ABSTRACT
Keratin 17 (KRT17; K17), a non-lamin intermediate filament protein, was recently found to occur in the nucleus. We report here on K17-dependent differences in nuclear morphology, chromatin organization, and cell proliferation. Human tumor keratinocyte cell lines lacking K17 exhibit flatter nuclei relative to normal. Re-expression of wild-type K17, but not a mutant form lacking an intact nuclear localization signal (NLS), rescues nuclear morphology in KRT17-null cells. Analyses of primary cultures of skin keratinocytes from a mouse strain expressing K17 with a mutated NLS corroborated these findings. Proteomics screens identified K17-interacting nuclear proteins with known roles in gene expression, chromatin organization and RNA processing. Key histone modifications and LAP2β (an isoform encoded by TMPO) localization within the nucleus are altered in the absence of K17, correlating with decreased cell proliferation and suppression of GLI1 target genes. Nuclear K17 thus impacts nuclear morphology with an associated impact on chromatin organization, gene expression, and proliferation in epithelial cells.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Chromatin, Gene expression, Intermediate filament, Keratin, Nucleus, Proliferation
Highlighted Article: Keratin 17 (K17) is a non-lamin IF protein that functions in the cell nucleus. K17 is here shown to regulate nuclear morphology, chromatin organization, LAP2 localization and cell proliferation.
INTRODUCTION
Intermediate filament (IF) proteins are encoded by a superfamily of >70 genes and assemble into cytoskeletal elements that fulfill key roles towards cellular structure and mechanics and many other cellular processes (Pan et al., 2013; Bouameur and Magin, 2017). Keratins, the largest subset of IFs, comprise 28 type I (acidic) and 26 type II (basic) genes and proteins, and form IFs as obligate heteropolymers in the cytoplasm of epithelial cells (Schweizer et al., 2006; Jacob et al., 2018). We and others recently discovered that select keratins and other non-lamin IF proteins can localize to the nucleus, contain functional importin α-dependent nuclear localization signal (NLS) sequences, and participate in nuclear processes, such as transcriptional control of gene expression, cell cycle progression, and protection from senescence (Escobar-Hoyos et al., 2015; Hobbs et al., 2015, 2016; Kumeta et al., 2013; Zhang et al., 2018; Zieman et al., 2019).
Before these findings, the generally accepted notion was that, with the exception of type V nuclear lamins, IF proteins strictly reside in the cytoplasm (Hobbs et al., 2016). Similar views were once held for other cytoskeletal proteins that have since been found to occur and function in the nucleus (Pereira et al., 1998; Yeh et al., 2004; Bettinger et al., 2004; Pederson, 2008; Akoumianaki et al., 2009; Castano et al., 2010). For example, actin adopts novel conformations in the nucleus, where it participates in chromatin remodeling, transcription, binding to nascent mRNA transcripts and mRNA export (Bettinger et al., 2004; Schoenenberger et al., 2005; Visa and Percipalle, 2010; Kapoor et al., 2013). There is evidence suggesting that the ancestral IF protein was a nuclear lamin-like protein from which all modern-day cytoplasmic IF proteins are derived (Erber et al., 1998). Accordingly, it could be that some modern-day keratin proteins and non-lamin IF proteins are performing some of the ‘ancient’ roles associated with IF proteins. This is not an unheard-of concept given nuclear actin (Kapoor et al., 2013).
