House mice from 4 replicate lines selectively bred for 61 generations for voluntary wheel-running behavior were compared with 4 non-selected control lines using multiple genome-wide analytical techniques on both haplotype and single nucleotide polymorphism data......
Keywords: artificial selection, behavior, complex traits, experimental evolution, population differentiation
Abstract
The biological basis of exercise behavior is increasingly relevant for maintaining healthy lifestyles. Various quantitative genetic studies and selection experiments have conclusively demonstrated substantial heritability for exercise behavior in both humans and laboratory rodents. In the “High Runner” selection experiment, four replicate lines of Mus domesticus were bred for high voluntary wheel running (HR), along with four nonselected control (C) lines. After 61 generations, the genomes of 79 mice (9–10 from each line) were fully sequenced and single nucleotide polymorphisms (SNPs) were identified. We used nested ANOVA with MIVQUE estimation and other approaches to compare allele frequencies between the HR and C lines for both SNPs and haplotypes. Approximately 61 genomic regions, across all somatic chromosomes, showed evidence of differentiation; 12 of these regions were differentiated by all methods of analysis. Gene function was inferred largely using Panther gene ontology terms and KO phenotypes associated with genes of interest. Some of the differentiated genes are known to be associated with behavior/motivational systems and/or athletic ability, including Sorl1, Dach1, and Cdh10. Sorl1 is a sorting protein associated with cholinergic neuron morphology, vascular wound healing, and metabolism. Dach1 is associated with limb bud development and neural differentiation. Cdh10 is a calcium ion binding protein associated with phrenic neurons. Overall, these results indicate that selective breeding for high voluntary exercise has resulted in changes in allele frequencies for multiple genes associated with both motivation and ability for endurance exercise, providing candidate genes that may explain phenotypic changes observed in previous studies.
MOST traits of interest in biology are complex, modulated by numerous genetic and environmental factors, and comprised of multiple lower-level (subordinate) traits that often influence higher-level traits in nonintuitive ways (Garland et al. 2016; Sella and Barton 2019). Examples of complex traits include human height, which is influenced by >9500 quantitative trait loci (QTL) (Wood et al. 2014), as well as one’s susceptibility to various psychological diseases (Horwitz et al. 2019).
One complex trait of great interest to medicine is exercise behavior. Exercise has been linked to numerous health benefits, including muscle and bone strength, weight control, reduced cardiac disease, and improved mental health (Manley 1996; Lightfoot et al. 2018). Nonetheless, the majority of Americans are not getting sufficient exercise and this problem is common worldwide (Guthold et al. 2018). Not only does insufficient exercise contribute to such health issues as obesity and diabetes (Booth et al. 2002; Cornier et al. 2008; Myers et al. 2017), but it also increases healthcare costs in the United States, e.g., by >US $100 billion annually between the years of 2006 and 2011 (Carlson et al. 2015). Conversely, higher levels of physical activity promote physical fitness and cardiovascular health, while lowering risk for depression, anxiety-related disorders, obesity, type 2 diabetes, and mortality (Blair and Morris 2009; Matta Mello Portugal et al. 2013; Mok et al. 2019).
The health benefits of exercise occur by various mechanisms (Neufer et al. 2015), as do the adverse effects of a lack of exercise (Booth et al. 2012). Acute exercise can have beneficial effects on immune function (Sellami et al. 2018) and cognition (Park and Etnier 2019). Chronic exercise training can cause changes in muscle fiber type composition that benefit regulation of energy metabolism and other metabolic pathways (Fan et al. 2013). Furthermore, exercise has been linked to lower blood pressure by reducing systemic vascular resistance (Cornelissen and Fagard 2005). Reduced blood pressure, in turn, reduces risk of cardiac disease (Benjamin et al. 2019). The release of endorphins and vascular endothelial growth factors have shown promise as explanations for the growth of new neurons in the brain, which may be the cause of reduces symptoms of neurological diseases such as depression (Ernst et al. 2006).
Identifying genetic determinants of exercise behavior could potentially lead to drug targets that would help promote motivation for exercise and/or benefits derived from exercise. Additionally, by identifying genetic causes of motivation for exercise we may also gain insight regarding higher-level structures or pathways that control this motivation. A variety of human studies have been conducted to determine the genes or chromosomal regions that modulate various components of exercise behavior, including both motivation and/or capability to exercise (Lightfoot et al. 2018). Many of these studies use observational methods to compare humans who engage in either frequent and/or strenuous exercise with those who are less active (Kostrzewa and Kas 2014; Lin et al. 2017). Historically, the most common approach to measuring human exercise levels was by use of questionnaires, which can be of dubious reliability, but an increasing number of studies use accelerometers (Prince et al. 2008; Dyrstad et al. 2014). Detecting QTL in these studies is generally done with genome-wide association studies (GWAS), which rely on phenotypic and genetic data from many individuals within a population and can identify particularly strong correlations between the phenotype and key genetic markers and loci.
Various QTL identified in humans are associated with motivation, e.g., dopaminergic regulation. Dopamine is a well-established modulator of exercise motivation or reward (Garland et al. 2011b). Various genes associated with the dopamine pathway are associated with exercise behavior in humans (Simonen et al. 2003; Loos et al. 2005; De Moor et al. 2009). The large body of evidence that dopamine signaling is a major component of exercise motivation dwarfs other motivational systems that have been associated with exercise, including serotonin and endocannabinoids (Dietrich 2004; Cordeiro et al. 2017), though serotonin has been implicated in GWAS of hyperactivity disorders (Aebi et al. 2016).
Other human studies have detected QTL associated with physical traits related to exercise abilities, including maximal oxygen consumption (VO2max) (Williams et al. 2017), bone density (Herbert et al. 2019), and more (Lin et al. 2017). The list of possible biological traits affiliated with exercise and their associated QTL is extensive (Sarzynski et al. 2016; Lightfoot et al. 2018).
Observational studies of human exercise behavior are limited by measurement error and environmental cofactors that cannot always be accounted for in statistical models (Garland et al. 2011b; Lightfoot et al. 2018). One alternative is to use animal models derived from selective breeding experiments (Garland and Rose 2009). Selective breeding will alter the proportions of alleles that affect a trait of interest, thus allowing for easier detection of such alleles (Britton and Koch 2001; Konczal et al. 2016). Finding the genetic factors that underlie a complex trait is also facilitated by reducing environmental variation (“noise”), as is possible with laboratory colonies of rodents (Parker and Palmer 2011).
To elucidate the biological basis of voluntary aerobic exercise behavior, a selection experiment was begun in 1993 using a base population of outbred Hsd:ICR mice. Four replicate lines have been bred for high voluntary wheel-running behavior and another four bred without regard to their wheel running as controls for founder effects and random genetic drift (Swallow et al. 1998). Since the beginning of this experiment, over 150 papers have been published that document a variety of phenotypic differences between the High Runner (HR) and Control (C) lines. These previous studies establish morphological and physiological differences in bone, kidney, heart, skeletal muscle, brain, and other organs and systems (Rhodes et al. 2005; Swallow et al. 2005; Kolb et al. 2013b; Wallace and Garland 2016), and, more generally, reaffirm the diversity of the systems involved in voluntary exercise behavior (Garland et al. 2011b; Lightfoot et al. 2018). The previous studies also give potential directions for informed analyses of the genome. For example, we would expect divergence in allele frequencies related to the reward system in the brain and to muscle function. The HR selection experiment is the world’s “largest” involving a behavioral trait in rodents in terms of the number of lines and generations. Therefore, addressing the genomic differences between the HR and C mice is expected to provide novel insights into the underpinnings of exercise behavior.
Previously, Xu and Garland (2017) used a mixed model (nested ANOVA) with minimum variance quadratic unbiased estimation (MIVQUE) to analyze medium-density single nucleotide polymorphism (SNP) data for the HR and control lines sampled from generation 61 (Xu and Garland 2017). This statistical method proved more powerful than the commonly used regularized F test and Generalized Linear Mixed Model (GLMM) methods when incorporating permutation-based multiple testing correction. The data used included 7–10 females from each of eight lines (four HR and four C). Genotypes were determined with the MegaMUGA SNP-chip (Morgan and Welsh 2015). After removing markers with missing data, 25,318 markers were analyzed with the mixed models, finding 152 markers to be significantly differentiated between the HR and C linetypes (i.e., test group). Although Xu and Garland (2017) demonstrated numerous SNP loci with evidence of differentiation between the HR and control lines, biological interpretations were not presented. Additionally, as demonstrated by the whole-genome sequence (WGS) data addressed in this paper, various differentiated loci were not detected in the previous SNP-chip analysis.
Here, we apply the mixed model with MIVQUE estimation method to WGS data obtained from the same individuals as in Xu and Garland (2017). We analyze both SNP and haplotype data to take full advantage of the information provided by each data type (Shim et al. 2009; Taliun et al. 2016). We also use simulations to explore some of the statistical properties of the MIVQUE estimation method for this application, and we implement procedures aimed at improving model fit and potentially statistical power. We identify numerous SNP and haplotype loci as potential candidates for functionally relevant genetic differentiation between the HR and C lines. Many of these can be tied to specific lower-level traits that should influence exercise behavior, through use of gene ontology terms and KO phenotype analyses of nearby genes.
Using information on known morphological and physiological differences between the HR and control lines, we were able to perform both broad and directed strategies to detecting significantly differentiated loci. We show that the method of Xu and Garland (2017) can be improved by allowing for different among- and within-line variance structures. We identified several potentially differentiated genes associated with bone, heart, and brain morphology. We also identified a few candidates with potential large-scale influences on the HR mice, including Sorl1, Dach1, and Cdh10.
Materials and Methods
HR mouse model
As described previously (Swallow et al. 1998; Careau et al. 2013), 112 males and 112 females of the outbred Hsd:ICR strain were purchased from Harlan Sprague Dawley in 1993. These mice were randomly bred in our laboratory for two generations. Then 10 males and 10 females were then randomly chosen as founders for each of eight closed lines (generation 0). Four of these lines were randomly picked to be HR lines, in which mice would be selected for breeding based on voluntary wheel running. The remaining four lines were used as C lines, without any selection. At ∼6–8 weeks of age, all mice were given access to wheels for 6 days. The amount of running (total revolutions) on days 5 and 6 was used as the selection criterion. For the nonselected C lines, one male and one female from each of 10 families were chosen as breeders to propagate the line. For the HR lines, the highest-running male and female from within each of 10 families were chosen as breeders (within-family selection). Sib-mating was disallowed in all lines (Swallow et al. 1998).
Whole-genome sequencing
DNA was collected from 80 mice (10 from each line), from generation 61, via phenol-chloroform extraction and sequenced on an Illumina HiSeq 2500 1T platform. Libraries were constructed using Nextera kit and reads were trimmed and aligned to the GRCm38/mm10 mouse genome assembly as described in Didion et al. (2016). This generated an average read depth of 12X per mouse. SNPs were filtered based on genotype quality (“GQ”) >5, read depth >3, MAF <0.0126 for all samples, and Mapping Quality (“MQ”) >30. Of the 80 mice, 1 was excluded due to likely contamination as in Xu and Garland (2017), leaving 79 for the following analyses. SNPs not found to be present in at least 2 of the 80 mice were also removed from analysis. Although Xu and Garland (2017) had identified these as females, they were in fact all males with exception of one female from line 5.
Heterozygosity calculations
Individual mouse heterozygosity (multi-locus heterozygosity) was calculated by dividing the number of heterozygous loci for each mouse by the total number of segregating loci across all 80 mice (n = 5,932,124). Heterozygosity per line is the average of the heterozygosity of all sequenced mice within that line.
SNP analysis
Individual SNPs were initially analyzed using a mixed model approach with the Minimum Variance Quadratic Unbiased Estimation of variance (MIVQUE) method of estimating variance parameters as described in Xu and Garland (2017). However, rather than removing loci or mice (which had been necessary in the Xu and Garland paper, resulting in 7–10 mice per line analyzed) with missing data, code was modified to remove only the missing values themselves. The MIVQUE analysis provides a P-value for each locus for rejecting the null hypothesis of no differentiation between the HR and C lines. Xu and Garland had performed the analysis using two different encoding schemes to represent genotypes as 0, 0.5 and 1 vs. as twin vectors of 0–0, 0–1 and 1–1. We have since determined that the twin vectors encoding was preferable, and we report only those results (File S7).
Multi-model analysis of SNP data from WGSs
The analyses performed in Xu and Garland (2017) used a single statistical model in R for all loci (our comparable SAS model being “Simple” in Table 1). This model did not allow for several possibilities that might be expected a priori and that were in fact observed, such as differing variances among the five replicate HR and C lines (designated “SepVarLines” in Table 1), as is the case for wheel-running behavior (Garland et al. 2011a). Beyond this, the amount of variation among individual mice within the replicate lines might differ for the HR and C lines (“Full” model). Interpretation of these different models is presented in the Discussion. In total, we applied four alternate models to the data for each locus, and followed a model selection procedure for the one with the lowest the Aikake Information Criterion (AIC), corrected for small sample sizes (AICc), and retained the P-value for its linetype effect (differentiation between the HR and C lines). All Multi-Model analyses were performed in SAS using PROCEDURE MIXED with the mivque0 method (File S10). We elected to prioritize SAS over R for its performance gains over large number of loci. For a direct comparison, we reanalyzed the MegaMUGA data in Xu and Garland (2017) the multi-model method (Supplemental Material, Figures S1 and S2).
Table 1. Summary of covariance models.
| Model | df. | Covariance parameters | Description | HR and C different among-line variance | HR and C different within-line variance | HR and C same among-line variance | HR and C same within-line variance | SAS Code |
|---|---|---|---|---|---|---|---|---|
| Full | 6 | 4 | Random effects for replicate line within selection treatment (linetype) and for mouse within line and linetype, allowing for separate variance estimates for both lines within linetype and mouse within line and linetype | x | x | proc mixed data = locus method = mivque0; | ||
| class pop sub mouse; | ||||||||
| model COL1 = pop/solution; | ||||||||
| random sub(pop) /group = pop; | ||||||||
| random mouse(sub pop) /group = pop; | ||||||||
| SepVarLines | 6 | 3 | Random effects for replicate line within selection treatment (linetype) and for mouse within line and linetype, allowing for separate variance estimates for line within linetype | x | x | proc mixed data = locus method = mivque0; | ||
| class pop sub mouse; | ||||||||
| model COL1 = pop/solution; | ||||||||
| random sub(pop) /group = pop; | ||||||||
| random mouse(sub pop); | ||||||||
| SepVarInd | 6 | 3 | Random effects for replicate line within selection treatment (linetype) and for mouse within line and linetype, allowing for separate variance estimates for mouse within line and linetype | x | x | proc mixed data = locus method = mivque0; | ||
| class pop sub mouse; | ||||||||
| model COL1 = pop/solution; | ||||||||
| random sub(pop); | ||||||||
| random mouse(sub pop) /group = pop; | ||||||||
| Simple | 6 | 2 | Random effects for replicate line within selection treatment (linetype) and for mouse within line and linetype (as used by Xu and Garland (2017)) | x | x | proc mixed data = locus method = mivque0; | ||
| class pop sub mouse; | ||||||||
| model COL1 = pop/solution; | ||||||||
| random sub(pop); | ||||||||
| random mouse(sub pop); |
Multiple models used to analyze the allelic SNP data (two values per mouse) for whole-genome sequences from 79 mice. For each model, we used SAS Procedure Mixed with MIVQUE estimation (Xu and Garland 2017) to obtain the test statistic (F), significance level (P), and AICc (df. method was containment). For some loci, the within-line variance was zero for all eight lines. In those cases, we used direct enumeration to calculate a significance level, i.e., the probability of observing the pattern vs. the 23 possible combinations. See text for further details.
Loci that contained no within-line variance (i.e., each line was fixed for one allele or the other) could not be analyzed with the foregoing procedures. We analyzed these loci by counting the net number of alternatively fixed lines among the HR and C linetypes. Those loci with greater difference in allele frequency between the HR and C linetypes are regarded as being more “significant.”
Multiple testing correction
Permutations for MegaMUGA data:
This approach is based on the permutation method used by Xu and Garland (2017), but modified to account for the multiple models. All permutations were performed using SAS PROC MIXED as described above in the section on multi-model approach. The mouse IDs, line, and linetype were randomly permuted as a block to break their original associations with the allelic data but not with each other. The permuted data for each locus were then analyzed with each of the four models listed in Table 1 (i.e., for the MegaMUGA SNP data, 4 × 25,332 analyses were performed). For each of the four models, the AICc was recorded, and the corresponding F-statistics were retained. From these 25,332 loci (for the MegaMUGA data), the F-statistic corresponding to the model with the lowest AICc was saved. The foregoing process was repeated 5000 times, the resulting F-statistics were sorted from largest to smallest, and the 250th largest F-statistic was used to establish the critical value for the 5% FWER.
