Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces a wide range of disease severity, ranging from asymptomatic infection to a life-threating illness, particularly in the elderly population and individuals with comorbid conditions. Among individuals with serious coronavirus 2019 (COVID-19) disease, acute respiratory distress syndrome (ARDS) is a common and often fatal presentation. Animal models of SARS-CoV-2 infection that manifest severe disease are needed to investigate the pathogenesis of COVID-19–induced ARDS and evaluate therapeutic strategies. We report two cases of ARDS in two aged African green monkeys (AGMs) infected with SARS-CoV-2 that had pathological lesions and disease similar to severe COVID-19 in humans. We also report a comparatively mild COVID-19 phenotype characterized by minor clinical, radiographic, and histopathologic changes in the two surviving, aged AGMs and four rhesus macaques (RMs) infected with SARS-CoV-2. Notable increases in circulating cytokines were observed in three of four infected, aged AGMs but not in infected RMs. All the AGMs had increased levels of plasma IL-6 compared with baseline, a predictive marker and presumptive therapeutic target in humans infected with SARS-CoV-2. Together, our results indicate that both RMs and AGMs are capable of modeling SARS-CoV-2 infection and suggest that aged AGMs may be useful for modeling severe disease manifestations, including ARDS.
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the development of coronavirus 2019 (COVID-19) disease is accompanied by a mild respiratory disease for most individuals. However, a small subset progress to develop severe respiratory disease, which, in some cases, is fatal.1 The most severely affected individuals often present with a fever, cough, dyspnea, and bilateral radiographic opacities, which, in most critically ill patients, progress to acute respiratory distress syndrome (ARDS).2 The onset of ARDS is often associated with an increase in circulating proinflammatory cytokines often referred to as a cytokine storm.3,4 Worsening of disease can be seen in the context of declining viral loads and markedly elevated cytokines, suggesting a role for these inflammatory responses in disease progression and immunopathology.5 Research into the causes and mechanisms of the most severe manifestations of COVID-19 is needed to facilitate the development of prophylactic and therapeutic approaches that can prevent this life-threatening outcome.
Nonhuman primates (NHPs) are ideally suited to model respiratory human viral infections because of the similarities to human respiratory anatomy and immunologic responses when compared with other animal species. Several NHP species have been successfully used to model pathogenesis6, 7, 8, 9, 10 and test vaccine candidates11, 12, 13, 14 for SARS-CoV-2. These prior studies have found that NHPs are susceptible to infection and develop mild to moderate disease, but none has been able to recapitulate the rapid clinical deterioration seen in individuals with severe disease and ARDS. Age is a well-established risk factor for severe disease and death in humans infected with SARS-CoV-22,15,16; therefore, older rhesus macaques (RMs) and African green monkeys (AGMs) were challenged with SARS-CoV-2 via two routes (aerosol and mulitroute).
This report describes the sudden and rapid health deterioration of two of the four aged AGMs experimentally infected with SARS-CoV-2. The two affected animals developed ARDS and elevated circulating cytokines similar to the complications reported in 5% to 13% of patients with COVID-19.17
Materials and Methods
Data Availability and Study Approval
The raw data supporting the findings and figures have been placed in the public data repository Figshare (https://figshare.com/articles/dataset/Raw_Data_Spreadsheets/12449654; last accessed October 30, 2020). Material requests can be made to the Tulane National Primate Research Center. Approved requests for materials will be released after completion of a material transfer agreement. The Institutional Animal Care and Use Committee of Tulane University reviewed and approved all the procedures for this experiment. The Tulane National Primate Research Center is fully accredited by the American Association for Accreditation of Laboratory Animal Care. All animals were cared for in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals.18 The Tulane University Institutional Biosafety Committee approved the procedures for sample handling, inactivation, and removal from BSL3 containment.
Virus
The virus used for experimental infection was SARS-CoV-2; 2019-nCoV/USA-WA1/2020 (https://www.ncbi.nlm.nih.gov/nuccore; accession number MN985325.1).19 Virus stock was prepared in Vero E6 cells and sequence confirmed by deep sequencing. Plaque assays were performed in Vero E6 cells. Vero E6 cells were acquired from ATCC (Manassas, VA).
Animals and Procedures
A total of eight animals, four aged (approximately 16 years of age), wild-caught AGMs (two male and two female) and four adult (13 to 15 years of age) RMs (3 male and 1 female) were used in this study. Animals (n = 4) were exposed to SARS-CoV-2 by small particle aerosol20 or multiroute combination. The four animals (AGM1, AGM4, RM3, and RM4) exposed by aerosol received an inhaled dose of approximately 2 × 103 median tissue culture infectious dose. The other four animals (AGM2, AGM3, RM1, and RM2) were exposed by inoculating a cumulative dose of 3.61 × 106 plaque-forming units through multiple routes (oral, 1 mL; nasal, 1 mL; intratracheal, 1 mL; and conjunctival, 50 μL per eye). Animals were observed for up to 28 days, including twice-daily monitoring. Pre-exposure and postexposure samples included blood, cerebrospinal fluid, feces, urine, bronchioalveolar lavage, and mucosal swabs (buccal, nasal, pharyngeal, rectal, vaginal, and bronchial brush). Blood was collected at post-exposure days −14, 1, 3 (aerosol) or 4 (multiroute), 7, 14, 21, and at necropsy. Cerebrospinal fluid, feces, urine, bronchioalveolar lavage, and mucosal swabs were collected at post-exposure days −14, 7, 14, 21, and at necropsy. Physical examination, plethysmography, and imaging (radiography and positron emission tomography/computed tomography) occurred 7 days before exposure and then weekly thereafter. Animals were euthanized for necropsy after 3 weeks postexposure or when humane end points were reached. Samples from the left cranial and caudal lung lobes were collected fresh and in media for further processing. All right lung lobes were infused and stored in fixative for microscopic evaluation. The remainder of the necropsy was performed routinely with collection of tissues in media, fixative, or fresh frozen.