Kumeta et al. (2013) conducted an unbiased screen to identify the molecular components of nuclear scaffolds and reported on the presence of keratins in the nucleus (e.g. GFP-tagged keratins 7, 8, 17, 18; also, endogenous K8), with most appearing as a soluble, non-filamentous haze. We have shown that keratin 17 (K17) and the transcriptional regulator AIRE colocalize in the nucleus and physically associate with NF-κB (e.g. p65 subunit, also known as RELA) consensus binding sites at the promoter regions of genes associated with inflammation (Hobbs et al., 2015). Escobar-Hoyos et al. (2015) has reported that K17 promotes cell cycle progression by binding to a G0/G1-to-S-phase transition inhibitor (p27KIP1; encoded by CDKN1B) in the nucleus and facilitating its nuclear export and degradation. We recently reported that another keratin protein, K16, localizes to the nucleus of keratinocytes (Zieman et al., 2019). The type IV IF protein nestin also localizes to the nucleus, where it binds to and stabilizes lamin A/C, thereby protecting cells from senescence (Zhang et al., 2018). In the case of K17 and nestin, functional importin α-dependent bipartite NLS sequences have been identified (Escobar-Hoyos et al., 2015; Hobbs et al., 2015, 2016; Zhang et al., 2018). This said, we are still in the early stages of understanding the function(s) and significance of non-lamin IF proteins in the nucleus. Here, we provide evidence that nuclear-localized K17 regulates nuclear morphology and chromatin organization.
RESULTS AND DISCUSSION
Loss of K17 alters nuclear morphology
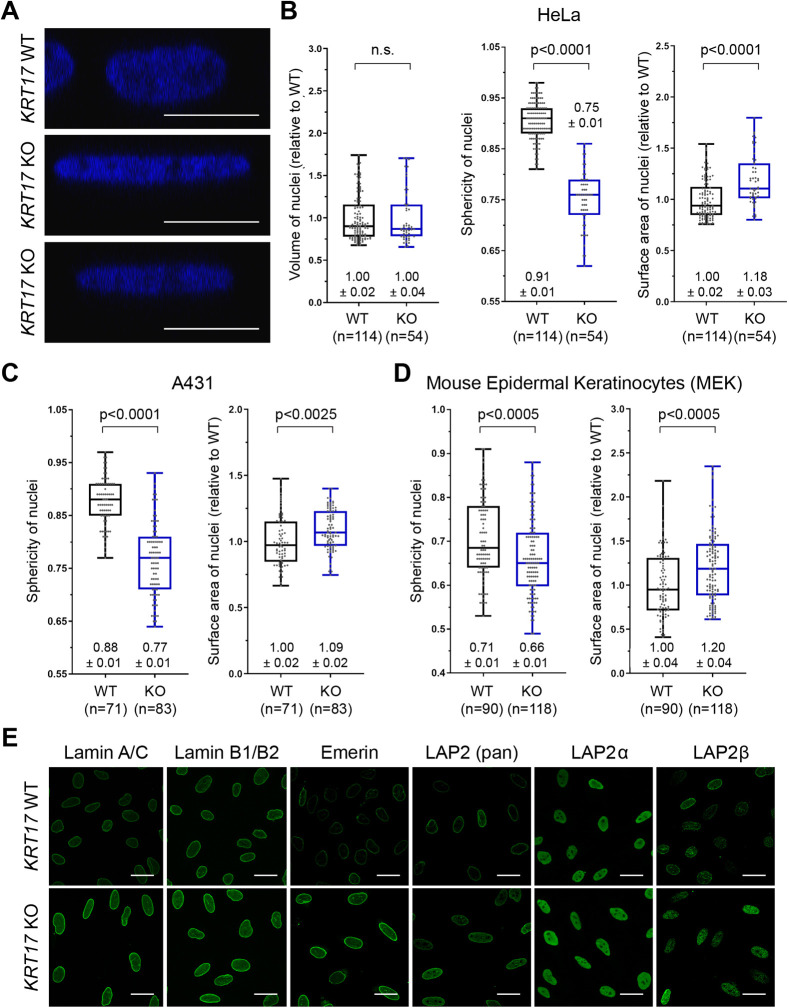
To build on our knowledge of the function(s) of nuclear K17, we utilized CRISPR/Cas9 genomic editing technology to generate KRT17 knockout (KO) clones in the tumor-derived HeLa and A431 cell lines (see Materials and Methods). We observed a correlation between K17 expression and nuclear size and shape. From 2D images, we found that loss of K17 correlated with larger nuclear area in HeLa (Fig. S1A,B) and A431 (Fig. S1C) human cells, and in mouse epidermal keratinocytes (MEKs) in primary culture (Fig. S1D). From confocal imaging, we determined that fewer optical sections were required to span the height of the nucleus (z-axis) in KRT17 KO relative to parental cells (Fig. 1A). The nuclei of KRT17 KO HeLa (Fig. 1B), A431 (Fig. 1C) and MEK (Fig. 1D) cells exhibited decreased sphericity and increased total surface area, but no change in volume (Fig. 1B).
