Abstract
T-cells play key roles in immunity to COVID-19 as well as the development of severe disease. T-cell immunity to COVID-19 is mediated through differentiated CD4+ T-cells and cytotoxic CD8+ T-cells, although their differentiation is often atypical and ambiguous in COVID-19 and single cell dynamics of key genes need to be characterized. Notably, T-cells are dysregulated in severe COVID-19 patients, although their molecular features are still yet to be fully revealed. Importantly, it is not clear which T-cell activities are beneficial and protective and which ones can contribute to the development of severe COVID-19. In this article, we examine the latest evidence and discuss the key features of T-cell responses in COVID-19, showing how T-cells are dysregulated in severe COVID-19 patients. Particularly, we highlight the impairment of FOXP3 induction in CD4+ T-cells and how the impaired FOXP3 expression can lead to the differentiation of abnormally activated (hyperactivated) T-cells and the dysregulated T-cell responses in severe patients. Furthermore, we characterise the feature of hyperactivated T-cells, showing their potential contribution to T-cell dysregulation and immune-mediated tissue destruction (immunopathology) in COVID-19.
1. Text
T-cells are required to induce immune responses specific to SARS-CoV-2 by recognizing viral antigens through their antigen receptor, T-cell receptor (TCR) [1]. Since TCR is highly variable due to the random recombination of the TCR genes, each antigen can only be recognized by a small number of T-cells [2,3]. Since T-cells recognize antigens as peptides bound to Major Histocompatibility Complex (MHC), T-cells can recognize not only structural proteins such as spike (S) and nucleocapsid (N) proteins but also non-structural proteins including ORF3a and ORF7 [1]. Once recognizing a viral antigen, CD4+ T-cells are activated and can differentiate into helper T-cell subsets through the activities of transcription factors and cytokines specific to each subset. CD4+ T-cell help promotes the maturation of B-cells, which undergo affinity maturation and class-switching of virus-specific antibodies through the action of activation-induced cytidine deaminase (AID) [4]. Meanwhile, CD8+ T-cells can get primed with the help of CD4+ T-cells and differentiate into cytotoxic T-cells, which produce cytotoxic molecules such as granzymes and perforins upon recognizing antigen and thereby induce the apoptosis of virus-infected cells [1,5]. Therefore, T-cells play central roles in viral infections including COVID-19, and thus, it is not surprising that T-cells are dysregulated particularly in severe COVID-19 patients. This article will show the evidence of T-cell dysregulation in severe COVID-19 disease and discuss underlying molecular mechanisms.
1.1. Lymphopenia and T-cell reduction in COVID-19
Severe COVID-19 patients show the reduction of all lymphocyte subsets including CD4+ and CD8+ T-cells, NK cells, and B cells (i.e. lymphopenia) [[6], [7], [8]], while monocytes and granulocytes increase in circulation [8]. COVID-19 patients show the increase of serum cortisol [9], which is suggested to be a cause of lymphopenia in SARS [10], because corticosteroid treatment can also transiently reduce lymphocyte numbers while increasing neutrophils and monocytes in circulation [11,12]. In addition, T-cells in severe COVID-19 patients highly express activation markers as discussed below. Thus, it is likely that other factors also contribute to the T-cell reduction in COVID-19.
T-cell numbers are regulated by proliferation and apoptosis during homeostasis [13], and accordingly, T-cell reduction in COVID-19 can be due to either or both of increased apoptosis and reduced proliferation rates. While Fas expression is increased in T-cells from COVID-19 patients [14], T-cell data in Zhu et al. showed that Fas, FasL, and Caspase-3 [15], which play key roles of T-cell apoptosis, were not significantly increased in COVID-19 patients [16]. Interleukin (IL)-7 is a key cytokine for T-cell homeostasis, sustaining the naïve T-cell pool [17]. However, serum IL-7 levels are increased in severe COVID-19 patients [18], indicating that the IL-7-mediated compensatory mechanism is operating normally. IL-15 is important for maintaining the size of the CD8+ T-cell and memory T-cell pool [17] and could play a role in T-cell homeostasis in COVID-19, although data for IL-15 in COVID-19 is limited. Interestingly, T-cell numbers are negatively correlated with the serum concentration of cytokines including IL-6 and IL-10 in COVID-19 patients [7]. IL-6 is primarily produced by macrophages, dendritic cells (DCs), B-cells, and T-cells and can promote the proliferation of T-cells in inflammatory conditions [19]. IL-10 is produced by a wide range of cells including DCs, macrophages, B-cells, and T-cells including T-helper type 2 (Th2) and regulatory T-cells (Treg). IL-10 can suppress the proliferation of CD4+ and CD8+ T-cells in some contexts [20] while enhancing T-cell proliferation in the presence of other γ-chain cytokines i.e. IL-2, IL-4, IL-7, and IL-15 [21]. Given the increased cytokine production in severe COVID-19 patients, it is unlikely that the elevated IL-10 levels is the cause of T-cell reduction. These collectively suggest that T-cell reduction is a consequence of the activation of various innate and adaptive immune cells, which inevitably leads to the activation of T-cells as well.
1.2. Interferon and cytokine responses to COVID-19
SARS-CoV-2-infected epithelial cells can detect viral RNA by cytosolic sensors including RIG-1 and MDA5 and produce type-I interferons (IFNs), which induces IFN-mediated anti-viral responses [22]. Notably, in comparison to other respiratory viruses, SARS-CoV-2 induces lower levels of type-I IFNs (IFN-α, IFN-β) and type-III IFN (IFN-λ) and higher chemokine expression in primary human bronchial epithelial cells in comparison to other common respiratory viruses [23]. Still, IL-1β and type I and type III IFNs are modestly induced in active COVID-19, while persisting and even increased during the recovery phase [24].
