Abstract
Background
Circ_0072995 is a novel identified circRNA and has been identified to involve in the metastasis of breast cancer. However, the detailed function and mechanism of circ_0072995 in the biological property of breast cancer cell remain vague.
Materials and Methods
The expression of circ_0072995, microRNA (miR)-149-5p and serine hydroxymethyltransferase 2 (SHMT2) mRNA was detected using quantitative real-time polymerase chain reaction. Western blot was used to detect the protein levels of SHMT2, hexokinase-2 (HK-2), lactate dehydrogenase a chain (LDHA), and glucose transporter 1 (GLUT1). Cell proliferation, apoptosis, migration, and invasion were analyzed using cell counting kit-8 assay, flow cytometry, caspase-3 activity analysis, cell adhesion assay and transwell assay, respectively. Glucose metabolism was calculated by measuring glucose uptake, lactate production, and adenosine triphosphate (ATP) levels. The interaction between miR-149-5p and circ_0072995 or SHMT2 was confirmed by dual-luciferase reporter assay. In vivo tumorigenesis was performed using the murine xenograft model.
Results
Circ_0072995 and SHMT2 were up-regulated in breast cancer tissues and cell lines, and knockdown of circ_0072995 or SHMT2 suppressed cell malignant properties and anaerobic glycolysis; importantly, SHMT2 overexpression attenuated the anticancer action of circ_0072995 knockdown in breast cancer. Besides, we also found circ_0072995 directly targeted miR-149-5p, thereby regulating its downstream gene SHMT2 by competitively binding to miR-149-5p. Additionally, xenograft analysis showed circ_0072995 silencing suppressed tumor growth via regulating SHMT2 and miR-149-5p in vivo.
Conclusion
This study demonstrated that circ_0072995 promoted cell malignant phenotypes and anaerobic glycolysis in breast cancer via up-regulating SHMT2 through sponging miR-149-5p, indicating a promising molecular target for breast cancer treatment.
Keywords: circ_0072995, miR-149-5p, SHMT2, breast cancer
Introduction
On a global scale, breast cancer is the most frequently diagnosed female malignancy, which seriously threatens the health of women in the world.1 As the first leading cause among female malignancy, the overall occurrence and mortality of patients with breast cancer have not significantly reduced despite the great improvements in diagnosis and therapeutic technology.2,3 Hence, it is paramount to investigate the mechanism underlying the progression of breast cancer for managing the clinical treatments.
Circular RNAs (circRNAs) are a special subclass of endogenous noncoding RNA molecules with covalently closed-loop structures, which make them resistant to exonuclease degradation relative to their linear RNAs decay.4 Previous studies have reported that circRNAs are important modulators in multiple cancer types via affecting various cellular processes, including cell proliferation, invasion, migration, drug resistance and metabolism,5–8 thus involving in the progression and development of cancers. In breast cancer, multiple functions of circRNAs have been identified to exert various influences on the tumorigenesis of cancer cells. For example, hsa_circ_0001982 interacted with miR-143 to promote tumor cell proliferation and invasion in breast cancer, thus contributing to the progression of cancer.9 CircRNA hsa_circ_0052112 induced breast cancer cell migration and invasion by targeting miR-125a-5p.10 CircRNA hsa_circRNA_002178 promoted cell viability, tube formation and energy metabolism via up-regulating COL1A1 through miR-328-3p.11 circ_0072995 is a novel identified circRNA, a recent study found circ_0072995 was elevated in breast cancer cells and promoted cell metastasis by binding to miR-30c-2-3p in vitro.12 Nevertheless, the detailed function and mechanism of circ_0072995 in breast cancer remain vague.
Serine hydroxymethyltransferase 2 (SHMT2), a mitochondrial SHMT, can reversibly and simultaneously catalyze the conversions of L-serine to glycine, playing significant roles in cellular one-carbon pathways.13,14 An adequate supply of glycine is strongly responsible for rapidly proliferating cancer cells, and glycine consumption and catabolism contribute to the carcinogenesis and malignancy.15,16 Silencing of SHMT2 is shown to be a key target for therapeutic intervention of cancer. However, little work has shown the role of SHMT2 in cell tumorigenesis in breast cancer.
In this study, we aimed to investigate the function and molecular mechanisms of circ_0072995 and SHMT2 in the malignant biological behaviors and glucose metabolism of breast cancer cells, explored whether SHMT2 involved in the action of circ_0072995 in the progression of breast cancer.
Materials and Methods
Clinical Specimens
Tumor tissues and matched non-tumor tissue (normal) samples from 70 breast cancer patients who underwent surgical resection at The Central Hospital of Enshi Tujia and Miao Autonomous Prefecture were collected, and then immediately frozen in liquid nitrogen at −80°C until subsequent analysis. All patients were diagnosed by histopathological assessment and none of them received preoperative treatment. The clinicopathological features of breast cancer patients are displayed in Table 1. We had obtained the approval of the Ethics Committee of The Central Hospital of Enshi Tujia and Miao Autonomous Prefecture and all subjects had provided written informed consents. The RNA-seq data of 1104 breast cancer and 113 adjacent non-tumor (normal) tissues and corresponding clinical data were obtained from the starBase database.
Table 1.
The Clinicopathological Features for Breast Cancer Patients
| Clinicopathological Features | Number of Cases |
|---|---|
| Age (years) | |
| <50 | 47 |
| ≥50 | 23 |
| Breast cancer type | |
| Invasive ductal carcinoma | 49 |
| Mucinous adenocarcinoma | 21 |
| TNM stages | |
| I and II | 34 |
| III | 36 |
| Distant metastasis | |
| Yes | 26 |
| No | 44 |
| ER status | |
| Positive | 39 |
| Negative | 31 |
| PR status | |
| Positive | 61 |
| Negative | 9 |
| HER2 status | |
| Positive | 22 |
| Negative | 48 |
Abbreviations: TNM, tumor-node-metastasis; ER, estrogen receptor; PR, progesterone receptor; Her2, human epidermal growth factor receptor type 2.