Fig. 1.
Loss of K17 alters nuclear morphology. (A) Orthogonal-view reconstruction of multi-plane confocal micrographs of individual nuclei from HeLa KRT17 WT versus KRT17 knockout (KO) cells. Nuclei are stained with DAPI (blue). Scale bars: 10 μm. (B–D) Quantification of nuclear volume, sphericity and surface area in (B) KRT17 WT versus KO cells HeLa and (C) A431 cells, and in (D) primary MEK cells. Numbers above or below each box-and-whisker plot designate the mean±s.e.m. In the box-and-whisker plots, the box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum and maximum values. P-values shown were calculated with a Student's t-test with two-tailed distribution, and two-sample equal variance (homoscedastic); n=cells counted. n.s., not significant. (E) Confocal micrograph maximum intensity projections (MIPs) of nuclei from KRT17 WT and KRT17 KO HeLa cells, immunostained with antibodies against nuclear lamina-associated proteins (green). Scale bars: 10 μm.
We hypothesized that differences in nuclear shape and surface area may entail changes in inner nuclear membrane (INM) proteins, including lamins A, C and B1/B2 and LEM-domain-containing proteins [LAP2α, LAP2β and LAP2γ (all encoded by TMPO) and emerin]. Immunoblotting of whole-cell lysates from KRT17 WT and KO cells revealed significant increases in the steady-state levels of these proteins, except for LAP2β, in cells lacking K17 (Fig. S1E,F). The observed increases (∼15–50%) were proportional to the increase in surface area (∼10–20%) in KRT17 KO nuclei relative to control (Fig. 1B–D). Consistent with these findings, indirect immunofluorescence indicated a significant increase in signal intensity for each analyzed protein in KRT17 KO cells compared to WT (Fig. 1E; quantification in Fig. S1G). Again, LAP2β behaved differently (see below).
Impact of K17 on nuclear morphology requires an intact NLS
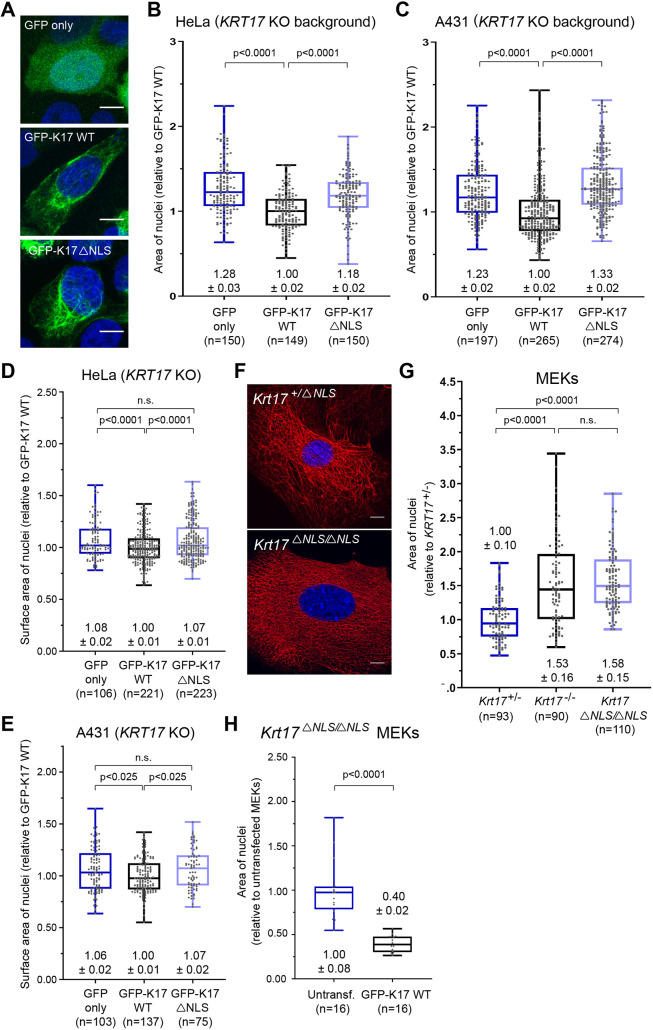
We next assessed whether nuclear-localized K17 impacts nuclear dimensions. We transiently transfected KRT17 KO cells (HeLa and A431) with either GFP (control), GFP–K17 WT or GFP–K17ΔNLS (in which K399A and K400A mutations impair NLS function; Escobar-Hoyos et al., 2015; Hobbs et al., 2015) (Fig. 2A) and assessed the size and shape of nuclei from 2D and 3D images. Expression of GFP–K17 WT, but not GFP–K17ΔNLS, restored normal dimensions to the nucleus, including decreased area (2D; Fig. 2B,C), increased sphericity (3D; Fig. S2A,B), and decreased surface area (3D; Fig. 2D,E). Expression of GFP–K17 ΔNES, in which L194A, L197A and L199A mutations impair the nuclear export sequence (NES) function of K17 (thus favoring nuclear retention; Escobar-Hoyos et al., 2015) (Fig. S2C), also rescues nuclear shape defects in KRT17 KO A431 keratinocytes. Expression of GFP–K14 WT (Fig. S2D) is ineffective in doing so, suggesting specificity for K17. Next, we used proximity ligation assays (PLAs) with two different host-species antibodies against K17 [allows the visualization of the K17 nuclear pool without Leptomycin B treatment (Hobbs et al., 2015)] in HeLa KRT17 KO cells expressing GFP–K17 WT or GFP–K17ΔNLS. Both constructs gave rise to well-developed filamentous arrays in the cytoplasm of transfected cells (Fig. 2A). However, GFP–K17ΔNLS gave rise to a markedly reduced nuclear signal relative to GFP–K17 WT (Fig. S2E; quantification in Fig. S2F). These data suggest that, in transfected human A431 and HeLa cells, a nuclear pool of K17 is responsible for the impact on nuclear morphology, independently of the property of fostering a dense network of keratin filaments in the perinuclear cytoplasm.
Fig. 2.
Impact of K17 on nuclear morphology requires an intact NLS. (A) Confocal micrograph MIPs of HeLa KRT17 KO cells transiently transfected (48 h) with GFP, GFP–K17 WT or GFP–K17ΔNLS (green). Nuclei are stained with DAPI (blue). Scale bars: 10 μm. (B,C) Quantified area (2-dimensional; 2D) of nuclei from (B) HeLa KRT17 KO and (C) A431 KRT17 KO cells expressing GFP, GFP–K17 WT or GFP–K17ΔNLS. (D,E) Quantified surface area (3-dimensional; 3D) of nuclei from (D) HeLa KRT17 KO and (E) A431 KRT17 KO cells expressing GFP, GFP–K17 WT or GFP–K17ΔNLS. (F) Confocal MIPs for K17 (red) in MEKs from Krt17+/ΔNLS and Krt17ΔNLS/ΔNLS mice. Nuclei are stained with DAPI (blue). Scale bars: 10 μm. (G) Quantified area of nuclei from primary MEKs from Krt17+/−, Krt17−/− and Krt17ΔNLS/ΔNLS mice. (H) Quantified area of nuclei from Krt17ΔNLS/ΔNLS MEKs untransfected or transfected with GFP-K17 WT. For B–E, G, H, numbers above or below each box-and-whisker plot represent the mean±s.e.m. In the box-and-whisker plots, the box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum and maximum values. P-values shown were calculated with a Student's t-test with two-tailed distribution, and two-sample equal variance (homoscedastic). n.s., not significant.