Histopathologic Scoring
Pulmonary pathology was scored using two separate random forest tissue segmentation algorithms trained by a veterinary pathologist (R.V.B.) in an unblinded fashion to recognize fibrin and edema and cellular inflammation using HALO software version 3.1 (Indica Labs, Albuquerque, NM). Tissue sections from each of the right lung lobes was segmented using the trained algorithms to quantify the percentage of tissue effected by fibrin and edema or cellular inflammation. The percentage of inflammation was converted to a pathology score based on the following scoring system: fibrin and edema score of 0, 0% to 2%; 1, 2% to 5%; 2, 5% to 15%; 3, 15% to 30%; and 4, >30%; and cellular inflammation score of 0, 0% to 0.5%; 1, 0.5% to 3%; 2, 3% to 6%; 3, 6% to 12%; and 4, >12%. The histopathology score was made by summating the fibrin and edema and cellular inflammation scores for each lobe.
Semiquantitative scores were generated by a veterinary pathologist (R.V.B.) for lesions within other tissues and specific tissue compartments within the lung. Lesions were scored based on severity as lacking a lesion (−) or being minimally (+), mildly (++), moderately (+++), or severely (++++) affected.
Quantification of Swab Viral RNA
Swab and bronchial brush samples were collected in 200 μL of DNA/RNA Shield 1× (catalog number R1200; Zymo Research, Irvine, CA) and extracted for viral RNA using the Quick-RNA viral kit (catalog number R1034/5; Zymo Research). The Viral RNA Buffer was dispensed directly to the swab in the DNA/RNA Shield. A modification to the manufacturers' protocol was made to insert the swab directly into the spin column to centrifugate, allowing all the solution to cross the spin column membrane. The viral RNA was then eluted (45 μL) from which 5 μL was added in a 0.1-mL fast 96-well optical microtiter plate format (catalog number 4346906; Thermo Fisher Scientific, Waltham, MA) for a 20-μL real-time quantitative RT-PCR (RT-qPCR) reaction. The RT-qPCR reaction used TaqPath 1-Step Multiplex Master Mix (catalog number A28527; Thermo Fisher Scientific) along with the 2019-nCoV RUO Kit (catalog number 10006713; IDTDNA, Coralville, IA), a premix of forward and reverse primers and a FAM-labeled probe targeting the N1 amplicon of the N gene of SARS2-nCoV19 (https://www.ncbi.nlm.nih.gov/nuccore; accession number MN908947). The reaction master mix was added using an X-stream repeating pipette (Eppendorf, Hauppauge, NY) to the microtiter plates, which were covered with optical film (catalog number 4311971; Thermo Fisher Scientific), vortexed, and pulse centrifuged. The RT-qPCR reaction was subjected to RT-qPCR at a program of uracil-DNA glycosylase incubation at 25°C for 2 minutes, room temperature incubation at 50°C for 15 minutes, and an enzyme activation at 95°C for 2 minutes followed by 40 cycles of a denaturing step at 95°C for 3 seconds and annealing at 60°C for 30 seconds. Fluorescence signals were detected with a QuantStudio 6 Sequence Detector (Applied Biosystems, Foster City, CA). Data were captured and analyzed with Sequence Detector Software version 1.3 (Applied Biosystems). Viral copy numbers were calculated by plotting Cq values obtained from unknown (ie, test) samples against a standard curve that represented known viral copy numbers. The limit of detection of the viral RNA assay was 10 copies per reaction volume. A 2019-nCoV–positive control (catalog number 10006625; IDTDNA) was analyzed in parallel with every set of test samples to verify that the RT-qPCR master mix and reagents were prepared correctly to produce amplification of the target nucleic acid. A nontemplate control was included in the qPCR to ensure that there was no cross-contamination between reactions.
Immunohistochemistry
Five micrometer sections of formalin-fixed, paraffin-embedded lung were incubated for 1 hour with the primary antibodies [SARS-CoV-2 nucleoprotein, mouse IgG1 [catalog number 40143-MM08; Sino Biological, Wayne, PA]; ACE2, rabbit polyclonal [catalog number HPA000288; Millipore, Burlington, MA]; Iba-1, rabbit polyclonal [catalog number 019-19741; Wako, Richmond, VA]; or pancytokeratin, rabbit polyclonal [catalog number Z0622; Dako, Santa Clara, CA] diluted in NGS at a concentration of 1:200 and 1:100, respectively. Secondary antibodies tagged with Alexa Fluor fluorochromes and diluted 1:1000 in NGS were incubated for 40 minutes. DAPI was used to label the nuclei of each section. Slides were imaged with a digital slide scanner (Zeiss Axio Scan.Z1; Zeiss, White Plains, NY).
Cytokine Production in Plasma
Plasma was collected by spinning and was thawed before use. Cytokines were measured using Mesoscale Discovery using a V-Plex Proinflammatory Panel 1, 10-Plex [interferon (IFN)-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, tumor necrosis factor-α] (catalog number K15049D; Mesoscale Discovery, Rockville, MD) following the instructions of the kit. The plate was read on a MESO Quick Plex SQ120 machine.
Heatmaps were generated using the pheatmap package in R (https://rdrr.io/cran/pheatmap/, last accessed July 22, 2020). Data were normalized by dividing raw values at week 1 and necropsy by baseline values for each animal, followed by the application of log2. Values below the limit of detection were replaced with the lowest limit of detection value based on the standard curve for each run or with the lowest value detected during the run, whichever was smaller. Polar coordinate plots were generated using the ggplot2 package in R,21 using the same normalized data shown in the heatmap. Scatterplots were drawn using raw data points and display the Pearson correlation coefficients.
Detection of Binding IgG Antibody in Plasma
Serum samples collected before infection and weekly after infection were tested for binding IgG antibodies against SARS-CoV-2 S1/S2 proteins using an enzyme-linked immunosorbent assay kit (catalog number SP864C; XpressBio, Frederick, MD). The assays were performed per directions of the manufacturer. The absorbance of the colorimetric reaction was read at 405 nm. Samples were considered positive if the difference between the absorbance on the positive viral antigen well and the absorbance on the negative control antigen well was ≥0.300.