Cell Culture and Transfection
Human breast cancer cell lines (SKBR3, HCC38, MCF-7, MDA-MB-231 and BT-549) and MCF-10A nonmalignant breast epithelial cells were purchased from Shanghai Academy of Life Science (Shanghai, China). All cells were grown in the Dulbecco’s modified Eagle’s medium (DMEM) harboring with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco, Carlsbad, CA, USA) with 5% CO2 at 37°C.
The miR-149-5p mimic (miR-149-5p), mimic control (miR-NC), small interfering RNA (siRNA) targeting circ_0072995 (si-circ#1, si-circ#2, si-circ#3), siRNA targeting SHMT2 (si-SHMT2#1, si-SHMT2#2, si-SHMT2#3), siRNA negative control (si-NC), pcDNA3.1 circ_0072995 overexpression vector (circ_0072995), negative plasmid (pcDNA), pcDNA3.1 SHMT2 overexpression vector (SHMT2), empty vector (vector), lentiviral particles stably expressing either short hairpin RNA (shRNA)-targeting circ_0072995 (sh-circ) or a scrambled control sequence (sh-NC) were designed and synthesized by Invitrogen (Carlsbad, CA, USA). Then, the transfection of oligonucleotides or vectors was performed using Lipofectamine 2000 (Invitrogen).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Whole-RNA extracts from tissues and cells were conducted by using TRIzol reagent (Invitrogen). Then, extracted RNA was reversely transcribed into complementary DNA (cDNA) using the Prime Script RT Master Mix (Takara, Dalian, China), and the synthesized cDNA template was detected using SYBR Premix Ex Taq (Takara). After that, the expression levels were measured by the 2−ΔΔCt method with glyceraldehyde 3-phosphate dehydrogenase (GADPH) or U6 small nuclear B noncoding RNA (U6) using as an internal reference. The same experiment was repeated three times, and the average was taken. The primer sequences are listed in Table 2.
Table 2.
The Sequences of Primers Used in This Study
| Forward (5ʹ-3ʹ) | Reverse (5ʹ-3ʹ) | Amplicon Size | |
|---|---|---|---|
| circ_0072995 | |||
| Divergent | CAGGCCAGAGGAAAGAACAG | CAGGCCCCATCTCATAGCCA | 102 |
| Convergent | GATGGGGCCTGGCTAAACTT | AGCTTGGAGTGTCCTTCACG | 127 |
| SHMT2 | CCCTTCTGCAACCTCACGAC | TGAGCTTATAGGGCATAGACTCG | 135 |
| miR-149-5p | CTGGCTCCGTGTCTTCACTC | CAGCTGCCCCAGCACAG | 72 |
| GAPDH | ACAACTTTGGTATCGTGGAAGG | GCCATCACGCCACAGTTTC | 101 |
| U6 | GCAAAACGCACCACGTGAC | GGAAATAGGCCCTCTTCCTGC | 72 |
Actinomycin D Assay
BT-549 cells were treated with 2 μg/mL Actinomycin D (Sigma-Aldrich, St. Louis, MO, USA) to block new RNA synthesis for 0, 4, 8, 12, and 16 h, and then, total RNA was extracted and qRT-PCR was carried out.
Western Blot
Total proteins were isolated using RIPA buffer (Beyotime, Shanghai, China). Subsequently, proteins were separated by the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto the polyvinylidene difluoride (PVDF) membranes. After blocking with 5% non-fat milk for 2 h, the membranes were incubated with the primary antibodies against SHMT2 (1:1000, ab180786, Abcam, Cambridge, MA, USA), hexokinase-2 (HK-2) (1:5000, ab104836, Abcam), lactate dehydrogenase A chain (LDHA) (1:3000, ab135366, Abcam), glucose transporter 1 (GLUT1) (1:10,000, ab40084, Abcam), and then incubated with the secondary HRP-conjugated antibody (1:1000, ab9482, Abcam). Protein bands were visualized using ECL detection reagent and normalized by β-actin (1:1000, 4967, Cell Signaling Technology, Inc., Beverly, MA, USA). Triplicate individual experiments were performed in this study.
Cell Proliferation Assay
Transfected SKBR3 and BT-549 cells were seeded in 96-well cell culture plates for 24 h, then per well was incubated with 10 μL cell counting kit-8 (CCK-8) solution (Beyotime, Shanghai, China) for about 2 h. Subsequently, the optical density was measured by a spectrophotometric microplate reader at 450 nm. The results represent as the average of three independent replicates.
Cell Apoptosis Analysis
Cell apoptosis was analyzed by flow cytometry with Annexin V-FITC apoptosis detection kit (BD Biosciences, San Jose, CA, USA). Transfected SKBR3 and BT-549 cells were collected and resuspended in binding buffer, followed by interaction with Annexin V-FITC (10 μL) and propidium iodide (PI) (10 μL) for 15 min. The apoptosis rate of cells was analyzed by flow cytometer. The experiment was repeated three times.
Measurement of Caspase-3 Activity
The caspase-3 activity was detected by a colorimetric assay kit (Sigma, St Louis, MO, USA) in SKBR3 and BT-549 cells following the standard procedure. Finally, the absorbance was measured by a microplate reader at 450 nm. Experiments were performed three times.
Cell Adhesion Assay
Transfected SKBR3 and BT-549 (1×105) were seeded into the 96-well culture plates pre-coated with 1:3 diluted Matrigel (Corning, NY, USA) for 30 min. After discarding the corresponding medium with non-adhered cells, cells were gently washed with PBS for two times to remove loosely adhered cells. After that, adhered cells were stained with 0.1% crystal violet, and then fixed with methanol for 30 minutes. Finally, adherent cells in six random fields were counted by a microscope at 100× magnification. Experiments were performed three times.
Cell Migration and Invasion Assay
Transwell chambers were used to detect cell migration and invasion. For invasion assay, the transwell chamber was pre-coated with Matrigel, while cell migration analysis did not need Matrigel. Then, SKBR3 and BT-549 cells transfected with the assigned vector were seeded in the upper chambers with 200 μL serum-free DMEM, and 600 μL of medium fixed with FBS was added to the lower chambers. Twenty-four hours later, cells on the lower face of the membranes were fixed and stained, and counted using an inverted light microscope in five random fields. The results represent as the average of three independent replicates.
