Abstract
The magnitude and the quality of humoral responses against SARS-CoV-2 have been associated with clinical outcome. Although the elicitation of humoral responses against different viral proteins is rapid and occurs in most infected individuals, its magnitude is highly variable among them and positively correlates with COVID-19 disease severity. This rapid response is characterized by the almost concomitant appearance of virus-specific IgG, IgA and IgM antibodies that contain neutralizing antibodies directed against different epitopes of the Spike glycoprotein. Of particularly interest, the antibodies against domain of the Spike that interacts with the cellular receptor ACE2, known as the receptor binding domain (RBD), are present in most infected individuals and are block viral entry and infectivity. Such neutralizing antibodies protect different animal species when administered before virus exposure; therefore, its elicitation is the main target of current vaccine approaches and their clinical use as recombinant monoclonal antibodies (mAbs) is being explored. Yet, little information exists on the duration of humoral responses during natural infection. This is a key issue that will impact the management of the pandemic and determine the utility of seroconversion studies and the level of herd immunity. Certainly, several cases of reinfection have been reported, suggesting that immunity could be transient, as reported for other coronaviruses. In summary, although the kinetics of the generation of antibodies against SASR-CoV-2 and their protective activity have been clearly defined, their role in COVID-19 pathogenesis and the length of these responses are still open questions.
Keywords: B cells, Antibodies, Vaccines, Therapy, Pathogenesis
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) zoonosis causing Coronavirus disease-19 (COVID-19) emerged in late 2019 in China and rapidly spread worldwide, altering the established societal, economic and scientific priorities [1]. Scientific efforts to understand and control this new infection have not only focused on the discovery of optimal treatments and vaccines to reduce the clinical impact and spread of the disease [2,3], but also on the understanding of the interplay between the new virus and immune system [4]. This is a key piece of evidence that will inform about pathogenesis, transmission and vaccine development.
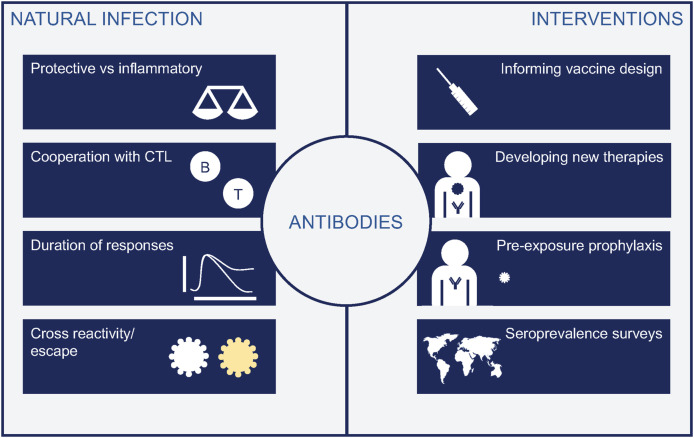
Key contributors to immunity are CD4+ T-cells that coordinate immune responses; B-cells that produce antibodies to target virus and infected cells; and CD8+ T-cells, that contribute to infected-cell killing. All three arms of adaptive immunity are relevant and work coordinately with innate cells to protect against infections [5]. Given the key role of antibodies in protection from viral diseases, in the development of serological tests and vaccines, but also their controversial role in immunopathogenesis of severe COVID-19 [4], we focused this review on humoral responses. We have recapitulated current knowledge on antibodies against SARS-CoV-2, their implications in our understanding of disease pathogenesis, and the development of antibody-based therapies and diagnostic tools or vaccines (Fig. 1 ).
Fig. 1.
Relevance of antibodies in our understanding of COVID-19. Left panel show main questions related to natural infection, including the still unknown duration of responses, while right panel enumerates the different tools developed thanks to our knowledge of antibody responses, that include vaccines, mAbs and diagnostic tools.