Samples collected before infection and weekly after infection until necropsy were tested for detection of binding IgG antibodies against SARS-CoV-2 nucleoprotein by MFIA COVID-Plex (Charles River Laboratories, Wilmington, MA). The assays were performed per directions of the manufacturer. Plates were read on a Bio-Plex 200 System (Bio-Rad Laboratories, Hercules, CA). MFIA scores were calculated using Bio-Plex Manager Software version 6.2 (Bio-Rad) as indicated by Charles River Laboratories. Samples were considered positive if the MFIA score was ≥3.0.
Statistical Analysis
Statistical tests were performed with GraphPad Prism software, version 8.4.3 (GraphPad Software, San Diego, CA). The Mann-Whitney U-test was used to compare viral load between species and route of exposure. The Pearson correlation test was used to test correlation between cytokines and viral load.
Results
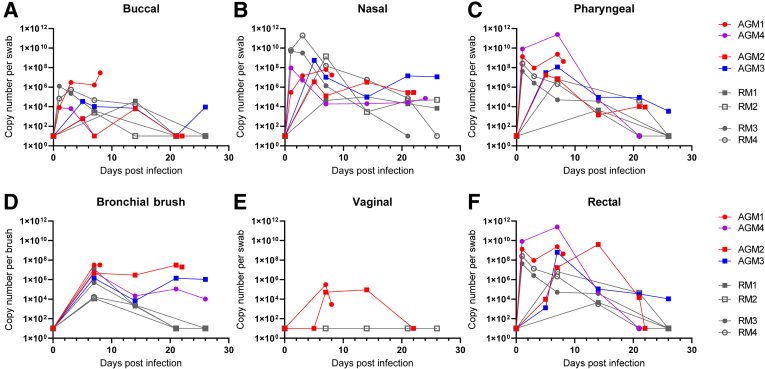
Four, aged AGMs and four RMs 13 to 15 years of age were exposed by two routes to SARS-CoV-2 isolate USA-WA1/2020. Four animals (AGM1, AGM4, RM3, and RM4) were exposed via small particle aerosol, and four animals (AGM 2, AGM3, RM1, and RM2) were exposed via multiple route installation (Table 1). SARS-CoV-2 RNA was detectable in swabs obtained from mucosal sites in all eight animals (Figure 1). The highest levels of viral RNA were detected in the pharynx and nasal cavity (Figure 1, B and C). Rectal swabs contained high viral RNA loads similar to reports in humans (Figure 1F).22,23 Despite the nearly three-log difference in exposure dose, no significant difference was observed in the viral RNA loads or kinetics based on the route of exposure.
Table 1.
Species, Source, Route of Exposure, Demographic Information, and Clinical Outcome from Each Animal in the Study
| Animal | Species | Source | Age, years | Sex | Weight, kg | Exposure (dose) | Outcome |
|---|---|---|---|---|---|---|---|
| AGM1 | Chlorocebus aethiops sabaeus | Wild caught, St. Kitts | 16 | F | 4.3 | Aerosol (1 × 104 PFU) | ARDS (8 DPI) |
| AGM2 | Chlorocebus aethiops sabaeus | Wild caught, St. Kitts | 16 | F | 3.9 | Multiroute (3.61 × 106 PFU) | ARDS (22 DPI) |
| AGM3 | Chlorocebus aethiops sabaeus | Wild caught, St. Kitts | 16 | M | 6.9 | Multiroute (3.61 × 106 PFU) | Reached study end point (26 DPI) |
| AGM4 | Chlorocebus aethiops sabaeus | Wild caught, St. Kitts | 16 | M | 7.5 | Aerosol (1 × 104 PFU) | Reached study end point (24 DPI) |
| RM1 | Macaca mulatta | Born at TNPRC | 14 | M | 16.7 | Multiroute (3.61 × 106 PFU) | Reached study end point (27 DPI) |
| RM2 | Macaca mulatta | Born at TNPRC | 13 | F | 6.9 | Multiroute (3.61 × 106 PFU) | Reached study end point (27 DPI) |
| RM3 | Macaca mulatta | Born at TNPRC | 13 | M | 11.6 | Aerosol (1 × 104 PFU) | Reached study end point (28 DPI) |
| RM4 | Macaca mulatta | Born at TNPRC | 15 | M | 11 | Aerosol (1 × 104 PFU) | Reached study end point (28 DPI) |
AGM, African green monkey; ARDS, acute respiratory distress syndrome; DPI, days post infection; PFU, plaque-forming units; RM, rhesus macque; TNPRC, Tulane National Primate Research Center.
Figure 1.
Quantification of viral loads from mucosal swabs. A–F; All animals [four African green monkeys (AGMs) and four rhesus macaques (RMs)] had detectable viral RNA at mucosal sites, including buccal (A), nasal (B), pharyngeal (C), bronchial brush (D), and rectal swabs (F). D: No significant differences are seen in viral load between species and route of exposure (Mann-Whitney U-test). Animals with acute respiratory disease syndrome (ARDS) trend to high viral loads in bronchial brush samples. E: Viral RNA is detected in vaginal swabs from both female AGMs but not the only female RM. Circles indicate aerosol exposure; squares, multiroute exposure; gray, RMs; red, developed ARDS; purple, increased cytokines without ARDS; and blue, no cytokine increase or ARDS.
After SARS-CoV-2 exposure, animals were followed up to 4 weeks after infection with regular clinical assessment that included physical examination, pulse oximetry, and plethysmography. Clinical findings during the study consisted of mild transient changes in temperature, oxygen saturation as measured by pulse oximetry, and appetite for all animals except on 8 (AGM1) and 22 (AGM2) days post-infection (DPI) (Supplemental Figure S1). The day before (7 and 21 DPI) all animals underwent a complete physical evaluation and an extensive sample collection protocol, including fluid, stool, swab, and bronchial brush collection; no remarkable findings were noted at that time in any of the animals. In the 24 hours after sample collection (8 DPI for AGM1 and 22 DPI for AGM2), both animals developed mild tachypnea that progressed to severe respiratory distress that included dyspnea, tachypnea, hypothermia, and an oxygen saturation as measured by pulse oximetry of 77% (Supplemental Figure S1). No significant clinical findings were observed in any of the remaining animals after 22 DPI.