A CRISPR/Cas9-based strategy was used to produce a mouse strain carrying K399A and K401A mutations at the Krt17 locus in its germline (Fig. S3A,B; and see Materials and Methods). Mice homozygous for this allele (Krt17ΔNLS/ΔNLS) are viable, fertile and show no obvious phenotype at baseline (Fig. S3C and data not shown). The skin of young adult Krt17ΔNLS/ΔNLS mice show normal histology relative to WT (Fig. S3D) and a normal pattern of immunostaining for K17 in hair follicles (Fig. S3E). In primary culture, Krt17ΔNLS/ΔNLS keratinocytes show a normal array of K17-containing filaments in the cytoplasm (Fig. S2F). PLAs using two different host-species antibodies for K17 coupled to single-plane confocal microscopy revealed a markedly reduced frequency of nuclear-localized signal in Krt17ΔNLS/ΔNLS mutant keratinocytes relative to WT (data not shown; similar to Figs. S2E, S2F). Measurements revealed that, in spite of the presence of a robust network of K17 filaments in their cytoplasm (Fig. 2F), the nuclei of newborn Krt17ΔNLS/ΔNLS keratinocytes show an area that is indistinguishable from Krt17−/− keratinocytes but markedly larger than Krt17+/− control (Fig. 2G). Transfection-mediated re-expression of GFP–K17 WT, but not GFP–K17ΔNLS, restored the nuclear dimensions in Krt17ΔNLS/ΔNLS keratinocytes in primary culture (Fig. 2H). Such findings extend the observations made in cultured human cell lines and markedly strengthen the connection between nuclear-localized K17 and nuclear shape in skin keratinocytes.
K17 interacts with proteins involved in chromatin organization and gene expression
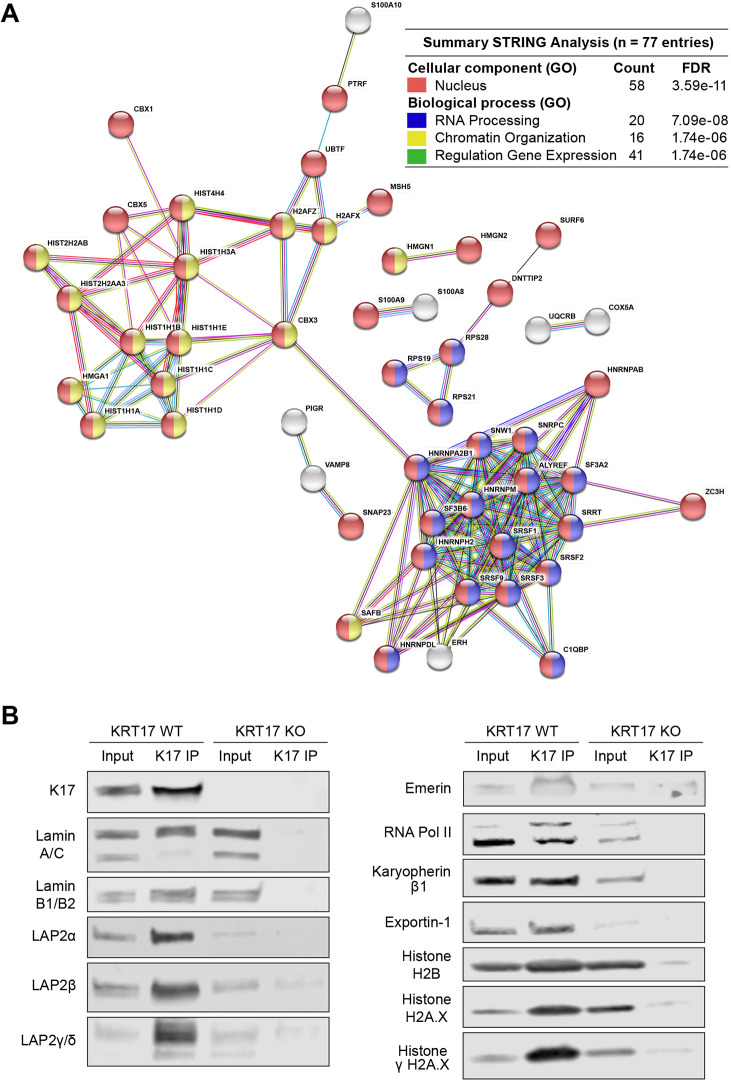
We next conducted an unbiased screen to identify proteins that interact with K17 in the nucleus. We immunoprecipitated K17 from a soluble fraction enriched for nuclear proteins and identified co-precipitated proteins by mass spectrometry (MS). Data filtering (e.g. 127N/130N ratio >1.2; 127N/130N count >4; see Materials and Methods) yielded 77 proteins that are enriched by K17 immunoprecipitation (IP) in KRT17 WT cells over KRT17 KO cells (see Table S1). Of the 77 proteins, 58 (75%) had at least one defined nuclear role according to Gene Ontology terms in the Universal Protein Resource (UniProt) database (https://www.uniprot.org/; Uniprot Consortium, 2019). Network