Abstract
Since the dawn of the 20th century, the fruit fly Drosophila melanogaster has been used as a model organism to understand the nature of genes and how they control development, behavior, and physiology. One of the most powerful experimental approaches employed in Drosophila is the forward genetic screen. In the 21st century, genome-wide screens have become popular tools for identifying evolutionarily conserved genes involved in complex human diseases. In the accompanying article “Amyotrophic Lateral Sclerosis Modifiers in Drosophila Reveal the Phospholipase D Pathway as a Potential Therapeutic Target,” Kankel and colleagues describe a forward genetic modifier screen to discover factors that contribute to the severe neurodegenerative disease amyotrophic lateral sclerosis (ALS). This primer briefly traces the history of genetic screens in Drosophila and introduces students to ALS. We then provide a set of guided reading questions to help students work through the data presented in the research article. Finally, several ideas for literature-based research projects are offered as opportunities for students to expand their appreciation of the potential scope of genetic screens. The primer is intended to help students and instructors thoroughly examine a current study that uses forward genetics in Drosophila to identify human disease genes.
Keywords: neurodegeneration, ALS, Drosophila, genetic screen, education
ONE of the most powerful approaches for discovering the functions of genes is the forward genetic screen. Screens in model organisms like Drosophila melanogaster can identify evolutionarily conserved genes involved in complex human diseases. As an example, in the accompanying article “Amyotrophic Lateral Sclerosis Modifiers in Drosophila Reveal the Phospholipase D Pathway as a Potential Therapeutic Target,” Kankel and colleagues describe a forward genetic screen to discover factors that contribute to the severe neurodegenerative disease amyotrophic lateral sclerosis (ALS). This primer guides students through the data in the article and offers ideas for literature-based research projects to expand students’ appreciation of genetic screens.
Model Organisms
It is impossible to imagine the successes of modern medicine without the contributions from so-called “model organisms.” Because fundamental genetic and cell biological processes are common to all life on Earth, profound insight into human biology can be gained by studying species simpler than our own. Research on organisms as distant from us as bacteria and their phages have helped provide the thorough and extensive understanding we have today of the genetic material, the code by which it stores information, and the process by which this information is decoded by cells. That foundational knowledge, built in model organisms, laid the groundwork for the molecular biology revolution of the mid-20th century, which in turn underpins the modern biomedical enterprise. Likewise, animal studies in rats, mice, chicks, frogs, fish, and invertebrates have been providing insight into the evolutionarily conserved mechanisms of our own development, physiology, and behavior, and related pathologies, for well over a century. In their recent article, Kankel and coauthors report on a genetic study using the fruit fly, D. melanogaster, as a model organism (Kankel et al. 2020).
Drosophila melanogaster
D. melanogaster has a particularly long history as a model organism, having first achieved fame in the early 20th century for its role in establishing the chromosome theory of inheritance. Because of its long history, its resurgence in popularity starting in the mid-20th century, and its “user-friendly” attributes, including short life-cycle, small size, high fecundity, and relatively compact genome, Drosophila has become a research platform with a plethora of experimental tools (Hales et al. 2015). These tools, coupled with the fact that more than half of Drosophila genes have orthologs in humans, have made the fruit fly a leading model system for studying the mechanisms of human biology and disease (Bellen and Yamamoto 2015). Additionally, its complex brain and behavioral repertoire make it a particularly useful model for understanding neurobiology (Bellen et al. 2010; McGurk et al. 2015).
Genetic Screens
After early experiments in Drosophila helped elucidate the rules of inheritance and the nature of genes, studies in this organism turned to the question of how genes direct biology. By examining what happens when genes are mutated, scientists have been able to tease apart how unmutated wild-type alleles contribute to the normal functioning of an organism. Since the 1960s, forward genetic screening has been used to identify novel genes involved in myriad biological processes (St Johnston 2013). Forward genetic screens begin by creating de novo mutations randomly throughout the genome, so that any gene has a chance of being altered (“hit”) and the researcher is unbiased toward any specific gene or class of genes (St Johnston 2013; Hales et al. 2015). Mutants then are surveyed for distinct phenotypic alterations as compared to the wild type. When applied on a large scale, this powerful method has the potential to capture any, and theoretically all, genes involved in a particular process (Nüsslein-Volhard and Wieschaus 1980; St Johnston 2002). In contrast to “reverse” genetics approaches, in which a specific gene or group of genes is targeted a priori, forward genetic screens are uniquely poised to discover pathways previously unknown to be involved in the process of interest. Some of the earliest screens centered on stereotyped behaviors, thereby demonstrating that even biological functions as complex as neurobiology and behavior are regulated by the activity of specific genes (e.g., Benzer 1967; Hotta and Benzer 1969; Pak et al. 1969; Jan and Jan 2008).