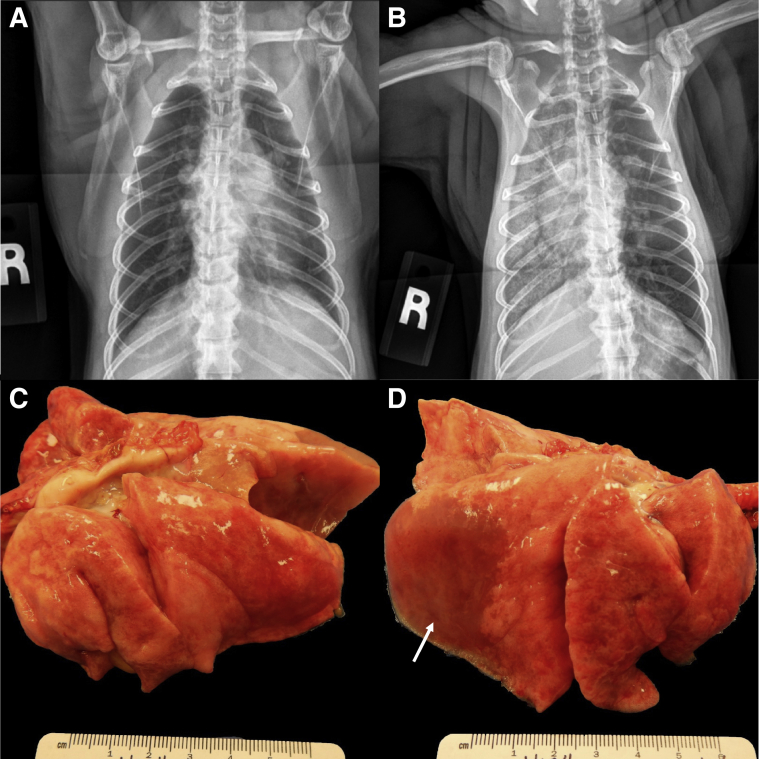
Thoracic radiographs for AGM1 and AGM2 revealed a diffuse alveolar pattern throughout the right lung fields and a lobar sign in the caudal dorsal lung field. In AGM2 the left caudal lung lobe also contained a mild alveolar pattern. These findings were in stark contrast to the radiographs from the day before highlighting the rapid disease progression (Figure 2, A and B). Because of their rapidly declining clinical condition, AGM1 and AGM2 were euthanized at 8 and 22 DPI, respectively. All remaining animals (2 AGM and 4 RM) were euthanized at the study end point between 3 and 4 weeks post-infection. A complete necropsy was performed on all animals.
Figure 2.
Radiographic and gross pathologic changes in SARS-CoV-2 exposed African green monkey (AGM) 1. A and B: Radiographs 22 hours before (A) and at the time of necropsy (B) showing the rapid development of alveolar lung opacities throughout the right lung lobes. C: The left lung lobes fail to collapse. D: There is extensive consolidation of the right lower lung lobe with pulmonary edema (arrow). The right middle and anterior lobes are less affected. On cut surface, all lobes ooze copious fluid.
Gross postmortem examination of AGM1 and AGM2 revealed severe consolidation and edema in the right caudal lung lobe with generalized failure to collapse of the remaining lobes, consistent with a bronchointerstitial pneumonia (Figure 2, C and D and Supplemental Figure S2). AGM3 had multifocal pleural adhesions between the left caudal lung lobe and the diaphragm. RM1 had a focal pulmonary scar in the right caudal lung lobe surrounded by acute hemorrhage (Supplemental Figure S2E). The lungs of the remaining animals (AGM4, RM2, RM3, and RM4) were grossly normal.
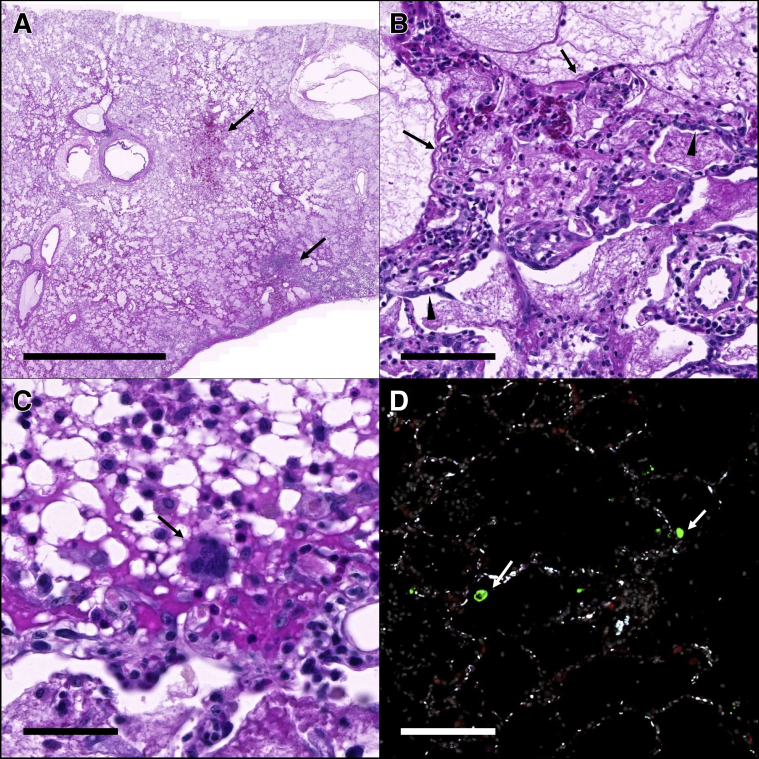
Histopathologic findings in the lungs of AGM1 and AGM2 were similar and characterized by alveoli that were filled with fibrin, hemorrhage, and proteinaceous fluid (Figure 3A). Alveoli were multifocally lined by hyaline membranes and/or type II pneumocytes, consistent with diffuse alveolar damage (Figure 3B). Alveoli contained rare multinucleated syncytial cells (Figure 3C). Fluorescent immunohistochemistry identified low numbers of SARS-CoV-2–infected cells in AGM1 but not AGM2 (Figure 3D). The animals that survived to study end point exhibited minimal to mild interstitial inflammation (Supplemental Figures S3 and S4). Three of four RMs (RM1, RM2, and RM4) had microscopic evidence of aspiration pneumonia characterized by foreign plant material within bronchioles. Histopathologic lesions in other tissues were mild and interpreted as not significant in all eight animals (Supplemental Table S1).
