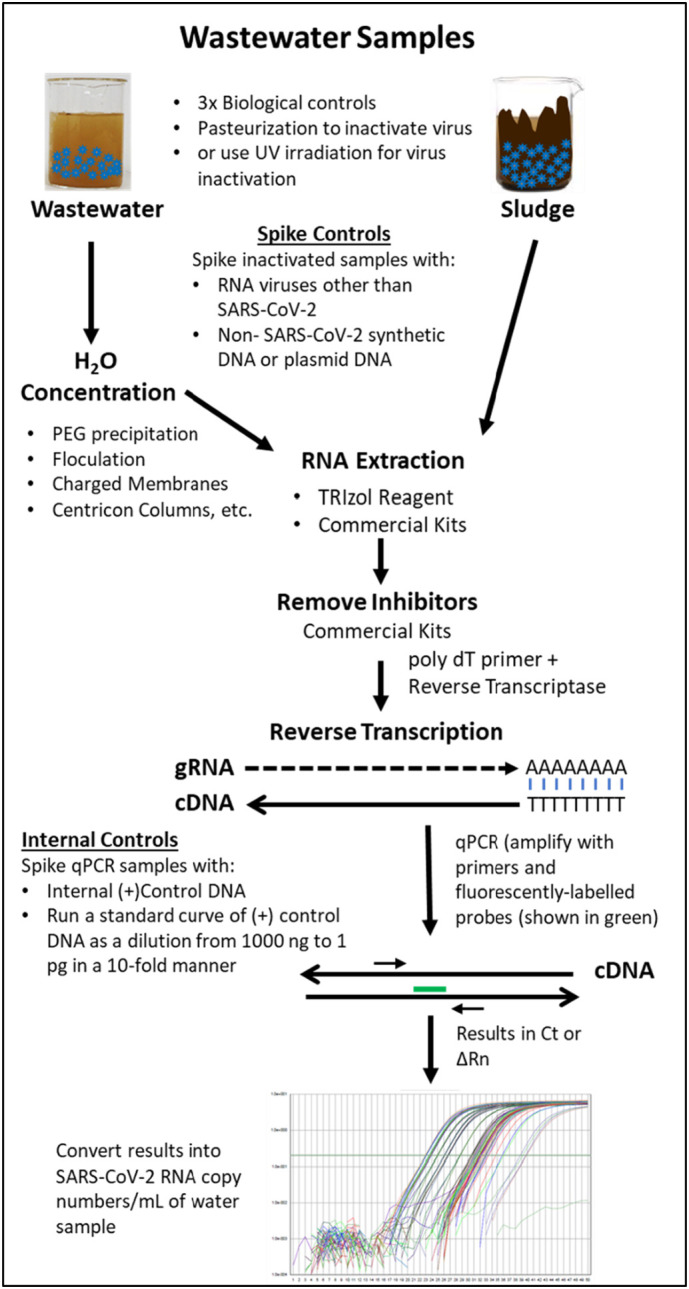
Schematic illustration of the general strategy being employed currently to collect wastewater or sludge samples, process for virus inactivation, virus concentration, and RNA isolation, followed by viral RNA detection using RT qPCR. Each step is shown systematically with key information provided next to the arrows.
3.7. Technical controls
Given the high variability observed in the recovery of virus depending upon the concentration and detection method used, as mentioned above, it is important to use quality control measures to increase confidence in the results obtained. Furthermore while PCR-based techniques are very powerful and sensitive for detection of low level of virus genetic material in the environment, its successful quantitation and correlation with virus burden in the communities requires critical controls that must be included to validate and generate biologically and clinically-relevant data for successful WBE (Ahmed et al., 2020a). First, all sewage samples should be tested in triplicates, irrespective of whether they are grab (one time) or composite (multiple sampling over a day) in nature. The controls should be added at different stages of sample preparation and virus detection to ensure that when a result is negative, it is not due to any technical issues. Some of these technical issues can include: 1) improper storage or handling of the samples (most samples are stored at 4 °C till processed though one study used −80 °C as well), 2) inappropriate concentration of the test sample during the early stages of sample concentration, 3) problems with RNA extraction such as residual phenol or ethanol remaining in samples, 4) presence of PCR inhibitors that can potentially prevent further steps of amplification, resulting in false negative results, etc. (Fig. 5). These controls can further allow investigators to estimate the percent recovery of the spiked-in control, allowing proper normalization of results to reduce intra-sample variability, a valuable aspect that has largely been ignored. Depending upon the study, different groups have used different types of controls for this purpose with some using purified plasmid or synthesized DNA (Ahmed et al., 2020a; Bar-Or et al., 2020; Medema et al., 2020b; Peccia et al., 2020; F. Wu et al., 2020; Wurtzer et al., 2020a, Wurtzer et al., 2020b), while others have used RNA viruses such as bacteriophages (bacterial viruses), mengovirus, other surrogate coronaviruses etc. (Fig. 5) (Balboa et al., 2020; Kocamemi et al., 2020a, Kocamemi et al., 2020b; Medema et al., 2020a, Medema et al., 2020b; Randazzo et al., 2020a, Randazzo et al., 2020b).
In addition to these, other controls are also needed to ensure accurate and specific amplification of the viral cDNA by adding a known non-SARS-CoV-2 DNA target in the test amplification reactions. Such internal controls have been designed by Centers for Disease Control and Prevention (CDC) for qPCR that detect the human RNase P that can be used for this purpose or any other customized qPCR assay (CDC, 2019). Finally, for quantification of copy numbers of viral RNA detected in wastewater, one must include independent samples that contain dilutions of a known amount of DNA to create a standard curve to estimate viral copy numbers. Most of the studies have included both these controls for a quantitative copy number analysis of the RNA detected, though some have presented more qualitative data without taking into account a standard curve (Medema et al., 2020a, Medema et al., 2020b; Rimoldi et al., 2020).
A framework for virus quantification in wastewater samples has previously been proposed (Haramoto et al., 2018). At the minimum, whole-process controls (positive and negative controls) should be employed to determine the overall efficiency of the virus concentration/detection process. More quality control measure should be implemented should a full-scale WBE investigation is undertaken such as listed below based on review of the literature provided in Table 1 :
-
•
Use of sterile sample containers and equipment
-
•
Sample storage preferably at 4 °C before concentration, while concentration within 24 h of storage. Storage at −20 to −80 °C is also acceptable.
-
•
The use of negative control samples, either by relying on bio-banked samples collected before the pandemic or by disinfecting a portion of the collected sample
-
•
Use of positive control samples by adding viral surrogates to lysed sewage samples to confirm virus concentration method recovery
-
•
Using biological replicates (wastewater sample replicates) to report the overall variability of reported results
-
•
Comparing results from more than one virus concentration method applied to the same sample
-
•
Use of PCR inhibitor removal resins to remove environmental inhibitors
-
•
Sample dilution to ensure removal of environmental inhibitors
-
•
Relying on different qPCR assays (Table 1) to compare the detection of viral RNA
-
•
Technical replicates (duplicates or triplicates for qPCR analysis) to ensure the reproducibility and reliability of the results.
Table 1.
Experimental characteristics of studies on detection of SARS-CoV-2 in domestic wastewater.