Permutations for haplotype data:
Permutations done for haplotypes were performed separately for 2-allele haplotype blocks and 3-allele blocks, using 1000 permutations to keep computational times manageable. As in the unpermuted haplotype analyses, blocks with three alleles (n = 5869) were analyzed with two dummy variables, each individual dummy variable was tested using the multi-model method, and the two P-values generated were combined using Fisher’s method (Fisher 1925). However, some permutations of the 3-allele blocks produced erroneous low P-values (apparently due to numerical issues), which, if included in subsequent calculations would have caused an artifactual reduction of the critical value needed to obtain the true 5% FWER. The permutations of the 2-allele blocks (n = 11,032) did not produce any artifactually low P-values. Given the problems with the 3-allele haplotype permutations, we elected to apply the MeguMUGA permutation threshold (P < 0.00526) to the haplotype blocks because of their similar sample size (MegaMUGA = 25,332; Haplotypes = 16,901) and the fact that they should be highly correlated.
Local maxima selection for WGS data:
In the original paper, which analyzed 25,332 SNPs from a commercial chip, a permutation procedure was used to control the family-wise Type I error rate (FWER) at 5% (Xu and Garland 2017). Those procedures were not computationally practical for the 5,932,124 SNPs from the WGSs, nor are linked SNPs within a haplotype block truly independent from each other. Accordingly, significant loci were chosen via a combination of −logP cutoff and local maximum (LM) determination, the latter acting as a filter to focus on actual selected loci over their hitchhikers. Similar methods have been previously described (Nicod et al. 2016). Briefly, suggestive loci with −logP >3.0 were clustered with a maximum gap of 1 Mbp. For each such cluster, the global peak, and a set of local maxima were determined for every 500 kbp spanned by the cluster. The set of local maxima were chosen as peaks separated by dips in the signal below the median −logP in the cluster. These LM SNPs were annotated using R libraries GenomicFeatures and VariantAnnotation, with the mm10 knownGene.sqlite database provided by the Genome Browser team at the University of California, Santa Cruz.
Haplotype determination
From the WGSs, haplotypes were determined using JMP 11 and JMP Scripting Language (SAS Institute Inc., Cary, NC). To construct haplotypes, we first defined the genomic block segments as consecutive 20 kbp windows that did not transition between homozygous and heterozygous states. For each block region, we performed a hierarchical clustering analysis using SNP genotype data (of homozygous regions only) as input. Preliminary haplotype analysis showed that the HR population at generation 61 rarely had more than three alleles in a given haplotype. Therefore, the analysis was restricted to a maximum of three clusters (haplotype alleles) per block (File S5).
Haplotype analysis
As for the SNP data, haplotype data were analyzed using the multi-model method described above. Haplotype blocks with only two alleles (n = 11,032) were analyzed the same way as for the SNP data (File S10). Blocks with three alleles (n = 5869) were analyzed with two dummy variables, with the base allele chosen as the most common one, and then two dummy variables coding for presence of the other two alleles. Each individual dummy variable was tested using the multi-model method. The two P-values generated from the two dummy variables were combined using Fisher’s method (Fisher 1925). Different models potentially were used for each dummy variable based on AICc, allowing for up to two models to contribute to the final P-value of a locus (File S6).
SNPs fixed in one treatment but polymorphic in the other
As noted previously with the SNP chip data (Xu and Garland 2017), we observed no loci that were fixed for one allele in all four HR lines while being fixed for the alternate allele in all four C lines (see Results). We did, however, observe loci fixed for a given allele in all four HR lines, which is symptomatic of a complete selective sweep (caused by directional selection) as described by Burke (2012), while remaining polymorphic in all four C lines. All loci that were fixed in the HR mice and simultaneously polymorphic in all C lines (FixedHR/PolyC) were extracted from the multi-model results and grouped such that those fixed loci that were within 100,000 bp of other fixed loci would be part of the same group. This process was then repeated for loci fixed in the Control lines but polymorphic in all HR lines (FixedC/PolyHR).
General ontology analysis
Transcribed regions (N = 56, as indicated in Table 2) found to contain LM based on the WGS analyses were analyzed using The Gene Ontology (GO) resource. GO analyses were performed based on biological process, molecular function, and cellular component. Ontologies reported as significant at raw P < 0.05 for any of these three categories are reported here. Analysis of these genes was also performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID). The results of these analyses did not vary greatly from the GO results.
Table 2. Basic descriptive statistics for the primary analyses.
| Dataset | Total loci | Significant loci | Critical threshold | Significant GENES |
|---|---|---|---|---|
| MegaMUGA | 25,332 | 162a | P < 0.00526 (5% FWER) | 174b |
| Whole-Genome SNPs | 5,932,124 | 84 | P < 0.001 (Local Maximum) | 27 |
| Haplotypes | 16,901 | 102c (28 regions) | P < 0.00526 (See text) | 154b |
| All HR Fixed, All C Polymorphic | 5,932,124 | 2562 (46 regions) | See text | 135b |
In Xu and Garland (2017), 152 SNPs were identified as statistically significant with a single model and the MIVQUE procedure. after use of a permutation procedure to control the family-wise Type I error rate (FWER) at 5% (P < 0.00343).
These are not genes that SNPs fell into. These are genes close to significant SNPs or haplotypes.
From 28 closely linked groups.
Targeted ontology analysis
Previous papers show that the HR lines of mice have diverged from the C lines for many different phenotypes (reviews in Rhodes et al. 2005; Garland et al. 2011b; Wallace and Garland 2016). Many of these phenotypes can be tied to specific neurobiological or physiological functions. In such cases, a logical approach is to analyze separately some candidate genes known to be affiliated with relevant functions and find differentiated SNPs for those genes. We used this approach for several ontologies. Specifically, lists of genes affiliated with dopamine, serotonin, brain, bone, cardiac muscle, and skeletal muscle were extracted from the Mouse Genome Informatics website. SNPs found within these genes were separated from the full WGS data and the most differentiated among these were recorded.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Any additional intermediary or results file are available upon request. Supplemental files are available at FigShare. File S1 contains supplemental figures and brief descriptions of all other supplemental files and tables. File S2 contains allelic SNP data. File S3 contains mouse data with line and lintype. File S4 contains all results for analyses of individual SNPs. File S5 contains all haplotype data. Files S6 contains all results for analyses of haplotype data. File S7 contains justification for use of allelic coding of alleles. File S8 includes simulations of Type I error rates for Mixed Model analyses using MIVQUE variance estimation. File S9 expands on the discussion of genes in consistent regions (see Results). File S10 includes all R and SAS code used for the SNP and haplotype analyses. File S11 includes a comprehensive list of genes containing SNPs and rankings by P-value. Table S1 includes local maxima associated genes. Table S2 contains groups of loci fixed in all lines of one lintype but polymorphic in all lines of the other. Table S3 includes heterozygosity for each individual mouse. Table S4 includes top 10 genes for each of the targeted ontologies analyses. Table S5 includes allele frequency by line of each loci identified as a local maximum. Table S6 includes genomic regions identified as suggestive (P < 0.001) by the SNP analyses. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12436649.
Results
Variation in genetic diversity
After 61 generations of the HR mouse selection experiment, and based on a sample of 79 mice, we found SNPs segregating at 5,932,124 loci (∼2.2 SNPs per kbp or 0.22%) across the entire set of lines (i.e., at least two mice containing an alternate allele were found across the 79 mice sequenced) with at least 1.5% minor allele frequency. Individual lines contained 2.04–2.82M SNPs (34%–48% of the total diversity) (Table 3), with no appreciable loss in diversity for the HR lines compared to the C replicates (Mann–Whitney U test, W = 6; P-value = 0.6857). SNP heterozygosity ranged from 10.3% to 20.6% among individual mice (Table S3) and averaged 12.7%–18.1% per line (Table 3).
Table 3. Summary of polymorphism and heterozygosity by line.
| Line | Polymorphic SNP loci | SNP % | Polymorphic haplotypes | Haplotype % | SNP Het | Haplotype Het |
|---|---|---|---|---|---|---|
| C1 | 2,333,951 | 39.3% | 7773 | 46.0% | 14.7% | 17.8% |
| C2 | 2,436,225 | 41.1% | 7652 | 45.3% | 13.7% | 16.6% |
| C3 | 2,602,007 | 43.9% | 7841 | 46.4% | 15.8% | 17.8% |
| C5 | 2,102,405 | 35.4% | 7160 | 42.4% | 12.7% | 16.5% |
| HR3 | 2,819,828 | 47.5% | 8717 | 51.6% | 18.1% | 19.6% |
| HR6 | 2,220,487 | 37.4% | 7060 | 41.8% | 13.5% | 16.2% |
| HR7 | 2,042,309 | 34.4% | 6304 | 37.3% | 13.0% | 14.7% |
| HR8 | 2,226,282 | 37.5% | 7315 | 43.3% | 14.4% | 16.6% |
Initial haplotype analysis demonstrated that there were rarely more than three alleles for any given haplotype block (region with little to no discernable recombination events within the 79 mice analyzed). Therefore, for the final haplotype analysis, hierarchical clustering was performed with a limit of three clusters. Of these blocks, 16,901 remained variable across the eight lines in generation 61. As would be expected, the number of haplotypes that have not gone to fixation in each line appears to be proportional to the number of SNPs that have not gone to fixation (Table 3). Heterozygosity for the haplotypes ranged from 12.2% to 25.5% for individual mice (Table S3), and 14.7%–19.6% when averaged per line (Table 3). Heterozygosity for the haplotype data were not significantly different between HR and C lines (Mann–Whitney U test, W = 8; P-value = 1.0 and W = 6; P-value = 0.6857, respectively).
Multi-model vs. single-model comparisons
As expected, we found that many, indeed most, loci were better fit by models other than the “Simple” model used by Xu and Garland (2017). Generally, the “Full” model was the most preferred, followed by the “Simple” model (Table 4). In general, differences between the P-values determined by the single and multi-model methods were negligible (Figure S2).
Table 4. Model preference by data set, test, and allele counts.
| Model | MegaMUGAa | WGSa | Hap 2-alleleb | Hap 3-alleleb |
|---|---|---|---|---|
| Full | 9875 (39.0%) | 2,441,601 (41.2%) | 4512 (40.9%) | 5510 (46.9%) |
| SepVarLine | 3105 (12.3%) | 504,946 (8.5%) | 1052 (9.5%) | 1583 (13.5%) |
| SepVarInd | 2983 (11.8%) | 716,265 (12.1%) | 726 (6.6%) | 748 (6.4%) |
| Simple | 8654 (34.2%) | 2,186,803 (36.9%) | 4594 (41.6%) | 3615 (30.8%) |
| # with no within-line variance | 715 (2.8%) | 82,533 (1.4%) | 148 (1.3%) | 282 (2.4%) |
Number of SNPs whose lowest AICc match the indicated model.
Number of haplotype blocks whose lowest AICc match the indicated model (one for each dummy variable for 3-allele blocks).
When analyzing data generated under the null hypothesis, the mixed models with MIVQUE estimation for both single and multi-model produced a deflated Type I error rate for α = 0.05 (File S8). The multi-model approach helped to correct this, but the Type I error rate did not improve greatly with the multi-model approach alone. We attempted to utilize the Kenward Rogers method of determining degrees of freedom to correct this low Type I error rate, but this did not bring Type I error rate to 0.05 and effectively dropped the nested line effect for many loci. We did not want to drop the nested line effect because this ignores the fundamental experimental design of the selection experiment. However, the permutation and local maxima methods of determining loci of interest are robust to this deflated Type I error rate (File S8), so we proceeded with our analyses using conservative results produced by the MIVQUE variance estimation method.
Three major analyses
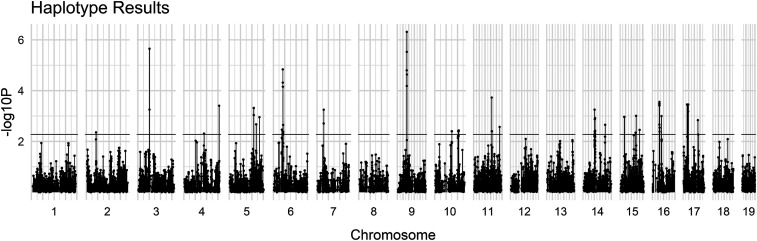
Whole-genome haplotype:
No haplotypes were identified as being fixed in all HR lines for one allele and fixed in all C lines for the opposite allele. The multi-model haplotype analysis produced 102 blocks of significant differentiation at the P < 0.005 (permutations) level. Significant blocks could be found on 13 chromosomes (Figure 1). We consider haplotype blocks within 1,000,000 bp of each other to be linked and therefore part of the same haplotype group: 28 such groups were determined (Table 5). These groups include a total of 154 transcribed sequences recognized by the Panther database for gene ontology. The largest of these groups was found on chromosome 14:52,100,155–54,334,868 bp (Table 5).
Figure 1.
Manhattan plot for haplotype data. Dashed horizontal line indicates P-value <0.005 (see Materials and Methods), which yielded 28 haplotype groups (see Table 5).
Table 5. Significant haplotype groups.
| Group | Chr | Start (bp) | End (bp) | Size (bp) | P-value |
|---|---|---|---|---|---|
| 1 | 2 | 43,100,041 | 43,214,647 | 114,606 | 4.42E-03 |
| 2 | 3 | 51,580,020 | 51,659,891 | 79,871 | 2.25E-06 |
| 3 | 4 | 89,300,145 | 89,357,884 | 57,739 | 4.92E-03 |
| 4 | 4 | 155,480,343 | 155,654,426 | 174,083 | 3.94E-04 |
| 5 | 5 | 108,000,623 | 108,679,807 | 679,184 | 4.85E-04 |
| 6 | 5 | 118,824,587 | 119,299,787 | 475,200 | 2.15E-03 |
| 7 | 5 | 132,540,807 | 133,720,551 | 1,179,744 | 1.12E-03 |
| 8 | 6 | 37,440,411 | 37,659,588 | 219,177 | 3.47E-03 |
| 9 | 6 | 41,584,862 | 43,431,434 | 1,846,572 | 1.47E-05 |
| 10 | 7 | 29,640,243 | 29,697,093 | 56,850 | 5.67E-04 |
| 11 | 9 | 41,240,184 | 42,275,833 | 1,035,649 | 4.90E-07 |
| 12 | 10 | 75,061,742 | 75,456,261 | 394,519 | 3.99E-03 |
| 13 | 10 | 103,363,232 | 104,139,953 | 776,721 | 3.94E-03 |
| 14 | 10 | 105,220,041 | 105,699,704 | 479,663 | 3.72E-03 |
| 15 | 11 | 79,724,263 | 81,409,849 | 1,685,586 | 1.89E-04 |
| 16 | 11 | 114,466,946 | 114,489,018 | 22,072 | 2.69E-03 |
| 17 | 14 | 52,100,155 | 54,334,868 | 2,234,713 | 5.62E-04 |
| 18 | 14 | 98,380,090 | 98,679,965 | 299,875 | 2.22E-03 |
| 19 | 15 | 18,960,135 | 19,759,996 | 799,861 | 1.09E-03 |
| 20 | 15 | 69,120,025 | 70,219,737 | 1,099,712 | 4.53E-03 |
| 21 | 15 | 71,480,090 | 71,559,595 | 79,505 | 9.91E-04 |
| 22 | 15 | 86,541,805 | 86,599,823 | 58,018 | 3.55E-03 |
| 23 | 16 | 31,540,757 | 33,178,952 | 1,638,195 | 2.79E-04 |
| 24 | 16 | 40,742,298 | 41,357,426 | 615,128 | 1.01E-03 |
| 25 | 17 | 18,020,933 | 18,039,390 | 18,457 | 3.54E-04 |
| 26 | 17 | 20,700,046 | 20,939,819 | 239,773 | 3.54E-04 |
| 27 | 17 | 23,000,233 | 23,599,776 | 599,543 | 3.54E-04 |
| 28 | 17 | 65,458,617 | 65,738,255 | 279,638 | 1.46E-03 |
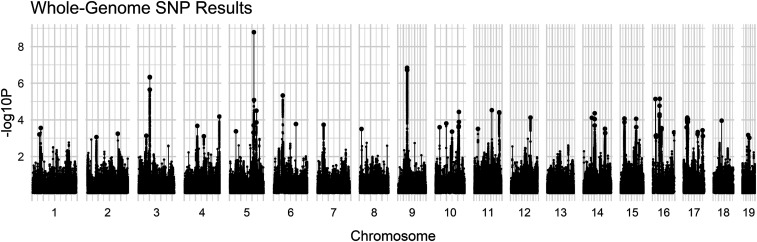
Whole-genome SNP:
Similarly to haplotypes, no individual SNPs were identified as being fixed in alternative alleles across all HR on one hand and all C lines on the other. At the P < 8.4E-09 critical level (Bonferroni-corrected), only two SNPs in chromosome 5 were identified to be significantly differentiated across the entire genome (Figure 2), both in an intron of an uncharacterized gene (GM34319). The syntenic/orthologous region of both the human and cat genomes correspond to a coding region (exon 3) of the MYL5 gene (Myosin light chain 5). Due to the small number of significant SNPs under Bonferroni and the computational difficulties of using permutations with the multi-model method, we focus on local maxima SNPs.