2. SARS-CoV-2 entry, tropism and pathogenesis
The pathogenesis of SARS-CoV-2 infection is directly related to its cellular tropism and the mechanisms of virus entry into target cells. Entry of SARS-CoV-2 into cellular targets is governed by the interactions of the Spike protein exposed on the viral particle with cellular receptors. This is a two-step process that relies on Spike binding to the cellular receptor Angiotensin-Converting Enzyme 2 (ACE2) [6]. This first step is mediated by a Spike domain close to its apex that is transiently exposed during conformational movements, this domain is known as the receptor binding domain (RBD) [7]. ACE2 is also exploited by other beta-coronaviruses such as SARS-CoV [8]. Once bound to ACE2, the Spike needs to be cleaved by proximal cellular proteases to facilitate viral fusion with the cellular membranes. This second step can take place on the plasma membrane if host proteases such as the Transmembrane Serine Protease 2 (TMPRSS2) are expressed on the cellular surface. Moreover, SARS-CoV-2 among other human beta-coronaviruses, uses other TMPRSS family members such as TMPRSS4 [9] or Furin [7]. However, in cells with low membrane-expressed proteases, alternative entry mechanisms take place via endocytic routes that accumulate viruses in early endosomes, where a plethora of cellular proteases such as cathepsins can prime the Spike protein to mediate fusion within these endosomal compartments [6,10].
Overall, the cellular tropism for SARS-CoV-2 is complex and mostly determined by the co-expression of ACE2 and host proteases [11] and allows SARS-CoV-2 to infect a wide range of cell types from the lung and different epithelia. Importantly, viral entry has clear implications for the design of effective therapies. The endosomal viral entry route is absent in pulmonary cells. Thus, TMPRSS inhibitors such as camostat may be the primary option to block viral entry [6]. At later stages, however, if SARS-CoV-2 reaches other extrapulmonary tissues, the endocytic route could complicate disease progression. Thus, to fully achieve viral suppression, effective inhibitors (including antibodies) need to block both plasma membrane and endosomal fusion to counteract infection in the diverse tissues where SARS-COV-2 has been found [12].
3. Humoral responses against SARS-CoV-2
A large amount of relevant information on antibody responses to SARS-CoV-2 has been generated, excellently reviewed by Vabret et al. [4], allowing for the definition of COVID-19 pathogenesis and the development of serodiagnosis tools for designing seroprevalence studies.
An early study of 173 patients with SARS-CoV-2 infection showed seroconversion in 93% of patient with an average time of 11 days [13]. Similar results are reported in other serological studies showing high seroconversion rates between 10 and 14 days after symptoms onset [[14], [15], [16], [17]]. Antibody response peak between the second and third-week after infection declining afterwards [13,18] and is characterized by the presence of IgA, IgM and IgG in plasma and saliva [13,14,18,19]. Although IgM is the first line of ng humoral response, one particularity of SARS-CoV-2 infection is that all three isotypes can be detected in a narrow time frame at seroconversion [13,18,20,21]. Actually, IgG and IgA can be frequently detected even before than IgM [18,21], indicating that the initial IgM response may be weak, that specific IgG or IgA B-cell precursor exist in the memory B-cell compartment or that class-switching occur rapidly after antigen encounter. Accordingly, specific IgG (mainly IgG1 and IgG3) can be detected 4–6 days after symptoms onset in some individuals [20]. Therefore, the detection of IgG or IgA may show higher sensitivity than IgM in early stages of infection [20,21]. No clear correlation has been observed between the levels of antibodies and the sex, age, or viral load [13], however they positively correlate with disease severity. Severe hospitalized patients present higher titter of IgG and IgA than mild cases, in which lower or even undetectable antibody levels have been reported [13,14,18,22].
Antibody response can be directed against all viral proteins, although Spike and nucleocapsid are considered the main targets of humoral response [13,18,23,24]. Antibodies against RBD, appear earlier in the course of infection than those antibodies against nucleocapsid [23]. Moreover, anti-RBD antibodies may provide a higher sensitivity and specificity for diagnosis than anti-nucleocapsid responses. RBD seroconversion in patients is frequent and fast with low cross-reactivity with SARS-CoV [20,24].