Glycolysis Analysis
After transfection, SKBR3 and BT-549 cells were placed in a 6-well plate, then supernatants of cell culture media were collected to analyze the levels of glucose and lactate by using a glucose or lactate assay kit (Sigma). The relative glucose uptake and lactate production were assessed based on the standard curve and normalized by the percentage of the control group. All experiments were repeated three times independently.
Levels detection of ATP in transfected SKBR3 and BT-549 cells were performed using an ATP Assay Kit (Sigma). Briefly, transfected cells were lysed, and then ATP reaction mix was added into the lysate and incubated for 30 min. Subsequently, the absorbance at 570 nm was detected by a microplate reader. All experiments were repeated three times independently.
Dual-Luciferase Reporter Assay
The circ_0072995 or SHMT2 3ʹUTR sequences with wild type (WT) or mutant (MUT) binding sites of miR-149-5p were amplified and cloned into the downstream region of firefly luciferase cassette in pGL3 vector (Promega, Shanghai, China). SKBR3 and BT-549 cells infected with miR-149-5p mimic or miR-NC, and then co-transfected with 50 ng constructed pGL3-UTR and 10 ng pRL-TK by using Lipofectamine 2000 (Invitrogen). Forty-eight hours later, firefly luciferase activities were examined using a Dual-Luciferase reporter assay kit (Promega) and normalized to Renilla luciferase activities. Each group was run in triplicate in 24-well plates.
Xenograft Experiments in vivo
Female BALB/c nude mice (5 weeks old, N=6) were used to establish xenograft models. We have obtained the approval of the Animal Research Committee of The Central Hospital of Enshi Tujia and Miao Autonomous Prefecture and performed this experiment in line with the guidelines of the National Animal Care and Ethics Institution. Six mice were divided into 2 groups for stable injection with BT-549 cells that were transfected with sh-NC or sh-circ. Tumor volume was recorded every week. At day 28, mice were killed and tumor masses were weighed and harvested for further molecular analysis.
Statistical Analysis
All analyses were conducted on GraphPad Prism 7 software. Data from at least three experiments were expressed in terms of means ± standard deviation (SD). Statistical difference between different groups was analyzed by Student’s t-test or one-way analysis of variance (ANOVA), and a P-value less than 0.05 was considered statistically significant.
Results
Identification of Circ_0072995 in Breast Cancer Cells
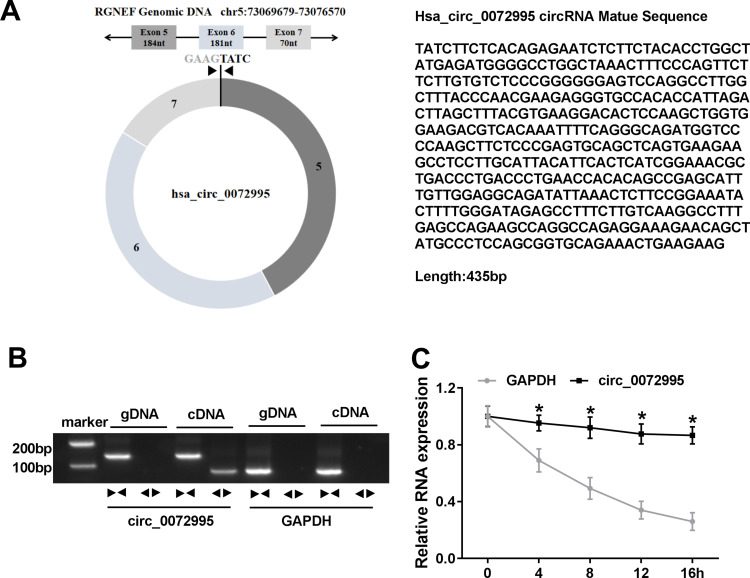
First, the characterization of circ_0072995 was investigated. Circ_0072995, arose from the RGNEF gene, is located on chromosome 5, the genomic length is 435 base pairs (bp), and comprised exons 5, 6 and 7 of its parental gene (Figure 1A). To confirm the formation of circ_0072995, convergent primers RGNEF mRNA and divergent primers to circ_0072995 were designed using cDNA and gDNA as templates, and results showed circ_0072995 could be amplified only in cDNA, and no products were detected in the extracted gDNA (Figure 1B), indicating that circ_0072995 could be formatted. Additionally, the stability of circ_0072995 was validated adopting Actinomycin D, an inhibitor of transcription, and the results showed that circ_0072995 had a longer half-life compared to linear GAPDH in BT-549 cells, revealing that circ_0072995 indeed circular (Figure 1C).
Figure 1.
Identification of circ_0072995 in breast cancer cells. (A) Schematic diagram of the genomic location and formation of circ_0072995 (435 bp). (B) The existence validation of circ_0072995 in BT-549 cells using RT-PCR. (C) qRT-PCR analysis of the abundances of circ_0072995 and GAPDH mRNA in BT-549 cells at the indicated time points after treatment with Actinomycin D. *P<0.05.