Type-I IFNs not only induce cell-intrinsic anti-viral states in infected and neighbouring cells, but also promote innate immune responses. In viral infections, DCs, particularly plasmacytoid DCs (pDCs), play key roles in inducing innate immune responses following IFN responses, as pDCs can rapidly produce large amounts of type-I IFNs upon viral infection [25]. Importantly, pDCs can produce type-I IFNs not only when they are infected but also when adjacent cells are infected and produce type-I IFNs. However, Laing et al. showed that pDCs were markedly reduced in PBMCs from severe COVID-19 patients in comparison to moderate patients and healthy controls [26].
In addition, type-I IFN signalling promotes the maturation of DCs, increasing the expression of MHC and CD80/86 [27], the latter of which enhances the activation of T-cells as discussed below. In severe COVID-19 patients, type-I IFN response is impaired in peripheral mononuclear blood cells (PBMCs) and BAL fluid, showing reduced type-I IFN production [28,29]. Similarly, in SARS-CoV-1, delayed type-I IFN responses can promote the development of a severe disease through the accumulation of inflammatory monocytes-macrophages, which leads to impaired T-cell responses [30]. In fact, circulating monocytes from severe patients express more IL-1β and TNF-α [31]. In addition, macrophages in bronchoalveolar lavage (BAL) fluids from severe COVID-19 patients express higher levels of IL-1β, IL-6 and TNF-α than moderate patients [32]. Intriguingly, the chemokine CCL3 (MIP1A) is increased in both serum and BAL fluid-derived T-cells from severe COVID-19 patients [18,33]. Thus, the combination of the impaired production of type-I IFNs and the increased production of proinflammatory cytokines may contribute to the development of the severe disease [22].
Collectively, the delayed IFN responses in COVID-19 can result in the increased proinflammatory cytokine responses, changing the dynamics of antigen presentation and cytokine production in innate immune cells, which may contribute to the dysregulation of T-cell responses.
1.3. T-cell activation in COVID-19
Antigen presenting cells such as mature DCs present antigens to T-cells on their MHCs, and highly express CD80/CD86, which interacts with CD28 on T-cells [34]. Then, antigen-specific T-cells receive TCR signals and costimulatory signals, resulting in their activation. Activated T-cells produce IL-2 while expressing the IL-2 receptor, which is composed of α (CD25), β (CD122), and γ chains [35]. CD25 is particularly important as an inducible protein to produce the high-affinity IL-2 receptor. Thus, CD25-expressing activated T-cells receive IL-2 signalling, which further promotes their proliferation and differentiation.
Interestingly, soluble CD25 is increased in COVID-19 patients [[24], [69]], and a study showed the increase of IL-2 as well [18]. Since both IL-2 and IL-2 receptors are expressed predominantly by T-cells, it is possible that the positive feedback loop for IL-2 signalling in T-cells is established in some patients. However, it is not known whether this is protective or contributes to the pathology of COVID-19. In normal situations, IL-2 signalling can induce FOXP3, which stops the IL-2 response as a negative feedback mechanism [36], but this FOXP3 induction is impaired in severe COVID-19 as discussed below. Importantly, therapeutic administration of recombinant IL-2 in human patients can induce vascular leak syndrome, which is characterized by multiple organ failure due to increased vascular permeability [37].
Activated CD4+ and CD8+ T-cells also increase the expression of the proliferation marker Ki67 and the activation markers CD38 and HLA-DR. Such T-cells will be identified in non-naïve T-cell fractions, which include activated, effector, and memory T-cells. Interestingly, circulating non-naïve CD4+ and CD8+ T-cells show higher expressions of Ki67, HLA-DR, and CD38 in COVID-19 patients in comparison to both recovered patients and healthy donors [38]. This indicates that a significant proportion of circulating T-cells react to the infection, which suggests that, in addition to the clonal expansion of SARS-CoV-2-specific T-cells, a broad range of the T-cell repertoire including self-reactive T-cells may be expanded in COVID-19. Currently, methods are limited for the analysis of the TCR-repertoire, but key resources may be obtained by single cell TCR-sequencing [39] of COVID-19 patients and the investigation of complementarity-determining region 3 (CDR3) of TCRs in each MHC haplotype [40] to understand the dynamics of virus-specific T-cells and self-reactive T-cells.
1.4. T-cell differentiation in COVID-19
Th1 cells, which produce IFN-γ and TNF-α [41], are important for viral infections, although the role of Th1 response in COVID-19 is not clear. SARS-CoV-2 spike (S) protein-specific T-cells are induced during the acute phase of COVID-19 and these cells produce IFN-γ [42]. However, another report showed that CD4+ T-cells had impaired IFN-γ and TNF-α production in severe COVID-19 patients [43], although severe COVID-19 patients show higher levels of serum IL-12 [8], which potently induces Th1 differentiation [41]. Lucas et al. showed that severe COVID-19 patients had higher serum levels of IFN-γ and IL-17A, the latter of which is produced by Th17 cells [8]. Th17 plays a key role in autoimmunity and infections including COVID-19 [45]. Notably, IL-6 signalling induces Th17 differentiation through activating STAT3 and inducing Th17 transcription factors including ROR-γt and AHR [46]. Intriguingly, T-cells from COVID-19 patients express higher levels of the IL-6 receptor than those from Influenza patients and healthy controls [16]. In addition, some T-cells from COVID-19 patients are identified as non-classical Th1 cells that highly express the Th17 markers CD161 and IL-1 receptor type I (IL-1RI) [24]. De Biasi et al. showed that not only IFN-γ but also Th2 cytokines including IL-4, IL-10, and IL-13 were increased in COVID-19 patients with pneumonia [45], while Schultheiβ et al. showed only marginal increase of IFN-γ in serum from COVID-19 patients [24]. Kalfaoglu et al. showed that both of the transcription factors TBX21 (T-bet) and GATA3, which induce Th1 and Th2 differentiation, respectively, were induced in activated CD4+ T-cells in BAL fluids from COVID-19 patients. In addition, IL4R and MAF, which are required for Th2 differentiation, were more highly induced in BAL-derived CD4+ T-cells from severe COVID-19 patients [33]. Collectively, it is likely that T-cells show mixed Th1, Th2, and Th17 responses in COVID-19, and it is yet to be revealed which Th response is the most protective.