association analysis (STRING) under a high stringency setting (0.7) categorized these 77 MS-identified proteins as overrepresented in nuclear processes including RNA processing (n=20), chromatin organization (n=16) and regulation of gene expression (n=41). RNA processing and chromatin organization each form a large hub in the network of functionally interconnected candidate nuclear protein interactors for K17 (Fig. 3A).
Fig. 3.
Screen for K17-interaction proteins in the nucleus. (A) STRING-db online analysis (v10.5; http://string-db.org/) of the 77 most-enriched MS-identified co-immunoprecipitated proteins in A431 nuclear extracts; 58 (75%) have at least one known function in the nucleus according to UniProt Gene Ontology (GO) Biological Process terms. Each circle or node represents one of the 77 candidate interactors, labeled with gene names. Lines connecting nodes indicate a known functional protein–protein interaction; line thickness reflects strength of the data available [only proteins with at least one connection are shown (n=52), explaining why K17 is not part of this network]. Red color reflects known nuclear localization; blue reflects a role in RNA processing; yellow reflects a role in chromatin organization; and green reflects a role in regulation of gene expression (multi-coloring reflects >1 role). Inset table (upper right) highlights key findings. Count, number of entries for a particular GO term (out of 77); FDR, false discovery rate. (B) Western blots of proteins (e.g. nuclear lamina-associated proteins, RNA polymerase II, nuclear import/export machinery, and histones) that co-immunoprecipitate with K17 in subcellular fractions enriched for nuclear proteins. Blots are representative of at least two, typically three, independent experiments.
We also conducted a target-specific survey that confirmed some of the MS results and identified several new K17-interacting proteins with well-defined roles in nuclear lamina architecture and chromatin organization. These included lamins A, B1 and B2, but not lamin C. Also identified were the LAP2 α, β, γ and Δ isoforms, emerin, histones H2B, H2A.X and γH2A.X, RNA Polymerase II, and the nucleo-cytoplasmic shuttling proteins exportin-1 and karyopherin β1 (Fig. 3B). Similar results were obtained in HeLa and A431 cells.