| Study | Location | Sample source | Volume used (mL) | Removal of large debris and suspended solids | Concentration method | Detection ratea | qPCR assay | |
|---|---|---|---|---|---|---|---|---|
| (Medema et al., 2020b) | The Netherlands | WWTP Influent | 100–200 | Centrifugation | Ultrafiltration: Centricon® Plus-70 | 14/18 | CDC N1, N2, N3, E_Sarbeco | |
| (F. Wu et al., 2020) | Boston, MA, USA | WWTP Influent | 40 | Filtration | Precipitation-flocculation: PEG 8000 + NaCl + 0.22 μm Filter | 10/10 | CDC N1, N2, N3 | |
| (Wurtzer et al., 2020b) | Paris, France | WWTP Influent | 11 | None | Ultracentrifugation | 18/18 | E_Sarbeco | |
| WWTP Treated | 6/8 | |||||||
| (Ahmed et al., 2020a) | Brisbane, Australia | WWTP Influent | 100–200 | None | Electronegative membrane filtrationb | 1/8 | N_Sarbeco, NIID_2019-nCoV | |
| Sewer Network | 0/1 | |||||||
| WWTP Influent | Centrifugation | Ultrafiltration: Centricon® Plus-70 | 1/8 | |||||
| Sewer Network | 0/1 | |||||||
| (Nemudryi et al., 2020) | Bozeman, MT, USA | WWTP Influent | 500 | Sequential filtration (20, 5, 0.45 μm filters) | Ultrafiltration: Corning® Spin-X® UF concentrators | 5/5 | CDC N1, N2 | |
| WWTP Influent | 2/2 | |||||||
| (Randazzo et al., 2020a) | Valencia, Spain | WWTP Influent | 200 | None | Precipitation-flocculationb: AlCl3 | 12/14 | CDC N1, N2 | |
| WWTP Treated | 0/9 | |||||||
| (Bar-Or et al., 2020) | Israel | WWTP Influent | 400 | Centrifugation | Precipitation-flocculation + Ultrafiltration | PEG 8000 + 0.22 μm filter + Amicon® Ultra-15 | 3/17 | E_Sarbeco |
| Sewer Network | 200 | 7/9 | ||||||
| WWTP Influent | 400 | Alum +0.22 μm filter + Amicon® Ultra-15 | 1/2 | |||||
| Sewer Network | 200 | 1/3 | ||||||
| (Rimoldi et al., 2020) | Milan, Italy | WWTP Influent | 500 | Sequential filtration (0.7, 0.2 μm filters) | None | 4/8 | Liferiver, N, E_Sarbeco, ORF1ab, | |
| WWTP Treated | 0/4 | |||||||
| Surface water | 3/4 | |||||||
| (Kocamemi et al., 2020a) | Istanbul, Turkey | WWTP Influent | 250 | Centrifugation + Sequential filtration (0.45, 0.22 μm filters) | Precipitation-flocculationb: PEG 8000 + NaCl | 5/7 | RdRp | |
| Sewer Network | 2/2 | |||||||
| (Wurtzer et al., 2020a) | Paris, France | WWTP Influent | 11 | None | Ultracentrifugation | 30/30 | E_Sarbeco, RdRp | |
| (La Rosa et al., 2020) | Milan & Rome, Italy | WWTP Influent | 250 | Centrifugation | Precipitation-flocculation: PEG 8000 + Dextran | 6/12 | ORF1ab, Spike | |
| (Kocamemi et al., 2020b) | Istanbul, Turkey | WWTP Sludge | 250 | Centrifugation + Sequential filtration (0.45, 0.22 μm filters) | Precipitation-flocculationb: PEG 8000 + NaCl | 9/9 | RdRp | |
| (Randazzo et al., 2020b) | Murcia, Spain | WWTP Influent | 200 | None | Precipitation-flocculationb: AlCl3 + Beef extract | 35/43 | CDC N1, N2, N3 | |
| WWTP Treated | 3/30 | |||||||
| (Medema et al., 2020a) | The Netherlands | WWTP Influent | 36–150 | Centrifugation | Ultrafiltration: Centricon® Plus-70 | 20/22 | CDC N1, N2, N3, E_Sarbeco | |
| (Peccia et al., 2020) | New Haven, CT, USA | WWTP Sludge | 2.5 | None | None | 43/43 | CDC N1, N2 | |
| (Green et al., 2020) | Syracuse, NY, USA | WWTP Influent | 20 | None | Ultracentrifugation: sucrose cushion (50% sucrose in TNE buffer) | 6/12 | RdRp (IP2, IP4) | |
| Sewer Network | 20 | 7/10 | ||||||
| (Balboa et al., 2020) | Ourense, Spain | WWTP Influent | 100 | Centrifugation | Ultrafiltration: Amicon® Ultra-15 | 5/5 | Allplex N, E_Sarbeco, RdRp | |
| WWTP Treated | 1/10 | |||||||
| WWTP Sludge | 50 | None | Precipitation-flocculation + Ultrafiltration: PEG 8000 + Glycin buffer +0.45 μm PES membrane + NaCl |
14/35 | ||||
| (Haramoto et al., 2020) | Yamanashi, Japan | WWTP Influent | 200 | None | Adsorption-elution: MgCl2 + 0.8 μm cellulose-ester membrane + elution buffer +0.45 μm filter + Centriprep YM-50 | 0/5 | CDC N1, N2, N_Sarbeco, NIID_2019-nCoV | |
| WWTP Treated | 5000 | 1/5 | ||||||
| Surface Water | 5000 | 0/3 | ||||||
| WWTP Influent | 200 | Electronegative filtration: MgCL2 + 0.8 μm cellulose-ester membrane | 0/5 | |||||
| WWTP Treated | 5000 | 0/5 | ||||||
| Surface Water | 5000 | 0/3 | ||||||
| (Sharif et al., 2020) | Pakistan | Sewer Network | 500 | Centrifugation | Precipitation-flocculation: PEG 8000 + dextran + chloroform | 21/78 | E_Sarbeco, ORF1ab, N-gene | |
| (Sherchan et al., 2020) | Louisiana, USA | WWTP Influent | 250 | None | Ultrafiltration: Centricon® Plus-70 | 2/2 | CDC N1, N2 | |
| WWTP Treated | 250 | 0/1 | ||||||
| WWTP Effluent | 250 | 0/1 | ||||||
| WWTP Influent | 100 | Adsorption-elution: MgCl2 + 0.45 μm electronegative membrane + H2SO4 + NaOH + Centriprep YM-50 | 0/2 | |||||
| WWTP Treated | 750 | 0/1 | ||||||
| WWTP Effluent | 750 | 0/1 | ||||||
| (Kumar et al., 2020) | Ahmedabad, India | WWTP Influent | 50 | Centrifugation + filtration (0.22 μm) | Precipitation-flocculation: PEG 9000 + NaCl | 2/2 | ORF1ab, N-gene, S-gene | |
| WWTP Treated | 50 | 0/2 | ||||||
Only samples collected after the first case are counted.
pH adjustment.
4. Lessons learnt from recent studies on prevalence of SARS-CoV-2 in wastewater
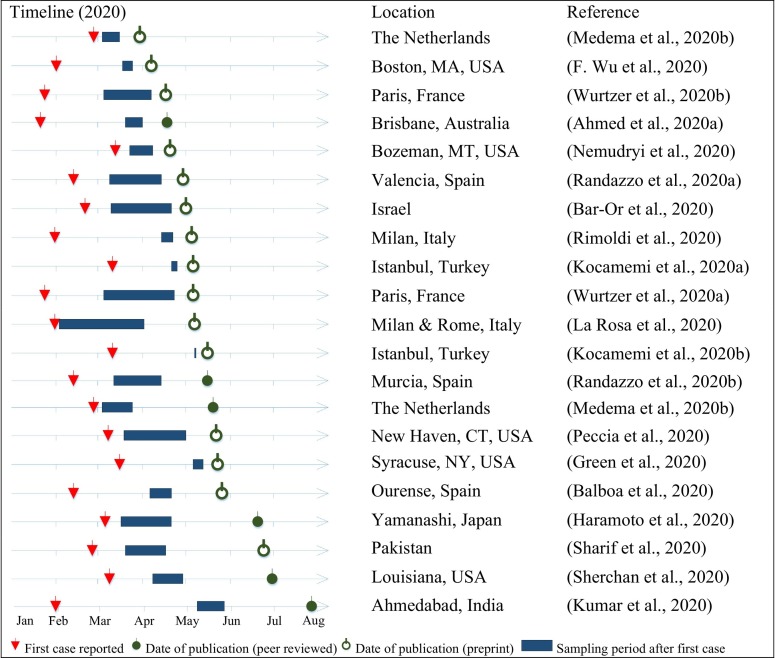
The earliest study on wastewater analysis for detecting SARS-CoV-2 appeared on 30th of March 2020 (Medema et al., 2020b). Since then more than a dozen studies from around the world were published, indicating a move towards mainstreaming wastewater surveillance for assessing the spread of SARS-CoV-2 in different cities. Fig. 6 reports the timeline of the reviewed studies against the reporting of the first patient case of SARS-CoV-2 in the respective study area. It also shows the duration of the sampling period, which varied considerably between the different studies. Some studies had shorter sampling durations, but samples were collected from several locations during the same period. Only few of the published studies were peer-reviewed. In general, all the published studies followed different technical procedures for wastewater sampling, sample concentration, and RNA detection (Table 1). It is of critical importance to compare the performance of the different analytical techniques as noted in earlier studies (Ahmed et al., 2015; Hjelmsø et al., 2017). The following sections focus on the technical differences between the different studies.