CD4+ T-cell responses to S-protein correlated with the concentration of the anti-SARS-CoV-2 IgG and IgA titres in convalescent patients [1], showing the importance of the T-cell help for B-cell maturation in recovery from COVID-19. This is most likely mediated by a subtype of CD4+ T-cells, T-follicular helper cells (Tfh), which provide the T-cell help that is required for B-cell differentiation and the affinity maturation of antibodies [47]. Tfh cells differentiate from antigen-reactive T-cells in lymph nodes. Tfh are identified by the expression of CXCR5 and PD-1 and are typically found in germinal centres (GCs). Some Tfh can be found in circulation (circulating Tfh, cTfh) and are identified as CXCR5+CD45RA− CD4+ T-cells (CD45RA− indicates antigen experienced T-cells). In COVID-19, cTfh cells are induced in COVID-19 patients [38,48] including severe patients [49]. Interestingly, cTfh upregulate CCR6, a marker of Th17 [50], upon stimulation with the S-protein of SARS-CoV-2, thus showing Th17-like cTfh features [51]. On the other hand, Kaneko et al. showed that GCs were not formed in lymph nodes and that BCL-6-expressing Tfh were reduced in COVID-19 [52]. In addition, Kalfaoglu et al. showed that BCL6 and Th17 genes (including RORC, IL17A, and CCR6) were repressed in BAL fluid-derived CD4+ T-cells from severe COVID-19 [33]. Collectively, these suggest that uncharacterised T-cell differentiation, which partially resemble Th17 and Tfh, may be important to control the infection.
CD8+ T-cells can be primed in lymph nodes and kill virus infected cells through the release of cytotoxic molecules such as perforin and granzyme B [53]. Interestingly, after the recovery from COVID-19, patients with mild disease sustain SARS-CoV-2-specific CD8+ T-cells than patients with severe disease, suggesting that the induction of potent CD8+ T-cell responses is protective [54]. However, during active disease, CD8+ T-cells can produce higher levels of perforin and granzyme B in severe COVID-19 patients than in mild patients and healthy donors [[43], [49],55]. Interestingly, Westmeier et al. showed that CD8+ T-cells from aged COVID-19 patients failed to produce perforin and granzymes [56], although further studies are required to confirm the result. Notably, CD8+ T-cells in severe COVID-19 patients had higher frequencies of PD-1+ cell subsets [7,49]. Although PD-1 is often used as a marker to identify exhausted T-cells, PD-1+CXCR5+CD8+ T-cells are important anti-viral effectors in the lymphocytic choriomeningitis virus (LCMV) infection model [57]. Further studies are required to determine whether PD-1+CD8+ T-cells in COVID-19 are protective or contribute to the development of severe disease.
1.5. FOXP3 repression and hyperactivated T-cells in COVID-19
FOXP3 is widely recognized as the Treg-specific transcription factor, and FOXP3-expressing T-cells can suppress T-cell responses [58,59]. In humans, ∼5% of CD4+ T-cells highly express FOXP3 [60,61]. Notably those FOXP3high Treg highly express CD25 and downregulate IL-7 receptor α (CD127) [61]. The majority of such naturally-occurring FOXP3+ Treg recognize self-antigens or microbiome antigens [62]. In fact, FOXP3+ Treg regularly receive TCR signals in vivo [63] and show higher Ki67 expression, indicating their activated and proliferating status [61]. The expression of both CD25 and FOXP3 is dynamically regulated in vivo [60,63] and is controlled by IL-2 and TCR signals [64].
When TCR and TGF-β signals are available, IL-2 signalling in activated T-cells promotes not only their survival and proliferation but also the expression of the FOXP3 gene [65]. FOXP3 represses IL2 transcription, inducing hyporesponsiveness (anergy) to TCR signals [66]. Importantly, natural FOXP3+ Treg also further upregulate FOXP3 transcription through its autoregulatory transcriptional loop during inflammation, increasing the expression of Treg-associated molecules including CD25 and CTLA-4, which is required for their suppressive function [67]. Thus, FOXP3 works as a negative feedback mechanism for suppressing T-cell activation whether in Treg and in non-Treg (Fig. 1 ).
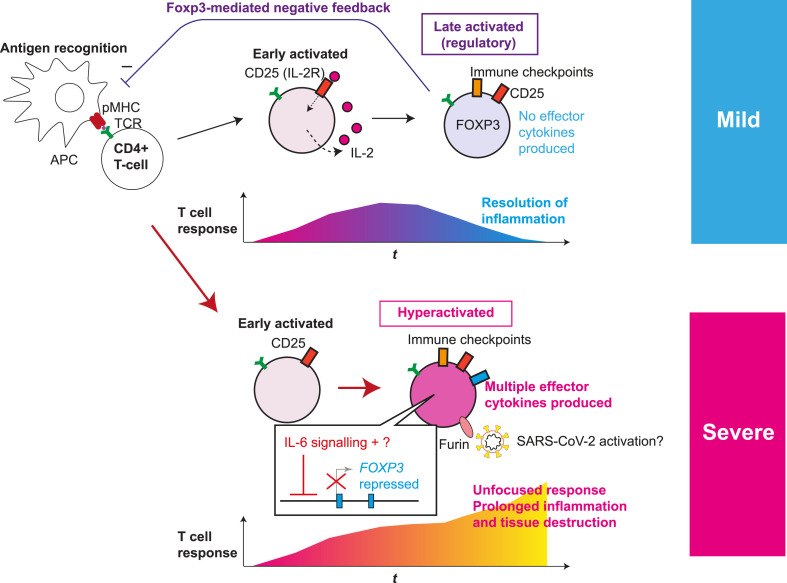
Fig. 1.