Dominant Modifiers
One of the most obvious physical features of the D. melanogaster fly is its eye—a large, complex, and beautiful neurological organ. Since the very first described Drosophila mutant with white eye color instead of red, some of the most notable forward genetic screens have looked for mutations that perturb the eye (Morgan 1910; St Johnston 2002). A number of these screens take advantage of the fact that genes in the same pathway have a tendency to be sensitive to each other’s dosage (e.g., Rogge et al. 1991; Simon et al. 1991). Because changes to the sequence of nucleotide bases in a gene often reduce the functionality of the encoded gene product (protein or RNA), many mutant alleles are loss of function. In diploid organisms, including Drosophila and humans, a single wild-type allele typically produces enough gene product to maintain normal activity; hence, most loss of function mutant alleles are recessive and must be homozygous to affect phenotype. However, if another component of the same pathway is altered in its genetic dosage, loss of function mutant alleles can exacerbate (enhance) or ameliorate (suppress) the phenotype as heterozygotes, and thus appear dominant. In this way, forward genetic modifier screens can be performed by changing the dosage of one pathway component, thereby creating a sensitized background, generating novel mutations randomly throughout the genome, and then screening for those second-site alleles that are dominant modifiers of the starting phenotype. This enables identification of genes that act in the same pathway as the original component, with the added advantage that the screen can be performed with only one generation of flies, as the dominant modifiers do not need to be homozygous to manifest the altered phenotype. This strategy is especially valuable because mutant alleles that are otherwise homozygous lethal can be identified as heterozygotes, with survival of the organism permitted. Furthermore, the eye, being a nonessential organ, is an ideal context in which to study development and physiology without impacting survival or fertility (Baker et al. 2014). In their study, Kankel et al. (2020) perform a genetic screen for dominant modifiers of a mutant eye phenotype.
Expressing Human Genes in Drosophila
The experimental range of Drosophila was expanded vastly by transgenic technology. Once the tools of molecular biology made it possible to isolate individual genes from the genome and engineer them in bacterial plasmids, methods were developed to deliver any gene of interest into the genome of another organism, of the same or different species (Rubin and Spradling 1982; Spradling and Rubin 1982). With this technology in hand, human genes now could be introduced into Drosophila (e.g., Jowett et al. 1991; Luo et al. 1992). In the decades since, this procedure has been used to demonstrate time and again the strong evolutionary conservation between flies and humans, with human proteins capable of rescuing mutant phenotypes conferred by loss of their fly counterparts. Furthermore, expressing human disease alleles in flies often mimics disease pathology, again highlighting the robust underlying conservation of genetic pathways and cellular networks (e.g., Jackson et al. 1998; Warrick et al. 1998). Kankel et al. (2020) use this approach of expressing human disease alleles in the fly eye, which creates a visible mutant phenotype.
GAL4-UAS
In order for an exogenous transgene to be expressed in the host organism, it must include regulatory sequences for transcription and translation. Early transgenes were engineered with specific Drosophila regulatory elements, but the GAL4-UAS system introduced a great technological improvement by separating the gene of interest from its regulation (Brand and Perrimon 1993). In this system, Drosophila transgenes are engineered downstream of an upstream activation sequence (UAS) from the Saccharomyces cerevisiae genome that binds the GAL4 transcription factor. Because the Drosophila genome does not endogenously encode GAL4, UAS-driven transgenes are effectively not expressed, thereby protecting the transgenic organisms from expression of deleterious transgenes. A vast array of separate transgenic Drosophila stocks has been generated and shared within the scientific community, each carrying the yeast GAL4 transcription factor expressed in a tissue- or cell-specific pattern under the control of an endogenous regulatory element (Hales et al. 2015). Thus, any UAS-driven transgene now can be expressed in almost any tissue or cell type by simply mating to a GAL4 stock. Kankel et al. (2020) use the GMR-GAL4 driver, which produces GAL4 protein in cells of the eye, to induce expression of UAS-controlled human disease alleles.
Genome-Wide Mutant Collections
By the last decade of the 20th century, the fly research community was inspired to generate new collections of mutants that could be used for rapid forward genetic screening and subsequent mapping (Cooley et al. 1988; Spradling et al. 1995). Collections aiming to represent every gene in the genome made use of mobile transposable elements that could integrate throughout the genome at random, creating loss of function mutations by disrupting local regulatory and/or coding sequences (Artavanis-Tsakonas 2004; Thibault et al. 2004; Bellen et al. 2011). In this study, Kankel et al. (2020) screen one of these collections of Drosophila insertion mutations, the Exelixis collection. These insertions can be mapped immediately by isolation of the inserted element along with its neighboring genomic DNA, greatly reducing labor- and time-intensive procedures for mapping hits from screens. The utility of collections like these has been augmented further by whole genome sequencing, which revolutionized the study of biology at the turn of the 21st century (Adams et al. 2000). Today, scientists have at their fingertips nearly the entire Drosophila genome sequence, along with libraries of mutants disrupting a substantial proportion of protein-coding loci, making possible rapid genetic screening, identification, and functional analysis. The availability of the human genome sequence, together with the revelation that 60%–70% of human genes have orthologs in Drosophila, and that this percentage is even higher (∼75%) for disease genes, has prompted the scientific community to use the powerful tools of Drosophila genetics to understand human disease (Lander et al. 2001; Venter et al. 2001; McGurk et al. 2015; Wangler et al. 2017; Johnston 2020).