Fig. 6.
Timeline of published articles on the detection of SARS-CoV-2in wastewater against the date of first reported case of SARS-CoV-2 in patients.
4.1. Sample collection
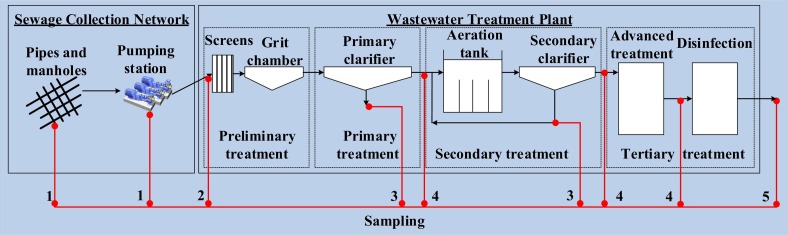
Most of the studies investigating the prevalence of SARS-CoV-2 in the wastewater focused on sampling the raw influent of wastewater into the WWTP (Fig. 7 ). Raw influent sampling is done routinely at any wastewater treatment facility and would thus not add any extra sampling effort to the operators. In addition, to mainstream WBE, it is important to rely on sample locations that have not been disturbed yet by treatment processes. Some studies have added other locations in the wastewater stream to investigate the variation of virus concentrations. Random network locations (manholes or pumping stations) were selected in some studies, samples from these locations gave low virus detection results (Ahmed et al., 2020a; Bar-Or et al., 2020, p.). Other studies have selected network locations that are close to a SARS-CoV-2 patient isolation center, a cruise, or a hospital (Ahmed et al., 2020c; Bar-Or et al., 2020; Kocamemi et al., 2020a). Sampling from such locations can be useful in verifying the virus quantification protocol, particularly when correlating to the number of infected individuals, which can be obtained from hospital records. Comparing results from high concentration locations (such as locations close to hospitals) with results from other locations in the network can provide a better understanding of the fate of the virus in the network and the impact of hydraulic and environmental factors on its concentration. From a risk assessment point of view, some studies investigated the concentration of the virus after sequential treatment at the WWTP, up to the effluent of secondary and tertiary treatment processes (before disinfection) and even in the water of receiving rivers (Balboa et al., 2020; Haramoto et al., 2020; Kumar et al., 2020; Randazzo et al., 2020a, Randazzo et al., 2020b; Rimoldi et al., 2020; Sherchan et al., 2020; Wurtzer et al., 2020b). Only very few of these locations resulted in positive results, which confirms that treatment processes implemented at WWTPs are highly effective in reducing the virus concentration. Finally, some studies were interested in investigating virus concentration in WWTPs sludge (Balboa et al., 2020; Kocamemi et al., 2020b; Peccia et al., 2020). The hypothesis was that since the WWTP processes focus mainly on concentrating solids in the wastewater stream, and since enveloped viruses have affinity to biosolids, then virus concentration would be higher in the sludge resulting from these treatment processes (Balboa et al., 2020). Thickened and primary sludge were identified as the locations with the highest virus concentration compared to WWTP influent (Balboa et al., 2020).
Fig. 7.
Reported wastewater sampling locations in the reviewed articles. 1) Sewer network, 2) WWTP influent, 3) WWTP sludge, 4) WWTP treated, and 5) WWTP effluent in addition to surface water.
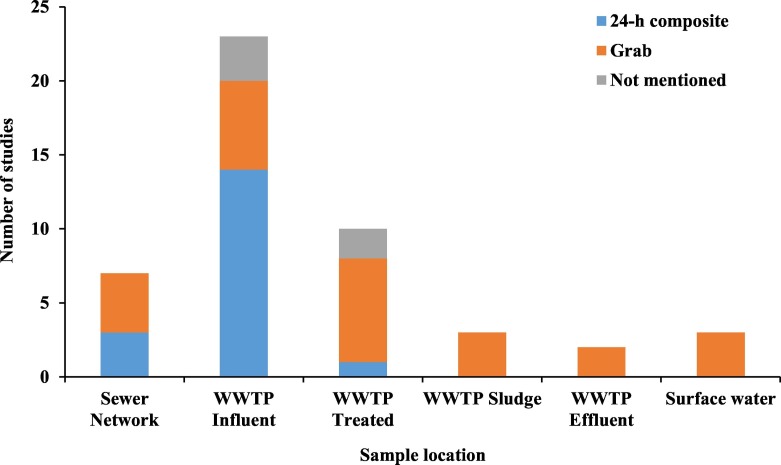
Composite sampling (24-h composite, flow proportional) was used in most studies investigating the concentration of SARS-CoV-2 in wastewater (Fig. 8 ). Composite samples are more homogenous and offer a better representation of the virus concentration in the wastewater (Matrajt et al., 2018). This kind of sampling is easier to apply at routine sampling locations in a WWTP, where the operator has control over the sampling process and an autosampler can be easily used. Composite sampling may not be possible at other locations in the sewer network, or sludge sampling, or even environmental (surface water) sampling. An alternative that was used by some studies is grab sampling, which is easier to do. Nevertheless, studies have shown that virus detection results from grab samples exhibited high variability compared to composite samples (Nemudryi et al., 2020). One way to reduce the variability of grab samples is to rely on grab-composite samples, where grab samples are taken at different periods throughout the day and a composite sample is created by mixing collected samples from any specific location (Ahmed et al., 2020a). The collected sample volume varies depending on the collection method. Only few of the reviewed studies reported on the collected sample volume and the sample storage method. Reported sample volumes ranged between 250 mL to 1000 mL, this is presumed to be the sample extracted from the composite volume. The volume used in virus concentration is different than the sample volume and will be addressed in Section 4.2. Typical unprocessed sample storage was at 4 °C; however, some studies reported storage at −20 to −80 °C (Bar-Or et al., 2020; La Rosa et al., 2020; Peccia et al., 2020). Most studies processed the samples upon receipt in the laboratory and only stored the concentrated viral RNA at −80 °C until further extraction and quantification.
Fig. 8.
Common wastewater sampling locations and sample type in recently published studies on the detection and quantification of SARS-CoV-2 in wastewater systems.
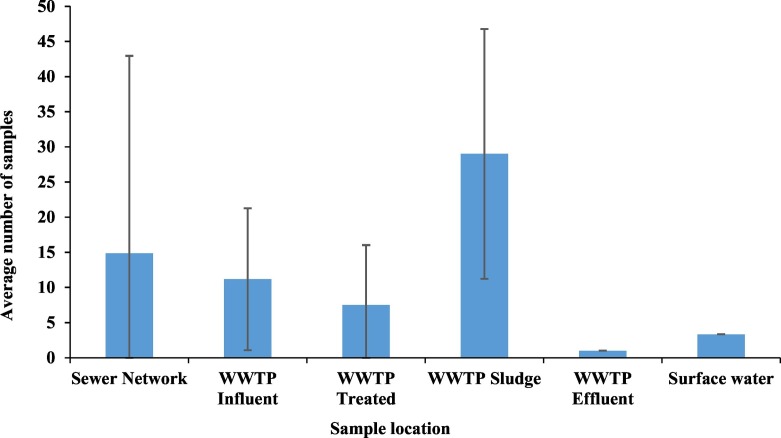
Virus concentration and quantification is a complex, time consuming, and often expensive undertaking. This is perhaps why the studies published on the quantification of SARS-CoV-2 in wastewater, till the date of this review, have relied on a few samples before a proof-of-concept is published. The number of samples analyzed varied considerably among all the studies (Fig. 9 ). Given the high uncertainty in virus concentration and detection methods (Ahmed et al., 2020a), it is imperative that future studies rely on a larger results dataset before conclusions are drawn. The objectives of the analysis would then dictate the sampling protocol, frequency, and the total number of samples. Some studies focused on collecting samples frequently from few locations for a time longer than a month to observe the changes in viral RNA load with time, and compared it against the temporal changes in the number of reported cases near the sample locations (Peccia et al., 2020; Wurtzer et al., 2020b). Other studies focused on sampling from more locations to identify points in the wastewater treatment stream with the most representative viral RNA load could be observed (Balboa et al., 2020; Bar-Or et al., 2020; Randazzo et al., 2020b; Rimoldi et al., 2020). Most studies, however, focused on the repeatability of the virus detection protocol at different WWTPs with few samples from each location (Ahmed et al., 2020a; Kocamemi et al., 2020a, Kocamemi et al., 2020b; La Rosa et al., 2020; Medema et al., 2020a, Medema et al., 2020b; Randazzo et al., 2020a; Wurtzer et al., 2020a). Only two studies relied on a few samples from the influent of one WWTP (Nemudryi et al., 2020; F. Wu et al., 2020).