Roles of T-cell hyperactivation in the lung of severe COVID-19 patients. The fates of activated T-cells in mild and severe COVID-19 patients are depicted. Antigen-presenting cells (APC) present antigens as peptide-MHC complex (pMHC) to CD4+ T-cells, which triggers T-cell receptor (TCR) signalling and subsequent activation and differentiation processes. Initially, early activated T-cells start to produce CD25 and IL-2, establishing a positive feedback loop for T-cell activation and proliferation. Subsequently the fates of activated T-cells are different in mild and severe COVID-19 patients. (Upper panel) In normal conditions and mild patients, IL-2 signalling enhances FOXP3 transcription in a part of activated T-cells. In addition, the expression of immune checkpoint molecules such as CTLA-4 is increased. Meanwhile, some T-cells can differentiate into Tfh and Th cells (not shown). CD25+CTLA-4+FOXP3+ T-cells can consume and occupy immunological resources including IL-2 and CD28 signalling, and thereby suppress T-cell activation, leading to resolution of inflammation. (Lower panel) In severe COVID-19, activated T-cells fail to express FOXP3 while further enhancing the expression of CD25 and immune checkpoint molecules, producing multiple Th cytokines and showing the features of hyperactivated T-cells. FOXP3 repression is likely due to the increased IL-6 availability and undefined factors. In addition, CD25+ hyperactivated T-cells produce FURIN, which may further enhance the viral infection, promoting prolonged inflammation and tissue destruction. Figure modified from Ref. [33].
FOXP3 expression in circulating CD4+ T-cells is higher in convalescent COVID-19 patients than unexposed individuals [68]. On the other hand, the frequency of Treg in circulation is reduced in severe patients [69], although results are variable between studies [26]. This suggests that, paradoxically, FOXP3 induction is favourable to the outcome in some patients, suggesting a role of FOXP3 in anti-viral T-cell immunity. Intriguingly, a significant percentage of CD4+ T-cells highly express CD25 while FOXP3 expression is repressed in the lung of severe COVID-19 patients [33]. Those CD25+ FOXP3- T-cells highly express CTLA-4, GITR, and other activation/Treg markers, suggesting that their differentiation into Treg is prematurely blocked (Fig. 1). In addition, Jeannet et al. showed that severe patients in Intensive Care Unit (ICU) showed sustained reduction of all lymphocyte subsets including T-cells, while the expression of CD25, CTLA-4, and PD-1 in CD4+ T-cells was increased, although their study did not analyse FOXP3 expression [6].
Although the mechanism of FOXP3 repression in severe COVID-19 is yet to be revealed, IL-6 produced by macrophages and monocytes may contribute to the repression. IL-6 signalling activates STAT3, which antagonises STAT5 activities and thereby represses IL-2-mediated FOXP3 transcription [70]. As discussed above, T-cells in severe COVID-19 show higher IL-6 receptor expression and thus are considered to be sensitive to IL-6 in the microenvironments. In addition, our scRNA-seq analysis showed that IL-2 was not detected in BAL fluid-derived CD4+ T-cells from severe COVID-19 patients [33], and critically ill patients show reduced serum IL-2 [71]. These suggest that IL-6 overproduction and IL-2 deprivation contribute to FOXP3 repression in severe COVID-19.
FOXP3 inhibition generally induces inflammation, although this may not be beneficial in infections [36]. Rather, the inhibition of FOXP3 induction can lead to unfocused T-cell responses that induce immunopathology (i.e. immune-mediated tissue destruction) and ineffective antiviral T-cell response. For example, in a mouse model of herpes simplex infection, the depletion of FOXP3+ Treg induces strong inflammation and prolongs the infection [72]. Similarly, Treg depletion results in increased mortality in a murine model of coronavirus-induced acute encephalitis [73].
In addition, depletion of FOXP3+ Treg can induce T-cell responses to self-antigens, which lead to the development of autoimmunity [59]. Interestingly, severe COVID-19 patients show B-cell repertoire features previously described in active systemic lupus erythematosus (SLE) patients, a systemic autoimmune disease [74]. Here it is worthwhile to note that active SLE patients have reduced FOXP3high Treg in circulation [61]. These collectively suggest that the impairment of FOXP3 induction in severe COVID-19 induces autoimmune-like T-cell responses to self-antigens, which deplete immunological resources that could have been used by virus-specific T-cells.
Interestingly, CD25+ T-cells in severe COVID-19 patients specifically express the protease FURIN [33]. FURIN and the serine protease TMPRSS2 can cleave spike (S) protein of SARS-CoV-2 and enhance the viral entry into human cells [75]. FURIN is expressed by Treg and a knockout study showed that FURIN expression is required for Treg-mediated immune suppression [76]. In addition, FURIN expression is induced in T-cells by TCR signalling in vivo and in vitro [33], and therefore, FOXP3+ Treg and highly activated CD25+CD4+ T-cells could potentially enhance the activation of S protein in inflammatory tissues. Since a larger proportion of macrophages produce FURIN and the number of FOXP3−CD25+CD4+ T-cells is small in each individual, the contribution of FURIN expression in those T-cells towards enhancing viral entry, if any, could be mediated through either SARS-CoV-2 infection of T-cells themselves or the enhancement of viral entry into DCs with which antigen-specific T-cells may intimately interact [77]. Although T-cells do not express ACE2, it is possible that SARS-CoV-2 enters non-ACE2 expressing cells using alternative receptors [78], and further investigation is needed.