Loss of K17 alters histone modifications that are relevant to chromatin status and gene expression
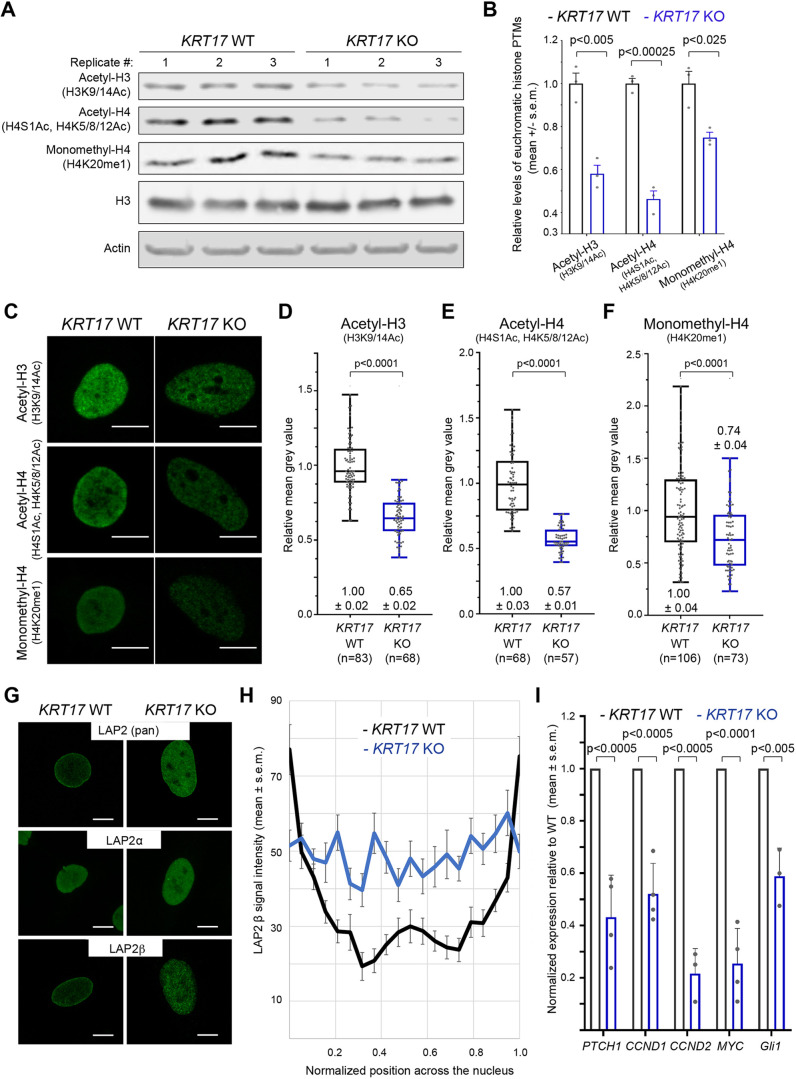
Changes in nuclear size, shape or lamina composition can lead to re-organization of chromatin-nuclear lamina interactions with an impact on gene expression (Mukherjee et al., 2016). Besides, the differential distribution of post-translational modifications (PTMs) on histone proteins are major determinants of chromatin and gene expression (Lawrence et al., 2016). We next tested whether histone PTMs are regulated by K17 expression status. Immunoblotting of whole-cell protein lysates from KRT17 WT and KO HeLa cells revealed marked decreases in several histone PTMs, including acetyl-histone H3 (at Lys-9 and/or Lys-14), acetyl-histone H4 (at Ser-1, Lys-5, Lys-8 and/or Lys-12) and monomethyl-histone H4 (at Lys-20) in the absence of K17 (Fig. 4A; quantification in Fig. 4B). Confocal microscopy analyses afford an opportunity to assess individual cells rather than cell populations and confirmed reduced signal intensities of these histone PTMs in KRT17 KO (human) cells relative to WT controls (Fig. 4C–F). Altogether, K17 expression is positively correlated with more spherical nuclei and higher levels of histone PTMs associated with transcriptionally active euchromatin, which suggest that K17 may regulate gene expression epigenetically, e.g. through modifying the histone code.