4. Neutralizing humoral response
Neutralizing antibodies are considered a major correlate of protective immunity and vaccine success [[25], [26], [27]]. In SARS-CoV-2 infection, these antibodies recognize several regions within the Spike glycoprotein, mainly but not exclusively the RBD, and inhibit viral infectivity by several mechanisms including the blockade of initial Spike binding to ACE2 [[28], [29], [30]]. Two main regions of vulnerability have been identified in the Spike, the RBD and the adjacent N-terminal domain (NTD) [28,31,32]. SARS-CoV and SARS-CoV-2 have a 80% homology and share approximately 75% of the spike glycoprotein sequence [7,33]. Although, few antibodies that exhibit cross-neutralizing activity between SARS-CoV and SARS-CoV-2 have been identified [34], the existence of potentially cross-reactive antibodies opens new avenues for the potential development of a pan-neutralizing vaccines against various coronaviruses [31].
Neutralizing antibodies are detected in approximately 40%–70% of infected individuals, depending on the criteria and the cohort studied. At least 30% of patients have no detectable antibody levels and less than 15% reach high titers of neutralization in vitro [[34], [35], [36], [37]]. An association between neutralizing antibody titer and severity of COVID-19 disease has been observed and those who have mild symptoms or are asymptomatic are more reluctant to generate a neutralizing response [38]. The kinetics of elicitation of neutralizing antibodies is similar to that reported for seroconversion. Approximately 6–15 days after symptoms onset neutralization activity is detected, although with a wide range of titers [20,36,37]. Then, the level of neutralizing antibodies gradually decreases over a period of 3 months [36] with an estimated half-life of 26 days [38], although these data should be confirmed in larger longitudinal analyses.
Neutralizing monoclonal antibodies (mABs) protect from SARS-CoV-2 lung infection and inflammation and weight loss in mice, rhesus macaques and other animal models [[39], [40], [41], [42], [43]]. Also, current vaccines induce high titers of neutralizing antibodies in most vaccinated individuals [[44], [45], [46], [47], [48]]; however, their efficacy and extent of protection are still open questions.
5. Role of antibodies in pathogenesis and protection
As mentioned above, neutralizing antibodies are protective as demonstrated in different animal models [[39], [40], [41], [42], [43]]. Accordingly, the administration of convalescent plasma to COVID19+ individuals improve their clinic status, at least in those treated with High neutralizing plasma during early disease stages [[49], [50], [51], [52]]. Actually, no clinical benefit was observed in those patients requiring invasive mechanical ventilation [50]. An additional indirect evidence about the antiviral activity of neutralizing antibodies in vivo is the identification of escape mutations that avoid the binding of neutralizing antibodies [53]. The studies performed with mAbs have also highlighted that the humoral response encompasses antibodies with different specificities that can work synergistically, opening the gateway to the use of rationally-designed antibody cocktails for therapy [30,54,55].
Beyond their neutralizing activity, antibodies develop additional functions depending on their isotype. Antibody-dependent cellular cytotoxicity (ADCC); antibody-dependent cellular phagocytosis (ADCP); and complement-dependent cytotoxicity (CDC) are important Fc-dependent functions that associate with protection in several infectious diseases [56]. Interestingly, S2M11 and S2E12, two recently described SARS-CoV-2 neutralizing antibodies, show ADCC and ADCP activities, respectively [30]. However, in addition to the potential protective role, antibodies can also be deleterious, promoting infection itself or causing the antibody-dependent enhancement (ADE) of the disease. This phenomenon has been documented for other pathogens (dengue or RSV) [57,58] and is mediated by immune receptors (FcRs) and the complement system, both promoting the infection or the progression of the disease. The fact that higher titers of total and neutralizing antibodies are observed in severe cases of SARS-CoV and SARS-CoV-2 infection [13,59,60], suggests that ADE might contribute to severity. Although in vitro and in vivo evidence exist for a role of ADE during SARS-CoV infection [[61], [62], [63], [64]], current observations do not support a strong contribution of ADE to COVID-19 severity: 1) the infusion of convalescent plasma in COVID19 patients have not reveled any adverse effect [[49], [50], [51], [52]]; 2) non-human primates develop antibodies against SARS-CoV-2 and are resistant to reinfection [65]; and 3) vaccinated animals develop antibodies and do not show signs of ADE after challenge [66,67].