Collectively, FOXP3−CD25+CD4+ T-cells in severe COVID-19 patients are considered to be abnormally activated (hyperactivated), failing to differentiate into specific T-cell subsets. Their activation and differentiation may also be prematurely stopped at an early activation stage and contribute to the infection and pathogenesis (Fig. 1).
2. Perspective and conclusion
FOXP3-mediated negative regulatory mechanisms of T-cell activation are impaired and CD25+ hyperactivated T-cells are induced in severe COVID-19 patients. In addition, atypical T-cell differentiation seems to occur in COVID-19, producing T-cells that partially resemble Th1, Th2, Th17 and Tfh but lack their cardinal features. SARS-CoV-2 can induce abnormal responses of macrophages and DCs through infection, which control the differentiation of hyperactivated T-cells, possibly through the impaired type-I IFN response and the production of cytokines including IL-6.
Key outstanding questions include: (1) how the impaired FOXP3 induction can lead to the development of severe COVID-19, in which dysregulated T-cell responses dominate protective immune responses and induce immunopathology; (2) which T-cell subsets are protective, and which genes are important for effective T-cell immunity against COVID-19; (3) in addition to CD25+ hyperactivated T-cells, which T-cell subsets are pathogenic in the severe disease and which molecules can be targeted to block the aberrant T-cell responses (4) whether and how impaired IFN responses lead to T-cell dysregulation. Single cell-level analysis, particularly the analysis of TCR repertoires and antigen-specific T-cells will be important for revealing the dynamics of virus-specific and tissue-specific T-cell activities. Furthermore, mechanistic understanding of protective T-cell immunity to COVID-19 and their dysregulation will be key to improved prevention and treatment strategies for COVID-19.
Acknowledgements
MO is supported by the MRC project grant MR/S000208/1 and the Kakenhi 19H05426 from the Japan Society for the Promotion of Science (JSPS). In addition, this research was supported in part by grants from the Japan Agency for Medical Research and Development (grant numbers JP20jm0210074 to YS and MO; JP20fk0410023 and JP19fm0208012 to YS).
References
- 1.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J., Zeng X., Sigal N., Lund P.J., Su L.F., Huang H., Chien Y.-h., Davis M.M. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:E1890–E1897. doi: 10.1073/pnas.1602488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reiss S., Baxter A.E., Cirelli K.M., Dan J.M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J.-P., Havenar-Daughton C., Crotty S., Kaufmann D.E. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PloS One. 2017;12 doi: 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honjo T., Kinoshita K., Muramatsu M. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 5.Wherry E.J., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeannet R., Daix T., Formento R., Feuillard J., François B. Severe COVID-19 is associated with deep and sustained multifaceted cellular immunosuppression. Intensive Care Med. 2020;46:1769–1771. doi: 10.1007/s00134-020-06127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Muenker M.C., Fournier J.B., Campbell M., Odio C.D., Casanovas-Massana A., Obaid A., Lu-Culligan A., Nelson A., Brito A., Nunez A., Martin A., Watkins A., Geng B., Kalinich C., Harden C., Todeasa C., Jensen C., Kim D., McDonald D., Shepard D., Courchaine E., White E.B., Song E., Silva E., Kudo E., DeIuliis G., Rahming H., Park H.-J., Matos I., Nouws J., Valdez J., Fauver J., Lim J., Rose K.-A., Anastasio K., Brower K., Glick L., Sharma L., Sewanan L., Knaggs L., Minasyan M., Batsu M., Petrone M., Kuang M., Nakahata M., Campbell M., Linehan M., Askenase M.H., Simonov M., Smolgovsky M., Sonnert N., Naushad N., Vijayakumar P., Martinello R., Datta R., Handoko R., Bermejo S., Prophet S., Bickerton S., Velazquez S., Alpert T., Rice T., Khoury-Hanold W., Peng X., Yang Y., Cao Y., Strong Y., Herbst R., Shaw A.C., Medzhitov R., Schulz W.L., Grubaugh N.D., Dela Cruz C., Farhadian S., Ko A.I., Omer S.B., Iwasaki A., Yale I.T. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan T., Khoo B., Mills E.G., Phylactou M., Patel B., Eng P.C., Thurston L., Muzi B., Meeran K., Prevost A.T., Comninos A.N., Abbara A., Dhillo W.S. Association between high serum total cortisol concentrations and mortality from COVID-19. Lancet Diabetes Endocrinol. 2020;8:659–660. doi: 10.1016/S2213-8587(20)30216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panesar N.S., Lam C.W.K., Chan M.H.M., Wong C.K., Sung J.J.Y. Lymphopenia and neutrophilia in SARS are related to the prevailing serum cortisol. Eur. J. Clin. Invest. 2004;34:382–384. doi: 10.1111/j.1365-2362.2004.01347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrillo J.E., Fauci A.S. Mechanisms of corticosteroid action on lymphocyte subpopulations. III. Differential effects of dexamethasone administration on subpopulations of effector cells mediating cellular cytotoxicity in man. Clin. Exp. Immunol. 1978;31:116–125. [PMC free article] [PubMed] [Google Scholar]
- 12.Fauci A.S. Mechanisms of corticosteroid action on lymphocyte subpopulations. I. Redistribution of circulating T and b lymphocytes to the bone marrow. Immunology. 1975;28:669–680. [PMC free article] [PubMed] [Google Scholar]