Figure 2.
Manhattan plot for WGS SNP data. Large dots represent local maxima (N = 84).
In the local maxima (LM) analyses, the suggestive cutoff (−logP > 3.0) produced 38,065 SNPs for analysis. A total of 44 clusters were found, ranging in size from 1 SNP to 3787 SNPs (Chr9: 41,303,824–42,478,817 bp). The largest single group in terms of genome spanned is on chr17: 17,846,983–23,586,163 bp (Table 6). From these groups, a total of 84 LM were determined; 31 of these SNPs were associated with 27 unique transcribed regions. Of the 27 genes, 26 could be utilized for GO analysis. Although chromosome 3 had no LM fall into specific genes (despite clear significance based on the Manhattan plot), the cluster on chr3 (chr3:51,190,735–52,498,029 bp) includes ∼10 validated coding genes and various predicted genes, but none of the LMs fall in these. However, all three LMs in this group are upstream of Setd7, a methyltransferase.
Table 6. Top 5 largest suggestive regions.
| Chr | Start (BP) | End (BP) | Size | Lowest P |
|---|---|---|---|---|
| 17 | 17,846,983 | 23,586,163 | 5,739,180 | 7.54E-05 |
| 10 | 103,429,623 | 105,529,701 | 2,100,078 | 3.73E-05 |
| 16 | 31,440,034 | 33,128,268 | 1,688,234 | 7.05E-06 |
| 15 | 18,958,730 | 20,635,226 | 1,676,496 | 8.49E-05 |
| 16 | 16,235,542 | 17,805,005 | 1,569,463 | 7.04E-04 |
The most significant SNPs with no within-line variance fell into three regions. One of these regions is on chromosome 5 [105–109 million base pair (mbp)], which is close to the LM identified in this chromosome. Another is on chromosome 16 (44 mbp), ∼2.5 mbp from the LM on chromosome 16 containing Lsamp, a gene which codes for a neuron-associated membrane protein. However, the last region falls in chromosome 7 (115 mbp), a chromosome which contained no LM. This location is downstream of Sox6, a developmental regulator broadly associated with muscle fiber type composition (van Rooij et al. 2009), hematopoiesis, bone growth, and heart function (Smits et al. 2001).
SNPs fixed in one treatment and polymorphic in the other:
SNPs that were fixed in all HR lines and polymorphic in all C lines (FixedHR/PolyC) were grouped into 95 regions, based on their being separated by at least 100 kbp (Table S2). Here, we were more strict on the definition of a group than for the haplotype groups (1 mbp) to limit the potential for single SNPs to greatly expand the size of a group by their spacing, whereas haplotypes, being made up of several SNPs, are naturally resilient to such inflation. Some of these regions are probably not independently segregating (e.g., chr17: 17,895,909–22,546,405 bp), and might therefore be combined further. Regions varied in size from 1 to 1,626,783 bp. These regions include or are proximal to (in the case of 1 bp regions) 135 transcribed regions, including genes, miRNA, and predicted genes. SNPs that were fixed in all C lines and polymorphic in all HR lines (FixedC/PolyHR) were combined into 64 regions. The size of each region varies from 1 to 753,066 bp. We expect the 1 bp loci may be spurious but chose to include them in results for completeness, especially given that the mini-muscle locus involves only a single base pair (Kelly et al. 2013). These regions include or are proximal to 63 transcribed regions, again including genes, miRNA, and predicted genes. FixedHR/PolyC regions were also identified in haplotypes. These haplotype blocks overlapped with the SNP regions identified by FixedHR/PolyC; however, some of the single unlinked loci that met these criteria were not identified using haplotypes.
Ontology analyses
General ontology:
GO analysis of biological process for the haplotype data reveal “sensory perception of chemical stimulus” to be a major term of interest (Table 7). This appears to be caused by various clusters of olfactory and vomeronasal genes. Many of the most prominent terms appear to be correlated to these olfactory and vomeronasal gene clusters. Although a single, large group of closely linked olfactory genes may over-represent olfactory’s role in selection, we were able to identify two distinct genomic regions of vomeronasal genes and three such regions of olfactory genes.
Table 7. Top biological process terms from GO analysis for haplotype.
| GO term | Total genes | Input genes | Expected | Fold enrichment | Raw P-value |
|---|---|---|---|---|---|
| Detection of chemical stimulus involved in sensory perception of smell | 3 | 1 | 0.02 | 47.88 | 2.74E-02 |
| Sensory perception of smell | 1128 | 27 | 7.85 | 3.44 | 2.46E-08 |
| Sensory perception of chemical stimulus | 1228 | 34 | 8.55 | 3.98 | 5.71E-12 |
| Sensory perception | 1641 | 36 | 11.42 | 3.15 | 7.12E-10 |
| Detection of chemical stimulus involved in sensory perception | 59 | 7 | 0.41 | 17.04 | 3.65E-07 |
| Detection of stimulus involved in sensory perception | 136 | 8 | 0.95 | 8.45 | 7.40E-06 |
| Detection of stimulus | 236 | 9 | 1.64 | 5.48 | 5.40E-05 |
| Detection of chemical stimulus | 85 | 7 | 0.59 | 11.83 | 3.53E-06 |
| G protein-coupled receptor signaling pathway | 1853 | 37 | 12.9 | 2.87 | 4.86E-09 |
| Regulation of systemic arterial blood pressure by aortic arch baroreceptor feedback | 1 | 1 | 0.01 | >100 | 1.38E-02 |
| System process | 2594 | 42 | 18.06 | 2.33 | 2.12E-07 |
| Multicellular organismal process | 7307 | 74 | 50.87 | 1.45 | 1.43E-04 |
| Nervous system process | 2085 | 39 | 14.51 | 2.69 | 9.97E-09 |
| Sensory perception of sour taste | 5 | 1 | 0.03 | 28.73 | 4.08E-02 |
| Sensory perception of taste | 71 | 7 | 0.49 | 14.16 | 1.15E-06 |
| Detection of chemical stimulus involved in sensory perception of bitter taste | 47 | 6 | 0.33 | 18.34 | 1.74E-06 |
| Sensory perception of bitter taste | 51 | 6 | 0.36 | 16.9 | 2.69E-06 |
| Detection of chemical stimulus involved in sensory perception of taste | 51 | 6 | 0.36 | 16.9 | 2.69E-06 |
Within groups of GO terms, items are listed in order of increasing generality.
The biological process GO terms for LM include many results that are consistent with our previous findings involving the HR mice, including cardiac- and myoblast-related terms (Table 8). Regulation of locomotion is among the most statistically significant GO terms.
Table 8. Top biological process terms from GO analysis for LM.
| GO term | Total genes | Input genes | Expected | Fold enrichment | Raw P-value |
|---|---|---|---|---|---|
| Locomotory exploration behavior | 16 | 1 | 0.02 | 53.6 | 1.96E-02 |
| Locomotory behavior | 240 | 4 | 0.28 | 14.29 | 1.72E-04 |
| Behavior | 685 | 6 | 0.8 | 7.51 | 1.17E-04 |
| Positive regulation by host of viral release from host cell | 5 | 1 | 0.01 | >100 | 6.97E-03 |
| Positive regulation of viral release from host cell | 15 | 1 | 0.02 | 57.17 | 1.85E-02 |
| Regulation of viral release from host cell | 31 | 1 | 0.04 | 27.66 | 3.66E-02 |
| Regulation of locomotion | 1040 | 7 | 1.21 | 5.77 | 1.47E-04 |
| Negative regulation of cardiac muscle cell proliferation | 17 | 2 | 0.02 | >100 | 2.20E-04 |
| Negative regulation of cell population proliferation | 684 | 3 | 0.8 | 3.76 | 4.46E-02 |
| Negative regulation of cardiac muscle tissue growth | 29 | 2 | 0.03 | 59.14 | 5.94E-04 |
| Regulation of cardiac muscle tissue growth | 74 | 2 | 0.09 | 23.18 | 3.53E-03 |
| Regulation of cardiac muscle tissue development | 98 | 2 | 0.11 | 17.5 | 6.02E-03 |
| Regulation of striated muscle tissue development | 160 | 2 | 0.19 | 10.72 | 1.52E-02 |
| Regulation of muscle tissue development | 163 | 2 | 0.19 | 10.52 | 1.57E-02 |
| Regulation of muscle organ development | 164 | 2 | 0.19 | 10.46 | 1.59E-02 |
| Regulation of heart growth | 80 | 2 | 0.09 | 21.44 | 4.09E-03 |
| Regulation of organ growth | 114 | 2 | 0.13 | 15.04 | 8.02E-03 |
| Negative regulation of cardiac muscle tissue development | 40 | 2 | 0.05 | 42.88 | 1.09E-03 |
| Negative regulation of striated muscle tissue development | 64 | 2 | 0.07 | 26.8 | 2.67E-03 |
| Negative regulation of muscle organ development | 66 | 2 | 0.08 | 25.99 | 2.83E-03 |
| Negative regulation of muscle tissue development | 67 | 2 | 0.08 | 25.6 | 2.92E-03 |
| Negative regulation of heart growth | 29 | 2 | 0.03 | 59.14 | 5.94E-04 |
| Bundle of His cell-Purkinje myocyte adhesion involved in cell communication | 6 | 1 | 0.01 | >100 | 8.13E-03 |
| Bundle of His cell to Purkinje myocyte communication | 13 | 1 | 0.02 | 65.96 | 1.62E-02 |
| Cell communication involved in cardiac conduction | 32 | 1 | 0.04 | 26.8 | 3.78E-02 |
| Multicellular organismal signaling | 109 | 2 | 0.13 | 15.73 | 7.37E-03 |
| Cardiac muscle cell-cardiac muscle cell adhesion | 7 | 1 | 0.01 | >100 | 9.28E-03 |
| Cell-cell adhesion | 389 | 3 | 0.45 | 6.61 | 1.04E-02 |
| Cell adhesion | 789 | 6 | 0.92 | 6.52 | 2.50E-04 |
| Biological adhesion | 799 | 6 | 0.93 | 6.44 | 2.68E-04 |
| Negative regulation of cellular extravasation | 8 | 1 | 0.01 | >100 | 1.04E-02 |
| Negative regulation of leukocyte migration | 41 | 2 | 0.05 | 41.83 | 1.14E-03 |
| Regulation of leukocyte migration | 209 | 2 | 0.24 | 8.21 | 2.49E-02 |
| Regulation of cell migration | 912 | 5 | 1.06 | 4.7 | 3.71E-03 |
| Regulation of cell motility | 963 | 5 | 1.12 | 4.45 | 4.67E-03 |
| Negative regulation of cell migration | 276 | 4 | 0.32 | 12.43 | 2.91E-04 |
| Negative regulation of cell motility | 289 | 4 | 0.34 | 11.87 | 3.46E-04 |
| Negative regulation of cellular component movement | 323 | 4 | 0.38 | 10.62 | 5.24E-04 |
| Definitive hemopoiesis | 21 | 2 | 0.02 | 81.67 | 3.25E-04 |
Within groups of GO terms, items are listed in order of increasing generality.
The FixedHR/PolyC GO analyses indicate terms: complement-receptor-mediated signaling pathway and response to pheromone. These terms were significant with a false discovery rate (FDR) correction (FDR < 0.05), P = 7.11E-04 and P = 2.40E-07, respectively) (Table 9). For FixedC/PolyHR, no GO terms were significantly enriched with FDR correction, some novel GO terms were deemed most significant. Included in these results is also CDP-choline pathway, which had also been implicated in the haplotype data. The full list of regions for both FixedHR/PolyC and FixedC/PolyHR can be found in (Table S2).
Table 9. Top GO results for FixedHR/PolyC implicated genes.
| GO term | Total genes | Input genes | Expected | Fold enrichment | Raw P-value |
|---|---|---|---|---|---|
| Response to pheromone | 104 | 8 | 0.63 | 12.7 | 3.93E-07 |
| Complement receptor mediated signaling pathway | 13 | 4 | 0.08 | 50.82 | 2.81E-06 |
| Phospholipase C-activating G protein-coupled receptor signaling pathway | 91 | 5 | 0.55 | 9.07 | 2.89E-04 |
| Exocytic insertion of neurotransmitter receptor to postsynaptic membrane | 8 | 3 | 0.05 | 61.93 | 3.40E-05 |
| Regulation of postsynaptic membrane neurotransmitter receptor levels | 62 | 3 | 0.38 | 7.99 | 7.09E-03 |
| Neurotransmitter receptor transport to postsynaptic membrane | 20 | 3 | 0.12 | 24.77 | 3.46E-04 |
| Neurotransmitter receptor transport to plasma membrane | 21 | 3 | 0.13 | 23.59 | 3.93E-04 |
| Vesicle-mediated transport to the plasma membrane | 90 | 3 | 0.54 | 5.51 | 1.87E-02 |
| Neurotransmitter receptor transport | 40 | 3 | 0.24 | 12.39 | 2.21E-03 |
| Establishment of protein localization to postsynaptic membrane | 21 | 3 | 0.13 | 23.59 | 3.93E-04 |
| Protein localization to postsynaptic membrane | 44 | 3 | 0.27 | 11.26 | 2.85E-03 |
| Protein localization to synapse | 76 | 3 | 0.46 | 6.52 | 1.21E-02 |
| Receptor localization to synapse | 51 | 3 | 0.31 | 9.72 | 4.23E-03 |
| Calcium ion import across plasma membrane | 9 | 2 | 0.05 | 36.7 | 1.91E-03 |
| Calcium ion import into cytosol | 10 | 2 | 0.06 | 33.03 | 2.28E-03 |
| Calcium ion transport into cytosol | 69 | 3 | 0.42 | 7.18 | 9.40E-03 |
| Positive regulation of cytosolic calcium ion concentration | 292 | 7 | 1.77 | 3.96 | 2.26E-03 |
| Regulation of cytosolic calcium ion concentration | 340 | 8 | 2.06 | 3.89 | 1.25E-03 |
| Cellular calcium ion homeostasis | 446 | 10 | 2.7 | 3.7 | 4.48E-04 |
| Calcium ion homeostasis | 463 | 10 | 2.8 | 3.57 | 5.95E-04 |
Within groups of GO terms, items are listed in order of increasing generality.
Targeted ontology:
The gene search for specific ontologies produced 45–820 genes and 7315–143,507 SNPs associated with each search (Table 10). The top 10 genes were chosen based on the most significant SNP within the gene (Table S4). The most significantly differentiated SNPs were generally found in genes associated with the brain, followed by bone and muscle related genes. Surprisingly, the reward-related ontologies (dopamine and serotonin) did not contain as strong evidence for differentiation as the others.
Table 10. Summary of ontology search.
| Search term | Total genes | Total SNPs | Top genes | Top P-value |
|---|---|---|---|---|
| Dopamin* | 254 | 43,890 | Gnb1, Fpra, Adora2a | 1.33E-04 |
| Serotonin | 45 | 7,315 | Htr7, Chrm2, Btbd9 | 9.33E-03 |
| Osteo* | 491 | 56,091 | Noct, Nf1, Mmp14 | 3.76E-05 |
| Cardiac | 820 | 143,507 | Myh11, Tbx5, Dlg1 | 7.25E-06 |
| “Skeletal Muscle” | 295 | 39,383 | Kel, Foxp1, Nf1 | 5.23E-06 |
| Brain | 667 | 123,416 | Sorl1, Gak, Fbxo45 | 1.92E-07 |
Genes are listed from most significant to least significant by SNP with lowest P-value.
Includes: Fpr1, Fpr2, Fpr3, Fpr-rs4 (all closely linked).
Consistent regions identified across multiple analyses
The major analyses (LM, haplotype, and FixedHR/PolyC) individually implicate ∼80, 24, and 46 differentiated genomic regions, respectively. Combined, 61 unique regions across the genome are indicated, including at least one region on every chromosome. Of these 61 regions, 12 are found in all three analyses (Table 11). These 12 consistent regions span just over 27.4 mbp and include 300 validated and predicted genes. Of the 300 genes, 77 are either olfactory or vomeronasal genes, which are predominantly located in two large regions on chromosomes 14 and 17. Surprisingly, many of these regions do not contain many of the most differentiated SNPs according to the multi-model MIVQUE analyses, but do have at least one SNP with P ≤ 0.001 by the LM criteria.