6. Duration of responses, implications
Considering the relatively short time from SARS-CoV-2 zoonosis, short longitudinal information of immune responses is available. Several studies indicate that 20 days after symptoms onset the levels of specific IgM and IgA against SARS-CoV-2 progressively decline over 3–5 months after infection [68,69]. Similar declines were also reported for MERS infection [70] or other human seasonal coronavirus, whose protective immunity seems to be short-lasting [71]. The absence of germinal centers and the lack of Bcl-6+ TFH cells after acute SARS-CoV-2 infection [72], provides an explanation for the low level of somatic hypermutation detected in anti-SARS-CoV-2 antibodies [73] and predicts a short-lasting response.
Vaccinated non-human primate and serological studies in humans indicate that the levels of neutralizing antibodies correlate with protection from SARS-CoV-2 [66,74]. Therefore, an obvious risk associated with a fast decay or a poor elicitation of neutralizing antibodies is the possibility of reinfection. Although animal models suggests that infection induces protective immunity [65], several cases of reinfection have been reported in humans [75,76], one of them clearly associated with lack of seroconversion after the initial infection [76]. A close surveillance of reinfection events accompanied by serological surveys will inform on the relevance of this phenomenon. Further dangers of low neutralization titers are linked to incomplete antibody mediated protection and risk of ADE or inflammatory clinical complications [77]. However, individuals with mild-symptomatic infection elicit low/undetectable neutralizing activity [78,79]. A potential explanation for this paradox could be found in virus-specific T-cells, which have been identified in mild COVID-19 cases, individuals with close contacts with COVID-19 patients and in healthy donors sampled before 2019. Thus, T-cells primed by other human coronaviruses may control SARS-CoV-2 replication, making unnecessary a large activation of the B-cell immune arm [80,81].
7. Conclusions and future directions
The large efforts aimed at understanding the interplay between SASR-CoV-2 and the immune system have paved the way for an extraordinary fast vaccine development and the design of clinically tested mAbs as new therapeutic tools. However, our deep knowledge of early responses contrasts with the lack of information on the protective efficacy of antibodies overtime. This is particularly relevant, because the answer will strongly impact the course of the pandemics. Additional questions, such as the reasons behind the large diversity in antibody levels and the lack of seroconversion in some individuals, and the role of T cells are still being investigated. A comprehensive analysis of all immune processes involved in virus control will be required to fully control the pandemic.
Declaration of competing interest
Outside the submitted work JB and JC are founders and shareholders of AlbaJuna Therapeutics, S.L. BC is founder and shareholder of AlbaJuna Therapeutics, S.L and AELIX Therapeutics, S.L.
Acknowledgements
Work at IrsiCaixa is funded by Grifols S. A. (Spain), Generalitat de Catalunya (grant DSL0016 and Grant DSL015), the Spanish Carlos III Institute (Grants PI17/01518 and PI18/01332) and the crowdfunding initiatives #joemcorono, BonPreu/Esclat and Correos (Spain). E.Pradenas was supported by a doctoral grant from ANID, Chile: Grant 72180406.
References
- 1.Science in the time of coronavirus. Nat. Methods. 2020;17:355. doi: 10.1038/s41592-020-0807-y. [DOI] [PubMed] [Google Scholar]
- 2.Thorlund K., Dron L., Park J., et al. A real-time dashboard of clinical trials for COVID-19. Lancet. Digit. Heal. 2020;2:e286–e287. doi: 10.1016/S2589-7500(20)30086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020 doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 4.Vabret N., Britton G.J., Gruber C., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020 doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walls A.C., Park Y.-J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W., Moore M.J., Vasllieva N., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003 doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zang R., Gomez Castro M.F., McCune B.T., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons G., Gosalia D.N., Rennekamp A.J., et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiao J., Li W., Bao J., et al. The expression of SARS-CoV-2 receptor ACE2 and CD147, and protease TMPRSS2 in human and mouse brain cells and mouse brain tissues. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanley B., Naresh K.N., Roufosse C., et al. Histopathological findings and viral tropism in UK patients with severe fatal COVID-19: a post-mortem study. The Lancet. Microbe. 2020 doi: 10.1016/S2666-5247(20)30115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J., Yuan Q., Wang H., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lou B., Li T.-D., Zheng S.-F., et al. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hueston L., Kok J., Guibone A., et al. The antibody response to SARS-CoV-2 infection. Open Forum Infect. Dis. 2020;7 doi: 10.1093/ofid/ofaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H., Sun B., Fang Z., et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 2020;56:2001526. doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellam P., Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. 