- 13.Yates A., Saini M., Mathiot A., Seddon B. Mathematical modeling reveals the biological program regulating lymphopenia-induced proliferation. J. Immunol. 2008;180:1414. doi: 10.4049/jimmunol.180.3.1414. [DOI] [PubMed] [Google Scholar]
- 14.Bellesi S., Metafuni E., Hohaus S., Maiolo E., Marchionni F., D’Innocenzo S., La Sorda M., Ferraironi M., Ramundo F., Fantoni M., Murri R., Cingolani A., Sica S., Gasbarrini A., Sanguinetti M., Chiusolo P., De Stefano V. Increased CD95 (Fas) and PD-1 expression in peripheral blood T lymphocytes in COVID-19 patients. Br. J. Haematol. 2020;191:207–211. doi: 10.1111/bjh.17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maher S., Toomey D., Condron C., Bouchier-Hayes D. Activation-induced cell death: the controversial role of Fas and Fas ligand in immune privilege and tumour counterattack. Immunol. Cell Biol. 2002;80:131–137. doi: 10.1046/j.1440-1711.2002.01068.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhu L., Yang P., Zhao Y., Zhuang Z., Wang Z., Song R., Zhang J., Liu C., Gao Q., Xu Q., Wei X., Sun H.-X., Ye B., Wu Y., Zhang N., Lei G., Yu L., Yan J., Diao G., Meng F., Bai C., Mao P., Yu Y., Wang M., Yuan Y., Deng Q., Li Z., Huang Y., Hu G., Liu Y., Wang X., Xu Z., Liu P., Bi Y., Shi Y., Zhang S., Chen Z., Wang J., Xu X., Wu G., Wang F.-S., Gao G.F., Liu L., Liu W.J. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53:685–696. doi: 10.1016/j.immuni.2020.07.009. e683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surh C.D., Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Jones L.L., Geiger T.L. IL-6 promotes T cell proliferation and expansion under inflammatory conditions in association with low-level RORγt expression. J. Immunol. 1950;201:2934–2946. doi: 10.4049/jimmunol.1800016. Baltimore, Md. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maynard C.L., Weaver C.T. Diversity in the contribution of interleukin-10 to T-cell-mediated immune regulation. Immunol. Rev. 2008;226:219–233. doi: 10.1111/j.1600-065X.2008.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geginat J., Sallusto F. A. Lanzavecchia cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4+ T cells. J. Exp. Med. 2001;194:1711–1720. doi: 10.1084/jem.194.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008737. e1008737-e1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultheiß C., Paschold L., Simnica D., Mohme M., Willscher E., von Wenserski L., Scholz R., Wieters I., Dahlke C., Tolosa E., Sedding D.G., Ciesek S., Addo M., Binder M. Next-Generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53:442–455. doi: 10.1016/j.immuni.2020.06.024. e444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reizis B. Plasmacytoid dendritic cells: development, regulation, and function. Immunity. 2019;50:37–50. doi: 10.1016/j.immuni.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laing A.G., Lorenc A., del Molino del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., Kamdar S., Joseph M., Davies D., Davis R., Jennings A., Zlatareva I., Vantourout P., Wu Y., Sofra V., Cano F., Greco M., Theodoridis E., Freedman J., Gee S., Chan J.N.E., Ryan S., Bugallo-Blanco E., Peterson P., Kisand K., Haljasmägi L., Chadli L., Moingeon P., Martinez L., Merrick B., Bisnauthsing K., Brooks K., Ibrahim M.A.A., Mason J., Lopez Gomez F., Babalola K., Abdul-Jawad S., Cason J., Mant C., Seow J., Graham C., Doores K.J., Di Rosa F., Edgeworth J., Shankar-Hari M., Hayday A.C. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 27.Simmons D.P., Wearsch P.A., Canaday D.H., Meyerson H.J., Liu Y.C., Wang Y., Boom W.H., Harding C.V. Type I IFN drives a distinctive dendritic cell maturation phenotype that allows continued class II MHC synthesis and antigen processing. J. Immunol. 1950;188:3116–3126. doi: 10.4049/jimmunol.1101313. Baltimore, Md. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.-A., Merkling S.H., Treluyer J.-M., Veyer D., Mouthon L., Blanc C., Tharaux P.-L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., Deczkowska A., Zhang S., Schwikowski B., Zhang Z., Amit I. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488. doi: 10.1016/j.cell.2020.05.006. e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.-S., Jeong S.J., Lee H.K., Park S.H., Park S.-H., Choi J.Y., Kim S.-H., Jung I., Shin E.-C. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Science Immunology. 2020;5 doi: 10.1126/sciimmunol.abd1554. eabd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 33.Kalfaoglu B., Almeida-Santos J., Tye C.A., Satou Y., Ono M. T-cell hyperactivation and paralysis in severe COVID-19 infection revealed by single-cell analysis. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.589380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orabona C., Grohmann U., Belladonna M.L., Fallarino F., Vacca C., Bianchi R., Bozza S., Volpi C., Salomon B.L., Fioretti M.C., Romani L., Puccetti P. CD28 induces immunostimulatory signals in dendritic cells via CD80 and CD86. Nat. Immunol. 2004;5:1134–1142. doi: 10.1038/ni1124. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu A., Kondo S., Sabe H., Ishida N., Honjo T. Structure and function of the interleukin 2 receptor: affinity conversion model. Immunol. Rev. 1986;92:103–120. doi: 10.1111/j.1600-065x.1986.tb01496.x. [DOI] [PubMed] [Google Scholar]