The Expression of Circ_0072995 and SHMT2 is Elevated in Breast Cancer
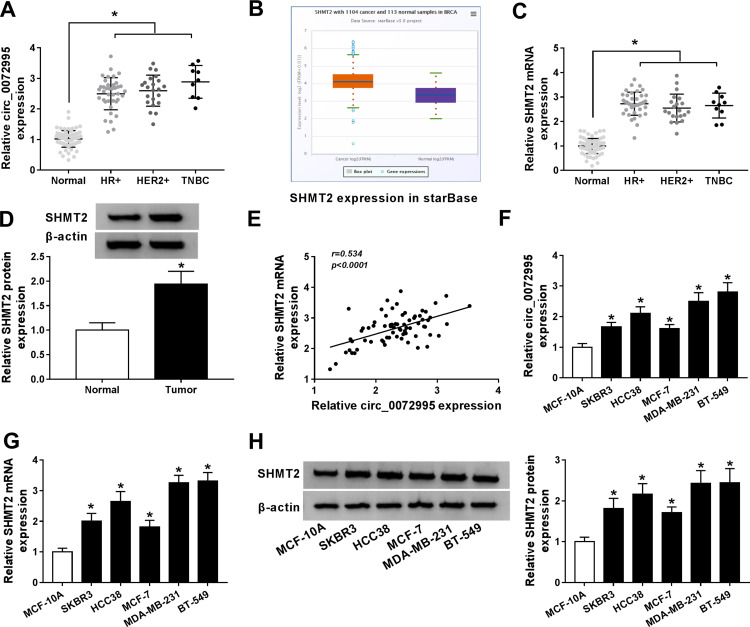
Next, the expression profile of circ_0072995 in breast cancer was investigated. Results showed that circ_0072995 expression was higher in breast cancer tissues (HR +, HER2+ and triple negative) than that in normal tissues (Figure 2A). To ensure whether SHMT2 was co-overexpressed with circ_0072995, analysis of RNA-seq data of 1104 breast cancer tissues and 113 adjacent normal tissues obtained from starBase suggested that SHMT2 was highly expressed in breast cancer tissues compared with normal tissues (Figure 2B). Furthermore, SHMT2 expression was determined in 70 pairs of clinical breast cancer samples, and qRT-PCR analysis also showed an up-regulation of SHMT2 in breast cancer tissues (Figure 2C and D). Importantly, a positive correlation between circ_0072995 and SHMT2 mRNA expression in breast cancer tissues was observed (Figure 2E). Similarly, levels of circ_0072995 and SHMT2were also elevated in breast cancer cells (SKBR3, HCC38, MCF-7, MDA-MB-231 and BT-549) compared with MCF-10A nonmalignant breast epithelial cells (Figure 2F–H). These results indicated that circ_0072995 and SHMT2 elevation might be associated with the initiation and development of breast cancer.
Figure 2.
The expression of circ_0072995 and SHMT2 is elevated in breast cancer. (A) qRT-PCR of circ_0072995 expression in breast cancer tissues and matched normal tissues. (B) The SHMT2 expression in starBase breast cancer tissues and normal tissues. (C and D) qRT-PCR and Western blot analysis of SHMT2 expression in breast cancer tissues and matched normal tissues. (E) Correlation analysis of circ_0072995 and SHMT2 mRNA expression in breast cancer tissues using Pearson correlation analysis. (F–H) Levels detection of circ_0072995 and SHMT2 in breast cancer cells (SKBR3, HCC38, MCF-7, MDA-MB-231 and BT-549) and MCF-10A nonmalignant breast epithelial cells using qRT-PCR and Western blot. *P<0.05.
Circ_0072995 Knockdown Inhibits Cell Malignant Phenotypes and Anaerobic Glycolysis in Breast Cancer
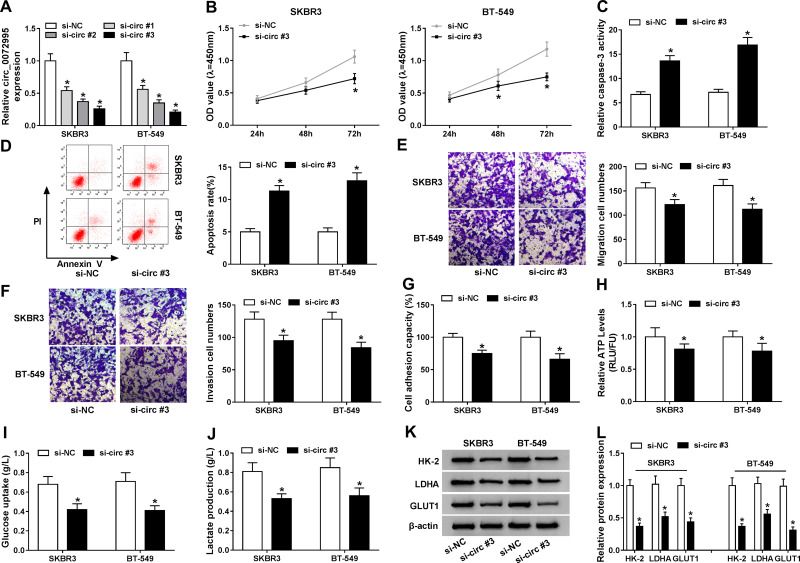
The detailed functions of circ_0072995 in the progression of breast cancer were investigated. Circ_0072995 was knocked down by transfecting constructed si-circ_0072995 plasmid into SKBR3 and BT-549 cells. Subsequently, we found si-circ_0072995 transfection significantly reduced the expression of circ_0072995 relative to the si-NC group in SKBR3 and BT-549 cells (Figure 3A), and si-circ#3 was selected for subsequent experiments because it was the most effective siRNA. By contrast with si-NC group, circ_0072995 knockdown inhibited SKBR3 and BT-549 cell proliferation (Figure 3B), but induced SKBR3 and BT-549 cell apoptosis, evidenced by the increase of caspase-3 activity and apoptotic cells (Figure 3C and D). Besides, transwell migration and invasion assays demonstrated that circ_0072995 down-regulation significantly inhibited both the cell migratory and invasive capabilities of SKBR3 and BT-549 cells (Figure 3E and F), and cell adhesion assay showed that circ_0072995 knockdown attenuated cell adhesion ability (Figure 3G). Additionally, glucose metabolism was calculated, results showed circ_0072995 silencing decreased ATP levels (Figure 3H), glucose uptake (Figure 3I), and lactate production (Figure 3J) in SKBR3 and BT-549 cells. Western blot analysis indicating levels of HK-2, LDHA, GLUT1 also were down-regulated by circ_0072995 knockdown in SKBR3 and BT-549 cells (Figure 3K and L). Thus, circ_0072995 silence inhibited cell anaerobic glycolysis in breast cancer. Afterwards, the effects of circ_0072995 on non-tumor cells was detected. The transfection of si-circ_0072995 significantly reduced the expression of circ_0072995 in MCF-10A cells (Figure S1A), then we found that circ_0072995 knockdown had no statistically significant effect on the growth (Figure S1B–D), migration, invasion (Figure S1E–G) and anaerobic glycolysis (Figure S1H–K) of MCF-10A cells. Taken together, circ_0072995 knockdown suppressed breast cancer progression.