2020;101:791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long Q.-X., Liu B.-Z., Deng H.-J., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 19.Ceron J., Lamy E., Martinez-Subiela S., et al. Use of saliva for diagnosis and monitoring the SARS-CoV-2: a general perspective. J. Clin. Med. 2020 doi: 10.3390/jcm9051491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suthar M.S., Zimmerman M.G., Kauffman R.C., et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Reports Med. 2020;1:100040. doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H., Zeng W., He H., et al. Serum IgA, IgM, and IgG responses in COVID-19, cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu W.T., Howell J.C., Ozturk T., et al. Antibody profiles according to mild or severe SARS-CoV-2 infection, atlanta, Georgia, USA. Emerg. Infect. Dis. 2020;(2020) doi: 10.3201/eid2612.203334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Premkumar L., Segovia-Chumbez B., Jadi R., et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc8413. eabc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chia W.N., Tan C.W., Foo R., et al. Serological differentiation between COVID-19 and SARS infections, Emerg. Microb. Infect. 2020;9:1497–1505. doi: 10.1080/22221751.2020.1780951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plotkin S.A. Vaccines: correlates of vaccine-induced immunity. Clin. Infect. Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 26.Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amanna I.J., Messaoudi I., Slifka M.K. Protective immunity following vaccination: how is it defined? Hum. Vaccine. 2008;4:316–319. doi: 10.4161/hv.4.4.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi X., Yan R., Zhang J., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 84. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brouwer P.J.M., Caniels T.G., van der Straten K., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;80 doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tortorici M.A., Beltramello M., Lempp F.A., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;80(3354):29–31. doi: 10.1029/2002GC000367. ISSN":"15252027","author":[{"family":"Hodell","give. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ju B., Zhang Q., Ge J., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 32.Liu L., Wang P., Nair M.S., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020 doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 33.Ou X., Liu Y., Lei X., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Guo X., Xin Q., et al. Neutralizing antibody responses to severe acute respiratory syndrome coronavirus 2 in coronavirus disease 2019 inpatients and convalescent patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu F., Liu M., Wang A., et al. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in shanghai, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K., Long Q.-X., Deng H.-J., et al. Longitudinal dynamics of the neutralizing antibody response to SARS-CoV-2 infection. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu F., Wang A., Liu M., et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. SSRN Electron. J. 2020:2020. doi: 10.2139/ssrn.3566211. 03.30.20047365. [DOI] [Google Scholar]
- 38.Legros V., Denolly S., Vogrig M., et al. A longitudinal study of SARS-CoV-2 infected patients shows high correlation. MedRxiv. 2020:2020. doi: 10.1101/2020.08.27.20182493. 08.27.20182493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassan A.O., Case J.B., Winkler E.S., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alsoussi W.B., Turner J.S., Case J.B., et al. A potently neutralizing antibody protects mice against SARS-CoV-2 infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers T.F., Zhao F., Huang D., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;(80-):963. doi: 10.1126/science.abc7520. eabc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Y., Su B., Guo X., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi R., Shan C., Duan X., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 44.Sadoff J., Le Gars M., Shukarev G., et al. Safety and immunogenicity of the Ad26.COV2.S COVID-19 vaccine candidate: interim results of a phase 1/2a, double-blind, randomized, placebo-controlled trial. MedRxiv. 2020:2020. doi: 10.1101/2020.09.23.20199604. 09.23.20199604. [DOI] [Google Scholar]
- 45.Folegatti P.M., Ewer K.J., Aley P.K., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson L.A., Anderson E.J., Rouphael N.G., et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/nejmoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y.-J., Zeng G., Pan H.-X., et al. MedRxiv; 2020. Immunogenicity and Safety of a SARS-CoV-2 Inactivated Vaccine in Healthy Adults Aged 18-59 Years: Report of the Randomized, Double-Blind, and Placebo-Controlled Phase 2 Clinical Trial. [Google Scholar]