Table 11. Genomic regions implicated by LM, haplotype, and FixedHR/PolyC analyses.
| Chr | First bp | Last bp | Included genes |
|---|---|---|---|
| 5 | 108,000,623 | 108,679,807 | Tmed5, Ccdc18, Pigg, Mfsd7a, Gak, Tmem175, Slc26a1 |
| 6 | 41,584,862 | 41,918,440 | Trpv5, Trpv6, Ephb6, Kel, Llcfc1, Olfr459 |
| 7 | 29,603,841 | 29,697,093 | Catsperg2 |
| 9 | 41,240,184 | 42,275,833 | Sorl1, Mir100hg, Mir100, Mir125b-1, Mirlet7a-2, Tbcela |
| 11 | 79,724,263 | 80,090,780 | Atad5, Suz12, Utp6, Crlf3 |
| 11 | 112,227,183 | 114,489,018 | BC006965, Sox9 |
| 14 | 52,072,148 | 53,779,979 | Olfrb, Travb |
| 14 | 97,645,171 | 98,679,965 | Dach1 |
| 15 | 18,960,135 | 20,609,074 | Cdh10, Gm35496 |
| 15 | 71,023,429 | 71,559,595 | Fam135b |
| 16 | 31,540,757 | 33,178,952 | Gm536, Rnf168, Ubxn7, Fbxo45, Tnk2, Tnk2os |
| 17 | 17,895,909 | 22,396,753 | Vmn2rb |
Tbcel is most differentiated gene in genome based on median P-value.
Several genes in this gene family were represented in this region.
Discussion
Variation in genetic diversity
For the present sample of 79 mice from generation 61, based on the polymorphic SNPs within each line (Table 2), each of the lines continues to retain ∼34%–48% of the total diversity across all three lines. Such a drop in genetic diversity would be expected after 61 generation with ∼10 breeding pairs per generation per each line. We found no evidence that HR and C lines had differing levels of genetic diversity, averaged across the whole genome.
Consistent regions from multiple analyses
Many of the identified regions span too many genes to allow ready identification of a candidate. However, a few of the regions contain a limited number of genes for which the reported functions make sense in the context of directional selection for high voluntary wheel-running behavior (from first principles of physiology and neurobiology) and/or given previously identified differences between the HR and C lines (see Introduction). Given the rich phenotyping literature on the HR mouse selection experiment (>150 publications), we discuss a relatively large number of genes. Additional regions are covered in Supplemental Material (File S9).
The region identified on chromosome 5 includes 16 genes (excluding predicted and noncoding), three of which were previously identified as differentially expressed in the striatum of the HR and C mice (Saul et al. 2017). These genes include Tmed5, Gak, and Mfsd7a. Tmed5 is a trafficking protein associated with cell proliferation and WNT7B expression in HeLa cells (Yang et al. 2019). Mice knockouts in Gak are generally lethal to adult and developing mice causing various abnormal symptoms, including altered brain development (Lee et al. 2008). Mfsd7a (aka Slc49a3) has been associated with ovarian cancer, but much remains unknown about this gene (Khan and Quigley 2013).
The region on chromosome 6 includes Trpv5 and Kel, both of which are associated with KO phenotypes that may be tied to known differences between the HR and C lines. Trpv5 KO is associated with phenotypes related to structural changes in the femur and kidney physiology (Hoenderop et al. 2003; Loh et al. 2013), both of which differ between HR and C lines (Swallow et al. 2005; Castro and Garland 2018). Trpv5 is also associated with calcium homeostasis (Hoenderop et al. 2003; Loh et al. 2013). Kel is a blood group antigen with KO phenotypes affiliated with weakness, gait, and motor coordination, neurological development, and heart function (Zhu et al. 2009, 2014). Previous experiments have shown the HR and C mice to have differences in heart physiology (Kolb et al. 2013a), gait and motor coordination (Claghorn et al. 2017), and brain development (Kolb et al. 2013b).
The region on chromosome 9 contains various predicted genes and miRNA, but also one large gene of interest, Sorl1 (aka SorlA). This gene is also implicated in our targeted search for genes related to the brain (Table 10). Sorl1 codes for a sorting receptor that has been associated with various neural and metabolic diseases (Schmidt et al. 2017). Although some of the associated phenotypes, such as obesity, may have some correlation to phenotypic differences between HR and C mice, such as difference in body fat (Swallow et al. 2001; Vaanholt et al. 2008; Hiramatsu and Garland 2018), this does not directly answer the question of how Sorl1 influences running behavior. Mouse knockouts in this gene have not shown changes in running gait (Rohe 2008), whereas differences in gait do exist between HR and C mice (Claghorn et al. 2017). However, these treadmill tests do not address exercise motivation, which might be influenced by such a neurobiologically relevant gene. Additionally, a more significantly differentiated haplotype can be found over 150,000 bp downstream of Sorl1, containing various predicted genes and miRNA. Therefore, further studies will be required to determine precisely the elements of this region that modulate wheel running. Although Tbcel is near this consistent region rather than included in it, it is the most differentiated gene in the genome (based on median P-value of included SNPs, P = 4.01E-07). This gene is known to regulate tubulin activity in sperm and the nervous system (Nuwal et al. 2012; Frédéric et al. 2013).
One region on chromosome 11 contains numerous genes of potential interest. One LM within this region is proximal to a handful of genes that may be influencing the HR phenotype, including: Tefm, Adap2, Crlf3, and Suz12. These genes are associated with KO phenotypes including enlarged heart and decreased body weight (Jiang et al. 2019), blood cell concentration (White et al. 2013), and brain morphology (Miro et al. 2009). All of these phenotypes have been found to differ between HR and C mice (Kolb et al. 2013b; Thompson 2017; Singleton and Garland 2019).
One region on chromosome 14 includes almost exclusively Dach1, which is an important regulator for various early developmental genes. Dach1 is a regulator of muscle satellite cell proliferation and differentiation (Pallafacchina et al. 2010). Although knockouts of Dach1 in mice do not appear to disrupt limb development (Davis et al. 2001), Dach1 mutants sometimes have stunted leg development in Drosophila (Mardon et al. 1994). Furthermore, Dach1 has been shown to localize around limb budding regions and interact with known limb patterning genes in both mice and poultry (Horner et al. 2002; Kida 2004; Salsi et al. 2008). Studies of skeletal muscle (Garland et al. 2002; Bilodeau et al. 2009) and of the peripheral skeleton show several differences between HR and C lines of mice (Garland and Freeman 2005; Kelly et al. 2006; Castro and Garland 2018; Schwartz et al. 2018). This gene has also been implicated in the development and function of the kidneys (Köttgen et al. 2010), which have been shown to be larger in the HR lines than C lines in some studies (Swallow et al. 2005).
A region on chromosome 15 includes Cdh10 among a few predicted genes. GO links Cdh10 to both “calcium ion binding” and “glutamatergic synapse,” terms that occasionally produced suggestive P-values for enrichment searches in our differentiation analyses (Table 7 and Table 9). These terms could have various implications for the HR mice. Cdh10 specifically is a cadherin with extensive expression in the brain (Liu et al. 2006; Matsunaga et al. 2015). This gene has been shown to have increased expression in phrenic neurons (Machado et al. 2014), potentially modulating diaphragm movement, and increased functionality of the diaphragm could partly underlie the elevated maximal rate of oxygen consumption during exercise (VO2max) observed in HR lines (Kolb et al. 2010; Hiramatsu et al. 2017; Singleton and Garland 2019). Cdh10 has also been shown to have increased expression in regions associated with olfactory system development (Akins et al. 2007) which could be corroborated by the other two consistent regions associated with olfactory and vomeronasal (see Results, General ontology). The other region detected on chromosome 15 currently only contains Fam135b among its annotations. Few studies have been conducted involving the function of Fam135b, but evidence indicates it has an important role in spinal motor neurons based on a >10,000-fold decrease in expression in spinal and bulbar muscular atrophy models (Sheila et al. 2019).
The region we identified on chromosome 16 contains various genes that may influence wheel running behavior. One example is Fbxo45, which has demonstrated itself essential for neuronal development (Saiga et al. 2009) and synaptic transmission (Tada et al. 2010). One gene that particularly caught our attention was Pcyt1a, which is an important modulator of the CDP-choline pathway, catalyzing the formation of CDP-choline (Andrejeva et al. 2020), also known as citicoline. Citicoline has been researched extensively for its clinical applications and has demonstrated capacity to stimulate dopamine synthesis in nigrostriatal areas (Drago et al. 1989, cited in Secades and Lorenzo (2006)), which are important for exercise and reward (Wise 2009). Additionally, CDP-choline has shown evidence of modulating dopamine receptors in the striatum (Giménez et al. 1991).
Ontology
General ontology:
The GO analyses in this paper serve two functions. The first includes determining pathways that have been influenced by the selective breeding protocol. Additionally, the vast publications and data on various morphological and physiological differences between the HR and C lines provide insight into differentiated biological processes.
The Haplotype and Fixed/Poly methods of identifying differentiated genes had considerable overlap between genes and regions identified, which seems to result in similar GO terms for these analyses. The term “sensory perception of chemical stimulus” is expected, given the large number olfactory and vomeronasal genes present in some of these regions. Selection for such genes is likely in response to how the mice are tested for wheel running. For logistical reasons, around two-thirds of the mice tested in a given generation were measured on wheels that had not been washed since the previous mouse was on that same wheel, although the attached cages were fresh (Dewan et al. 2019). The scent of the previous mouse would potentially elicit different running behavior, dependent on these vomeronasal and olfactory genes (e.g., see Drickamer and Evans 1996). We checked the Allen Brain Atlas for some of these genes (particularly those in the consistent region on chromosome 17) and found that only a few of these olfactory and vomeronasal genes had data. One of these includes Vmn2r107, with expression most consistent around the olfactory bulb. However, Olfr1509 had expression levels seemingly around the anterior cingulate cortex, a region associated with cognitive control of motor behavior (Holroyd et al. 2004). GO terms related to postsynaptic neurotransmitters were largely indicated by three genes. Cplx1 has been linked to severe ataxia and movement limitations in knockout rats (Xu et al. 2020), Dlg1 (aka SAP97) is a scaffolding protein that localizes glutamate receptors in postsynaptic membranes and has shown altered expression in rats exposed to cocaine (Caffino et al. 2018), and Shisa6 has been associated with the localization of AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptors (Klaassen et al. 2016), which have shown reduced expression after prolonged cocaine exposure (Cooper et al. 2017). Such terms are perhaps not surprising, given observations of the HR mice having larger midbrains and altered reward mechanisms (Belke and Garland 2007; Mathes et al. 2010; Garland et al. 2011b; Keeney et al. 2012; Kolb et al. 2013b; Thompson et al. 2017).
The local maxima GO results are generally quite different from the haplotype and Fixed/Poly analyses. This is partially attributable to less overlapping of identified genomic regions. Additionally, LM is useful for gene culling to reduce influence of hitchhiking genes in the GO analyses. Many of the top terms for LM genes are associated with heart development and function. Heart ventricle mass is greater in the HR mice (Kolb et al. 2013a; Kelly et al. 2017; Kay et al. 2019) and correlates with VO2max in both HR and C mice (Rezende et al. 2006). The genes most associated with cardiac development include Pkp2, Myh11, and Tbx5 (also a forelimb regulator). Forelimb development may be altered in the HR mice, while humerus sizes do not seem to differ (Copes et al. 2018), differences have been found in metatarsal and metacarpal lengths (Young et al. 2009).
Targeted ontology:
As the target ontologies were chosen based on structures and systems known to have been altered by the selective breeding regimen, we would expect to find at least one gene of each ontology that would contain a differentiated SNP. Of these ontologies, “serotonin” and “dopamine” are associated with some of our less impressive P-values (Table 10), with many of the top dopamine-related genes (Fpr1, Fpr2, Fpr3, and Fpr-rs4) being present potentially because of linkage to highly differentiated vomeronasal genes (Table 10). However, expression data from the Allen Brain Atlas implicates the Fpr-rs3 gene as being highly expressed in nucleus raphe obscurus. The nucleus raphe structure is well established for modulating serotonin (Walker and Tadi 2020) and the obscurus region itself has been implicated in modulating respiratory neurons (Lalley et al. 1997). As Fpr-rs3 is the most differentiated gene of the FPR family (median P = 0.000393 over six SNPs), it may be contributing to the selection signature of this genomic region rather than simply hitchhiking. The most significantly differentiated loci in a dopamine-related gene are in Gnb1, part of the Gβγ complex, which activates Girk2 in dopamine neuron membranes (Wang et al. 2016). We are surprised not to have found more impressive results for dopamine-related genes, given clear differences in dopamine function between the HR and C mice (Rhodes et al. 2001, 2005; Rhodes and Garland 2003; Bronikowski et al. 2004; Mathes et al. 2010). A possible explanation for this is that trans-regulating sites for these genes have been more influenced by the HR selection regime (Kelly et al. 2012; Nica and Dermitzakis 2013). Unfortunately, a limitation of the current study is it lacks the necessary expression data to identify trans-regulating SNPs (Kelly et al. 2012, 2014).
The remaining ontologies (bone, cardiac, skeletal muscle, and brain) all have at least one gene containing a SNP with P < 0.0001 (Table 10). Some of these are included with our LM genes, such as Myh11 (a myosin gene affiliated with the “cardiac” tag) and Sorl1 (“Brain” tag). However, some of these are not present among the LM list. Kel, described above as influencing various phenotypes relevant for high running behavior, may appear to be a confusing “miss” for the LM detection process, with a P-value = 1.49E-05. However, the region does have two local maxima, neither of which land in genes, but one is ∼15,000 bp upstream of Kel. This might be taken as evidence that the LM approach to determining affected genes ought to be modified to better catch nearby genes that could be affected.
The expression patterns of the top genes implicated by the “brain” targeted ontology were determined using the Allen Brain Atlas. The top four genes (Sorl1, Gak, Fbxo45, and Tbx3) showed interesting consistency in their expression patterns. Sorl1, Gak, and Fbxo45 all have increased expression around the hippocampus, which has been associate with spatial learning (Schiller et al. 2015) and may play a role in addiction (Koob and Volkow 2010). Sorl1, Gak, and Tbx3 have higher expression in the retrospenial area, which has also been suggested as a potential modulator of spatial memory (Vann et al. 2009), potentially in coordination with the hippocampus (Schiller et al. 2015). Gak and Tbx3 both have notable expression levels in the retrohippocampal region, particularly the entorhinal cortex, which is thought to modulate movement speed (Geisler et al. 2007; Kropff et al. 2015; Ye et al. 2018). Additionally, Gak, Fbxo45, and Tbx3 have high expression in olfactory regions.
The hippocampus has been linked to the regulation of speed during locomotor behavior in both mice and rats by theta (Li et al. 2012; Fuhrmann et al. 2015; Sheremet et al. 2019), gamma (Chen et al. 2011; Ahmed and Mehta 2012), and delta oscillations (Furtunato et al. 2020). Notably, the difference in daily running distance between HR and control lines is attributable mainly to an increase in average (and maximum) running speed, rather than the duration of running, especially in females (e.g., see Garland et al. 2011a; Claghorn et al. 2016, 2017; Copes et al. 2018; Hiramatsu and Garland 2018). Another consideration is the impact of physical activity on neurogenesis in the hippocampus (Rhodes et al. 2003b; Clark et al. 2010; Rendeiro and Rhodes 2018), which, perhaps, could create a sort of feedback loop relating to running speed.
Comparison with previous studies
Exercise behavior and the genetic factors that affect it have been the subject of various other GWAS and gene expression studies in mice, as well as comparisons of inbred strains (reviews in Kostrzewa and Kas 2014; Lightfoot et al. 2018). In general, these previous studies do not show strong agreement with each other. The primary exception is that several studies have implicated dopamine pathway genes (Bronikowski et al. 2004; Lightfoot 2011; Dawes et al. 2014; Roberts et al. 2017). This is of little surprise, as dopamine has been long recognized as a primary neurotransmitter involved with physical activity (Freed and Yamamoto 1985; Rhodes et al. 2005). As another example of consistencies across previous studies, Dawes et al. (2014) found differential gene expression in C57L/J (high running) and C3H/HeJ (low running) inbred strains for Mstn, a gene previously implicated by Lightfoot et al. (2010) using 41 inbred strains of mice to associate alleles with wheel running. Mstn is established as a regulator of skeletal muscle proliferation (Grobet et al. 1997; Amthor et al. 2007; Mosher et al. 2007). The present study contributes several new regions that have not been previously identified (see above). However, we can also identify examples of overlapping results.