Figure 3.
Circ_0072995 knockdown inhibits cell malignant phenotypes and anaerobic glycolysis in breast cancer. SKBR3 and BT-549 cells were transfected with si-circ_0072995 (si-circ#1, si-circ#2, and si-circ#3) or si-NC. (A) qRT-PCR analysis of circ_0072995 expression in SKBR3 and BT-549 cells after transfection. (B) The proliferation analysis of SKBR3 and BT-549 cells using CCK-8 assay. (C) Detection of caspase-3 activity in SKBR3 and BT-549 cells using a colorimetric assay kit. (D) Apoptosis analysis of SKBR3 and BT-549 cells using flow cytometry. (E and F) Analysis of SKBR3 and BT-549 cell migration and invasion with transwell assay (G) The analysis of cell adhesion ability in SKBR3 and BT-549 cells using cell adhesion assay. (H–J) The ATP levels, glucose uptake, and lactate production analyses in SKBR3 and BT-549 using an ATP, glucose, or lactate assay kit, respectively. (K and L) Levels detection of HK-2, LDHA, and GLUT1 protein using Western blot in SKBR3 and BT-549 cells. *P<0.05.
SHMT2 Knockdown Inhibits Cell Malignant Phenotypes and Anaerobic Glycolysis in Breast Cancer
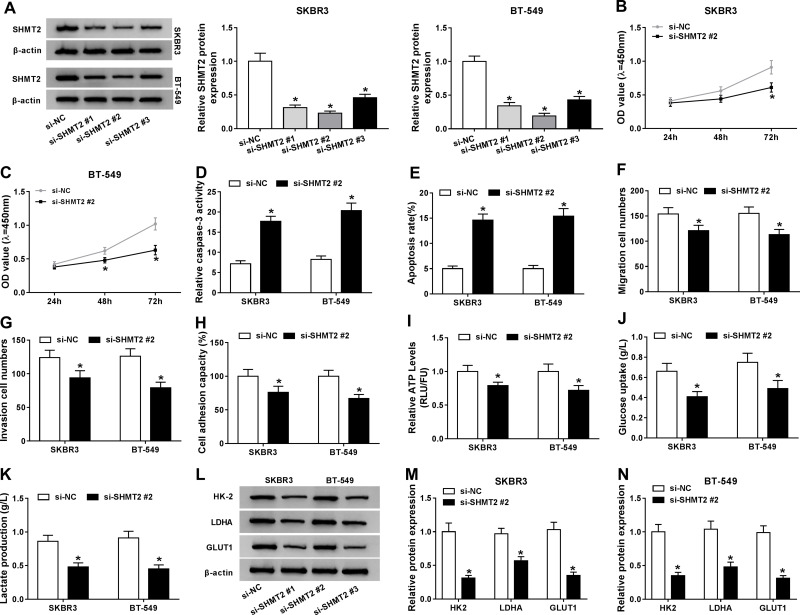
The function of SHMT2 in the progression of breast cancer was then explored. Three of si-SHMT2 were designed and used to knock down the expression of SHMT2 in SKBR3 and BT-549 cells. Western blot analysis showed si-SHMT2 significantly decreased the level of SHMT2 relative to the si-NC group in SKBR3 and BT-549 cells (Figure 4A), and si-SHMT2 #2 was selected for subsequent analyses due to its best interference rate. Then functional experiments were conducted. Results displayed that SHMT2 knockdown mediated proliferation inhibition (Figure 4B and C), apoptosis promotion (Figure 4D and E), and migration, invasion (Figure 4F and G) and adhesion (Figure 4H) abilities suppression in SKBR3 and BT-549 cells. Besides, we also found SHMT2 silencing repressed anaerobic glycolysis in SKBR3 and BT-549 cells, reflected by the decrease of ATP levels (Figure 4I), glucose uptake (Figure 4J), lactate production (Figure 4K), as well as reduction of HK-2, LDHA, GLUT1 levels (Figure 4L–N) in SKBR3 and BT-549 cells. These data suggested that SHMT2 knockdown also impeded the development of breast cancer.
Figure 4.
SHMT2knockdown inhibits cell malignant phenotypes and anaerobic glycolysis in breast cancer. SKBR3 and BT-549 cells were transfected with si-SHMT2 (si-SHMT2#1, si-SHMT2 #2, and si-SHMT2 #3) or si-NC.(A) Western blot analysis of SHMT2 expression in SKBR3 and BT-549 cells after transfection. (B and C) CCK-8 assay analysis of SKBR3 and BT-549 cell proliferation. (D) Caspase-3 activity analysis of SKBR3 and BT-549 cells using a colorimetric assay kit. (E) Apoptosis analysis of SKBR3 and BT-549 cells using flow cytometry. (F and G) The analysis of migratory and invasive capabilities of SKBR3 and BT-549 cells with transwell assay. (H) The analysis of cell adhesion ability in SKBR3 and BT-549 cells using cell adhesion assay. (I–K) Detection of ATP levels, glucose uptake, and lactate production in SKBR3 and BT-549 using an ATP, glucose, or lactate assay kit, respectively. (L–N) Levels detection of HK-2, LDHA, and GLUT1 protein using Western blot in SKBR3 and BT-549 cells. *P<0.05.