Figure 3.
Histopathology and fluorescent immunohistochemistry in African green monkey (AGM) 1. A: The right lower lung lobe is filled with fibrin and edema with areas of hemorrhage and necrosis (arrows). B: Alveoli are variably lined by hyaline membranes (arrows) and type II pneumocytes (arrowheads). C: Rare multinucleated syncytia (arrow) are scattered throughout the affected lungs; D: Fluorescent immunohistochemistry for SARS-CoV-2 nucleoprotein (green, arrows) and ACE2 (red) identified low numbers of SARS-CoV-2 positive cells within the affected lung lobes. White indicates DAPI/nuclei; green, CoV-2; red, ACE2; and blue, Empty. Scale bars: 5 mm (A); 100 μm (B and D); 50 μm (C).
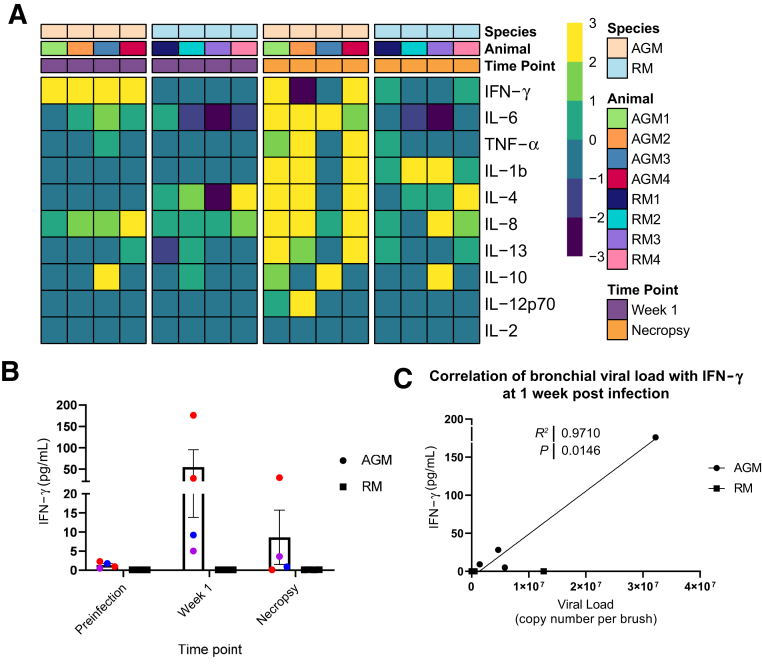
A group of cytokines similar to those observed in human COVID-19 were up-regulated in the two animals that progressed to ARDS (AGM1 and AGM2) at the time of necropsy compared with baseline levels (Figure 4A and Supplemental Figure S5). Elevated markers included IFN-γ, IL-6, IL-4/IL-13, IL-8, IL-1β, and tumor necrosis factor-α. indicating a cytokine storm. AGM4 did not develop ARDS; however, this animal had a similar increase in cytokine concentrations, but with only a mild elevation of IL-6. In contrast, RMs only had modest changes in cytokine expression at 1 week post-infection and necropsy, despite comparable peak viral loads. IFN-γ levels were high in AGM1 and AGM2 during acute infection and were associated with viral RNA in the bronchial brushes at the same time point (1 week) (Figure 4, B and C). Binding IgG antibodies were not detected in AGM1 (euthanized 8 DPI), but all remaining animals had detectable antibodies by 21 DPI (Supplemental Figure S6).
Figure 4.
Cytokine increase. Heat maps (A) separated by species (first row), animal (second row), and timepoint (third row) showing changes in the levels of 10 cytokines in plasma at week 1 (purple, first two heat maps) and necropsy (orange, third and fourth heat maps) with respect to the baseline in African green monkeys (AGMs) and rhesus macaques (RMs). AGMs at 1 week post-infection (heat map 1) show increased levels of interferon (IFN)-γ, which is not observed in RMs at the same timepoint (heat map 2). At necropsy, three of the four AGMs exhibit elevated levels of multiple plasma cytokines (heat map 3) in contrast to RMs (heat map 4). Data are normalized (log2). B: Levels of IFN-γ in plasma at baseline, week 1, and at necropsy. C: Association between IFN-γ levels at week 1 and viral load in bronchial brushes (Pearson test). TNF-α, tumor necrosis factor-α.
Discussion
Several recent studies have been published using AGMs6,9 and RMs7,8 to model SARS-CoV-2 infection and have found that both species are capable of modeling mild to moderate disease and are useful for testing prospective vaccines and therapeutics. However, none of these prior studies have been able to recapitulate the severe disease phenotype seen in a subset of individuals with COVID-19. Our results indicate that after infection with SARS-CoV-2, aged AGMs can develop ARDS and cytokine elevations similar to that reported in humans with severe COVID-19.24 Apart from the severe phenotype observed in two of the animals, our findings are otherwise consistent with prior studies, with the surviving AGMs having mild clinical disease, pathology, and prolonged viral shedding.6,9 The RMs in our study also exhibited mild clinical disease and pathology with shorter viral shedding from mucosal sites compared with the AGMs. These findings indicate that even in the absence of severe disease both AGMs and RMs have utility for testing SARS-CoV-2 vaccines and therapeutics.