Fig. 9.
Average number of samples analyzed in recently published studies on the detection and quantification of SARS-CoV-2 in wastewater systems.
4.2. Effect of concentration methods on the qPCR results
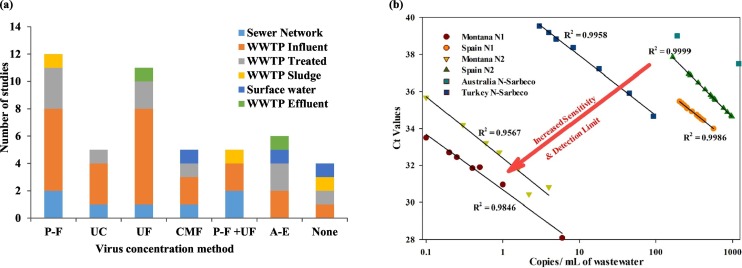
The main wastewater concentration methods could be summarized in four major categories as: electronegative membranes, ultrafiltration, precipitation, and ultracentrifugation. The advantages and disadvantages of different viral concentration methods for recovery of SARS-CoV-2 from wastewater were recently investigated and reviewed (Ahmed et al., 2020b; Lu et al., 2020). The study's results elucidated that the most efficient method for virus recovery is the adsorption-extraction method relying on charged membrane filtration with MgCl2 addition or without manipulation. Ultrafiltration using the Amicon® Ultra-15 exhibited a recovery efficiency close to that of the adsorption-extraction (Ahmed et al., 2020b). Fig. 10a shows that the most used concentration method, reported in the reviewed studies on SARS-CoV-2 recovery from wastewater, are precipitation-flocculation followed by ultrafiltration methods. Of the reviewed studies, only few reported on the recovery of the virus concentration methods implemented.
Fig. 10.
Virus concentration methods used for SARS-CoV-2 in literature. a) Different virus concentration methods applied in the reviewed studies (P-F = precipitation-flocculation, UC = ultracentrifugation, UF = ultrafiltration, CMF = charged membrane filtration, A-E = adsorption-elution) b) Gene copies/ mL of wastewater vs. Ct values at different locations. Locations correspond to the following studies: Montana (Nemudryi et al., 2020), Spain (Randazzo et al., 2020a), Australia (Ahmed et al., 2020a), Turkey (Kocamemi et al., 2020a).
(Note: the figure repeats the count of a study that uses different methods to the same sample location, or the same method for different sample locations.)
The viral RNA concentration method used in each study should have a direct impact on the sensitivity of the results. All the studies that measured SARS-CoV-2 RNA showed strong inverse correlations between cycle threshold (Ct) values and the number of copies /mL of wastewater (Fig. 10b). The lowest R2 reported equals 0.95 having an average slope of −1.3621 ± 0.33. At a low Ct, the method that generates the lowest number of gene copies per mL of wastewater will indicate the highest sensitivity of concentration method employed. It should be noted that multiple factors could affect the method sensitivity including the concentration method and the primer used. However, using different primers as shown in Fig. 10b within the same study did not result in a significant change in the method sensitivity (Kocamemi et al., 2020a; Randazzo et al., 2020a). This suggests that the concentration method is the most important factor in determining the sensitivity, which is in line with the findings of Lu et al. (2020). Based on the results shown in Fig. 10b, the concentration methods that resulted in the most sensitivity was reported as follows; 1) Ultrafiltration: Corning Spin-X UF concentrators with 100 kDa molecular weight cut-off down to 150–200 μL after pretreatment consisting of sequential filtration: 20 μm, 5 μm, then 0.45 μm (Nemudryi et al., 2020), 2) the second most sensitive method used was PEG precipitation preceded by centrifugation and sequential filtration 0.45 μm and 0.2 μm (Kocamemi et al., 2020a), and 3) the third most sensitive method was precipitation using AlCl3 without pretreatment (Randazzo et al., 2020a). Although the least sensitive method was found to be electronegative filtration, it was reported based on one data point in each study (Ahmed et al., 2020a; Haramoto et al., 2020). Availability of more data points specific to SARS-CoV-2 could change the ranking of the method sensitivity discussed above. Lu et al. (2020) recommended that the method used should depend on the sample volume, which will have an impact on the detection limit. It should be noted that the data in Fig. 10b does not include all the SARS-CoV-2 studies and includes only the ones with reported data points. Moreover, qPCR inhibition could cause a similar impact on the results shown in the figure as reported by Kocamemi et al. (2020a).
4.3. Primers and probes for real time qPCR
Early on in the COVID-19 pandemic, to assist with virus detection in the public health settings in the absence of positive control virus material, several groups such as an international group of scientists from Europe and China (Corman et al., 2020), Japan (Shirato et al., 2020), and the CDC, USA (CDC, 2019), developed primers and positive control material which is what most of these studies used (Ahmed et al., 2020a; Balboa et al., 2020; Bar-Or et al., 2020; Haramoto et al., 2020; Kocamemi et al., 2020a, Kocamemi et al., 2020b; Kumar et al., 2020; La Rosa et al., 2020; Medema et al., 2020a, Medema et al., 2020b; Nemudryi et al., 2020; Peccia et al., 2020; Randazzo et al., 2020a, Randazzo et al., 2020b; Rimoldi et al., 2020; Sharif et al., 2020; Sherchan et al., 2020; F. Wu et al., 2020; Wurtzer et al., 2020a, Wurtzer et al., 2020b). The real time PCR assays in these assays mostly targeted the N and E genes on the genomic RNA, though other regions of the genome were also targeted in some cases such as ORF1a and ORF1b (RdRp gene) and the S gene (Fig. 3A). Depending upon the study– the mode of virus concentration used, RNA isolation techniques employed, and amplification method used, different groups have observed different levels of sensitivity of these assays, making it hard to cross-compare them. Thus, Jung et al. (2020) have conducted a study that compared the relative efficiency of some of these now commonly-used assays under the same laboratory conditions and demonstrated that the 2019-nCoV-N2 and N3 primer-probes from CDC and ORF1ab primer-probe from China are the most sensitive combinations among the tested primer-probe pairs.
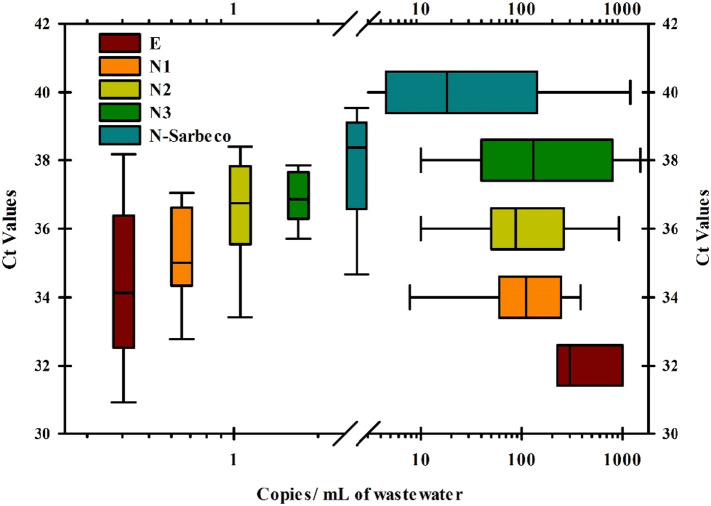
In an attempt to reanalyze data from many of these studies, RT qPCR was used and enough data sets were available for the N and E genes qPCR assays to determine which assay had more sensitivity. This was achieved by comparing the Ct values obtained compared to the estimated copy numbers of viral RNA detected per mL of wastewater and mapping the results obtained as box plots (Fig. 11 ). The same relative trend in sensitivity was observed among the various real time assays whether the box plots were mapped vertically or horizontally (N-Sarbeco > N3 > N2 > N1 > E). Thus, the qPCR assay for the E gene was the least sensitive since it showed the lowest Ct value (32) for the highest copy numbers (250–1000 copies/mL), unlike the assays based on N gene that gave high Ct values when detecting lower levels of RNA. The N-Sarbeco assay was the most sensitive with the ability to detect anywhere from 5.5–120 copies of viral RNA/mL at a Ct of 40, followed by N3 (40–1000 copies/mL) at a Ct of 38, N2 (50–300 copies/mL) at a Ct of 36, and lastly N1 which was similar to N2 with the ability to detect 70 to 250 copies/mL at a Ct of 34. These results somewhat agree with those of Jung et al. (2020), who also found the CDC-designed N2 and N3 were the most sensitive primers among the ten primer sets that they had tested. Unfortunately, they had not tested N-Sarbeco which is quite a commonly used primer set (Corman et al., 2020). Ahmed et al. (2020c) found out that N1 and N2 had better sensitivity than N-Sarbeco which might be attributed to nucleotide mutations. It is clear, that there is no consensus on the sensitivity of the different primers.