Organizing Screen Hits
A successful large-scale genome-wide genetic screen can identify hundreds of hits, genes whose mutation leads to interesting phenotypic alterations. To organize these hits, scientists often use publicly available gene ontology (GO) classification, which annotates each gene with molecular functions, biological processes, and cellular sites of activity (http://geneontology.org/). Annotations are based on data from many different model systems, as well as on computer-based analyses, and are curated by a large global consortium of scientists (Gaudet et al. 2017). This diversity of inputs, along with a controlled vocabulary that is clearly defined and adheres to a consistent logical framework, makes the GO classification structure universally applicable (Hastings 2017). GO classification can help group screen hits according to function and/or cellular localization, thereby illuminating biological pathways important for the process under investigation.
Amyotrophic Lateral Sclerosis
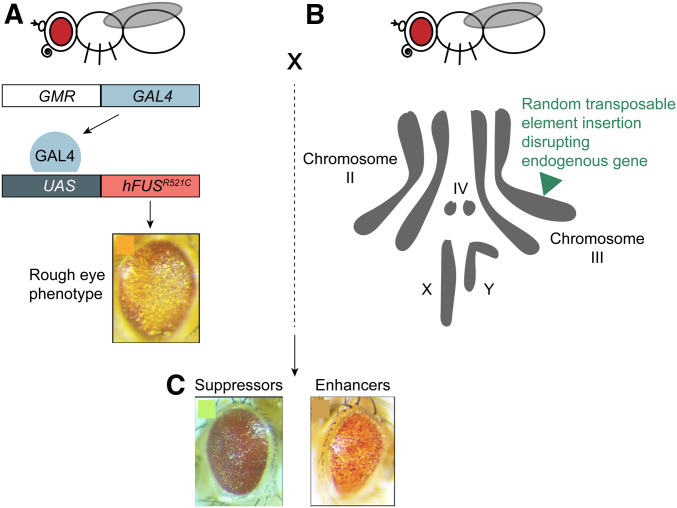
Human life expectancy has increased dramatically over the course of the last century due largely to improved nutrition, public health, and medical care, but with increased lifespan comes greater risk of age-associated diseases, including neurodegenerative disease (ND, https://www.nia.nih.gov/research/dbsr/global-aging). ND encompasses a range of conditions that cause the death of neurons and loss of neurological function, including Alzheimer’s disease, Lewy body diseases like Parkinson’s, polyglutamine diseases like Huntington’s disease, and amyotrophic lateral sclerosis (ALS). Despite much investigation, a comprehensive understanding of ND has been confounded by the complex contribution of a variety of genetic and environmental factors. ALS, commonly known as Lou Gehrig’s disease, is a severe ND in which motor neurons die and their target muscles atrophy (Brown and Al-Chalabi 2017). Although a modest proportion of ALS cases are clearly inherited (familial ALS, fALS), ∼80%–90% of ALS cases are sporadic (sALS), with only ∼20% of sporadic cases linked to specific genes (Vucic et al. 2014; Brown and Al-Chalabi 2017; Martin et al. 2017). This, together with the limited treatment options, creates a pressing need to understand better the genetic and cellular pathways underlying this serious disease. Because fALS and sALS bear high clinical and pathological similarity, and because the identified genetic loci are common to both, it is presumed that fALS and sALS share molecular and cellular etiologies (Brown and Al-Chalabi 2017; Martin et al. 2017). Numerous genes have been identified in fALS (https://alsod.ac.uk/), with four loci accounting for the majority of cases, namely FUS, TARDBP/TDP-43, C9orf72, and SOD1 (Brown and Al-Chalabi 2017; Hardiman et al. 2017). Interestingly, FUS and TDP-43 are both RNA-binding proteins, and abnormal RNA processing may contribute to disease progression (Ranganathan et al. 2020; Yerbury et al. 2020). Still, ALS-associated mutations in FUS or TDP-43 increase the tendency of the encoded proteins to misfold and aggregate abnormally, a property likewise observed for ALS-associated SOD1 mutants, suggesting that dysregulation of protein homeostasis (proteostasis) and resultant proteotoxicity is the cause of neurodegeneration (Martin et al. 2017; Yerbury et al. 2020). In support of this idea, TDP-43 aggregates are found in up to 97% of ALS patients, representing both sporadic and familial cases, including in patients with wild-type TDP-43 alleles (Hardiman et al. 2017). Toxic aggregates are also a feature of ALS linked to mutations in c9orf72. The protein encoded by c9orf72 is implicated in autophagy, an important cellular process for regulating proteostasis, but ALS-causing mutations derive from expansion of a hexanucleotide GGGGCC repeat in a noncoding intron of the gene. These repeated sequences are transcribed into RNAs that aggregate and sequester RNA binding proteins, and the repeat containing RNAs can be translated via a noncanonical protein translation pathway into dipeptide repeats that form toxic protein aggregates (Ranganathan et al. 2020). Much is still unknown about how ALS develops, including whether and how dysregulation of RNA processing and proteostasis lead to neurodegeneration, whether ALS-associated mutations cause disease by loss of their wild-type functions or gain of toxicity, and whether these four genes, as well as others, act in the same genetic and biochemical