Two of the AGMs in our study developed widespread radiographic opacities and severe respiratory distress (oxygen saturation as measured by pulse oximetry, 77%) that progressed during a 24-hour period. Taken together with the postmortem findings of diffuse alveolar damage and no evidence of congestive heart disease, these findings are consistent with a diagnosis of ARDS in both AGM1 and AGM2. Of note, both of the animals that developed ARDS did so within 24 hours of routine sampling procedures, which included anesthesia and bronchoalveolar lavage. It has been our experience that these procedures are well tolerated, and procedure-related complications are exceedingly rare (fatal complication within 48 hours of procedure after 2 of 11,431 procedures in animals ranging from 1 to 31 years of age; unpublished data). Furthermore, AGM1 and AGM2 previously underwent the same routine sampling procedures one and three times, respectively, without complication. In our previous experience, fatal complications have only occurred in animals that were severely debilitated at the time of the procedure, and even in these rare cases the pathologic lesions were distinct (no evidence of diffuse alveolar disease) from the two AGMs described herein. Therefore, in our experience, routine sampling procedures do not cause the severe COVID-19 phenotype observed in AGM1 and AGM2, even in the rare cases where fatal complications occur.
Furthermore, notable increases in plasma cytokines compatible with cytokine storm were found in the aged AGMs that progressed to ARDS. Proinflammatory cytokines, including tumor necrosis factor-α, IL-1β, IL-8, IL-6, granulocyte colony-stimulating factor, monocyte chemoattractant protein 1, and macrophage inflammatory protein 1, are elevated during the acute phases of acute lung injury.25 In human COVID-19, circulating IL-6 correlates with radiographic abnormalities of pneumonia.3 Indeed, overexpression of several of these cytokines were observed in both animals that progressed to ARDS. This finding differed from the cytokine profile in the AGMs and RMs, which reached study end point. Interestingly, at 7 DPI all four AGMs had increased levels of IFN-γ, with the two AGMs that progressed having the highest plasma concentration. Thus, elevated IFN-γ in plasma could be explored as a potential predictive biomarker for advanced disease in individuals.
Several factors may have contributed to the severe disease phenotype observed in the AGMs in our study. Age,2 weight,26 and sex2,27 have been identified as potential predisposing factors for developing severe disease in humans. All the AGMs included in our study were aged, with an estimated age of 16 years. Both animals that progressed to severe disease were also female and low weight. This differs from the findings in patients with COVID-19 in which male sex15,28 and obesity26 are associated with a higher prevalence of severe disease. The AGMs used in this infection study were also imported from nondomestic sources, and although the animals were housed for 10 months at the Tulane National Primate Research Center and deemed clinically healthy at the time of initiation of the study, historical factors may have predisposed them to enhanced COVID-19 disease.
This study found that after exposure to SARS-CoV-2 aged AGMs develop a spectrum of disease from mild to severe COVID-19, which in some cases progress to ARDS. The cytokine expression profile in the two animals that developed ARDS is similar to that seen in the severe human disease phenotype. Our data suggest that both RMs and AGMs are capable of modeling mild manifestations of SARS-CoV-2 infection and that aged AGMs may additionally be capable of modeling severe disease manifestations, including ARDS.
Acknowledgment
We thank Natalie Thornburg at the National Center for Immunization and Respiratory Diseases for her help in acquiring and characterizing the viral stock used in this infection study.
Footnotes
Supported by NIH grant P51 OD011104 59, NIH National Institute of Allergy and Infectious Diseases grant R24 AI120942 (S.C.W.), George Mason University, Emergent Ventures Fast Grants (T.F. and M.V.), a Joe and Dorothy Dorsett Brown Foundation grant (J.R.), and Tulane University.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2020.10.016.
Author Contributions
R.V.B., L.A.D.-M., C.J.R., K.R.-L., G.L., N.G., N.J.M., T.F., R.P.B., and J.R. designed the study. R.V.B., L.A.D.-M., C.J.R., C.J.M., X.Q., P.K.D., N.J.M., T.F., R.P.B., and J.R. wrote and edited the manuscript. R.V.B., M.V., C.J.M., B.B., B.T., T.P., C.A., M.B.B., P.K.D., N.J.M., R.P.B., and J.R. collected and analyzed the data. K.S.P., J.A.P., S.C.W. provided large preparations of deep sequenced virus from the WRCEVA collection and wrote the manuscript.
Supplemental Data
Clinical parameters of African green monkeys (AGMs) and rhesus macaques (RMs) after exposure to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). No significant differences in clinical parameters are found leading up to development of acute respiratory disease syndrome (ARDS) in the two animals that progressed (red). Progression to ARDS is associated with spike in physical examination scores, respiratory rate, and a notable decline in oxygen saturation as measured by pulse oximetry. Neither weight loss nor fever was associated with SARS-CoV-2 exposure in any of the eight animals. Circles indicate aerosol exposure; squares, multiroute exposure; gray, RMs; red, developed ARDS; purple, increased cytokines without ARDS; and blue, no cytokine increase or ARDS.
Gross pathology of the right lung lobes of severe acute respiratory syndrome coronavirus 2–infected African green monkeys (AGMs) and rhesus macaques (RMs). A and B: There is extensive consolidation of the right caudal lung lobes of AGM1 (A) and AGM2 (B), with lesser involvement of the middle and cranial lobes. C: AGM3 has focal area of hemorrhage along the caudodorsal margin of the right lower lobe. D: No gross abnormalities are visible in AGM4. E–H: RM1 (E) has a scar along the caudodorsal margin of the right caudal lobe surrounded by acute hemorrhage; RM2 (F) has red mottling of the dorsal margin of the right caudal and cranial lung lobes; and no gross abnormalities are visible in RM3 and RM4 (G and H). Arrows point to the lesion described for each animal. Scale bar = 1 cm.
Histopathology (hematoxylin and eosin) of the right caudal lung lobes of severe acute respiratory syndrome coronavirus 2 infected African green monkeys (AGMs) and rhesus macaques (RMs). Representative images of the histopathology of the right caudal lung lobe from the AGMs (top row, left to right AGM1 to AGM4) and RMs (bottom row, left to right RM1 to RM4). Scale bar = 250 μm.