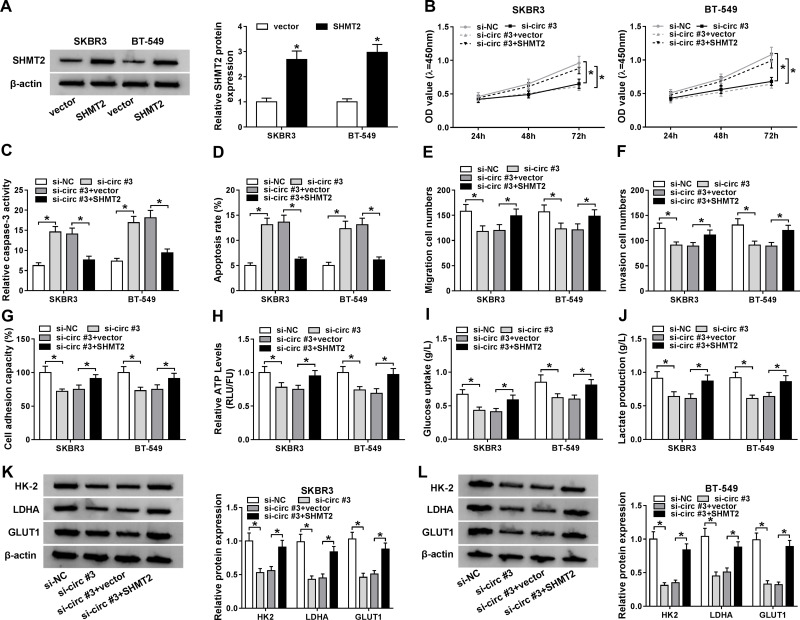
SHMT2 Overexpression Reverses the Anticancer Functions of si-circ_0072995 in Breast Cancer Cells
Given the positive correlation between circ_0072995 and SHMT2 expression in breast cancer, whether si-circ_0072995 exerted anticancer effects via SHMT2 was investigated. First, SKBR3 and BT-549 cells were transfected with SHMT2 or vector, and Western blot analysis showed SHMT2 transfection significantly elevated SHMT2 levels compared with vector transfection (Figure 5A). Next, SKBR3 and BT-549 cells were transfected with si-NC, si-circ#3, si-circ#3 + vector, or si-circ#3 + SHMT2 to perform rescue assay. We found SHMT2 overexpression attenuated si-circ_0072995-induced proliferation inhibition (Figure 5B), apoptosis promotion (Figure 5C and D), and suppression of migration (Figure 5E), invasion (Figure 5F) and adhesion (Figure 5G) abilities in SKBR3 and BT-549 cells. Moreover, the suppression of anaerobic glycolysis in SKBR3 and BT-549 cells induced by circ_0072995 knockdown also was rescued by SHMT2 re-expression (Figure 5H–L). Altogether, circ_0072995 contributed to the progression of breast cancer by regulating SHMT2.
Figure 5.
SHMT2 overexpression reverses the anticancer functions of si-circ_0072995 in breast cancer cells. (A) Western blot analysis of SHMT2 protein expression in SKBR3 and BT-549 cells transfected with SHMT2 or vector. SKBR3 and BT-549 cells were transfected with si-NC, si-circ#3, si-circ#3 + vector, or si-circ#3 + SHMT2. (B) CCK-8 assay analysis of SKBR3 and BT-549 cell proliferation. (C) Caspase-3 activity analysis of SKBR3 and BT-549 cells using a colorimetric assay kit. (D) The analysis of SKBR3 and BT-549 cell apoptosis using flow cytometry. (E and F) The analysis of the migratory and invasive capabilities of SKBR3 and BT-549 cells using transwell assay. (G) Cell adhesion ability analysis in SKBR3 and BT-549 cells using cell adhesion assay. (H–J) Detection of ATP levels, glucose uptake, and lactate production in SKBR3 and BT-549 using an ATP, glucose, or lactate assay kit, respectively. (K and L) Western blot analysis of HK-2, LDHA, and GLUT1 protein in SKBR3 and BT-549 cells. *P<0.05.
MiR-149-5p Was a Target of Circ_0072995 and Directly Target SHMT2 mRNA in Breast Cancer Cell
It has been reported that circRNAs can act as competitive endogenous RNAs (ceRNAs) for miRNAs, thus regulating the depression of miRNA targets.17 Therefore, we hypothesized that circ_0072995 knockdown-mediated anticancer function might operate via a ceRNA mechanism. Hence, six online databases (circBase, circInteractome, TargetScan, starbase, miRwalk and miRDB) were used to search for the potential target miRNA binding partners of circ_0072995 and SHMT2 mRNA, among which miR-149-5p was selected from the overlap between the databases (Figure 6A). The sequences of circ_0072995 or SHMT2 3ʹUTR containing binding sites of miR-149-5p are shown in Figure 6B. Subsequently, the reduction of luciferase activity in SKBR3 and BT-549 cells co-transfected with WT-circ_0072995 and miR-149-5p, or SHMT2 3ʹUTR-WT and miR-149-5p confirmed the interaction between miR-149-5p and circ_0072995 or SHMT2 (Figure 6C and D). Besides, we also found circ_0072995 knockdown enhanced miR-149-5p expression in SKBR3 and BT-549 cells (Figure 6E); moreover, after elevating the expression of miR-149-5p by transfecting with miR-149-5p mimic (Figure 6F), it was also discovered that miR-149-5p overexpression reduced the expression of SHMT2 in SKBR3 and BT-549 cells (Figure 6G). In all, these results suggested that circ_0072995 was a sponge of miR-149-5p and negatively regulated its expression; besides, miR-149-5p targetedly suppressed SHMT2 expression.
Figure 6.
MiR-149-5p directly binds to circ_0072995 or SHMT2 in breast cancer cells. (A) Schematic illustration exhibiting overlapping of the target miRNAs of circ_0072995 and SHMT2 predicted by online databases (circBase, circInteractome, TargetScan, starbase, miRwalk and miRDB), (B) The predicted binding sites of miR-149-5p on circ_0072995 or SHMT2 sequences. (C and D) Interaction analysis between miR-149-5p and circ_0072995 or SHMT2 in SKBR3 and BT-549 cells using the dual-luciferase reporter assay. (E) qRT-PCR analysis of miR-149-5p expression in SKBR3 and BT-549 cells transfected with si-NC or si-circ#3. (F) qRT-PCR analysis of miR-149-5p expression in SKBR3 and BT-549 cells transfected with miR-149-5p or miR-NC. (G) Western blot analysis of SHMT2 protein expression in SKBR3 and BT-549 cells transfected with miR-149-5p or miR-NC. *P<0.05.