We first compiled a list of genes from our study that contain at least one variable SNP (see Materials and Methods). For each gene, all of the SNPs within the transcribed or promotor region were accumulated and the lowest P-value and median P-value (from File S4) were recorded. These are presented in File S11. We then cross-reference these P-values (with emphasis on median P-value) against the regions and genes identified by previous studies. This method is limited by not addressing regulatory loci located outside the promotor and transcribed region. For the previous studies, we focused on regions, SNPs, and genes that were specifically associated with running distance, rather than speed or duration of running (if reported), as the HR mice were specifically bred for running distance.
Shimomura et al. (2001) performed an F2 cross between BALB/cJ and C57BL/6J and mapped daily running levels in constant darkness. Although the primary purpose of their study was to identify circadian QTL, two regions were associated directly with wheel-running distance. One of these regions is on chromosome 16 (97,608,543–97,608,688 bp, mm10), not far from one of our local maxima (96,795,226 bp, P = 4.97E-04).
A study involving a cross between high- and low-running inbred strains located several markers on both chromosome 9 and chromosome 13 (Lightfoot et al. 2008). Although none of these markers fall within our own significant region on chromosome 9 (∼41,000,000–42,000,000 bp), one of the markers identified by Lightfoot et al. (2008) on chromosome 9 is only ∼500,000 bp from the gene Leo1. For our sample of mice, only one SNP in this gene was polymorphic, and it was in the noncoding region (File S11: P = 0.00186)
Lightfoot et al. (2010) used haplotype association mapping to identify 12 QTL associated with wheel running among 41 inbred strains of mice. One of the regions they identified on chromosome 5 (114,584,508–117,669,848 bp after conversion to mm10) is intriguingly close to one of our own haplotype regions (118,824,587–119,299,787 bp, Table 5). Additionally, we detected a local maximum on chromosome 12 (88,919,735 bp, P = 7.54E-05) near their identified haplotype (88,113,842–88,220,086 bp, mm10). Lightfoot et al. (2010) also identified a region on chromosome 13 (95,477,271–95,863,515 bp, mm10), which coincides with a few of our FixedHR/PolyC loci (95,595,237–95,947,205 bp). Aside from these, the best example of similarity with the present study is a gene on chromosome 8 (Galntl6) that was found as suggestive in the current study (File S11, median P = 0.039, SNPs= 5925). Lightfoot et al. (2010) also identified a region on chromosome 12, ∼0.5 mbp upstream of Nrxn3. Both our LM and FixedHR/PolyC methods indicated this gene as a strong candidate, with a segment of intron 1 containing several low P-values (median P = 2.04E-04, SNPs = 195), but it was not listed as a consistent region because the haplotype results did not produce a significant haplotype near Nrxn3. Nrxn3 is a single-pass transmembrane protein found in presynaptic terminals and functions as a cell adhesion molecule (Stoltenberg et al. 2011; Kasem et al. 2018). Nrxn3 creates particular interest in that it is associated with various addictive behaviors (Zheng et al. 2018), which is consistent with evidence that the HR mice are to some extent addicted to running (Rhodes et al. 2005; Kolb et al. 2013b). Previous work has associated Nrxn3 with addictive behaviors involving nicotine (Wolock et al. 2013) and opioids (Lachman et al. 2007), predominantly through association and expression studies (Kasem et al. 2018). Exercise addiction is not a new concept, but remains controversial (Nogueira et al. 2018).
QTL mapping of the G4 intercross of C57BL/6J with one of the four HR lines implicated a region on chromosome 7 (101–130 mbp) that contains numerous olfactory/vomeronasal genes (Kelly et al. 2010). We identified FixedHR/PolyC SNPs within that region at 127,385,309–127,947,542 bp. We also identified vomeronasal genes on chromosome 17. (Kelly et al. (2010) reported other QTL associated with running on the first 2 days of wheel exposure, but this phenotype may reflect variation in neophobia more than exercise motivation or ability.)
Saul et al. (2017) performed expression analysis using the striatum of the HR and C lines from generation 66. The mice were sampled after several hours of wheel deprivation, which is believed to induce high expression of motivation-related genes (Rhodes et al. 2003a). Some of their highlighted differentially expressed genes include: Htr1b, Slc38a2, Tmed5, 5031434O11Rik, Gak, Mfsd7a, and Gpr3. Tmed5, Gak, and Mfsd7a are all found within a highly differentiated region in our SNP data (median P = 4.85E-04 for all three genes, SNPs = 671, File S11). Although 5031434O11Rik and the associated Setd7 are not found within our consistent regions (due to no FixedHR/PolyC SNPs), they both contain many of the most differentiated loci of individual SNP analyses (median P = 3.78E-05, SNPs = 4). Knockouts of Setd7 (aka Set9) have been associated with altered lung development and morphology (Elkouris et al. 2016). Lung differences in the HR and C lines have not been greatly explored. Three studies have reported no statistical difference in lung mass (Meek et al. 2009; Kolb et al. 2010; Dlugosz et al. 2013), but an unpublished study of males from generation 21 found that HR lines tended to have higher pulmonary diffusion capacity and capillary surface area determined via morphometry (T. Garland, and S. F. Perry, personal communication), and a study of females from generation 37 reported a trend for HR mice to have higher dry lung mass (Meek et al. 2009; Kelly et al. 2017). We are uncertain of what Setd7 may be doing in the brain. However, the Allen Brain Atlas does indicate increased expression levels of Setd7 in the sensory regions of the midbrain, motor related regions of the medulla, and the cerebellar cortex, which has been associated with motor function and reward (Doya 2000). Furthermore, Setd7 has been shown to modulate pain and inflammation following nerve injury, potentially enabling an individual to proceed to exercise despite injury (Shen et al. 2019).
Overall, studies attempting to identify the genetic underpinnings of exercise behavior in rodents have produced a wide variety of results. We can offer several reasons for such inconsistencies. First, some of these studies address gene expression (Bronikowski et al. 2003, 2004; Dawes et al. 2014; Saul et al. 2017) and eQTL (Kelly et al. 2012, 2014), which will commonly implicate different genetic factors for complex traits than studies looking at genetic variants, likely as a result of complex interactions between genetic variants and gene expression (Bouchard 2015; Parker et al. 2016). Second, some studies compare inbred strains (Lightfoot et al. 2008, 2010; Dawes et al. 2014) with very different genetic histories and likely different biologically significant alleles available to them than in the Hsd:ICR mice that formed the basis for the present selection experiment. Furthermore, a trait as complex as voluntary exercise (Lightfoot et al. 2018) would be expected to have numerous underlying subordinate traits, which, in turn, could have innumerable potential genetic factors modulating them (Garland et al. 2016; Sella and Barton 2019). Finally, in the current study, we sought to detect specifically those factors that are shared across all four HR lines, which likely does not reflect all of the exercise-relevant loci that vary among the replicate HR lines. However, those alleles implicated by all four HR lines arguably provide the strongest evidence for biologically significant regions in this selection experiment and also for the Hsd:ICR base population.
Mini-muscle allele
The mini-muscle phenotype was discovered in the HR selection experiment and is associated with alterations in various organs, especially skeletal muscle, but also including heart, kidney, and overall body mass of the mice (Swallow et al. 2005; Meek et al. 2009; Kolb et al. 2013a; Talmadge et al. 2014; Kay et al. 2019) as well as behaviors (Kelly et al. 2006; Singleton and Garland 2019). This phenotype is caused by a single recessive SNP mutation located in an Myh4 (myosin heavy polypeptide 4) gene (Kelly et al. 2013). Mice expressing the mini-muscle phenotype have often been found to run faster and sometimes for longer distances than other HR mice (Kolb et al. 2013a). This polymorphism was lost, presumably via random genetic drift, from all lines except for HR lines 3 (where it went to fixation) and line 6 (where it remains polymorphic with the wildtype allele). Population-genetic analyses indicate that the allele was under positive selection in the HR lines (Garland et al. 2002). The current WGS data show (generation 61) that the mutation is still only present in lines 3 (fixed) and 6, with allele frequency of 0.65 in line 6. As the mini-muscle phenotype appears to enable faster overall running on wheels at the cost of running duration, it has been regarded as an alternative “solution” to the selection criterion (Garland et al. 2011a), not unlike the concept of “private” alleles (Martin et al. 1996). Such a mutation is expected to change the genetic background of line 3 (and to a lesser extent, line 6) giving rationale to analyzing these lines separately for possible QTL, in future studies.
Allele frequency implications
The general pattern of allele frequencies across the replicate lines can be used to infer patterns of selection. Table 12 includes some of the potential profiles that could possibly be observed and (for the most part) were observed in the WGS data.
Table 12. Potential fixation profiles.
| Test Group 1 | Test Group 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Profile | Rep 1 | Rep 2 | Rep 3 | Rep 4 | Rep 1 | Rep 2 | Rep 3 | Rep 4 |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| 3 | Het | Het | Het | Het | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 |
Profile 1:
No observed genetic variation. For our 79 mice, this accounts for ∼99.8% of the genome (Table 2).
Profile 2:
Fixation for alternate alleles in the two selection treatments would imply opposing directional selection, as might occur in experiments with replicate lines selected for high vs. low values of a trait. The HR mouse selection experiment includes high-selected and control treatments, but not a low-selected treatment. Thus, fixation for alternate alleles in the HR and C lines would not necessarily be expected, and indeed was never observed for either the WGS data or the MegaMUGA data reported previously (Xu and Garland 2017). Importantly, even data from selection experiments that include high- and low-selected treatments are not showing much evidence of fixation for alternate alleles (Burke et al. 2010; Lillie et al. 2019).
Profile 3:
Stabilizing selection or random drift for one group and directional selection for the other. This was the focus of the scans for loci fixed in all lines of one linetype and polymorphic in all lines of the other (Fixed/Poly) in our own haplotype and WGS data and produced several prospective regions of interest. The fixed allele can either be entirely the reference (0) or alternative (1).
Profile 4:
Selection for test group 2 but evidence of drift for group 1 (likely caused by little to no selection). Some of the loci of the WGS SNP data meet this profile. For example, Chromosome 11: 96,332,082 bp (P = 0.051).
Profile 5:
Random genetic drift for both test groups. Such loci will be among those analyzed, but this pattern of differentiation is unlikely to result from the selective breeding regimen.
In general, as with any population that is relatively well adapted to the prevailing environmental conditions, breeding colonies of laboratory house mice maintained under standard vivarium housing conditions should experience continuing stabilizing selection at many loci. Under standard housing conditions, an allele with a strong positive influence on wheel running, or activity in cages without wheels, might be disfavored if it were negatively associated with such aspects of the life history as litter size or maternal care. In contrast, under the conditions of the HR mouse selection experiment, an allele with a strong positive influence on wheel running might be expected to go to fixation rapidly in all HR lines in a manner consistent with a “complete sweep” (Burke 2012). Thus, to fix an allele, directional selection in the HR lines must be strong enough to overcome a presumed prevailing background of stabilizing selection and possibly negative selection. Regions that are FixedHR/PolyC (profile 3) should, therefore, be indicative of relatively strong directional selection in the HR lines.
Alternatively, some loci may have come under stabilizing selection in the HR lines, e.g., due to heterozygote advantage or epistatic interactions with other loci, preventing them from going to fixation. Hence, we also examined loci polymorphic in all HR lines but fixed in all C lines (FixedC/PolyHR). The GO analyses of the included genes in these regions were consistently less significant (raw P ≥ 0.0026 for all implicated terms). However, such terms as “synapse assembly” and those related to glycerolipids emerged may merit further exploration.
Interpretation of the four models
The four models in the multi-model analysis were included to allow for different variance structures within and between the HR and C linetypes. The within-line variance is the variability of allele frequency among the ∼10 mice within each line. This variance is zero when a line is fixed for one allele or another, but maximized when five mice within each line are homozygous for one allele while five mice are homozygous for the other. The among-line variance indicates how different the replicate lines within a linetype are from each other. This variance component is minimized when all four lines within a linetype are fixed for the same allele, but maximized when two lines are fixed for one allele while two lines are fixed for the other.
In principle, both the within-line and among-line variances can differ between the two selection treatments (linetypes); hence, the Full model includes separate estimates of both within- and among-line variances. For wheel running in later generations of the selection experiment, a full model has been shown to fit well (Garland et al. 2011a). The SepVarInd model includes only the within-line variance. The SepVarLine model includes only the among-line variance. Lastly, the Simple model does not include either of these two variances, and corresponds to the single model used by Xu and Garland (2017).
As expected, we found many loci that were better fit by models other than the Simple model used by Xu and Garland (2017) (Table 4). Figure 3 gives examples. In A, the Full model is implemented because C lines exhibit very little within- and among-line variance while HR lines exhibit both. In B, the SepVarInd model is used because C lines have high within-line variance (while HR lines are comparatively low), but both have similar among-line variance. In C, SepVarLines model is used because nearly all lines contain very little within-line variance (6 are fixed for a single allele), but C lines, being fixed for opposing alleles, creates different among-line variance. D identifies a Simple model locus because these variances are roughly the same for the different linetypes. E represents a locus with no within-line variance and thus could not be analyzed with the mixed model ANOVA like other loci. However, use of multiple models did not increase the number of loci identified as statistically significant based on repeat analyses of the MEGAMuga data with both methods (Figure 1).
Figure 3.
These are images of different variance structures depicted by actual examples from the MegaMUGA data (Xu and Garland 2017). This includes example data that were best fit by the “Full” model, marker = JAX00157775, P = 0.1008 (A), “SepVarInd” model, marker = UNC25816949, P = 0.006754 (B), “SepVarLines” model, marker = JAX00170011, P =0.04885 (C), the “Simple” model, marker = UNC080521812, P = 0.2687 (D), or has no within line variance, thus not fitting any model, marker = backupJAX00272176 (E).
Summary, Limitations, and Future Directions
Exercise, or the lack of exercise, has far-reaching medical and financial implications (Manley 1996; Booth et al. 2012; Carlson et al. 2015). Numerous studies have provided strong evidence for the existence of genetic underpinnings of exercise behavior and physical activity (Kostrzewa and Kas 2014; Lightfoot et al. 2018), including in the HR mouse selection experiment (Bronikowski et al. 2003, 2004; Careau et al. 2013; Saul et al. 2017; Xu and Garland 2017). Here, we used three different analytical methods with WGS data to address the genetic basis of the threefold increase in daily running distances observed in the four replicate selectively bred HR lines of mice. These methods include haplotype and SNP statistical analysis, as well as nonstatistical analysis of fixation patterns in HR and C lines.
The intersection of multiple analyses indicated 61 genomic regions of differentiation, with 12 identified as of particular interest. These regions include genes known to influence systems that have already been demonstrated to differ between HR and C mice, such as response to conspecific odors, brain development, body weight, and relative heart size. However, they also contain genes whose role in voluntary running behavior is as yet unclear.
This study does have the limitation of focusing on males, whereas exercise behavoir and much of the physiology and morphology related to exercise abilities differ between sexes in both rodents and humans (Eikelboom and Mills 1988; Thomas and Thomas 1988; Rowland 2016; Sheel 2016; Rosenfeld 2017; Thompson et al. 2017). A natural next step would then be to conduct similar analyses in females. This approach, however, can establish correlation but not causation. Therefore, studies of wheel-running behavoir of mice with knockouts or Cre modifications of genes in some of the genomic regions identified here may help to establish or dismiss causal relationships between the genes and phenotype. Furthermore, as the HR mouse experiment has complete pedigree information for all mice and lines (Careau et al. 2013, 2015), it will also be possible to use this information to better account for relatedness between mice in statistical analyses and so provide more informed estimates of loci acted upon by selection.
Importantly, none of the analytical approaches we used address the possibility of “private alleles” (Martin et al. 1996) in one or more of the HR lines that may influence exercise behavior, thus representing “multiple solutions” to the selective breeding regime (Garland et al. 2011a), but this will be an important possibility to consider in future studies. We already know of one private allele of major effect (mini-muscle) that has far-reaching effects on mouse muscle and organ development (Swallow et al. 2005; McGillivray et al. 2009; Kelly et al. 2013), as well as many other aspects of the phenotype, and has been favored by the selection protocol (Garland et al. 2002). Determination of such alleles will be an important area for future research.
Acknowledgments
Supported by National Science Foundation (NSF) grant DEB-1655362 to T.G.
Author contributions: Conceptualization, D.A.H., L.Y., F.P.M.dEV., D.P., S.X., F.C., T.G.; investigation, D.A.H., L.Y., G.M.W., F.P.M.dEV., D.P., A.S.F., F.C., T.G.; software, D.A.H., L.Y., S.X.; formal analysis, D.A.H., L.Y., A.S.F., S.X., F.C., T.G.; writing – original draft, D.A.H., L.Y., A.S.F., F.C., T.G.; writing – review and editing, D.A.H., L.Y., G.M.W., F.P.M.dEV., D.P., A.S.F., S.X., F.C., T.G.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12436649.