Histopathologic scoring of pulmonary inflammation. Whole slide images of tissue sections from each lung lobe were analyzed for fibrin/edema and cellular inflammation using HALO's Tissue Classifier module. Top row: Representative images of fibrin/edema scores 0 to 4. Middle row: Representative images of cellular inflammation scores 0 to 2 and 4. None of the animals within the study had cellular inflammation score of 3, so a representative image of this score is not available. Graphs illustrate the score of inflammation in each of the right lung lobes. The histopathologic score is the aggregate score of fibrin and edema and cellular inflammation. Scale bar = 250 μm. AGM, African green monkey; RM, rhesus macaque.
Serum cytokines in severe acute respiratory syndrome coronavirus 2–infected African green monkeys (AGMs) and rhesus macaques (RMs). AGMs exhibit higher levels of interferon (IFN)-γ at 1 week post-infection compared with RMs with the two animals (AGM1 and AGM2, red) that progressed showing the highest elevation. At necropsy, the two progressors had elevated IL-6, IL-1Β, IL-8, IL-13, and IL-4. AGM4 (purple) had a similar cytokine profile except with little elevation in IL-6. AGM3 (blue) had elevated IL-10 1 week post-infection and is notable for having the lowest pathology scores of the four AGMs. RMs had modest changes in serum cytokines at 1 week post infection and at necropsy. Circles indicate aerosol exposure; squares, multiroute exposure; gray, RMs; red developed acute respiratory disease (ARDS); purple, increased cytokines without ARDS; and blue, no cytokine increase or ARDS.
Kinetics of binding antibody responses in severe acute respiratory syndrome coronavirus 2–infected African green monkeys (AGMs) and rhesus macaques (RMs). Circles indicate aerosol exposure; squares, multiroute exposure; gray, RMs; red, developed acute respiratory disease (ARDS); purple, increased cytokines without ARDS; and blue, no cytokine increase or ARDS.
References
- 1.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chien J.Y., Hsueh P.R., Cheng W.C., Yu C.J., Yang P.C. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11:715–722. doi: 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. Adv Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolsey C., Borisevich V., Prasad A.N., Agans K.N., Deer D.J., Dobias N.S., Heymann J.C., Foster S.L., Levine C.B., Medina L., Melody K., Geisbert J.B., Fenton K.A., Geisbert T.W., Cross R.W. Establishment of an African green monkey model for COVID-19. and protection against re-infection. Nature Immunology. 2020;24:1–13. doi: 10.1038/s41590-020-00835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh D.K., Ganatra S.R., Singh B., Cole J., Alfson K.J., Clemmons E. SARS-CoV-2 infection leads to acute infection with dynamic cellular and inflammatory flux in the lung that varies across nonhuman primate species. bioRxiv. 2020 [Epub] doi:10.1101/2020.06.05.136481. [Google Scholar]
- 8.Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Perez-Perez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., Avanzato V.A., Rosenke R., Hanley P.W., Saturday G., Scott D., Fischer E.R., de Wit E. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartman A.L., Nambulli S., McMillen C.M., White A.G., Tilston-Lunel N.L., Albe J.R., Cottle E., Dunn M.D., Frye L.J., Gilliland T.H., Olsen E.L., O'Malley K.J., Schwarz M.M., Tomko J.A., Walker R.C., Xia M., Hartman M.S., Klein E., Scanga C.A., Flynn J.L., Klimstra W.B., McElroy A.K., Reed D.S., Duprex W.P. SARS-CoV-2 infection of African green monkeys results in mild respiratory disease discernible by PET/CT imaging and prolonged shedding of infectious virus from both respiratory and gastrointestinal tracts. PLoS pathogens. 2020;16 doi: 10.1371/journal.ppat.1008903. e1008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A., Schipper D., van Run P., Leijten L., Sikkema R., Verschoor E., Verstrepen B., Bogers W., Langermans J., Drosten C., Fentener van Vlissingen M., Fouchier R., de Swart R., Koopmans M., Haagmans B.L. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E.R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Perez-Perez L., Edwards N.J., Wright D., Bissett C., Gilbride C., Williamson B.N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P.W., Scott D., Saturday G., de Wit E., Gilbert S.C., Munster V.J. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586 doi: 10.1038/s41586-020-2608-y. 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erasmus J.H., Khandhar A.P., O'Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B., Draves K.E., Randall S., Guerriero K.A., Duthie M.S., Carter D., Reed S.G., Hawman D.W., Feldmann H., Gale M., Jr., Veesler D., Berglund P., Fuller D.H. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci Transl Med. 2020;12:eabc9396. doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudley J.P., Lee N.T. Disparities in age-specific morbidity and mortality from SARS-CoV-2 in China and the Republic of Korea. Clin Infect Dis. 2020;71:863–865. doi: 10.1093/cid/ciaa354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., Cai H., Hu J., Lian J., Gu J., Zhang S., Ye C., Lu Y., Jin C., Yu G., Jia H., Zhang Y., Sheng J., Li L., Yang Y. Epidemiological, clinical characteristics of cases of SARS-CoV-2 infection with abnormal imaging findings. Int J Infect Dis. 2020;94:81–87. doi: 10.1016/j.ijid.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y., Sun J., Dai Z., Deng H., Li X., Huang Q., Wu Y., Sun L., Xu Y. Prevalence and severity of corona virus disease 2019 (COVID-19): a systematic review and meta-analysis. J Clin Virol. 2020;127:104371. doi: 10.1016/j.jcv.2020.104371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Committee for the Update of the Guide for the Care and Use of Laboratory Animals . National Academies Press; Washington, DC: 2011. National Research Council: Guide for the Care and Use of Laboratory Animals: Eighth Edition. [Google Scholar]