- 36.Ono M., Tanaka R.J. Controversies concerning thymus-derived regulatory T cells: fundamental issues and a new perspective. Immunol. Cell Biol. 2016;94:3–10. doi: 10.1038/icb.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Assier E., Jullien V., Lefort J., Moreau J.-L., Vargaftig B.B., Silva J.R J.L.e., Thèze J. Constitutive expression of IL-2Rbeta chain and its effects on IL-2-induced vascular leak syndrome. Cytokine. 2005;32:280–286. doi: 10.1016/j.cyto.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., Manne S., Chen Z., Huang Y.J., Reilly J.P., Weisman A.R., Ittner C.A.G., Kuthuru O., Dougherty J., Nzingha K., Han N., Kim J., Pattekar A., Goodwin E.C., Anderson E.M., Weirick M.E., Gouma S., Arevalo C.P., Bolton M.J., Chen F., Lacey S.F., Ramage H., Cherry S., Hensley S.E., Apostolidis S.A., Huang A.C., Vella L.A., Betts M.R., Meyer N.J., Wherry E.J. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020 doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redmond D., Poran A., Elemento O. Single-cell TCRseq: paired recovery of entire T-cell alpha and beta chain transcripts in T-cell receptors from single-cell RNAseq. Genome Med. 2016;8:80. doi: 10.1186/s13073-016-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madi A., Shifrut E., Reich-Zeliger S., Gal H., Best K., Ndifon W., Chain B., Cohen I.R., Friedman N. T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity. Genome Res. 2014;24:1603–1612. doi: 10.1101/gr.170753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szabo S.J., Sullivan B.M., Peng S.L., Glimcher L.H. Molecular mechanisms RegulatinG Th1 immune responses. Annu. Rev. Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 42.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., Haagmans B.L., de Swart R.L., Sette A., de Vries R.D. Phenotype and kinetics of SARS-CoV-2–specific T cells in COVID-19 patients with acute respiratory distress syndrome. Science Immunology. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M., Paolini A., Menozzi M., Milić J., Franceschi G., Fantini R., Tonelli R., Sita M., Sarti M., Trenti T., Brugioni L., Cicchetti L., Facchinetti F., Pietrangelo A., Clini E., Girardis M., Guaraldi G., Mussini C., Cossarizza A. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutiérrez-Vázquez C., Quintana F.J. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity. 2018;48:19–33. doi: 10.1016/j.immuni.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B., Tong S.Y.C., Lewin S.R., Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe. COVID- 2020;19 doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., Kuthuru O., Apostolidis S.A., Bershaw L., Dougherty J., Greenplate A.R., Pattekar A., Kim J., Han N., Gouma S., Weirick M.E., Arevalo C.P., Bolton M.J., Goodwin E.C., Anderson E.M., Hensley S.E., Jones T.K., Mangalmurti N.S., Luning Prak E.T., Wherry E.J., Meyer N.J., Betts M.R. Comprehensive mapping of immune perturbations associated with severe COVID-19. Science Immunology. 2020 doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unutmaz D. RORC2: the master of human Th17 cell programming. Eur. J. Immunol. 2009;39:1452–1455. doi: 10.1002/eji.200939540. [DOI] [PubMed] [Google Scholar]
- 51.Juno J.A., Tan H.-X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., Gherardin N.A., Pymm P., Dietrich M.H., Scott N.E., Tham W.-H., Godfrey D.I., Subbarao K., Davenport M.P., Kent S.J., Wheatley A.K. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 52.Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., Bartsch Y.C., Bonheur N., Caradonna T.M., Chevalier J., Chowdhury F., Diefenbach T.J., Einkauf K., Fallon J., Feldman J., Finn K.K., Garcia-Broncano P., Hartana C.A., Hauser B.M., Jiang C., Kaplonek P., Karpell M., Koscher E.C., Lian X., Liu H., Liu J., Ly N.L., Michell A.R., Rassadkina Y., Seiger K., Sessa L., Shin S., Singh N., Sun W., Sun X., Ticheli H.J., Waring M.T., Zhu A.L., Alter G., Li J.Z., Lingwood D., Schmidt A.G., Lichterfeld M., Walker B.D., Yu X.G., Padera R.F., Pillai S. Loss of bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157. doi: 10.1016/j.cell.2020.08.025. e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang N., Bevan M.J. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., López-Camacho C., Slon-Campos J., Zhao Y., Stuart D.I., Paesen G.C., Grimes J.M., Antson A.A., Bayfield O.W., Hawkins D.E.D.P., Ker D.-S., Wang B., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., de Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.-L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B.M., Fry J.W., Schwabe N.F., Semple M.G., Baillie J.K., Moore S.C., Openshaw P.J.M., Ansari M.A., Dunachie S., Barnes E., Frater J., Kerr G., Goulder P., Lockett T., Levin R., Zhang Y., Jing R., Ho L.-P., Barnes E., Dong D., Dong T., Dunachie S., Frater J., Goulder P., Kerr G., Klenerman P., Liu G., McMichael A., Napolitani G., Ogg G., Peng Y., Salio M., Yao X., Yin Z., Kenneth Baillie J., Klenerman P., Mentzer A.J., Moore S.C., Openshaw P.J.M., Semple M.G., Stuart D.I., Turtle L., Cornall R.J., Conlon C.P., Klenerman P., Screaton G.R., Mongkolsapaya J., McMichael A., Knight J.C., Ogg G., Dong T., T.c.C. Oxford Immunology Network Covid-19 Response. Investigators I.C. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020 doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang C.K., Han G.-C., Kim M., Kim G., Shin H.M., Song K.-H., Choe P.G., Park W.B., Kim E.S., Kim H.B., Kim N.-J., Kim H.-R., Oh M.-D. Aberrant hyperactivation of cytotoxic T-cell as a potential determinant of COVID-19 severity. Int. J. Infect. Dis. 2020;97:313–321. doi: 10.1016/j.ijid.2020.05.