Fig. 11.
Comparative analysis of the Ct values obtained relative to viral copy numbers observed for different real time PCR assays used in SARS-CoV-2 RNA surveillance WBE. The Ct values are normalized to copies of SARS-CoV-2 RNA detected per milliliter of wastewater tested in each study.
4.4. Relating the concentration of genes copies in wastewater to cumulative infection
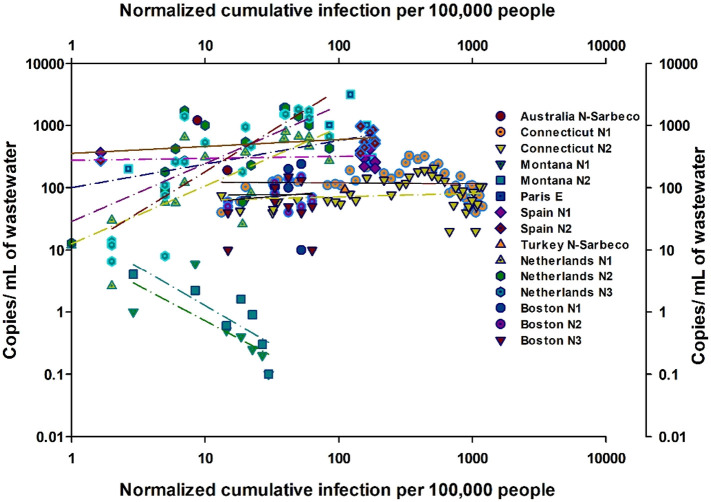
Fig. 12 shows the relationship between the cumulative infections in the area of each study normalized to 100,000 people in the study area with the number of copies/mL of wastewater. The location considered in each study varied depending upon the specificity of the reporting study in identifying the location of the sampling and the breakdown of the infection data. The number of infections and total population served was broken down by city level in the Netherlands and Spain (Medema et al., 2020a; Randazzo et al., 2020a), state level in Massachusetts, Montana, and Connecticut (Nemudryi et al., 2020; Peccia et al., 2020; F. Wu et al., 2020), and country level in Turkey, France, and Australia (Ahmed et al., 2020a; Kocamemi et al., 2020a; Wurtzer et al., 2020b). Within each study, the data was further broken down by the type of primer used. The relationship between both parameters (slope) varied greatly depending on the stage of the pandemic. The slope presented in Table 2 is calculated based on Fig. 12. The calculated slope is negative at early stages of the cumulative infection curve and keeps increasing with time along the cumulative curve. The correlation between the calculated slope and the average new cases per day during the analysis period showed a weak relationship (R2 = 0.5). Nevertheless, if data quality in future studies is improved and controlled protocols are applied, it is suggested that the qPCR results can be used to predict the number of new cases. Since wastewater surveillance accounts for infected cases, including those who did not display any symptoms (the asymptomatic cases), the slope was correlated with the average new cases per day, 8-days after the analysis period (Table 2). Thus, wastewater sampling results can help estimate the cumulative curve behavior as early as 8 days temporal shift (Wurtzer et al., 2020a). Using the average of new cases 8 days after the study period showed slightly improved correlation (R2 = 0.6).
Fig. 12.
Relationship between normalized cumulative infection per 100,000 people and gene copies /mL of wastewater measured plotted per primer at each location studied for the SARS-CoV-2 in wastewater. References of the locations in the figure are indicated in Table 2.
Table 2.
Relationship between qPCR results in copies/mL of waste, the normalized cumulative population, and the location on the cumulative infection curve.
| Location | Reference | Primer | Slope of curve in Fig. 12 | R2 | Average new cases per day during the analysis period | Average new cases per day 8-days after the analysis period | Sampling period in relation to cumulative curve |
|---|---|---|---|---|---|---|---|
| Montana | (Nemudryi et al., 2020) | N1 | −1.1292 | 0.49 | 18 | 9 | Very Early in the curve |
| N2 | −1.2436 | 0.66 | |||||
| Connecticut | (Peccia et al., 2020) | N1 | −0.0127 | 0.01 | 658 | 748 | Mid of the exponential growth |
| N2 | 0.0541 | 0.02 | |||||
| Boston | (F. Wu et al., 2020) | N1 | 0.0183 | 0.01 | 166 | 1223 | Early in the curve |
| N2 | 0.1809 | 0.04 | |||||
| N3 | −0.0127 | 0.01 | |||||
| Paris | (Wurtzer et al., 2020a) | E | 0.3771 | 0.50 | 874 | 2348 | The entire exponential growth period |
| Spain | (Randazzo et al., 2020a) | N1 | 0.0318 | 0.06 | 139 | 151 | Early to mid of the exponential growth |
| N2 | 0.1109 | 0.32 | |||||
| Netherlands | (Medema et al., 2020a) | N1 | 0.9317 | 0.58 | 265 | 579.2 | Early in the curve |
| N2 | 0.9279 | 0.55 | |||||
| N3 | 1.3142 | 0.70 |
5. Conclusions
With a new world adapting to the COVID-19 pandemic, wastewater is expected to play a major role in its surveillance and tracking. WBE can be used to detect the spread of pathogens. It is a cheaper and more efficient means of tracking infectious agents in communities, since it overcomes the need to test a large proportion of the population individually. WBE can help determine the spread of the pathogen and their hotspots in the community. WBE will work if the virus could survive the stomach acidity and be excreted in the feces. While WBE was proven to work with older human pathogens, such as poliovirus, new protocols need to be developed for SARS-CoV-2. In this study, a framework for WBE was proposed specifically for SARS-CoV-2. The framework incorporates lessons learnt from earlier studies on WBE. The implementation of this framework, can result in cost savings and better prediction of infections than the current practices.
Detection and quantification of viral SARS-CoV-2 RNA in wastewater was proven by multiple researchers all over the world. Among the studies, samples varied by location and collection methods. Some studies suggested that sampling from the wastewater treatment plant sludge was more appropriate than raw influent wastewater as it resulted in higher viral load. Moreover, sample handling, viral load concentration and extraction methods varied. Five different primers sets were primarily used for qPCR quantification. There were wide differences between the methods employed, resulting in inconsistent number of genes; sometimes even within the same study. Based on comparing the results of different primers extracted from multiple studies, N-Sarbeco appears to be the most sensitive primer. However, there is no general consensus on the most sensitive primer. A comparison between the different SARS-CoV-2 concentration methods was presented. The most sensitive methods were observed to be ultrafiltration followed by PEG precipitation. The analysis of SARS-CoV-2 detection focused on calculating the slope of the relation between the number of gene copies versus the cumulative number of infections normalized to the total population served. This slope was correlated to the average new cases within the sampling period. The resulting correlation suggests that qPCR results could be used to predict new infections. Since wastewater sampling accounts for all infections, regardless of being symptomatic or not, the correlation between the slope and the average new cases was improved if a lag period was introduced.
6. Recommendations
Implications of wastewater management on transmission of SARS-CoV-2
Defects in the wastewater plumbing system was identified as the transmission mode for SARS within a building in Hong Kong facilitated by the transport of “virus laden droplets” (WHO, 2003). Self-isolation, due to SARS-CoV-2, can lead to a greater number of infected people in a building and potential system overuse (Gormley et al., 2020). The use of hospital wards as quarantine areas is also a concern because of the interconnectedness of the plumbing system (Gormley et al., 2020). A study revealed that the estimated risks for the aggressive and extreme scenarios for the exposure of raw wastewater were likely to be above a WHO benchmark of tolerable risk used for virus infection, thus reinforcing the concern of sewage systems as a transmission pathway of SARS-CoV-2 (Zaneti et al., 2020). Several researchers called for the need of studies about the survival of the virus in sewage (Carducci et al., 2020; Qu et al., 2020). Until proved safe, fecal transmission routes of SARS-CoV-2 should be considered, and extra occupational safety precautions should be practiced in managing sewage collection and processing. The same should be applied during the implementation of a WBE program to assure the safety of workers involved in sample collection and analysis.