- 48.Zhu F.C., Guan X.H., Li Y.H., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dagotto G., Yu J., Barouch D.H. Approaches and challenges in SARS-CoV-2 vaccine development. Cell Host Microbe. 2020;28:364–370. doi: 10.1016/j.chom.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia X., Li K., Wu L., et al. Improved clinical symptoms and mortality among patients with severe or critical COVID-19 after convalescent plasma transfusion. Blood. 2020;136:755–759. doi: 10.1182/BLOOD.2020007079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen C., Wang Z., Zhao F., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hegerova L., Gooley T.A., Sweerus K.A., et al. Use of convalescent plasma in hospitalized patients with COVID-19: case series. Blood. 2020;136:759–762. doi: 10.1182/blood.2020006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baum A., Fulton B.O., Wloga E., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;80–:1018. doi: 10.1126/science.abd0831. eabd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Y., Wang F., Shen C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;(80-):1278. doi: 10.1126/science.abc2241. eabc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baum A., Fulton B.O., Wloga E., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu L.L., Suscovich T.J., Fortune S.M., et al. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katzelnick L.C., Gresh L., Halloran M.E., et al. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358(80):929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Openshaw P.J.M., Tregoning J.S. Immune responses and disease enhancement during respiratory syncytial virus infection. Clin. Microbiol. Rev. 2005;18:541–555. doi: 10.1128/CMR.18.3.541-555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho M.-S., Chen W.-J., Chen H.-Y., et al. Neutralizing antibody response and SARS severity. Emerg. Infect. Dis. 2005;11:1730–1737. doi: 10.3201/eid1111.040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee N., Chan P.K.S., Ip M., et al. Anti-SARS-CoV IgG response in relation to disease severity of severe acute respiratory syndrome. J. Clin. Virol. 2006;35:179–184. doi: 10.1016/j.jcv.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu L., Wei Q., Lin Q., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4 doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q., Zhang L., Kuwahara K., et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect. Dis. 2016;2:361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z.Y., Werner H.C., Kong W.P., et al. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. U. S. A. 2005;102:797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang S., Tseng S., Yen C., et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem. Biophys. Res. Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng W., Bao L., Liu J.J., et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;80–:369. doi: 10.1126/science.abc5343. 818 LP – 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu J., Tostanoski L.H., Peter L., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(80):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zang J., Gu C., Zhou B., et al. Immunization with the receptor-binding domain of SARS-CoV-2 elicits antibodies cross-neutralizing SARS-CoV-2 and SARS-CoV without antibody-dependent enhancement. Cell Discov. 2020;6:4–7. doi: 10.1038/s41421-020-00199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsueh P.R., Huang L.M., Chen P.J., et al. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004 doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He Z., Dong Q., Zhuang H., et al. Kinetics of severe acute respiratory syndrome (SARS) coronavirus-specific antibodies in 271 laboratory-confirmed cases of SARS. Clin. Diagn. Lab. Immunol. 2004 doi: 10.1128/CDLI.11.4.792-794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015 doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edridge A.W.D., Kaczorowska J., Hoste A.C.R., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020 doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 72.Kaneko N., Kuo H.H., Boucau J., et al. 2020. Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19, Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seydoux E., Homad L.J., MacCamy A.J., et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53:98–105. doi: 10.1016/j.immuni.2020.06.001. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Addetia A., Crawford K.H.D., Dingens A., et al. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gupta V., Bhoyar R.C., Jain A., et al. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.To K.K.-W., Hung I.F.-N., Ip J.D., et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wan Y., Shang J., Sun S., et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020;94 doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grzelak L., Temmam S., Planchais C., et al. SARS-CoV-2 serological analysis of COVID-19 hospitalized patients, pauci-symptomatic individuals and blood donors. MedRxiv. 2020:2020. doi: 10.1101/2020.04.21.20068858. 04.21.20068858. [DOI] [Google Scholar]
- 79.Chen X., Pan Z., Yue S., et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19, Signal Transduct. Target. Ther. 2020;5:180. doi: 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sekine T., Perez-Potti A., Rivera-Ballesteros O., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]