Communicating editor: A. Palmer
Literature Cited
- Aebi M., van Donkelaar M. M. J., Poelmans G., Buitelaar J. K., Sonuga-Barke E. J. S. et al. , 2016. Gene-set and multivariate genome-wide association analysis of oppositional defiant behavior subtypes in attention-deficit/hyperactivity disorder. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 171: 573–588. 10.1002/ajmg.b.32346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed O. J., and Mehta M. R., 2012. Running speed alters the frequency of hippocampal gamma oscillations. J. Neurosci. 32: 7373–7383. 10.1523/JNEUROSCI.5110-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins M. R., Benson D. L., and Greer C. A., 2007. Cadherin expression in the developing mouse olfactory system. J. Comp. Neurol. 501: 483–497. 10.1002/cne.21270 [DOI] [PubMed] [Google Scholar]
- Amthor H., Macharia R., Navarrete R., Schuelke M., Brown S. C. et al. , 2007. Lack of myostatin results in excessive muscle growth but impaired force generation. Proc. Natl. Acad. Sci. USA 104: 1835–1840 (erratum: Proc. Natl. Acad. Sci. USA 104: 4240). 10.1073/pnas.0604893104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrejeva G., Gowan S., Lin G., Wong Te Fong A.-C. L., Shamsaei E. et al. , 2020. De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy. Autophagy 16: 1044–1060. 10.1080/15548627.2019.1659608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belke T. W., and Garland T. Jr, 2007. A brief opportunity to run does not function as a reinforcer for mice selected for high daily wheel-running rates. J. Exp. Anal. Behav. 88: 199–213. 10.1901/jeab.2007.62-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin E. J., Muntner P., Alonso A., Bittencourt M. S., Callaway C. W. et al. , 2019. Heart disease and stroke statistics—2019 update: a report from the American heart association. Circulation 139: e56–e528 [Corrigenda: Circulation 141: e33 (2020)]. 10.1161/CIR.0000000000000659 [DOI] [PubMed] [Google Scholar]
- Bilodeau G. M., Guderley H., Joanisse D. R., and Garland T. Jr, 2009. Reduction of type IIb myosin and IIB fibers in tibialis anterior muscle of mini-muscle mice from high-activity lines. J. Exp. Zool. Part Ecol. Genet. Physiol. 311A: 189–198. 10.1002/jez.518 [DOI] [PubMed] [Google Scholar]
- Blair S. N., and Morris J. N., 2009. Healthy Hearts—and the universal benefits of being physically active: physical activity and health. Ann. Epidemiol. 19: 253–256. 10.1016/j.annepidem.2009.01.019 [DOI] [PubMed] [Google Scholar]
- Booth F. W., Chakravarthy M. V., Gordon S. E., and Spangenburg E. E., 2002. Waging war on physical inactivity: using modern molecular ammunition against an ancient enemy. J. Appl. Physiol. 93: 3–30. 10.1152/japplphysiol.00073.2002 [DOI] [PubMed] [Google Scholar]
- Booth F. W., Roberts C. K., and Laye M. J., 2012. Lack of exercise is a major cause of chronic diseases, Comprehensive Physiology, edited by Terjung R., John Wiley & Sons, Inc., Hoboken, NJ: 10.1002/cphy.c110025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C., 2015. Exercise genomics—a paradigm shift is needed: a commentary: Table 1. Br. J. Sports Med. 49: 1492–1496. 10.1136/bjsports-2015-095294 [DOI] [PubMed] [Google Scholar]
- Britton S. L., and Koch L. G., 2001. Animal genetic models for complex traits of physical capacity. Exerc. Sport Sci. Rev. 29: 7–14. 10.1097/00003677-200101000-00003 [DOI] [PubMed] [Google Scholar]
- Bronikowski A. M., Carter P. A., Morgan T. J., Garland T. Jr., Ung N. et al. , 2003. Lifelong voluntary exercise in the mouse prevents age-related alterations in gene expression in the heart. Physiol. Genomics 12: 129–138. 10.1152/physiolgenomics.00082.2002 [DOI] [PubMed] [Google Scholar]
- Bronikowski A. M., Rhodes J. S., Garland T. Jr., Prolla T. A., Awad T. A. et al. , 2004. The evolution of gene expression in mouse hippocampus in response to selective breeding for increased locomotor activity. Evolution 58: 2079–2086. 10.1111/j.0014-3820.2004.tb00491.x [DOI] [PubMed] [Google Scholar]
- Burke M. K., 2012. How does adaptation sweep through the genome? Insights from long-term selection experiments. Proc. Roy. Soc. B 279: 5029–5038. 10.1098/rspb.2012.0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M. K., Dunham J. P., Shahrestani P., Thornton K. R., Rose M. R. et al. , 2010. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467: 587–590. 10.1038/nature09352 [DOI] [PubMed] [Google Scholar]
- Caffino L., Messa G., and Fumagalli F., 2018. A single cocaine administration alters dendritic spine morphology and impairs glutamate receptor synaptic retention in the medial prefrontal cortex of adolescent rats. Neuropharmacology 140: 209–216. 10.1016/j.neuropharm.2018.08.006 [DOI] [PubMed] [Google Scholar]
- Careau V., Wolak M. E., Carter P. A., and Garland T. Jr, 2013. Limits to behavioral evolution: the quantative genetics of a complex trait under directional selection. Evolution 67: 3102–3119. 10.1111/evo.12200 [DOI] [PubMed] [Google Scholar]
- Careau V., Wolak M. E., Carter P. A., and Garland T. Jr, 2015. Evolution of the additive genetic variance–covariance matrix under continuous directional selection on a complex behavioural phenotype. Proc. Biol. Sci. 282: 1819 10.1098/rspb.2015.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson S. A., Fulton J. E., Pratt M., Yang Z., and Adams E. K., 2015. Inadequate physical activity and health care expenditures in the United States. Prog. Cardiovasc. Dis. 57: 315–323. 10.1016/j.pcad.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A. A., and Garland T. Jr, 2018. Evolution of hindlimb bone dimensions and muscle masses in house mice selectively bred for high voluntary wheel-running behavior. J. Morphol. 279: 766–779. 10.1002/jmor.20809 [DOI] [PubMed] [Google Scholar]
- Chen Z., Resnik E., McFarland J. M., Sakmann B., and Mehta M. R., 2011. Speed controls the amplitude and timing of the hippocampal gamma rhythm. PLoS One 6: e21408 10.1371/journal.pone.0021408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claghorn G. C., Fonseca I. A. T., Thompson Z., Barber C., and Garland T. Jr, 2016. Serotonin-mediated central fatigue underlies increased endurance capacity in mice from lines selectively bred for high voluntary wheel running. Physiol. Behav. 161: 145–154. 10.1016/j.physbeh.2016.04.033 [DOI] [PubMed] [Google Scholar]
- Claghorn G. C., Thompson Z., Kay J. C., Ordonez G., Hampton T. G. et al. , 2017. Selective breeding and short-term access to a running wheel alter stride characteristics in house mice. Physiol. Biochem. Zool. 90: 533–545. 10.1086/692909 [DOI] [PubMed] [Google Scholar]
- Clark P. J., Kohman R. A., Miller D. S., Bhattacharya T. K., Haferkamp E. H. et al. , 2010. Adult hippocampal neurogenesis and c-Fos induction during escalation of voluntary wheel running in C57BL/6J mice. Behav. Brain Res. 213: 246–252. 10.1016/j.bbr.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S., Robison A. J., and Mazei-Robison M. S., 2017. Reward circuitry in addiction. Neurotherapeutics 14: 687–697. 10.1007/s13311-017-0525-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copes L. E., Schutz H., Dlugsoz E. M., Judex S., and Garland T. Jr, 2018. Locomotor activity, growth hormones, and systemic robusticity: an investigation of cranial vault thickness in mouse lines bred for high endurance running. Am. J. Phys. Anthropol. 166: 442–458. 10.1002/ajpa.23446 [DOI] [PubMed] [Google Scholar]
- Cordeiro L. M. S., Rabelo P. C. R., Moraes M. M., Teixeira-Coelho F., Coimbra C. C. et al. , 2017. Physical exercise-induced fatigue: the role of serotonergic and dopaminergic systems. Braz. J. Med. Biol. Res. 50: e6432 10.1590/1414-431X20176432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen V. A., and Fagard R. H., 2005. Effects of endurance training on blood pressure, blood pressure–regulating mechanisms, and cardiovascular risk factors. Hypertension 46: 667–675. 10.1161/01.HYP.0000184225.05629.51 [DOI] [PubMed] [Google Scholar]
- Cornier M.-A., Dabelea D., Hernandez T. L., Lindstrom R. C., Steig A. J. et al. , 2008. The metabolic syndrome. Endocr. Rev. 29: 777–822. 10.1210/er.2008-0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. J., Shen W., Sandler Y. I., Amoui M., Purcell P. et al. , 2001. Dach1 mutant mice bear no gross abnormalities in eye, limb, and brain development and exhibit postnatal lethality. Mol. Cell. Biol. 21: 1484–1490. 10.1128/MCB.21.5.1484-1490.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes M., Moore-Harrison T., Hamilton A. T., Ceaser T., Kochan K. J. et al. , 2014. Differential gene expression in high- and low-active inbred mice. BioMed Res. Int. 2014: e12469. 10.1155/2014/361048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moor M. H. M., Liu Y.-J., Boomsma D. I., Li J., Hamilton J. J. et al. , 2009. Genome-wide association study of exercise behavior in Dutch and American adults. Med. Sci. Sports Exerc. 41: 1887–1895. 10.1249/MSS.0b013e3181a2f646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan I., Garland T. Jr., Hiramatsu L., and Careau V., 2019. I smell a mouse: indirect genetic effects on voluntary wheel-running distance, duration and speed. Behav. Genet. 49: 49–59. 10.1007/s10519-018-9930-2 [DOI] [PubMed] [Google Scholar]
- Didion J. P., Morgan A. P., Yadgary L., Bell T. A., McMullan R. C. et al. , 2016. R2d2 drives selfish sweeps in the house mouse. Mol. Biol. Evol. 33: 1381–1395. 10.1093/molbev/msw036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich A., 2004. Endocannabinoids and exercise. Br. J. Sports Med. 38: 536–541. 10.1136/bjsm.2004.011718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosz E. M., Schutz H., Meek T. H., Acosta W., Downs C. J. et al. , 2013. Immune response to a Trichinella spiralis infection in house mice from lines selectively bred for high voluntary wheel running. J. Exp. Biol. 216: 4212–4221. 10.1242/jeb.087361 [DOI] [PubMed] [Google Scholar]
- Doya K., 2000. Complementary roles of basal ganglia and cerebellum in learning and motor control. Curr. Opin. Neurobiol. 10: 732–739. 10.1016/S0959-4388(00)00153-7 [DOI] [PubMed] [Google Scholar]
- Drago F., Valerio C., D'Agata V., Spadaro F., Astuto C. et al. , 1989. Razionale farmacologico dell'impiego della CDP-colina nelle cerebrovasculopatie croniche. Ann. Ital. Med. Int. 4: 261–267. [Google Scholar]
- Drickamer L. C., and Evans T. R., 1996. Chemosignals and activity of wild stock house mice, with a note on the use of running wheels to assess activity in rodents. Behav. Processes 36: 51–66. 10.1016/0376-6357(95)00015-1 [DOI] [PubMed] [Google Scholar]
- Dyrstad S. M., Hansen B. H., Holme I. M., and Anderssen S. A., 2014. Comparison of self-reported vs. accelerometer-measured physical activity. Med. Sci. Sports Exerc. 46: 99–106. 10.1249/MSS.0b013e3182a0595f [DOI] [PubMed] [Google Scholar]
- Eikelboom R., and Mills R., 1988. A microanalysis of wheel running in male and female rats. Physiol. Behav. 43: 625–630. 10.1016/0031-9384(88)90217-X [DOI] [PubMed] [Google Scholar]
- Elkouris M., Kontaki H., Stavropoulos A., Antonoglou A., Nikolaou K. C. et al. , 2016. SET9-mediated regulation of TGF-β signaling links protein methylation to pulmonary fibrosis. Cell Rep. 15: 2733–2744. 10.1016/j.celrep.2016.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C., Olson A. K., Pinel J. P. J., Lam R. W., and Christie B. R., 2006. Antidepressant effects of exercise: evidence for an adult-neurogenesis hypothesis? J. Psychiatry Neurosci. 31: 84–92. [PMC free article] [PubMed] [Google Scholar]
- Fan W., Atkins A. R., Yu R. T., Downes M., and Evans R. M., 2013. Road to exercise mimetics: targeting nuclear receptors in skeletal muscle. J. Mol. Endocrinol. 51: T87–T100. 10.1530/JME-13-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1925. Statistical methods for research workers, pp. 25–235 in Biological monographs and manuals, edited by Crew F. A. E., and Cutler D. W.. Oliver and Boyd, Edinburgh. [Google Scholar]
- Frédéric M. Y., Lundin V. F., Whiteside M. D., Cueva J. G., Tu D. K. et al. , 2013. Identification of 526 conserved metazoan genetic innovations exposes a new role for cofactor E-like in neuronal microtubule homeostasis. PLoS Genet. 9: e1003804 10.1371/journal.pgen.1003804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed C., and Yamamoto B., 1985. Regional brain dopamine metabolism: a marker for the speed, direction, and posture of moving animals. Science 229: 62–65. 10.1126/science.4012312 [DOI] [PubMed] [Google Scholar]
- Fuhrmann F., Justus D., Sosulina L., Kaneko H., Beutel T. et al. , 2015. Locomotion, theta oscillations, and the speed-correlated firing of hippocampal neurons are controlled by a medial septal glutamatergic circuit. Neuron 86: 1253–1264. 10.1016/j.neuron.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Furtunato A. M. B., Lobão-Soares B., Tort A. B. L., and Belchior H., 2020. Specific increase of hippocampal delta oscillations across consecutive treadmill runs. Front. Behav. Neurosci. 14: 101 10.3389/fnbeh.2020.00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T. Jr, and Freeman P. W., 2005. Selective breeding for high endurance running increases hindlimb symmetry. Evolution 59: 1851–1854. 10.1111/j.0014-3820.2005.tb01832.x [DOI] [PubMed] [Google Scholar]
- Garland T. Jr, and Rose M. R., 2009. Experimental evolution: concepts, methods, and applications of selection experiments, University of California Press, Berkeley: 10.1525/9780520944473 [DOI] [Google Scholar]
- Garland T. Jr, Morgan M. T., Swallow J. G., Rhodes J. S., Girard I. et al. , 2002. Evolution of a small-muscle polymorphism in lines of house mice selected for high activity levels. Evolution 56: 1267–1275. 10.1111/j.0014-3820.2002.tb01437.x [DOI] [PubMed] [Google Scholar]
- Garland T. Jr, Kelly S. A., Malisch J. L., Kolb E. M., Hannon R. M. et al. , 2011a How to run far: multiple solutions and sex-specific responses to selective breeding for high voluntary activity levels. Proc. Roy. Soc. B 278: 574–581. 10.1098/rspb.2010.1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T. Jr, Schutz H., Chappell M. A., Keeney B. K., Meek T. H. et al. , 2011b The biological control of voluntary exercise, spontaneous physical activity and daily energy expenditure in relation to obesity: human and rodent perspectives. J. Exp. Biol. 214: 206–229. 10.1242/jeb.048397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T. Jr, Zhao M., and Saltzman W., 2016. Hormones and the evolution of complex traits: insights from artificial selection on behavior. Integr. Comp. Biol. 56: 207–224. 10.1093/icb/icw040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler C., Robbe D., Zugaro M., Sirota A., and Buzsaki G., 2007. Hippocampal place cell assemblies are speed-controlled oscillators. Proc. Natl. Acad. Sci. USA 104: 8149–8154. 10.1073/pnas.0610121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez R., Raïch J., and Aguilar J., 1991. Changes in brain striatum dopamine and acetylcholine receptors induced by chronic CDP-choline treatment of aging mice. Br. J. Pharmacol. 104: 575–578. 10.1111/j.1476-5381.1991.tb12471.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobet L., Royo Martin L. J., Poncelet D., Pirottin D., Brouwers B. et al. , 1997. A deletion in the bovine myostatin gene causes the double–muscled phenotype in cattle. Nat. Genet. 17: 71–74. 10.1038/ng0997-71 [DOI] [PubMed] [Google Scholar]
- Guthold R., Stevens G. A., Riley L. M., and Bull F. C., 2018. Worldwide trends in insufficient physical activity from 2001 to 2016: a pooled analysis of 358 population-based surveys with 1·9 million participants. Lancet Glob. Health 6: e1077–e1086. 10.1016/S2214-109X(18)30357-7 [DOI] [PubMed] [Google Scholar]