pathway. Moreover, despite the prevalence of RNA processing defects and proteotoxic aggregates in ALS, many other cellular activities are affected as well (Hardiman et al. 2017). The goal of this study is to link previously unassociated genes to ALS, in order to better understand how the disease progresses and to reveal potential therapeutic targets. Prior studies have demonstrated that expressing human disease variants of FUS or TDP-43 with the GAL4-UAS system in photoreceptor neurons of the Drosophila eye induces degeneration, mimicking the cellular pathology of the disease (Ritson et al. 2010; Lanson et al. 2011). The authors screen the Exelixis collection of Drosophila insertion mutations for dominant modifiers of this eye degeneration phenotype, to elucidate the molecular pathways leading from the human disease alleles to ALS pathology (Figure 1).
Figure 1.
To conduct their screen, the authors collected flies from (A) a Drosophila strain carrying an ALS-associated allele, for example hFUSR521C, under control of a UAS regulatory element along with the GMR-GAL4 driver. Expression of UAS-hFUSR521C with GMR-GAL4 causes a degenerative rough eye phenotype. These flies were crossed to (B) flies from the Exelixis collection, each carrying an individual mutation caused by random insertion of a transposable element in the genome (green triangle). F1 progeny (C) were examined for enhancement or suppression of the rough eye phenotype.
Unpacking the Work
In order to conduct genome-wide screens for ALS genes, the authors develop a strategy to cross transgenic Drosophila strains expressing ALS-associated human alleles to insertion mutations from the Exelixis collection and to examine the F1 progeny (Figure 1). Females were collected from Drosophila strains carrying either of two ALS-associated alleles, hFUSR521C or hTDP-43M337V, each controlled by a UAS regulatory element. Each strain also carries the GMR-GAL4 driver, which produces GAL4 protein in cells of the eye, thereby causing expression of the human ALS-associated proteins there. These females were mated to males from the Exelixis collection, each strain of which carries an individual insertion mutation.
All starting strains also carry balancer chromosomes [see (Hales et al. 2015) for a detailed description of balancer chromosomes], which allow stocks to be maintained as heterozygotes by bearing recessive lethal mutations and suppressing meiotic recombination with homologous chromosomes. The balancers are marked with the dominant wing phenotype Curly (Cy, curly wings instead of straight, on the balancer CyO), or the dominant larval phenotype Tubby (Tb, larvae are shorter and fatter than wild type, on the balancer TM6B). The second chromosome balancers in the strains expressing the human ALS alleles additionally carry the yeast gene GAL80 expressed in all cells. GAL80 protein inhibits GAL4 activity, preventing expression of the ALS alleles in the starting strains (St Johnston 2013).
Technical Glossary
This glossary provides some technical information to assist with understanding the experiments in the research article.
RNAi (RNA interference) uses a small double-stranded RNA molecule to target an endogenous mRNA for post-transcriptional silencing. To employ RNAi in vivo, a transgene is generated encoding an RNAi transcript under control of a UAS regulatory element, allowing directed expression by the GAL4-UAS system. The transgene is designed with sequence complementarity to an endogenous target mRNA and an inverted repeat structure, which facilitates folding of the RNAi transcript into a double-stranded RNA. The double-stranded RNA promotes destruction of the target mRNA with matching sequence, thereby inhibiting production of functional protein from the target gene. This process of post-transcriptional knockdown often produces similar phenotypes as loss of function alleles and circumvents the time and labor needed to create genomic mutations. Furthermore, the Drosophila research community has produced libraries of RNAi transgenes targeting every gene in the genome, which are readily available (Mohr and Perrimon 2012).
Imaginal discs are epithelial sacs of primordial cells that give rise to the external anatomical structures of the adult fly, also called the imago (Beira and Paro 2016). For example, the eye imaginal discs are the precursors of the adult eyes. Imaginal discs are specified during embryogenesis, are patterned and grow during larval development, and form their distinct morphologies during metamorphosis.