Implication of greywater management on transmission of SARS-CoV-2
Prior to the COVID-19 pandemic, a study investigated the risk of microbial quality using greywater as a transmission pathway (O'Toole et al., 2012). Enteric viruses were detected in 18% samples comprising enterovirus, norovirus genogroup GI, norovirus genogroup GII, and rotavirus (O'Toole et al., 2012). However, so far no study has examined the possibility of SARS-CoV-2 transmission through recycling of greywater. Although greywater does not include wastewater generated from toilet flushing, it could include the virus due to discharge of other bodily fluids via sink, laundry and shower wastewater. Transmission routes via greywater include inhalation of splashed greywater aerosols through toilet flushing and outdoor irrigation. For instance, SARS-CoV-2 RNA has been detected in non-potable irrigation water used in Paris (Connexion journalist, 2020). As a precaution, greywater needs to be disinfected before reuse until transmission of the virus in greywater is thoroughly investigated.
Study of the decay of SARS-CoV-2
Success of utilizing WBE to assess the circulation of SARS-CoV-2 in exposed communities depends largely on understanding the mechanisms affecting the transport of the viral RNA within the wastewater system. No study has been done yet to elucidate the mechanisms influencing the transport of the viral RNA in sewage. Studies are needed to assess viral decay due to biological and chemical activities as well as retardation of its transport due to sorption on biofilm and the pipe network material. The outcome of these studies will enable better prediction of the viral RNA level at the source.
Effectiveness of digital PCR
As indicated, qRT-PCR is currently the most extensively used technique for SARS-CoV-2 detection and the method has shown great success. Although digital PCR has not been widely adopted, but it has been tested and found to be valuable, especially when the viral loads are lower which happens during early stages of the virus spread in the community or when the virus is waning (Suo et al., 2020). Thus, there is a need for further studies to assess the effectiveness of digital PCR.
Method sensitivity
The sensitivity of the analytical method is very important in adopting a WBE program. The viral concentration method, as presented in this study, appears to be the most important factor affecting method sensitivity. Therefore, large scale comparative studies for specific concentration methods are needed to assure method reproducibility. Furthermore, comparative studies are needed to reach a consensus on the sensitivity of different primers used in the employed analytical methods.
Use of a standardized protocol
The call for additional studies to optimize the analytical method for detection of SARS-CoV-2 in wastewater will hopefully lead to the development of a standardized protocol that could be adopted globally. The protocol should provide a detailed description of sample collection, preservation, safe handling, pre-treatment and analysis steps. The use of a standardized analytical protocol is necessary to avoid improper interpretation of the results across different studies and among different laboratories. In fact, some researchers proposed a global effort to coordinate methodologies and data-sharing to ensure better evaluation of WBE for the current and future outbreaks of disease considering viral mutations (Bivins et al., 2020). It is recommended that future studies should consider a lag-period when conducting WBE for SARS-CoV-2 investigations.
Declaration of competing interest
No potential conflict of interest was reported by the authors.
Acknowledgments
Acknowledgement
This work was supported by the National Water Center at the United Arab Emirates University (COVID-19 seed grants).
Editor: Damia Barcelo
References
- Adcock N.J., Rice E.W., Sivaganesan M., Brown J.D., Stallknecht D.E., Swayne D.E. The use of bacteriophages of the family Cystoviridae as surrogates for H5N1 highly pathogenic avian influenza viruses in persistence and inactivation studies. J. Environ. Sci. Health A. 2009;44:1362–1366. doi: 10.1080/10934520903217054. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Harwood V.J., Gyawali P., Sidhu J.P.S., Toze S. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol. 2015;81:2042–2049. doi: 10.1128/AEM.03851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J., Simpson S., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020:139960. doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas D.S.C., K.V., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travelers. Journal of Travel Medicine. 2020 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdiouni H., Faouzi A., Fariat N., Hassar M., Soukri A., Nourlil J. Detection and molecular identification of human adenoviruses and enteroviruses in wastewater from Morocco. Lett. Appl. Microbiol. 2012;54:359–366. doi: 10.1111/j.1472-765X.2012.03220.x. [DOI] [PubMed] [Google Scholar]
- Apostol L.N.G., Imagawa T., Suzuki A., Masago Y., Lupisan S., Olveda R., Saito M., Omura T., Oshitani H. Genetic diversity and molecular characterization of enteroviruses from sewage-polluted urban and rural rivers in the Philippines. Virus Genes. 2012;45:207–217. doi: 10.1007/s11262-012-0776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino de Carvalho N., Stachler E.N., Cimabue N., Bibby K. Evaluation of Phi6 persistence and suitability as an enveloped virus surrogate. Environmental science & technology. 2017;51:8692–8700. doi: 10.1021/acs.est.7b01296. [DOI] [PubMed] [Google Scholar]
- Armanious A., Aeppli M., Jacak R., Refardt D., Sigstam T., Kohn T., Sander M. Viruses at solid–water interfaces: a systematic assessment of interactions driving adsorption. Environmental science & technology. 2016;50:732–743. doi: 10.1021/acs.est.5b04644. [DOI] [PubMed] [Google Scholar]
- Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. Jama. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodríguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. 2020. The Fate of SARS-CoV-2 in Wastewater Treatment Plants Points Out the Sludge Line as a Suitable Spot for Incidence Monitoring. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelo D. An environmental and health perspective for COVID-19 outbreak: meteorology and air quality influence, sewage epidemiology indicator, hospitals disinfection, drug therapies and recommendations. Journal of Environmental Chemical Engineering. 2020;8(4) doi: 10.1016/j.jece.2020.104006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E.F., Paitan Y., Bitkover E., Berchenko Y., Kushmaro A. 2020. Regressing SARS-CoV-2 Sewage Measurements onto COVID-19 Burden in the Population: A Proof-of-concept for Quantitative Environmental Surveillance. medRxiv 2020.04.26.20073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisseux M., Colombet J., Mirand A., Roque-Afonso A.-M., Abravanel F., Izopet J., Archimbaud C., Peigue-Lafeuille H., Debroas D., Bailly J.-L. Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: a one-year experiment in central France, 2014 to 2015. Eurosurveillance. 2018;23 doi: 10.2807/1560-7917.ES.2018.23.7.17-00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., de los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam Md.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Blomqvist S., El Bassioni L., Nasr E.M.E.M., Paananen A., Kaijalainen S., Asghar H., de Gourville E., Roivainen M. Detection of imported wild polioviruses and of vaccine-derived polioviruses by environmental surveillance in Egypt. Appl. Environ. Microbiol. 2012;78:5406–5409. doi: 10.1128/AEM.00491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A.B., Silverman A.I., Schriewer A., Goodwin K. Systematic review and meta-analysis of decay rates of waterborne mammalian viruses and coliphages in surface waters. Water Res. 2019;164 doi: 10.1016/j.watres.2019.114898. [DOI] [PubMed] [Google Scholar]
- Bofill-Mas S., Pina S., Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 2000;66:238–245. doi: 10.1128/aem.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer A.F., Eisenberg J.N., Pomeroy C.D., Shulman L.M., Hindiyeh M., Manor Y., Grotto I., Koopman J.S., Eisenberg M.C. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Natl. Acad. Sci. 2018;115:E10625–E10633. doi: 10.1073/pnas.1808798115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Nolan T. RT-qPCR testing of SARS-CoV-2: a primer. IJMS. 2020;21:3004. doi: 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A., Verani M., Battistini R., Pizzi F., Rovini E., Andreoli E., Casini B. Epidemiological surveillance of human enteric viruses by monitoring of different environmental matrices. Water Sci. Technol. 2006;54:239–244. doi: 10.2166/wst.2006.475. [DOI] [PubMed] [Google Scholar]
- Carducci A., Federigi I., Liu D., Thompson J.R., Verani M. Making waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L.M., Weaver S.R. Inactivation of an enveloped surrogate virus in human sewage. Environmental Science & Technology Letters. 2015;2:76–78. [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Outbreak of swine-origin influenza A (H1N1) virus infection-Mexico, March–April 2009. MMWR Morb. Mortal. Wkly Rep. 2009;58:467. [PubMed] [Google Scholar]
- CDC . 2012. First Global Estimates of 2009 H1N1 Pandemic Mortality Released by CDC-led Collaboration. [Google Scholar]
- CDC, N Real-time rRT-PCR panel primers and probes [WWW document]. Coronavirus disease 2019 (COVID-19) 2019. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html URL.