- Herbert A. J., Williams A. G., Hennis P. J., Erskine R. M., Sale C. et al. , 2019. The interactions of physical activity, exercise and genetics and their associations with bone mineral density: implications for injury risk in elite athletes. Eur. J. Appl. Physiol. 119: 29–47. 10.1007/s00421-018-4007-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu L., and Garland T. Jr, 2018. Mice selectively bred for high voluntary wheel-running behavior conserve more fat despite increased exercise. Physiol. Behav. 194: 1–8. 10.1016/j.physbeh.2018.04.010 [DOI] [PubMed] [Google Scholar]
- Hiramatsu L., Kay J. C., Thompson Z., Singleton J. M., Claghorn G. C. et al. , 2017. Maternal exposure to Western diet affects adult body composition and voluntary wheel running in a genotype-specific manner in mice. Physiol. Behav. 179: 235–245. 10.1016/j.physbeh.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenderop J. G. J., van Leeuwen J. P. T. M., van der Eerden B. C. J., Kersten F. F. J., van derKemp A. W. C. M. et al. , 2003. Renal Ca2+ wasting, hyperabsorption, and reduced bone thickness in mice lacking TRPV5. J. Clin. Invest. 112: 1906–1914. 10.1172/JCI200319826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holroyd C., Nieuwenhuis S., Mars R., and Coles M., 2004 Anterior cingulate cortex, selection for action, and error processing, pp. 219–231 in Cognitive neuroscience of attention, Guilford Press, New York. [Google Scholar]
- Horner A., Shum L., Ayres J. A., Nonaka K., and Nuckolls G. H., 2002. Fibroblast growth factor signaling regulates Dach1 expression during skeletal development. Dev. Dyn. 225: 35–45. 10.1002/dvdy.10132 [DOI] [PubMed] [Google Scholar]
- Horwitz T., Lam K., Chen Y., Xia Y., and Liu C., 2019. A decade in psychiatric GWAS research. Mol. Psychiatry 24: 378–389. 10.1038/s41380-018-0055-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Koolmeister C., Misic J., Siira S., Kühl I. et al. , 2019. TEFM regulates both transcription elongation and RNA processing in mitochondria. EMBO Rep. 20: e48101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasem E., Kurihara T., and Tabuchi K., 2018. Neurexins and neuropsychiatric disorders. Neurosci. Res. 127: 53–60. 10.1016/j.neures.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Kay J. C., Claghorn G. C., Thompson Z., Hampton T. G., and Garland T. Jr, 2019. Electrocardiograms of mice selectively bred for high levels of voluntary exercise: effects of short-term exercise training and the mini-muscle phenotype. Physiol. Behav. 199: 322–332. 10.1016/j.physbeh.2018.11.041 [DOI] [PubMed] [Google Scholar]
- Keeney B. K., Meek T. H., Middleton K. M., Holness L. F., and Garland T. Jr, 2012. Sex differences in cannabinoid receptor-1 (CB1) pharmacology in mice selectively bred for high voluntary wheel-running behavior. Pharmacol. Biochem. Behav. 101: 528–537. 10.1016/j.pbb.2012.02.017 [DOI] [PubMed] [Google Scholar]
- Kelly S. A., Czech P. P., Wight J. T., Blank K. M., and Garland T. Jr, 2006. Experimental evolution and phenotypic plasticity of hindlimb bones in high-activity house mice. J. Morphol. 267: 360–374. 10.1002/jmor.10407 [DOI] [PubMed] [Google Scholar]
- Kelly S. A., Nehrenberg D. L., Peirce J. L., Hua K., Steffy B. M. et al. , 2010. Genetic architecture of voluntary exercise in an advanced intercross line of mice. Physiol. Genomics 42: 190–200. 10.1152/physiolgenomics.00028.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. A., Nehrenberg D. L., Hua K., Garland T. Jr., and Pomp D., 2012. Functional genomic architecture of predisposition to voluntary exercise in mice: expression QTL in the brain. Genetics 191: 643–654. 10.1534/genetics.112.140509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. A., Bell T. A., Selitsky S. R., Buus R. J., Hua K. et al. , 2013. A novel intronic single nucleotide polymorphism in the myosin heavy polypeptide 4 gene is responsible for the mini-muscle phenotype characterized by major reduction in hind-limb muscle mass in mice. Genetics 195: 1385–1395. 10.1534/genetics.113.154476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. A., Nehrenberg D. L., Hua K., Garland T. Jr., and Pomp D., 2014. Quantitative genomics of voluntary exercise in mice: transcriptional analysis and mapping of expression QTL in muscle. Physiol. Genomics 46: 593–601. 10.1152/physiolgenomics.00023.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. A., Gomes F. R., Kolb E. M., Malisch J. L., and Garland T. Jr, 2017. Effects of activity, genetic selection and their interaction on muscle metabolic capacities and organ masses in mice. J. Exp. Biol. 220: 1038–1047. 10.1242/jeb.148759 [DOI] [PubMed] [Google Scholar]
- Khan A. A., and Quigley J. G., 2013. Heme and FLVCR-related transporter families SLC48 and SLC49. Mol. Aspects Med. 34: 669–682. 10.1016/j.mam.2012.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y., 2004. Chick Dach1 interacts with the Smad complex and Sin3a to control AER formation and limb development along the proximodistal axis. Development 131: 4179–4187. 10.1242/dev.01252 [DOI] [PubMed] [Google Scholar]
- Klaassen R. V., Stroeder J., Coussen F., Hafner A.-S., Petersen J. D. et al. , 2016. Shisa6 traps AMPA receptors at postsynaptic sites and prevents their desensitization during synaptic activity. Nat. Commun. 7: 10682. 10.1038/ncomms10682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb E. M., Kelly S. A., Middleton K. M., Sermsakdi L. S., Chappell M. A. et al. , 2010. Erythropoietin elevates but not voluntary wheel running in mice. J. Exp. Biol. 213: 510–519. 10.1242/jeb.029074 [DOI] [PubMed] [Google Scholar]
- Kolb E. M., Kelly S. A., and Garland T. Jr, 2013a Mice from lines selectively bred for high voluntary wheel running exhibit lower blood pressure during withdrawal from wheel access. Physiol. Behav. 112–113: 49–55. 10.1016/j.physbeh.2013.02.010 [DOI] [PubMed] [Google Scholar]
- Kolb E. M., Rezende E. L., Holness L., Radtke A., Lee S. K. et al. , 2013b Mice selectively bred for high voluntary wheel running have larger midbrains: support for the mosaic model of brain evolution. J. Exp. Biol. 216: 515–523. 10.1242/jeb.076000 [DOI] [PubMed] [Google Scholar]
- Konczal M., Koteja P., Orlowska-Feuer P., Radwan J., Sadowska E. T. et al. , 2016. Genomic response to selection for predatory behavior in a mammalian model of adaptive radiation. Mol. Biol. Evol. 33: 2429–2440. 10.1093/molbev/msw121 [DOI] [PubMed] [Google Scholar]
- Koob G. F., and Volkow N. D., 2010. Neurocircuitry of addiction. Neuropsychopharmacology 35: 217–238 (erratum: Neuropsychopharmacology 35: 1051). 10.1038/npp.2009.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrzewa E., and Kas M. J., 2014. The use of mouse models to unravel genetic architecture of physical activity: a review. Genes Brain Behav. 13: 87–103. 10.1111/gbb.12091 [DOI] [PubMed] [Google Scholar]
- Köttgen A., Pattaro C., Böger C. A., Fuchsberger C., Olden M. et al. , 2010. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 42: 376–384. 10.1038/ng.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropff E., Carmichael J. E., Moser M.-B., and Moser E. I., 2015. Speed cells in the medial entorhinal cortex. Nature 523: 419–424. 10.1038/nature14622 [DOI] [PubMed] [Google Scholar]
- Lachman H. M., Fann C. S. J., Bartzis M., Evgrafov O. V., Rosenthal R. N. et al. , 2007. Genomewide suggestive linkage of opioid dependence to chromosome 14q. Hum. Mol. Genet. 16: 1327–1334. 10.1093/hmg/ddm081 [DOI] [PubMed] [Google Scholar]
- Lalley P. M., Benacka R., Bischoff A. M., and Richter D. W., 1997. Nucleus raphe obscurus evokes 5-HT-1A receptor-mediated modulation of respiratory neurons. Brain Res. 747: 156–159. 10.1016/S0006-8993(96)01233-4 [DOI] [PubMed] [Google Scholar]
- Lee D., Zhao X., Yim Y.-I., Eisenberg E., and Greene L. E., 2008. Essential role of cyclin-G–associated kinase (auxilin-2) in developing and mature mice. Mol. Biol. Cell 19: 2766–2776. 10.1091/mbc.e07-11-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot J. T., 2011. Current understanding of the genetic basis for physical activity. J. Nutr. 141: 526–530. 10.3945/jn.110.127290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot J. T., Turner M. J., Pomp D., Kleeberger S. R., and Leamy L. J., 2008. Quantitative trait loci for physical activity traits in mice. Physiol. Genomics 32: 401–408. 10.1152/physiolgenomics.00241.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot J. T., Leamy L., Pomp D., Turner M. J., Fodor A. A. et al. , 2010. Strain screen and haplotype association mapping of wheel running in inbred mouse strains. J. Appl. Physiol. 109: 623–634. 10.1152/japplphysiol.00525.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot J. T., De Geus E. J. C., Booth F. W., Bray M. S., Den Hoed M. et al. , 2018. Biological/genetic regulation of physical activity level: consensus from GenBioPAC. Med. Sci. Sports Exerc. 50: 863–873. 10.1249/MSS.0000000000001499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Kuo T. B. J., Hsieh I.-T., and Yang C. C. H., 2012. Changes in hippocampal theta rhythm and their correlations with speed during different phases of voluntary wheel running in rats. Neuroscience 213: 54–61. 10.1016/j.neuroscience.2012.04.020 [DOI] [PubMed] [Google Scholar]
- Lillie M., Honaker C. F., Siegel P. B., and Carlborg Ö., 2019. Bidirectional selection for body weight on standing genetic variation in a chicken model. G3 (Bethesda) 9: 1165–1173. . 10.1534/g3.119.400038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Eaton C. B., Manson J. E., and Liu S., 2017. The genetics of physical activity. Curr. Cardiol. Rep. 19: 119 10.1007/s11886-017-0938-7 [DOI] [PubMed] [Google Scholar]
- Liu Q., Duff R. J., Liu B., Wilson A. L., Babb-Clendenon S. G. et al. , 2006. Expression of cadherin10, a type II classic cadherin gene, in the nervous system of the embryonic zebrafish. Gene Expr. Patterns 6: 703–710. 10.1016/j.modgep.2005.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh N. Y., Bentley L., Dimke H., Verkaart S., Tammaro P. et al. , 2013. Autosomal dominant hypercalciuria in a mouse model due to a mutation of the epithelial calcium channel, TRPV5. PLoS One 8: e55412 10.1371/journal.pone.0055412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos R. J. F., Rankinen T., Tremblay A., Pérusse L., Chagnon Y. et al. , 2005. Melanocortin-4 receptor gene and physical activity in the Québec Family Study. Int. J. Obes. 29: 420–428. 10.1038/sj.ijo.0802869 [DOI] [PubMed] [Google Scholar]
- Machado C. B., Kanning K. C., Kreis P., Stevenson D., Crossley M. et al. , 2014. Reconstruction of phrenic neuron identity in embryonic stem cell- derived motor neurons. Development 141: 784–794. 10.1242/dev.097188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley A. F., 1996. Physical activity and health: a report of the Surgeon General, DIANE Publishing, Darby, PA. [Google Scholar]
- Mardon G., Solomon N. M., and Rubin G. M., 1994. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120: 3473–3486. [DOI] [PubMed] [Google Scholar]
- Martin G. M., Austad S. N., and Johnson T. E., 1996. Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat. Genet. 13: 25–34. 10.1038/ng0596-25 [DOI] [PubMed] [Google Scholar]
- Mathes W. F., Nehrenberg D. L., Gordon R., Hua K., Garland T. Jr. et al. , 2010. Dopaminergic dysregulation in mice selectively bred for excessive exercise or obesity. Behav. Brain Res. 210: 155–163. 10.1016/j.bbr.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E., Nambu S., Oka M., and Iriki A., 2015. Complex and dynamic expression of cadherins in the embryonic marmoset cerebral cortex. Dev. Growth Differ. 57: 474–483. 10.1111/dgd.12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta Mello Portugal E., Cevada T., Sobral Monteiro-Junior R., Teixeira Guimarães T., da Cruz Rubini E. et al. , 2013. Neuroscience of exercise: from neurobiology mechanisms to mental health. Neuropsychobiology 68: 1–14. 10.1159/000350946 [DOI] [PubMed] [Google Scholar]
- McGillivray D. G., Garland T. Jr., Dlugosz E. M., Chappell M. A., and Syme D. A., 2009. Changes in efficiency and myosin expression in the small-muscle phenotype of mice selectively bred for high voluntary running activity. J. Exp. Biol. 212: 977–985. 10.1242/jeb.026625 [DOI] [PubMed] [Google Scholar]
- Meek T. H., Lonquich B. P., Hannon R. M., and Garland T. Jr, 2009. Endurance capacity of mice selectively bred for high voluntary wheel running. J. Exp. Biol. 212: 2908–2917. 10.1242/jeb.028886 [DOI] [PubMed] [Google Scholar]
- Miro X., Zhou X., Boretius S., Michaelis T., Kubisch C. et al. , 2009. Haploinsufficiency of the murine polycomb gene Suz12 results in diverse malformations of the brain and neural tube. Dis. Model. Mech. 2: 412–418. 10.1242/dmm.001602 [DOI] [PubMed] [Google Scholar]
- Mok A., Khaw K.-T., Luben R., Wareham N., and Brage S., 2019. Physical activity trajectories and mortality: population based cohort study. BMJ 365: 12323 10.1136/bmj.l2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., and Welsh C. E., 2015. Informatics resources for the Collaborative Cross and related mouse populations. Mamm. Genome 26: 521–539. 10.1007/s00335-015-9581-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher D. S., Quignon P., Bustamante C. D., Sutter N. B., Mellersh C. S. et al. , 2007. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 3: e79 10.1371/journal.pgen.0030079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A., Gibbons C., Finlayson G., and Blundell J., 2017. Associations among sedentary and active behaviours, body fat and appetite dysregulation: investigating the myth of physical inactivity and obesity. Br. J. Sports Med. 51: 1540–1544. 10.1136/bjsports-2015-095640 [DOI] [PubMed] [Google Scholar]
- Neufer P. D., Bamman M. M., Muoio D. M., Bouchard C., Cooper D. M. et al. , 2015. Understanding the cellular and molecular mechanisms of physical activity-induced health benefits. Cell Metab. 22: 4–11. 10.1016/j.cmet.2015.05.011 [DOI] [PubMed] [Google Scholar]
- Nica A. C., and Dermitzakis E. T., 2013. Expression quantitative trait loci: present and future. Philos. Trans. R. Soc. B Biol. Sci. 368: 20120362 10.1098/rstb.2012.0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicod J., Davies R. W., Cai N., Hassett C., Goodstadt L. et al. , 2016. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat. Genet. 48: 912–918. 10.1038/ng.3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira A., Molinero O., Salguero A., and Márquez S., 2018. Exercise addiction in practitioners of endurance sports: a literature review. Front. Psychol. 9: 1484 10.3389/fpsyg.2018.01484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwal T., Kropp M., Wegener S., Racic S., Montalban I. et al. , 2012. The Drosophila homologue of tubulin-specific chaperone E–like protein is required for synchronous sperm individualization and normal male fertility. J. Neurogenet. 26: 374–381. 10.3109/01677063.2012.731119 [DOI] [PubMed] [Google Scholar]
- Pallafacchina G., François S., Regnault B., Czarny B., Dive V. et al. , 2010. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. (Amst.) 4: 77–91. 10.1016/j.scr.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Parker C. C., and Palmer A. A., 2011. Dark Matter: are mice the solution to missing heritability? Front. Genet. 2: 32 10.3389/fgene.2011.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. C., Gopalakrishnan S., Carbonetto P., Gonzales N. M., Leung E. et al. , 2016. Genome-wide association study of behavioral, physiological and gene expression traits in outbred CFW mice. Nat. Genet. 48: 919–926. 10.1038/ng.3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., and Etnier J. L., 2019. Beneficial effects of acute exercise on executive function in adolescents. J. Phys. Act. Health 16: 423–429. 10.1123/jpah.2018-0219 [DOI] [PubMed] [Google Scholar]
- Prince S. A., Adamo K. B., Hamel M., Hardt J., Connor Gorber S. et al. , 2008. A comparison of direct vs. self-report measures for assessing physical activity in adults: a systematic review. Int. J. Behav. Nutr. Phys. Act. 5: 56 10.1186/1479-5868-5-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendeiro C., and Rhodes J. S., 2018. A new perspective of the hippocampus in the origin of exercise–brain interactions. Brain Struct. Funct. 223: 2527–2545. 10.1007/s00429-018-1665-6 [DOI] [PubMed] [Google Scholar]