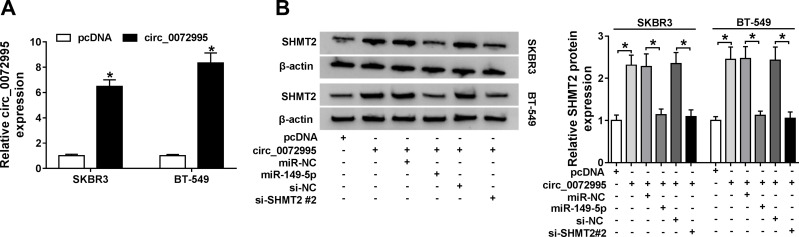
Circ_0072995 Regulates SHMT2 Expression via miR-149-5p
Given that circ_0072995 bound to miR-149-5p, and SHMT2 was a target of miR-149-5p, we then detected whether circ_0072995 regulated SHMT2 via miR-149-5p in breast cancer. First, SKBR3 and BT-549 cells were transfected with pcDNA or circ_0072995, and qRT-PCR analysis showed that circ_0072995 expression was obviously increased (Figure 7A). Next, SKBR3 and BT-549 cells were transfected with pcDNA, circ_0072995, circ_0072995 + miR-NC, circ_0072995 + miR-149-5p, circ_0072995 + si-NC, or circ_0072995 + si-SHMT2 #2 to perform rescue assay. Results indicated that the up-regulation of SHMT2 expression induced by circ_0072995 was abated by miR-149-5p increase or SHMT2 decrease in SKBR3 and BT-549 cells (Figure 7B). These data suggest that circ_0072995 promoted the expression of SHMT2 by competitively binding with miR-149-5p.
Figure 7.
Circ_0072995 regulates SHMT2 expression via miR-149-5p. (A) qRT-PCR analysis of circ_0072995 in SKBR3 and BT-549 cells transfected with pcDNA or circ_0072995. (B) Western blot analysis of SHMT2 in SKBR3 and BT-549 cells transfected with pcDNA, circ_0072995, circ_0072995 + miR-NC, circ_0072995 + miR-149-5p, circ_0072995 + si-NC, or circ_0072995 + si-SHMT2 #2. *P<0.05.
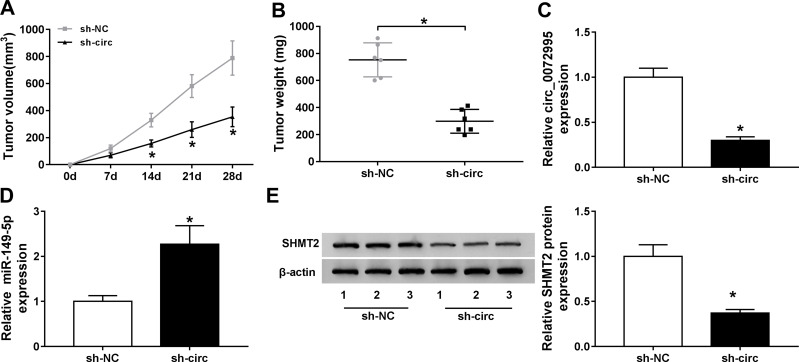
Circ_0072995 Impedes the Growth of Xenograft Tumors in vivo
Following on from the above findings, we further explored whether the miR-149-5p/SHMT2 pathway was implicated in the anticancer effects of circ_0072995 knockdown in the murine xenograft model. As shown in Figure 8A and B, circ_0072995 knockdown suppressed the tumor growth in vivo, demonstrated by the smaller of size and weight of xenograft tumors in sh-circ_0072995 groups compared with sh-NC groups. Then, molecular analysis showed the levels of circ_0072995 and SHMT2 were decreased while miR-149-5p was increased in tumors from sh-circ_0072995 groups (Figure 8C–E). Collectively, circ_0072995 knockdown inhibited tumor growth in vivo via miR-149-5p/SHMT2 axis.
Figure 8.
Circ_0072995 impedes the growth of xenograft tumors in vivo. (A and B) Measurement of the size and weight of xenograft tumors. (C and D) qRT-PCR analysis of circ_0072995 and miR-149-5p levels in tumor masses from each group. (E) Western blot analysis of SHMT2 protein levels in tumor masses. *P<0.05.
Discussion
Up to date, there is increasing evidence emerging to reveal that circRNAs have important roles in the progression of cancers. For example, circHIPK3 up-regulatedBDNF expression by competitively binding with miR-107, thus facilitating cell viability and migration in gastric cancer.18 CircMAT2B induced the glycolysis and malignancy in hepatocellular carcinoma via regulating PKM2 through sponging miR-338-3p.19 Therefore, circRNAs are involved in the regulation of cell malignant phenotypes and metabolism, thus affecting the occurrence and development of cancers.
The occurrence of breast cancer is a complex biological process with multiple genes and multiple factors. Recent evidence has identified the involvement of circRNAs in the progression and development of breast cancer.9,20 Circ_0072995 is a novel identified circRNA and has been found to induce cell metastasis in breast cancer in vitro.12 Thus, this study further investigated the detailed function and mechanism of circ_0072995 in breast cancer progression. We found circ_0072995 was elevated in breast cancer tissues and cell lines, and knockdown of circ_0072995 inhibited cell proliferation, migration, and invasion while induced cell apoptosis in vitro; importantly, xenograft tumors analysis showed circ_0072995 knockdown also impeded tumor growth in vivo. Currently, increasing researches have indicated that most cancer cells prefer to take adenosine triphosphate (ATP) through aerobic glycolysis rather than oxidative phosphorylation to meet their metabolic needs for sustained cell proliferation even in the presence of oxygen.21,22 Inhibition of aerobic glycolysis is an effective therapeutic strategy to hinder cancer malignancy.23 Therefore, glucose metabolism was calculated in this study, and we found circ_0072995 silencing decreased glucose uptake, lactate production, ATP levels, enzymes of HK-2 and LDHA, as well as GLUT1, thereby suppressing aerobic glycolysis in breast cancer cells. Taken together, suppression of circ_0072995 could repress breast cancer progression.
Recently, it was found that SHMT2 was highly expressed in breast cancer cells, SHMT2 was valuable independent prognostic markers in breast cancer, which high expression predicated poor overall survival.24 Besides, SHMT2 expression was positively associated with breast cancer grade.25 Li et al discovered that inhibition of SHMT2, the enzyme the mitochondrial serine and one-carbon pathway, suppressed tumor growth and lung metastasis in metastatic breast cancer.26 Thus, SHMT2 may be a potential target for the treatment and drug discovery of breast cancer. In this study, an elevation of SHMT2 expression was also observed in breast cancer tissues and cell lines, then functional experiments exhibited SHMT2 silencing inhibited cell malignant phenotypes and anaerobic glycolysis in breast cancer. Thus, we confirmed that SHMT2 promoted breast cancer progression.