- Cevik M., Bamford C., Ho A. COVID-19 pandemic–a focused review for clinicians. Clin. Microbiol. Infect. 2020;26(7):842–847. doi: 10.1016/j.cmi.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A., Chu J., Perera M., Hui K., Yen H.-L., Chan M., Peiris M., Poon L. 2020. Stability of SARS-CoV-2 in Different Environmental Conditions. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat. Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- Clemente-Casares P., Pina S., Buti M., Jardi R., Martín M., Bofill-Mas S., Girones R. Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 2003;9:449. doi: 10.3201/eid0904.020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connexion journalist . 2020. ‘No Risk’ as Corona Found in Paris Non-potable Water. The Connexion. [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Map [WWW Document] Johns Hopkins coronavirus resource center. 2020. https://coronavirus.jhu.edu/map.html URL.
- Curtis K., Keeling D., Yetka K., Larson A., Gonzalez R. 2020. Wastewater SARS-CoV-2 Concentration and Loading Variability from Grab and 24-hour Composite Samples. medRxiv 2020.07.10.20150607. [DOI] [Google Scholar]
- Daniel C., Talbot P. Physico-chemical properties of murine hepatitis virus, strain A59. Arch. Virol. 1987;96:241–248. doi: 10.1007/BF01320963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020:138149. doi: 10.1016/j.scitotenv.2020.138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dika C., Ly-Chatain M., Francius G., Duval J., Gantzer C. Non-DLVO adhesion of F-specific RNA bacteriophages to abiotic surfaces: importance of surface roughness, hydrophobic and electrostatic interactions. Colloids Surf. A Physicochem. Eng. Asp. 2013;435:178–187. [Google Scholar]
- Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Duan S., Zhao X., Wen R., Huang J., Pi G., Zhang S., Han J., Bi S., Ruan L., Dong X. Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomedical and environmental sciences: BES. 2003;16:246–255. [PubMed] [Google Scholar]
- Edition F. Guidelines for drinking-water quality. WHO chronicle. 2011;38:104–108. [PubMed] [Google Scholar]
- El Bassioni L., Barakat I., Nasr E., de Gourville E.M., Hovi T., Blomqvist S., Burns C., Stenvik M., Gary H., Kew O.M. Prolonged detection of indigenous wild polioviruses in sewage from communities in Egypt. Am. J. Epidemiol. 2003;158:807–815. doi: 10.1093/aje/kwg202. [DOI] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses. Springer; 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.-T., Lipp E.K. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005;69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S., Baric R., de Groot R., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley M., Aspray T.J., Kelly D.A. COVID-19: mitigating transmission via wastewater plumbing systems. Lancet Glob. Health. 2020;8:e643. doi: 10.1016/S2214-109X(20)30112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H., Wilder M., Middleton F.A., Collins M., Fenty A., Gentile K., Kmush B., Zeng T., Larsen D.A. 2020. Quantification of SARS-CoV-2 and Cross-Assembly Phage (crAssphage) From Wastewater to Monitor Coronavirus Transmission Within Communities. medRxiv 2020.05.21.20109181. [DOI] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ Virol. 2008;1:10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Hamza I.A., Bibby K. Critical issues in application of molecular methods to environmental virology. J. Virol. Methods. 2019;266:11–24. doi: 10.1016/j.jviromet.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737:140405. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller L., Mota C.R., Greco D.B. COVID-19 faecal-oral transmission: are we asking the right questions? Sci. Total Environ. 2020;138919 doi: 10.1016/j.scitotenv.2020.138919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan C., Brassey J., Jefferson T. COVID-19: what proportion are asymptomatic? The Centre for Evidence-Based Medicine develops, promotes and disseminates better evidence for healthcare. 2020. https://www.cebm.net/covid-19/covid-19-what-proportion-are-asymptomatic/ URL.
- Hjelmsø M.H., Hellmér M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsässer D., Aarestrup F.M., Löfström C., Bofill-Mas S., Abril J.F., Girones R., Schultz A.C. Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi T., Shulman L., Van der Avoort H., Deshpande J., Roivainen M., De Gourville E. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiology & Infection. 2012;140:1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- Jahne M.A., Brinkman N.E., Keely S.P., Zimmerman B.D., Wheaton E.A., Garland J.L. Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: implications for onsite reuse. Water Res. 2020;169 doi: 10.1016/j.watres.2019.115213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Flury M. Advances in Agronomy. Elsevier; 2002. Fate and transport of viruses in porous media; pp. 39–102. [Google Scholar]
- John D.E., Rose J.B. Review of factors affecting microbial survival in groundwater. Environmental Science & Technology. 2005;39:7345–7356. doi: 10.1021/es047995w. [DOI] [PubMed] [Google Scholar]
- Jung Y.J., Park G.-S., Moon J.H., Ku K., Beak S.-H., Kim S., Park E.C., Park D., Lee J.-H., Byeon C.W. 2020. Comparative Analysis of Primer-probe Sets for the Laboratory Confirmation of SARS-CoV-2. (BioRxiv) [DOI] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. 2020. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. medRxiv 2020.05.03.20089417. [DOI] [Google Scholar]
- Kocamemi B.A., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. 2020. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. medRxiv 2020.05.12.20099358. [DOI] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. 2020. First Detection of SARS-CoV-2 in Untreated Wastewaters in Italy. medRxiv 2020.04.25.20079830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago P.M., Gary H.E., Jr., Pérez L.S., Cáceres V., Olivera J.B., Puentes R.P., Corredor M.B., Jímenez P., Pallansch M.A., Cruz R.G. Poliovirus detection in wastewater and stools following an immunization campaign in Havana, Cuba. Int. J. Epidemiol. 2003;32:772–777. doi: 10.1093/ije/dyg185. [DOI] [PubMed] [Google Scholar]
- Lamarre A., Talbot P.J. Effect of pH and temperature on the infectivity of human coronavirus 229E. Can. J. Microbiol. 1989;35:972–974. doi: 10.1139/m89-160. [DOI] [PubMed] [Google Scholar]
- Li M.-Y., Li L., Zhang Y., Wang X.-S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infectious diseases of poverty. 2020;9:1–7. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Huang Z., Luo J., Zhang X., Sha S. Primary concentration – the critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: a mini-review. Sci. Total Environ. 2020;747:141245. doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor Y., Shulman L., Kaliner E., Hindiyeh M., Ram D., Sofer D., Moran-Gilad J., Lev B., Grotto I., Gamzu R. Intensified environmental surveillance supporting the response to wild poliovirus type 1 silent circulation in Israel, 2013. Eurosurveillance. 2014;19 doi: 10.2807/1560-7917.es2014.19.7.20708. [DOI] [PubMed] [Google Scholar]
- Mao K., Zhang K., Du W., Ali W., Feng X., Zhang H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Current Opinion in Environmental Science & Health. 2020;17:1–7. doi: 10.1016/j.coesh.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrajt G., Naughton B., Bandyopadhyay A.S., Meschke J.S. A review of the most commonly used methods for sample collection in environmental surveillance of poliovirus. Clin. Infect. Dis. 2018;67:S90–S97. doi: 10.1093/cid/ciy638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K., Ishikura M., Yoshida H., Nakayama T., Hasegawa S., Ando S., Horie H., Miyamura T., Kitamura T. Assessment of poliovirus eradication in Japan: genomic analysis of polioviruses isolated from river water and sewage in Toyama Prefecture. Appl. Environ. Microbiol. 2000;66:5087–5091. doi: 10.1128/aem.66.11.5087-5091.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. 2020. Presence of SARS-Coronavirus-2 in Sewage. medRxiv 2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Hamasaki M., Yoshitomi H., Ishibashi T., Yoshiyama C., Maeda E., Sera N., Yoshida H. Environmental surveillance of poliovirus in sewage water around the introduction period for inactivated polio vaccine in Japan. Appl. Environ. Microbiol. 2015;81:1859–1864. doi: 10.1128/AEM.03575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir J., Haumacher R., Ike A., Stumpf P., Böhm R., Marschang R.E. Long-term study on tenacity of avian influenza viruses in water (distilled water, normal saline, and surface water) at different temperatures. Avian Dis. 2010;54:720–724. doi: 10.1637/8754-033109-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. 2020. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. medRxiv 2020.04.15.20066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Delgado A. What do we know about the SARS-CoV-2 coronavirus in the environment? Sci. Total Environ. 2020;727 doi: 10.1016/j.scitotenv.2020.138647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okware S.I., Omaswa F.G., Zaramba S., Opio A., Lutwama J.J., Kamugisha J., Rwaguma E.B., Kagwa P., Lamunu M. An outbreak of Ebola in Uganda. Tropical Med. Int. Health. 2002;7:1068–1075. doi: 10.1046/j.1365-3156.2002.00944.x. [DOI] [PubMed] [Google Scholar]