The neuromuscular junction (NMJ) is the site of contact between motor neurons and muscle tissue. In the Drosophila larva, motor neuron axon terminals at the NMJ form synaptic boutons, round structures that contain the synaptic active zones. Each Drosophila motor neuron creates a stereotypical number of boutons, providing a quantitative measurement of the fidelity of NMJ development and maintenance (Menon et al. 2013). In ALS, motor neuron degeneration is accompanied by disassembly of the NMJ and atrophy of muscle tissue (Cappello and Francolini 2017).
A dominant negative allele is one that interferes with the activity of the wild-type allele. Dominant negative alleles are functionally dominant, i.e., they generate a mutant phenotype as heterozygotes, but because their mutant phenotypes result from reduced activity of the wild-type allele, they are loss of function.
Guided Reading Questions
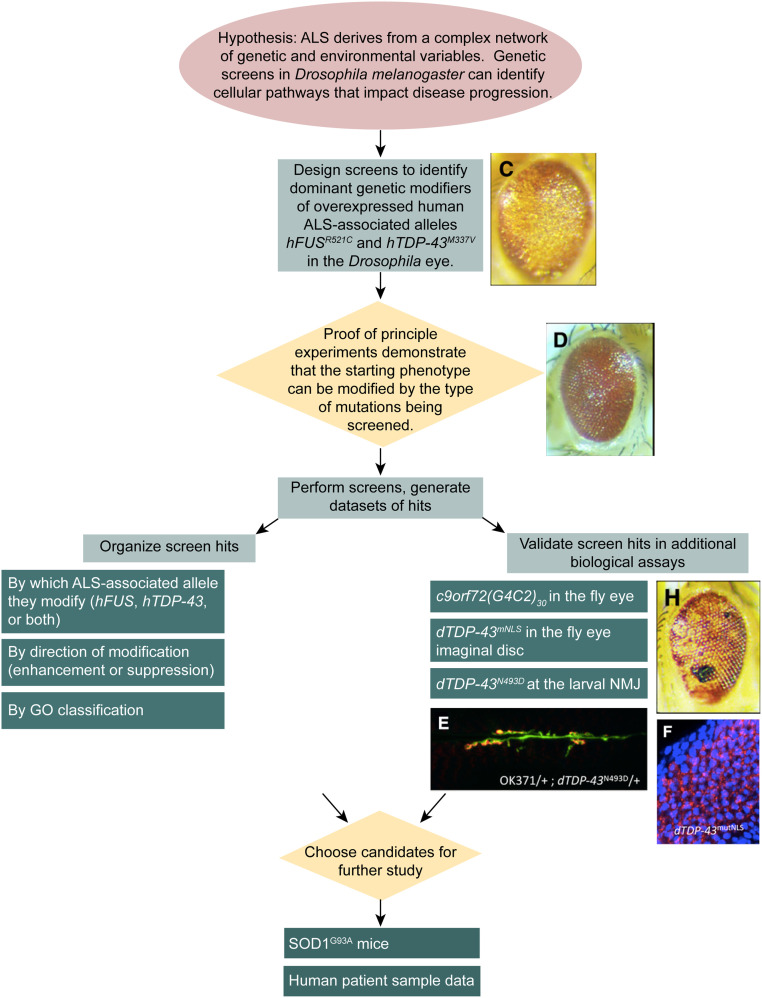
The guided reading questions below are intended to help students work through the results of Kankel et al. (2020). Students can be expected to spend ∼15–30 min on each figure, on average. Questions can be assigned as homework in preparation for class discussions or as in-class group work. The overall logic of the experiments presented in the article is diagrammed in Figure 2. Answers to the guided reading questions are available as Supplemental Material online.
Figure 2.
Workflow diagram illustrating the logic of the screening process. Genetic screens begin with a hypothesis, i.e., that genetic screens in Drosophila can be used to identify novel genes associated with ALS. Because human ALS-associated alleles hFUSR521C or hTDP-43M337V cause a visible degenerative eye phenotype in adult Drosophila, libraries of mutants like the Exelixis collection can be screened for dominant modifiers of this phenotype. Once the screens have been planned, proof-of-principle experiments are conducted to demonstrate that the screen has the potential to be successful, using selected Drosophila mutants of other ALS gene orthologs as positive controls. The screens are then performed, and a list of hits is generated. The list can be organized by which ALS allele is affected by each modifier, whether modifiers are enhancers or suppressors, and according to GO classification for \biological function and/or cellular localization. Screen hits also are validated experimentally by assaying whether they modify other ALS model systems, including c9orf72(G4C2)30-mediated degeneration of the adult Drosophila eye, dTDP-43mNLS aggregation in the larval eye imaginal disc, and dTDP-43N493D perturbation of the larval NMJ. Candidates that show activity in the secondary assays may be chosen for further study, which includes examination in an ALS model mouse and in human ALS patient data.