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westmeier J., Paniskaki K., Karaköse Z., Werner T., Sutter K., Dolff S., Overbeck M., Limmer A., Liu J., Zheng X., Brenner T., Berger M.M., Witzke O., Trilling M., Lu M., Yang D., Babel N., Westhoff T., Dittmer U., Zelinskyy G. Impaired cytotoxic CD8+ T cell response in elderly COVID-19 patients. mBio. 2020;11 doi: 10.1128/mBio.02243-20. e02243-02220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He R., Hou S., Liu C., Zhang A., Bai Q., Han M., Yang Y., Wei G., Shen T., Yang X., Xu L., Chen X., Hao Y., Wang P., Zhu C., Ou J., Liang H., Ni T., Zhang X., Zhou X., Deng K., Chen Y., Luo Y., Xu J., Qi H., Wu Y., Ye L. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature. 2016;537:412–416. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 58.Li M.O., Rudensky A.Y. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 2016;16:220–233. doi: 10.1038/nri.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakaguchi S., Yamaguchi T., Nomura T., Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. S0092-8674(08)00624-7 [pii] [DOI] [PubMed] [Google Scholar]
- 60.Fujii H., Josse J., Tanioka M., Miyachi Y., Husson F., Ono M. Regulatory T cells in melanoma revisited by a computational clustering of FOXP3+ T cell subpopulations. J. Immunol. 2016 doi: 10.4049/jimmunol.1402695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyara M., Yoshioka Y., Kitoh A., Shima T., Wing K., Niwa A., Parizot C., Taflin C., Heike T., Valeyre D., Mathian A., Nakahata T., Yamaguchi T., Nomura T., Ono M., Amoura Z., Gorochov G., Sakaguchi S. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30:899–911. doi: 10.1016/j.immuni.2009.03.019. S1074-7613(09)00202-7 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Harrison O.J., Powrie F.M. Regulatory T cells and immune tolerance in the intestine. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a018341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bending D., Martin P.P., Paduraru A., Ducker C., Marzaganov E., Laviron M., Kitano S., Miyachi H., Crompton T., Ono M. A timer for analyzing temporally dynamic changes in transcription during differentiation in vivo. J. Cell Biol. 2018 doi: 10.1083/jcb.201711048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ono M. Control of regulatory T-cell differentiation and function by T-cell receptor signalling and Foxp3 transcription factor complexes. Immunology. 2020;160:24–37. doi: 10.1111/imm.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freudenberg K., Lindner N., Dohnke S., Garbe A.I., Schallenberg S., Kretschmer K. Critical role of TGF-β and IL-2 receptor signaling in Foxp3 induction by an inhibitor of DNA methylation. Front. Immunol. 2018;9:125. doi: 10.3389/fimmu.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hori S., Nomura T., Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 67.Bending D., Ono M. From stability to dynamics: understanding molecular mechanisms of regulatory T cells through Foxp3 transcriptional dynamics. Clin. Exp. Immunol. 2018 doi: 10.1111/cei.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.liu j., Yang X., Wang H., Li Z., Deng H., jing L., Xiong S., He J., Guo C., Wang W., Zelinskyy G., Trilling M., Dittmer U., Lu M., Sutter K., Senff T., Menne C., Timm J., Zhang Y., Deng F., Feng X., Lu Y., Wu J., Yang D., Wang B., Zheng X. The analysis of the long-term impact of SARS-CoV-2 on the cellular immune system in individuals recovering from COVID-19 reveals a profound NKT cell impairment. medRxiv. 2020:2020. doi: 10.1101/2020.08.21.20179358. 2008.2021.20179358. [DOI] [Google Scholar]
- 69.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020 doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 71.Shi H., Wang W., Yin J., Ouyang Y., Pang L., Feng Y., Qiao L., Guo X., Shi H., Jin R., Chen D. The inhibition of IL-2/IL-2R gives rise to CD8+ T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia. Cell Death Dis. 2020;11:429. doi: 10.1038/s41419-020-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lund J.M., Hsing L., Pham T.T., Rudensky A.Y. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320:1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anghelina D., Zhao J., Trandem K., Perlman S. Role of regulatory T cells in coronavirus-induced acute encephalitis. Virology. 2009;385:358–367. doi: 10.1016/j.virol.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., Lee S., Suryadevara N., Case J.B., Bugrovsky R., Chen W., Estrada J., Morrison-Porter A., Derrico A., Anam F.A., Sharma M., Wu H.M., Le S.N., Jenks S.A., Tipton C.M., Staitieh B., Daiss J.L., Ghosn E., Diamond M.S., Carnahan R.H., Crowe J.E., Hu W.T., Lee F.E.-H., Sanz I. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020 doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pesu M., Watford W.T., Wei L., Xu L., Fuss I., Strober W., Andersson J., Shevach E.M., Quezado M., Bouladoux N., Roebroek A., Belkaid Y., Creemers J., O’Shea J.J. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature. 2008;455:246–250. doi: 10.1038/nature07210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rush C.M., Millington O.R., Hutchison S., Bryson K., Brewer J.M., Garside P. Characterization of CD4+ T-cell-dendritic cell interactions during secondary antigen exposure in tolerance and priming. Immunology. 2009;128:463–471. doi: 10.1111/j.1365-2567.2009.03124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh M., Bansal V., Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]