- O’Reilly K.M., Verity R., Durry E., Asghar H., Sharif S., Zaidi S.Z., Wadood M.Z.M., Diop O.M., Okayasu H., Safdar R.M. Population sensitivity of acute flaccid paralysis and environmental surveillance for serotype 1 poliovirus in Pakistan: an observational study. BMC Infect. Dis. 2018;18:176. doi: 10.1186/s12879-018-3070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole J., Sinclair M., Malawaraarachchi M., Hamilton A., Barker S.F., Leder K. Microbial quality assessment of household greywater. Water Res. 2012;46:4301–4313. doi: 10.1016/j.watres.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., Li P., Hu B., Wang J., Hu C. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 2020;115 doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Weinberger D.M., Omer S.B. 2020. SARS-CoV-2 RNA Concentrations in Primary Municipal Sewage Sludge as a Leading Indicator of COVID-19 Outbreak Dynamics. medRxiv 2020.05.19.20105999. [DOI] [Google Scholar]
- Pocock D., Garwes D. The influence of pH on the growth and stability of transmissible gastroenteritis virusin vitro. Arch. Virol. 1975;49:239–247. doi: 10.1007/BF01317542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powelson D.K., Simpson J.R., Gerba C.P. Virus transport and survival in saturated and unsaturated flow through soil columns. J. Environ. Qual. 1990;19:396–401. [Google Scholar]
- Pratelli A. Canine coronavirus inactivation with physical and chemical agents. Vet. J. 2008;177:71–79. doi: 10.1016/j.tvjl.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu G., Li X., Hu L., Jiang G. 2020. An Imperative Need for Research on the Role of Environmental Factors in Transmission of Novel Coronavirus (COVID-19) [DOI] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuan R., Domingo-Calap P., Sanchez G. 2020. Metropolitan Wastewater Analysis for COVID-19 Epidemiological Surveillance. medRxiv 2020.04.23.20076679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranta J., Hovi T., Arjas E. Poliovirus surveillance by examining sewage water specimens: studies on detection probability using simulation models. Risk Anal. 2001;21:1087–1096. doi: 10.1111/0272-4332.t01-1-216174. [DOI] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Moja L., Gismondo M.R., Salerno F. 2020. Presence and Vitality of SARS-CoV-2 Virus in Wastewaters and Rivers. medRxiv 2020.05.01.20086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos R. Center for Infectious Disease Research and Policy; Minneapolis, Minnesota: 2011. Study Puts Global 2009 H1N1 Infection Rate at 11–21% [Google Scholar]
- Sharif S., Ikram A., Khurshid A., Salman M., Mehmood N., Arshad Y., Ahmad J., Angez M., Alam M.M., Rehman L., Mujtaba G., Hussain J., Ali J., Akthar Ri., Malik M.W., Baig Z.I., Rana M.S., Usman M., Ali M.Q., Ahad A., Badar N., Umair M., Tamim S., Ashraf A., Tahir F., Ali N. 2020. Detection of SARS-Coronavirus-2 in Wastewater, Using the Existing Environmental Surveillance Network: An Epidemiological Gateway to an Early Warning for COVID-19 in Communities. (medRxiv) [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., Katoh H., Sakata M., Nakatsu Y., Mori Y. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;JJID-2020 doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- Sikka V., Chattu V.K., Popli R.K., Galwankar S.C., Kelkar D., Sawicki S.G., Stawicki S.P., Papadimos T.J. The emergence of Zika virus as a global health security threat: a review and a consensus statement of the INDUSEM Joint Working Group (JWG) J. Global Infect. Dis. 2016;8:3. doi: 10.4103/0974-777X.176140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139:105689. doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X. 2020. ddPCR: A more Sensitive and Accurate Tool for SARS-CoV-2 Detection in Low Viral Load Specimens. (medRxiv) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Wang H., Li Y., Xu A., Zhang Yong, Song L., Yoshida H., Xu Q., Yang J., Zhang Yan. Cocirculation of two transmission lineages of echovirus 6 in Jinan, China, as revealed by environmental surveillance and sequence analysis. Appl. Environ. Microbiol. 2011;77:3786–3792. doi: 10.1128/AEM.03044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M., Mustafa F., Rizvi T.A., Loney T., Suwaidi H.A., Al-Marzouqi A.H.H., Eldin A.K., Alsabeeha N., Adrian T.E., Stefanini C. SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12:526. doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Avoort H., Reimerink J., Ras A., Mulders M., Van Loon A. Isolation of epidemic poliovirus from sewage during the 1992–3 type 3 outbreak in The Netherlands. Epidemiology & Infection. 1995;114:481–491. doi: 10.1017/s0950268800052195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A., Ganesan H., Sudalaimuthu Raja S.S., Govindasamy V., Arunachalam M., Narayanasamy A., Sivaprakash P., Rahman P.K.S.M., Gopalakrishnan A.V., Siama Z., Vellingiri B. Novel wastewater surveillance strategy for early detection of COVID – 19 hotspots. Current Opinion in Environmental Science & Health. 2020 doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., Xiao W.-J., Yin J., Wei W., Wang G.-J., Si B., Guo B.-Z., Liu C., Ou G.-R., Wang M.-N., Fang T.-Y., Chao F.-H., Li J.-W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil M., Mandelboim M., Mendelson E., Manor Y., Shulman L., Ram D., Barkai G., Shemer Y., Wolf D., Kra-oz Z. Human enterovirus D68 in clinical and sewage samples in Israel. J. Clin. Virol. 2017;86:52–55. doi: 10.1016/j.jcv.2016.11.013. [DOI] [PubMed] [Google Scholar]
- WHO . 2003. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS) [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. The lancet Gastroenterology & hepatology. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N. 2020. SARS-CoV-2 Titers in Wastewater Are Higher than Expected From Clinically Confirmed Cases. (medRxiv) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics Through Viral Genome Quantification in Paris Wastewaters. medRxiv 2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Moulin L. 2020. Time Course Quantitative Detection of SARS-CoV-2 in Parisian Wastewaters Correlates With COVID-19 Confirmed Cases. (medRxiv) [Google Scholar]
- Xagoraraki I., O’Brien E. Women in Water Quality. Springer; 2020. Wastewater-based epidemiology for early detection of viral outbreaks; pp. 75–97. [Google Scholar]
- Xagoraraki I., Yin Z., Svambayev Z. Fate of viruses in water systems. J. Environ. Eng. 2014;140 [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P.D., Shete A.M., Kumar G.A., Sarkale P., Sahay R.R., Radhakrishnan C., Lakra R., Pardeshi P., Gupta N., Gangakhedkar R.R. Nipah virus sequences from humans and bats during Nipah outbreak, Kerala, India, 2018. Emerg. Infect. Dis. 2019;25:1003. doi: 10.3201/eid2505.181076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environmental science & technology. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Horie H., Matsuura K., Kitamura T., Hashizume S., Miyamura T. Prevalence of vaccine-derived polioviruses in the environment. J. Gen. Virol. 2002;83:1107–1111. doi: 10.1099/0022-1317-83-5-1107. [DOI] [PubMed] [Google Scholar]
- Zaneti R.N., Girardi V., Spilki F.R., Mena K., Westphalen A.P.C., Colares E.R. da C., Pozzebon A.G., Etchepare R.G. 2020. QMRA of SARS-CoV-2 for Workers in Wastewater Treatment Plants. medRxiv 2020.05.28.20116277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerda K.S., Gerba C.P., Hou K., Goyal S. Adsorption of viruses to charge-modified silica. Appl. Environ. Microbiol. 1985;49:91–95. doi: 10.1128/aem.49.1.91-95.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]