Figure 1. A genome-wide screening strategy for dominant modifiers of human ALS-associated allele induced eye degeneration in Drosophila.
What is the purpose of the screens described in this article? How were the screens conducted?
Explain each of the starting Drosophila strains used for the screens, shown in Figure 1A of Kankel et al. (2020). What genetic elements does each starting strain carry?
Draw Punnett squares representing the screen crosses. Which are the desired progeny and how will they be selected?
Which photomicrograph panel shows a normal fly eye? How do Figure 1, C and H compare to normal?
Describe what is shown in Figure 1, D–G and I–L in your own words. For each of the four genes shown, dSETX, dco, Hsc70Cb, and Ask, explain how it was tested and whether it is a suppressor or enhancer of hFUSR521C and hTDP-43M337V.
Figure 2. Dominant modifiers of human ALS-associated allele induced eye degeneration are identified in the screens.
How many insertion mutations were screened? How many crosses were established in the screens?
How old were the F1 flies when they were screened?
How many hits were recovered from the screens? What percentage of the total do the hits represent? Show your calculations. Is this percentage higher, lower, or on par with expectations?
How many hits affect both hFUSR521C and hTDP-43M337V transgenes? Do those common hits always affect the two transgenes in the same way? Give an example and propose a hypothesis to explain these results.
Figure 3. Validating screen hits using another ALS genetic model.
Explain the experiment shown in Figure 3. How does it differ from the original screens? What are the advantages and disadvantages of this experiment compared to the original screens?
What does the graph in Figure 3I show? How is each percentage calculated?
What percentage of genes tested shows effects in this experiment?
Figure 4. Validating screen hits using another ALS-associated phenotype.
How do the transgenes in the experiment shown in Figure 4 differ from those in the experiments shown in Figure 1?
Compare and contrast the proteins expressed from these transgenes with the original constructs. What additional disease-relevant property do these proteins show?
Is this additional property modified by the hits from the screens? What does that suggest about how this property relates to degeneration?
Figure 5. Validating screen hits in another cellular context.
In Figure 5, the authors switch their focus away from the eye. What anatomical structure is the focus of this figure? In which cells is OK371-GAL4 expressed? Why do the authors choose to examine this structure?
The authors use the OK371-GAL4 driver to express three different dTDP-43 variants. Among these three variants, which one has the most severe mutant phenotype and how do you know? How does this relate to the anatomical structure shown in the photomicrographs? What does this suggest about this variant?
The authors test three genes, SF2, lilli, and klp98A, for their ability to modify the phenotypes caused by OK371-GAL4 expression of dTDP-43. How were these three genes chosen?
Which of these three genes has a significant effect on the OK371-dTDP-43 phenotype? Does any of the three fail to affect the OK371-dTDP-43 phenotype? Propose a hypothesis to explain these results.
Figure 6. Phospholipase D is an important player in ALS.
From their screens, the authors discover the Phospholipase D (PLD) pathway (schematized in Figure 9). How do they test the importance of PLD in ALS in Figure 6? Describe five results shown in this figure that corroborate the importance of PLD.
What effect would you expect if Drosophila RalA expression were reduced instead of PLD in the same type of experiments?
Figure 7. The Phospholipase D pathway is important for ALS progression.
Is your expectation above (Figure 6, question 2) confirmed? Explain why or why not.
How do the results in Figure 7 further bolster the importance of the PLD pathway in ALS?
Based on the experiments presented in Figures 6 and 7, are PLD pathway effectors likely to function upstream or downstream of FUS, TDP-43, and c9orf72 in ALS disease development?
Figure 8. ValidatingPhospholipase D in another model organism.
What are the advantages of performing genetic screens in Drosophila melanogaster as opposed to mice? What are the advantages of testing genetic interactions in mice as opposed to Drosophila?
Compare the grip strength of wild-type mice, SOD1G93A-expressing mice, and SOD1G93A-expressing mice with mutations in PLD1, PLD2, or PLD1 and 2. How does mutating PLD modify the effects of SOD1G93A expression?
Are the results shown in Figure 8 consistent with those shown in Figure 6? Why or why not?
Putting It All Together
Other recent research articles have found that PLD pathway genes are upregulated in sALS patients with early-onset disease (Rabin et al. 2010; Kaplan et al. 2014). Is this finding consistent with the data presented in Figures 6–8? Explain.
Do you think the PLD pathway is a worthwhile therapeutic target for ALS? Why or why not?
Propose an experiment to follow up on the idea of PLD as a therapeutic target for ALS.
Student Projects
After reading the article and working through the guided reading questions, instructors may choose to give their students the opportunity to develop a project of their own on a related topic. Below are two different options for student-directed projects. We recommend giving students several weeks to complete the assignment and submit in midsemester or at the end of the term. Projects can be formatted as a written paper or as an oral presentation, with students working as individuals or in groups.