In our work, a positive correlation between SHMT2 and circ_0072995 expression in breast cancer was found, and SHMT2 up-regulation blocked the anticancer function of si-circ_0072995 in breast cancer cells. Previous studies have reported that circRNAs can act as ceRNAs for miRNAs, thereby abolishing the function of the corresponding miRNA and regulating the depression of miRNA targets.17,27 Thus, whether circ_0072995 knockdown-mediated anticancer function may operate via a ceRNA mechanism was explored, and we confirmed that circ_0072995 was a sponge of miR-149-5p and miR-149-5p directly targeted SHMT2 mRNA in this study; moreover, circ_0072995 positively regulated SHMT2 expression by binding to miR-149-5p. Therefore, a circ_0072995/miR-149-5p/SHMT2 network in the progression of breast cancer was identified.
In conclusion, these findings showed that circ_0072995 functioned as an oncogenic circRNA to promote breast cancer progression by inducing cell malignant phenotypes and anaerobic glycolysis through miR-149-5p/SHMT2 axis, suggesting a new insight into the pathogenesis of breast cancer and providing potential targets for the development of therapeutic strategies for breast cancer.
Funding Statement
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Seigel R, Naishadham D, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208 [DOI] [PubMed] [Google Scholar]
- 2.Fang J, Huang C, Ke J, et al. lncRNA TTN-AS1 facilitates proliferation, invasion, and epithelial-mesenchymal transition of breast cancer cells by regulating miR-139-5p/ZEB1 axis. J Cell Biochem. 2020. doi: 10.1002/jcb.29700 [DOI] [PubMed] [Google Scholar]
- 3.Goldvaser H, Amir E. Role of bisphosphonates in breast cancer therapy. Curr Treat Options Oncol. 2019;20(4):26. doi: 10.1007/s11864-019-0623-8 [DOI] [PubMed] [Google Scholar]
- 4.Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633. doi: 10.1038/bjc.2016.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9(9):1175–1188. [DOI] [PubMed] [Google Scholar]
- 8.Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90. doi: 10.1186/s13045-019-0776-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang YY, Zhao P, Zou TN, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36(11):901–908. doi: 10.1089/dna.2017.3862 [DOI] [PubMed] [Google Scholar]
- 10.Jiang LH, Hou JC, Zhong SL, et al. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed Pharmacother. 2018;107:1342–1353. doi: 10.1016/j.biopha.2018.08.030 [DOI] [PubMed] [Google Scholar]
- 11.Liu T, Ye P, Ye Y, Lu S, Han B. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA‐328‐3p‐mediated inhibition of COL1A1. J Cell Mol Med. 2020;24:2189–2201. doi: 10.1111/jcmm.14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Jiang L, Hou J, et al. Circular RNA hsa circ 0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10:1229–1242. doi: 10.2217/epi-2018-0002 [DOI] [PubMed] [Google Scholar]
- 13.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25(1):27–42. doi: 10.1016/j.cmet.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Salvo ML, Contestabile R, Paiardini A, Maras B. Glycine consumption and mitochondrial serine hydroxymethyltransferase in cancer cells: the heme connection. Med Hypotheses. 2013;80(5):633–636. doi: 10.1016/j.mehy.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 15.Jain M, Nilsson R, Sharma S, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–1044. doi: 10.1126/science.1218595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reina-Campos M, Diaz-Meco MT, Moscat J. The complexity of the serine glycine one-carbon pathway in cancerSerine biosynthesis in cancer. J Cell Biol. 2020;219(1):e201907022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John-Aryankalayil M, Palayoor ST, Makinde AY, et al. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat Res. 2012;178(3):105–117. doi: 10.1667/RR2703.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei J, Xu H, Wei W, et al. circHIPK3 promotes cell proliferation and migration of gastric cancer by sponging miR-107 and regulating BDNF expression. OncoTargets Ther. 2020;13:1613–1624. doi: 10.2147/OTT.S226300 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR‐338‐3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316. doi: 10.1002/hep.30671 [DOI] [PubMed] [Google Scholar]
- 20.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566. [PMC free article] [PubMed] [Google Scholar]
- 21.Xu XD, Shao SX, Jiang HP, et al. Warburg effect or reverse Warburg effect? A review of cancer metabolism. Oncol Res Treat. 2015;38(3):117–122. doi: 10.1159/000375435 [DOI] [PubMed] [Google Scholar]
- 22.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 23.Li C, Zhang G, Zhao L, Ma Z, Chen H. Metabolic reprogramming in cancer cells: glycolysis, glutaminolysis, and Bcl-2 proteins as novel therapeutic targets for cancer. World J Surg Oncol. 2016;14(1):15. doi: 10.1186/s12957-016-0769-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernhardt S, Bayerlová M, Vetter M, et al. Proteomic profiling of breast cancer metabolism identifies SHMT2 and ASCT2 as prognostic factors. Breast Cancer Res. 2017;19(1):112. doi: 10.1186/s13058-017-0905-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin K. Positive correlation between expression level of mitochondrial serine hydroxymethyltransferase and breast cancer grade. OncoTargets Ther. 2015;8:1069–1074. doi: 10.2147/OTT.S82433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li AM, Ducker GS, Li Y, et al. Metabolic profiling reveals a dependency of human metastatic breast cancer on mitochondrial serine and one-carbon unit metabolism. Mol Cancer Res. 2020;18(4):599–611. doi: 10.1158/1541-7786.MCR-19-0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Wang D, Wan D, et al. Circular RNA in invasive and recurrent clinical nonfunctioning pituitary adenomas: expression profiles and bioinformatic analysis. World Neurosurg. 2018;117:e371–e386. doi: 10.1016/j.wneu.2018.06.038 [DOI] [PubMed] [Google Scholar]