- Rezende E. L., Gomes F. R., Malisch J. L., Chappell M. A., and Garland T. Jr, 2006. Maximal oxygen consumption in relation to subordinate traits in lines of house mice selectively bred for high voluntary wheel running. J. Appl. Physiol. 101: 477–485. 10.1152/japplphysiol.00042.2006 [DOI] [PubMed] [Google Scholar]
- Rhodes J. S., and Garland T. Jr, 2003. Differential sensitivity to acute administration of Ritalin, apormorphine, SCH 23390, but not raclopride in mice selectively bred for hyperactive wheel-running behavior. Psychopharmacology (Berl.) 167: 242–250. 10.1007/s00213-003-1399-9 [DOI] [PubMed] [Google Scholar]
- Rhodes J. S., Hosack G. R., Girard I., Kelley A. E., Mitchell G. S. et al. , 2001. Differential sensitivity to acute administration of cocaine, GBR 12909, and fluoxetine in mice selectively bred for hyperactive wheel-running behavior. Psychopharmacology (Berl.) 158: 120–131. 10.1007/s002130100857 [DOI] [PubMed] [Google Scholar]
- Rhodes J. S., Garland T. Jr., and Gammie S. C., 2003a Patterns of brain activity associated with variation in voluntary wheel-running behavior. Behav. Neurosci. 117: 1243–1256. 10.1037/0735-7044.117.6.1243 [DOI] [PubMed] [Google Scholar]
- Rhodes J. S., van Praag H., Jeffrey S., Girard I., Mitchell G. S. et al. , 2003b Exercise increases hippocampal neurogenesis to high levels but does not improve spatial learning in mice bred for increased voluntary wheel running. Behav. Neurosci. 117: 1006–1016. 10.1037/0735-7044.117.5.1006 [DOI] [PubMed] [Google Scholar]
- Rhodes J. S., Gammie S. C., and Garland T. Jr, 2005. Neurobiology of mice selected for high voluntary wheel-running activity. Integr. Comp. Biol. 45: 438–455. 10.1093/icb/45.3.438 [DOI] [PubMed] [Google Scholar]
- Roberts M. D., Ruegsegger G. N., Brown J. D., and Booth F. W., 2017. Mechanisms associated with physical activity behavior: insights from rodent experiments. Exerc. Sport Sci. Rev. 45: 217–222. 10.1249/JES.0000000000000124 [DOI] [PubMed] [Google Scholar]
- Rohe, M., 2008 Role of SORLA in the brain and its relevance for Alzheimer disease. PhD Thesis, Freien Universität Berlin, 110 p. [Google Scholar]
- Rosenfeld C. S., 2017. Sex-dependent differences in voluntary physical activity. J. Neurosci. Res. 95: 279–290. 10.1002/jnr.23896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland T., 2016. Biologic regulation of physical activity, Human Kinetics Publishers, Champaign, IL. [Google Scholar]
- Saiga T., Fukuda T., Matsumoto M., Tada H., Okano H. J. et al. , 2009. Fbxo45 forms a novel ubiquitin ligase complex and is required for neuronal development. Mol. Cell. Biol. 29: 3529–3543. 10.1128/MCB.00364-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsi V., Vigano M. A., Cocchiarella F., Mantovani R., and Zappavigna V., 2008. Hoxd13 binds in vivo and regulates the expression of genes acting in key pathways for early limb and skeletal patterning. Dev. Biol. 317: 497–507. 10.1016/j.ydbio.2008.02.048 [DOI] [PubMed] [Google Scholar]
- Sarzynski M. A., Loos R. J. F., Lucia A., Pérusse L., Roth S. M. et al. , 2016. Advances in exercise, fitness, and performance genomics in 2015. Med. Sci. Sports Exerc. 48: 1906–1916. 10.1249/MSS.0000000000000982 [DOI] [PubMed] [Google Scholar]
- Saul M. C., Majdak P., Perez S., Reilly M., Garland T. Jr. et al. , 2017. High motivation for exercise is associated with altered chromatin regulators of monoamine receptor gene expression in the striatum of selectively bred mice. Genes Brain Behav. 16: 328–341. 10.1111/gbb.12347 [DOI] [PubMed] [Google Scholar]
- Schiller D., Eichenbaum H., Buffalo E. A., Davachi L., Foster D. J. et al. , 2015. Memory and space: towards an understanding of the cognitive map. J. Neurosci. 35: 13904–13911. 10.1523/JNEUROSCI.2618-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt V., Subkhangulova A., and Willnow T. E., 2017. Sorting receptor SORLA: cellular mechanisms and implications for disease. Cell. Mol. Life Sci. 74: 1475–1483. 10.1007/s00018-016-2410-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz N. L., Patel B. A., Garland T. Jr., and Horner A. M., 2018. Effects of selective breeding for high voluntary wheel-running behavior on femoral nutrient canal size and abundance in house mice. J. Anat. 233: 193–203. 10.1111/joa.12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secades J., and Lorenzo J., 2006. Citicoline: pharmacological and clinical review, 2006 update. Methods Find Exp Clin Pharmacol 28 (Suppl B): 1–56. [PubMed] [Google Scholar]
- Sella G., and Barton N. H., 2019. Thinking about the evolution of complex traits in the era of genome-wide association studies. Annu. Rev. Genomics Hum. Genet. 20: 461–493. 10.1146/annurev-genom-083115-022316 [DOI] [PubMed] [Google Scholar]
- Sellami M., Gasmi M., Denham J., Hayes L. D., Stratton D. et al. , 2018. Effects of acute and chronic exercise on immunological parameters in the elderly aged: can physical activity counteract the effects of aging? Front. Immunol. 9: 2187 10.3389/fimmu.2018.02187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheel A. W., 2016. Sex differences in the physiology of exercise: an integrative perspective. Exp. Physiol. 101: 211–212. 10.1113/EP085371 [DOI] [PubMed] [Google Scholar]
- Sheila M., Narayanan G., Ma S., Tam W. L., Chai J. et al. , 2019. Phenotypic and molecular features underlying neurodegeneration of motor neurons derived from spinal and bulbar muscular atrophy patients. Neurobiol. Dis. 124: 1–13. 10.1016/j.nbd.2018.10.019 [DOI] [PubMed] [Google Scholar]
- Shen Y., Ding Z., Ma S., Ding Z., Zhang Y. et al. , 2019. SETD7 mediates spinal microgliosis and neuropathic pain in a rat model of peripheral nerve injury. Brain Behav. Immun. 82: 382–395. 10.1016/j.bbi.2019.09.007 [DOI] [PubMed] [Google Scholar]
- Sheremet A., Kennedy J. P., Qin Y., Zhou Y., Lovett S. D. et al. , 2019. Theta-gamma cascades and running speed. J. Neurophysiol. 121: 444–458. 10.1152/jn.00636.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H., Chun H., Engelman C. D., and Payseur B. A., 2009. Genome-wide association studies using single-nucleotide polymorphisms vs. haplotypes: an empirical comparison with data from the North American Rheumatoid Arthritis Consortium. BMC Proc. 3: S35 10.1186/1753-6561-3-s7-s35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura K., Low-Zeddies S. S., King D. P., Steeves T. D., Whiteley A. et al. , 2001. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 11: 959–980. 10.1101/gr.171601 [DOI] [PubMed] [Google Scholar]
- Simonen R. L., Rankinen T., Pérusse L., Leon A. S., Skinner J. S. et al. , 2003. A dopamine D2 receptor gene polymorphism and physical activity in two family studies. Physiol. Behav. 78: 751–757. 10.1016/S0031-9384(03)00084-2 [DOI] [PubMed] [Google Scholar]
- Singleton J. M., and Garland T. Jr, 2019. Influence of corticosterone on growth, home-cage activity, wheel running, and aerobic capacity in house mice selectively bred for high voluntary wheel-running behavior. Physiol. Behav. 198: 27–41. 10.1016/j.physbeh.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Smits P., Li P., Mandel J., Zhang Z., Deng J. M. et al. , 2001. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell 1: 277–290. 10.1016/S1534-5807(01)00003-X [DOI] [PubMed] [Google Scholar]
- Stoltenberg S. F., Lehmann M. K., Christ C. C., Hersrud S. L., and Davies G. E., 2011. Associations among types of impulsivity, substance use problems and Neurexin-3 polymorphisms. Drug Alcohol Depend. 119: e31–e38. 10.1016/j.drugalcdep.2011.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow J. G., Garland T. Jr., Carter P. A., Zhan W.-Z., and Sieck G. C., 1998. Effects of voluntary activity and genetic selection on aerobic capacity in house mice (Mus domesticus). J. Appl. Physiol. 84: 69–76. 10.1152/jappl.1998.84.1.69 [DOI] [PubMed] [Google Scholar]
- Swallow J. G., Koteja P., Carter P. A., and Garland T. Jr, 2001. Food consumption and body composition in mice selected for high wheel-running activity. J. Comp. Physiol. B 171: 651–659. 10.1007/s003600100216 [DOI] [PubMed] [Google Scholar]
- Swallow J. G., Rhodes J. S., and Garland T. Jr, 2005. Phenotypic and evolutionary plasticity of organ masses in response to voluntary exercise in house mice. Integr. Comp. Biol. 45: 426–437. 10.1093/icb/45.3.426 [DOI] [PubMed] [Google Scholar]
- Tada H., Okano H. J., Takagi H., Shibata S., Yao I. et al. , 2010. Fbxo45, a novel ubiquitin ligase, regulates synaptic activity. J. Biol. Chem. 285: 3840–3849. 10.1074/jbc.M109.046284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliun D., Gamper J., Leser U., and Pattaro C., 2016. Fast sampling-based whole-genome haplotype block recognition. IEEE/ACM Trans. Comput. Biol. Bioinform. 13: 315–325. 10.1109/TCBB.2015.2456897 [DOI] [PubMed] [Google Scholar]
- Talmadge R. J., Acosta W., and Garland T. Jr, 2014. Myosin heavy chain isoform expression in adult and juvenile mini-muscle mice bred for high-voluntary wheel running. Mech. Dev. 134: 16–30. 10.1016/j.mod.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Thomas J. R., and Thomas K. T., 1988. Development of gender differences in physical activity. Quest 40: 219–229. 10.1080/00336297.1988.10483902 [DOI] [Google Scholar]
- Thompson Z., 2017. The neurobiological basis of voluntary exercise in selectively-bred high runner mice, University of California, Riverside. [Google Scholar]
- Thompson Z., Argueta D., Garland T. Jr., and DiPatrizio N., 2017. Circulating levels of endocannabinoids respond acutely to voluntary exercise, are altered in mice selectively bred for high voluntary wheel running, and differ between the sexes. Physiol. Behav. 170: 141–150. 10.1016/j.physbeh.2016.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaanholt L. M., Jonas I., Doornbos M., Schubert K. A., Nyakas C. et al. , 2008. Metabolic and behavioral responses to high-fat feeding in mice selectively bred for high wheel-running activity. Int. J. Obes. 32: 1566–1575. 10.1038/ijo.2008.136 [DOI] [PubMed] [Google Scholar]
- van Rooij E., Quiat D., Johnson B. A., Sutherland L. B., Qi X. et al. , 2009. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 17: 662–673. 10.1016/j.devcel.2009.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann S. D., Aggleton J. P., and Maguire E. A., 2009. What does the retrosplenial cortex do? Nat. Rev. Neurosci. 10: 792–802. 10.1038/nrn2733 [DOI] [PubMed] [Google Scholar]
- Walker, E. P., and P. Tadi, 2020 Neuroanatomy, Nucleus Raphe (updated 10 July 2020), In: StatPearls [Internet]. StatPearls Publishing, Treasure Island, FL. Available from: https://www.ncbi.nlm.nih.gov/books/NBK544359/. [PubMed] [Google Scholar]
- Wallace I. J., and Garland T. Jr, 2016. Mobility as an emergent property of biological organization: insights from experimental evolution. Evol. Anthropol. 25: 98–104. 10.1002/evan.21481 [DOI] [PubMed] [Google Scholar]
- Wang W., Touhara K. K., Weir K., Bean B. P., and MacKinnon R., 2016. Cooperative regulation by G proteins and Na+ of neuronal GIRK2 K+ channels. eLife 5: e15751 10.7554/eLife.15751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. K., Gerdin A.-K., Karp N. A., Ryder E., Buljan M. et al. , 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154: 452–464. 10.1016/j.cell.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. J., Williams M. G., Eynon N., Ashton K. J., Little J. P. et al. , 2017. Genes to predict VO2max trainability: a systematic review. BMC Genomics 18(Suppl 8): 831 10.1186/s12864-017-4192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise R. A., 2009. Roles for nigrostriatal—not just mesocorticolimbic—dopamine in reward and addiction. Trends Neurosci. 32: 517–524. 10.1016/j.tins.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock S. L., Yates A., Petrill S. A., Bohland J. W., Blair C. et al. , 2013. Gene × smoking interactions on human brain gene expression: finding common mechanisms in adolescents and adults. J. Child Psychol. Psychiatry 54: 1109–1119. 10.1111/jcpp.12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. R., Esko T., Yang J., Vedantam S., Pers T. H. et al. , 2014. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46: 1173–1186. 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., and Garland T., 2017. A mixed model approach to genome-wide association studies for selection signatures, with application to mice bred for voluntary exercise behavior. Genetics 207: 785–799. 10.1534/genetics.117.300102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhao X.-M., Liu J., Wang Y.-Y., Xiong L.-L. et al. , 2020. Complexin I knockout rats exhibit a complex neurobehavioral phenotype including profound ataxia and marked deficits in lifespan. Eur. J. Phys. 472: 117–133. 10.1007/s00424-019-02337-5 [DOI] [PubMed] [Google Scholar]
- Yang Z., Sun Q., Guo J., Wang S., Song G. et al. , 2019. GRSF1 -mediated MIR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy 15: 668–685. 10.1080/15548627.2018.1539590 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ye J., Witter M. P., Moser M.-B., and Moser E. I., 2018. Entorhinal fast-spiking speed cells project to the hippocampus. Proc. Natl. Acad. Sci. USA 115: E1627–E1636. 10.1073/pnas.1720855115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. M., Hallgrímsson B., and Garland T. Jr, 2009. Epigenetic effects on integration of limb lengths in a mouse model: selective breeding for high voluntary locomotor activity. Evol. Biol. 36: 88–99. 10.1007/s11692-009-9053-z [DOI] [Google Scholar]
- Zheng J.-J., Li W.-X., Liu J.-Q., Guo Y.-C., Wang Q. et al. , 2018. Low expression of aging-related NRXN3 is associated with Alzheimer disease: a systematic review and meta-analysis. Medicine (Baltimore) 97: e11343 10.1097/MD.0000000000011343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Cho E.-S., Sha Q., Peng J., Oksov Y. et al. , 2014. Giant axon formation in mice lacking Kell, XK, or Kell and XK. Am. J. Pathol. 184: 800–807. 10.1016/j.ajpath.2013.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Rivera A., Golub M. S., Peng J., Sha Q. et al. , 2009. Changes in red cell ion transport, reduced intratumoral neovascularization, and some mild motor function abnormalities accompany targeted disruption of the Mouse Kell gene (Kel). Am. J. Hematol. 84: 492–498. 10.1002/ajh.21453 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Any additional intermediary or results file are available upon request. Supplemental files are available at FigShare. File S1 contains supplemental figures and brief descriptions of all other supplemental files and tables. File S2 contains allelic SNP data. File S3 contains mouse data with line and lintype. File S4 contains all results for analyses of individual SNPs. File S5 contains all haplotype data. Files S6 contains all results for analyses of haplotype data. File S7 contains justification for use of allelic coding of alleles. File S8 includes simulations of Type I error rates for Mixed Model analyses using MIVQUE variance estimation. File S9 expands on the discussion of genes in consistent regions (see Results). File S10 includes all R and SAS code used for the SNP and haplotype analyses. File S11 includes a comprehensive list of genes containing SNPs and rankings by P-value. Table S1 includes local maxima associated genes. Table S2 contains groups of loci fixed in all lines of one lintype but polymorphic in all lines of the other. Table S3 includes heterozygosity for each individual mouse. Table S4 includes top 10 genes for each of the targeted ontologies analyses. Table S5 includes allele frequency by line of each loci identified as a local maximum. Table S6 includes genomic regions identified as suggestive (P < 0.001) by the SNP analyses. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12436649.