Choose a disease besides ALS that interests you. Does this disease have a genetic component? Has this disease been modeled in Drosophila or another model organism? Design a genetic screen to identify novel factors in this disease.
-
Using PubMed (https://pubmed.ncbi.nlm.nih.gov/), find another research article that describes a screen for ALS components. Compare and contrast with this article, using the questions below to help you.
What model organism or model system was used in your article?
How was ALS simulated in your article? What phenotypic aspects of ALS were examined?
What are the advantages and disadvantages of the model system in your article compared to the one used in Kankel et al. (2020)?
How was the screen conducted?
What were the outcomes of the screen?
Are there any screen hits in common between your article and Kankel et al. (2020)? Why might this be? What does this suggest?
Acknowledgments
We thank Elizabeth De Stasio, Molly Gallop, Rebecca Delventhal, Mark Kankel, Anindya Sen, and Spyros Artavanis-Tsakonas for helpful comments on the manuscript. The Steinhauer laboratory is supported by National Institutes of Health (NIH) grant R15-HD080511.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12889613.
Communicating editor: E. De Stasio
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D. et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., 2004. Accessing the Exelixis collection. Nat. Genet. 36: 207 10.1038/ng1316 [DOI] [PubMed] [Google Scholar]
- Baker N. E., Li K., Quiquand M., Ruggiero R., and Wang L. H., 2014. Eye development. Methods 68: 252–259. 10.1016/j.ymeth.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beira J. V., and Paro R., 2016. The legacy of Drosophila imaginal discs. Chromosoma 125: 573–592. 10.1007/s00412-016-0595-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., and Yamamoto S., 2015. Morgan’s legacy: fruit flies and the functional annotation of conserved genes. Cell 163: 12–14 (erratum: Cell 163: 772). 10.1016/j.cell.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Tong C., and Tsuda H., 2010. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci. 11: 514–522. 10.1038/nrn2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M. et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188: 731–743. 10.1534/genetics.111.126995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer S., 1967. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. USA 58: 1112–1119. 10.1073/pnas.58.3.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., and Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brown R. H., and Al-Chalabi A., 2017. Amyotrophic lateral sclerosis. N. Engl. J. Med. 377: 162–172. 10.1056/NEJMra1603471 [DOI] [PubMed] [Google Scholar]
- Cappello V., and Francolini M., 2017. Neuromuscular junction dismantling in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 18: 2092 10.3390/ijms18102092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Kelley R., and Spradling A., 1988. Insertional mutagenesis of the Drosophila genome with single P elements. Science 239: 1121–1128. 10.1126/science.2830671 [DOI] [PubMed] [Google Scholar]
- Gaudet P., Škunca N., Hu J. C., and Dessimoz C., 2017. Primer on the gene ontology. Methods Mol. Biol. 1446: 25–37. 10.1007/978-1-4939-3743-1_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales K. G., Korey C. A., Larracuente A. M., and Roberts D. M., 2015. Genetics on the fly: a primer on the Drosophila model system. Genetics 201: 815–842. 10.1534/genetics.115.183392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiman O., Al-Chalabi A., Chio A., Corr E. M., Logroscino G. et al. , 2017. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 3: 17071 (erratum: Nat. Rev. Dis. Primers 3: 17085). 10.1038/nrdp.2017.85 [DOI] [PubMed] [Google Scholar]
- Hastings J., 2017. Primer on ontologies. Methods Mol. Biol. 1446: 3–13. 10.1007/978-1-4939-3743-1_1 [DOI] [PubMed] [Google Scholar]
- Hotta Y., and Benzer S., 1969. Abnormal electroretinograms in visual mutants of Drosophila. Nature 222: 354–356. 10.1038/222354a0 [DOI] [PubMed] [Google Scholar]
- Jackson G. R., Salecker I., Dong X., Yao X., Arnheim N. et al. , 1998. Polyglutamine-expanded human Huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21: 633–642. 10.1016/S0896-6273(00)80573-5 [DOI] [PubMed] [Google Scholar]
- Jan Y. N., and Jan L., 2008. Retrospective: Seymour Benzer (1921-2007). Science 319: 45 10.1126/science.1154050 [DOI] [PubMed] [Google Scholar]