- 19.Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H.A., Wang L., Whitaker B., Lynch B., Gautam R., Schindewolf C., Lokugamage K.G., Scharton D., Plante J.A., Mirchandani D., Widen S.G., Narayanan K., Makino S., Ksiazek T.G., Plante K.S., Weaver S.C., Lindstrom S., Tong S., Menachery V.D., Thornburg N.J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartings J.M., Roy C.J. The automated bioaerosol exposure system: preclinical platform development and a respiratory dosimetry application with nonhuman primates. J Pharmacol Toxicol Methods. 2004;49:39–55. doi: 10.1016/j.vascn.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Wickham H. 2016. ggplot2: Elegant Graphics for Data Analysis, New York, NY: Springer. [Google Scholar]
- 22.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Grady N.P., Preas H.L., Pugin J., Fiuza C., Tropea M., Reda D., Banks S.M., Suffredini A.F. Local inflammatory responses following bronchial endotoxin instillation in humans. Am J Respir Crit Care Med. 2001;163:1591–1598. doi: 10.1164/ajrccm.163.7.2009111. [DOI] [PubMed] [Google Scholar]
- 26.Simonnet A., Chetboun M., Poissy J., Raverdy V., Noulette J., Duhamel A., Labreuche J., Mathieu D., Pattou F., Jourdain M., LICORN and the Lille COVID-19 and Obesity Study Group High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity (Silver Spring) 2020;28:1195–1199. doi: 10.1002/oby.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter L.A., McGregor A.J. Sex- and gender-specific observations and implications for COVID-19. West J Emerg Med. 2020;21:507–509. doi: 10.5811/westjem.2020.4.47536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin J.M., Bai P., He W., Wu F., Liu X.F., Han D.M., Liu S., Yang J.K. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical parameters of African green monkeys (AGMs) and rhesus macaques (RMs) after exposure to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). No significant differences in clinical parameters are found leading up to development of acute respiratory disease syndrome (ARDS) in the two animals that progressed (red). Progression to ARDS is associated with spike in physical examination scores, respiratory rate, and a notable decline in oxygen saturation as measured by pulse oximetry. Neither weight loss nor fever was associated with SARS-CoV-2 exposure in any of the eight animals. Circles indicate aerosol exposure; squares, multiroute exposure; gray, RMs; red, developed ARDS; purple, increased cytokines without ARDS; and blue, no cytokine increase or ARDS.
Gross pathology of the right lung lobes of severe acute respiratory syndrome coronavirus 2–infected African green monkeys (AGMs) and rhesus macaques (RMs). A and B: There is extensive consolidation of the right caudal lung lobes of AGM1 (A) and AGM2 (B), with lesser involvement of the middle and cranial lobes. C: AGM3 has focal area of hemorrhage along the caudodorsal margin of the right lower lobe. D: No gross abnormalities are visible in AGM4. E–H: RM1 (E) has a scar along the caudodorsal margin of the right caudal lobe surrounded by acute hemorrhage; RM2 (F) has red mottling of the dorsal margin of the right caudal and cranial lung lobes; and no gross abnormalities are visible in RM3 and RM4 (G and H). Arrows point to the lesion described for each animal. Scale bar = 1 cm.
Histopathology (hematoxylin and eosin) of the right caudal lung lobes of severe acute respiratory syndrome coronavirus 2 infected African green monkeys (AGMs) and rhesus macaques (RMs). Representative images of the histopathology of the right caudal lung lobe from the AGMs (top row, left to right AGM1 to AGM4) and RMs (bottom row, left to right RM1 to RM4). Scale bar = 250 μm.
Histopathologic scoring of pulmonary inflammation. Whole slide images of tissue sections from each lung lobe were analyzed for fibrin/edema and cellular inflammation using HALO's Tissue Classifier module. Top row: Representative images of fibrin/edema scores 0 to 4. Middle row: Representative images of cellular inflammation scores 0 to 2 and 4. None of the animals within the study had cellular inflammation score of 3, so a representative image of this score is not available. Graphs illustrate the score of inflammation in each of the right lung lobes. The histopathologic score is the aggregate score of fibrin and edema and cellular inflammation. Scale bar = 250 μm. AGM, African green monkey; RM, rhesus macaque.
Serum cytokines in severe acute respiratory syndrome coronavirus 2–infected African green monkeys (AGMs) and rhesus macaques (RMs). AGMs exhibit higher levels of interferon (IFN)-γ at 1 week post-infection compared with RMs with the two animals (AGM1 and AGM2, red) that progressed showing the highest elevation. At necropsy, the two progressors had elevated IL-6, IL-1Β, IL-8, IL-13, and IL-4. AGM4 (purple) had a similar cytokine profile except with little elevation in IL-6. AGM3 (blue) had elevated IL-10 1 week post-infection and is notable for having the lowest pathology scores of the four AGMs. RMs had modest changes in serum cytokines at 1 week post infection and at necropsy. Circles indicate aerosol exposure; squares, multiroute exposure; gray, RMs; red developed acute respiratory disease (ARDS); purple, increased cytokines without ARDS; and blue, no cytokine increase or ARDS.
Kinetics of binding antibody responses in severe acute respiratory syndrome coronavirus 2–infected African green monkeys (AGMs) and rhesus macaques (RMs). Circles indicate aerosol exposure; squares, multiroute exposure; gray, RMs; red, developed acute respiratory disease (ARDS); purple, increased cytokines without ARDS; and blue, no cytokine increase or ARDS.
Data Availability Statement
The raw data supporting the findings and figures have been placed in the public data repository Figshare (https://figshare.com/articles/dataset/Raw_Data_Spreadsheets/12449654; last accessed October 30, 2020). Material requests can be made to the Tulane National Primate Research Center. Approved requests for materials will be released after completion of a material transfer agreement. The Institutional Animal Care and Use Committee of Tulane University reviewed and approved all the procedures for this experiment. The Tulane National Primate Research Center is fully accredited by the American Association for Accreditation of Laboratory Animal Care. All animals were cared for in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals.18 The Tulane University Institutional Biosafety Committee approved the procedures for sample handling, inactivation, and removal from BSL3 containment.