- Editorial, 2020. Model organisms: nature’s gift to disease research. Genetics 214: 233–234. 10.1534/genetics.120.303050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett T., Wajidi M. F., Oxtoby E., and Wolf C. R., 1991. Mammalian genes expressed in Drosophila: a transgenic model for the study of mechanisms of chemical mutagenesis and metabolism. EMBO J. 10: 1075–1081. 10.1002/j.1460-2075.1991.tb08047.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel M. W., Sen A., Lu L., Theodorou M., Dimlich D. N. et al. , 2020. Amyotrophic lateral sclerosis modifiers in Drosophila reveal the phospholipase D pathway as a potential therapeutic target. Genetics 215: 747–766. 10.1534/genetics.119.302985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A., Spiller K. J., Towne C., Kanning K. C., Choe G. T. et al. , 2014. Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron 81: 333–348. 10.1016/j.neuron.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C. et al. ; International Human Genome Sequencing Consortium , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 (erratum: Nature 412: 565). 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lanson N. A., Maltare A., King H., Smith R., Kim J. H. et al. , 2011. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum. Mol. Genet. 20: 2510–2523. 10.1093/hmg/ddr150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Tully T., and White K., 1992. Human amyloid precursor protein ameliorates behavioral deficit of flies deleted for Appl gene. Neuron 9: 595–605. 10.1016/0896-6273(92)90024-8 [DOI] [PubMed] [Google Scholar]
- Martin S., Al Khleifat A., and Al-Chalabi A., 2017. What causes amyotrophic lateral sclerosis? F1000Res. 6: 371 10.12688/f1000research.10476.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk L., Berson A., and Bonini N. M., 2015. Drosophila as an in vivo model for human neurodegenerative disease. Genetics 201: 377–402. 10.1534/genetics.115.179457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon K. P., Carrillo R. A., and Zinn K., 2013. Development and plasticity of the Drosophila larval neuromuscular junction. Wiley Interdiscip. Rev. Dev. Biol. 2: 647–670. 10.1002/wdev.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S. E., and Perrimon N., 2012. RNAi screening: new approaches, understandings, and organisms. Wiley Interdiscip. Rev. RNA 3: 145–158. 10.1002/wrna.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H., 1910. Sex limited inheritance in Drosophila. Science 32: 120–122. 10.1126/science.32.812.120 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., and Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Pak W. L., Grossfield J., and White N. V., 1969. Nonphototactic mutants in a study of vision of Drosophila. Nature 222: 351–354. 10.1038/222351a0 [DOI] [PubMed] [Google Scholar]
- Rabin S. J., Kim J. M., Baughn M., Libby R. T., Kim Y. J. et al. , 2010. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum. Mol. Genet. 19: 313–328. 10.1093/hmg/ddp498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R., Haque S., Coley K., Shepheard S., Cooper-Knock J. et al. , 2020. Multifaceted genes in amyotrophic lateral sclerosis-frontotemporal dementia. Front. Neurosci. 14: 684 10.3389/fnins.2020.00684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritson G. P., Custer S. K., Freibaum B. D., Guinto J. B., Geffel D. et al. , 2010. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 30: 7729–7739. 10.1523/JNEUROSCI.5894-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge R. D., Karlovich C. A., and Banerjee U., 1991. Genetic dissection of a neurodevelopmental pathway: son of sevenless functions downstream of the sevenless and EGF receptor tyrosine kinases. Cell 64: 39–48. 10.1016/0092-8674(91)90207-F [DOI] [PubMed] [Google Scholar]
- Rubin G. M., and Spradling A. C., 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. 10.1126/science.6289436 [DOI] [PubMed] [Google Scholar]
- Simon M. A., Bowtell D. D., Dodson G. S., Laverty T. R., and Rubin G. M., 1991. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67: 701–716. 10.1016/0092-8674(91)90065-7 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., and Rubin G. M., 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. 10.1126/science.6289435 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Stern D. M., Kiss I., Roote J., Laverty T. et al. , 1995. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92: 10824–10830. 10.1073/pnas.92.24.10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., 2002. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3: 176–188. 10.1038/nrg751 [DOI] [PubMed] [Google Scholar]
- St Johnston D., 2013. Using mutants, knockdowns, and transgenesis to investigate gene function in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2: 587–613. 10.1002/wdev.101 [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A. et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J. et al. , 2001. The sequence of the human genome. Science 291: 1304–1351 (erratum: Science 292: 1838). 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Vucic S., Rothstein J. D., and Kiernan M. C., 2014. Advances in treating amyotrophic lateral sclerosis: insights from pathophysiological studies. Trends Neurosci. 37: 433–442. 10.1016/j.tins.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Wangler M. F., Yamamoto S., Chao H. T., Posey J. E., Westerfield M. et al. , 2017. Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics 207: 9–27. 10.1534/genetics.117.203067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick J. M., Paulson H. L., Gray-Board G. L., Bui Q. T., Fischbeck K. H. et al. , 1998. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell 93: 939–949. 10.1016/S0092-8674(00)81200-3 [DOI] [PubMed] [Google Scholar]
- Yerbury J. J., Farrawell N. E., and McAlary L., 2020. Proteome homeostasis dysfunction: a unifying principle in ALS pathogenesis. Trends Neurosci. 43: 274–284. 10.1016/j.tins.2020.03.002 [DOI] [PubMed] [Google Scholar]