Fig. 4.
Loss of K17 alters histone modifications and LAP2β localization. (A) Western blots of whole-cell lysates from KRT17 WT and KRT17 KO cells (n=3 biological replicates each). Primary antibody used shown at left. (B) Quantification (mean±s.e.m.) of band intensities in (A) normalized to β-actin loading control. P-values shown were calculated with a Student's t-test with two-tailed distribution, and two-sample equal variance (homoscedastic). (C) Confocal maximum intensity projections (MIPs) of KRT17 WT and KRT17 KO cells immunostained for acetylated histone H3 (H3K9/K14Ac), acetylated histone H4 (H4S1Ac, H4K5/K8/K12Ac) or monomethylated Histone H4 (H4K20me1) (green). Scale bars: 10 μm. (D–F) Quantification of mean signal intensity for (D) acetylated histone H3 (H3K9/K14Ac), (E) acetylated histone H4 (H4S1Ac, H4K5/K8/K12Ac), and (F) monomethylated histone H4 (H4K20me1) in KRT17 WT versus KRT17 KO cells. Numbers above or below each box-and-whisker plot are mean±s.e.m. relative to KRT17 WT. In the box-and-whisker plots, the box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum and maximum values. P-values shown were calculated with a Student's t-test with two-tailed distribution, and two-sample equal variance (homoscedastic). (G) Confocal MIPs of KRT17 WT and KRT17 KO HeLa cells immunostained for LAP2 (pan-isoform), LAP2α, and LAP2β (green). Scale bars: 10 μm. (H) Quantification of signal intensity across line scans through individual nuclei from KRT17 WT and KRT17 KO HeLa cells (mean±s.e.m.; n=20 nuclei per genotype). Signal at the peripheries of the x-axis (i.e. normalized positions 0.00–0.05 and 0.95–1.00) reflect LAP2β signal at the nuclear envelope. Signal in the interior of the x-axis (i.e. normalized positions 0.05–0.95) reflect LAP2β signal in nucleoplasm. (I) Normalized expression of select GLI1-target genes in KRT17 KO relative to KRT17 WT HeLa cells (mean±s.e.m.; n=3–4 biological replicates per mRNA). P-values shown were calculated with a Student's t-test with two-tailed distribution, and two-sample equal variance (homoscedastic).
Loss of K17 alters LAP2β localization, cell proliferation and GLI1 function
LAP2β stood out by many criteria in our survey of nuclear envelope-associated proteins (Fig. 1E; Fig. S1E–G). Quantitative confocal microscopy showed that LAP2β localization shifts from occurring in the INM and nucleoplasm of KRT17-positive cells to being in the nucleoplasm in KRT17 KO cells (Fig. 4G,H). Mirza et al. (2019) showed that GLI1, a terminal effector of Hedgehog signaling and powerful mitogenic transcription factor (Mazza et al., 2013), is subject to a two-step, LAP2-dependent regulation when inside the nucleus. After import, acetylated-GLI1 is initially docked to the INM in a LAP2β-dependent fashion and unable to drive expression of its target genes (Mirza et al., 2019), many of which drive cell proliferation (Mastrangelo and Milani, 2018). We surmised that the disruption of a potential interplay between K17, LAP2β and GLI1 may explain previously reported differences in the proliferation properties of Krt17-null keratinocytes (Hobbs et al., 2015; Depianto et al., 2010).
We found that KRT17-null HeLa and A431 tumor cells proliferate more slowly than parental controls, with the defect being more pronounced in HeLa cells (Fig. S4A). To assess for cell autonomy, we mixed and plated parental and KRT17-null HeLa cells in a 1:1 ratio and monitored growth over time. Reproducibly, parental HeLa cells became dominant over time (Fig. S4B). Use of mitomycin C (Blake et al., 2006) revealed that the two genotypes survived equally well (Fig. S4C), ruling out a role for differential cell death. These findings are supported by DepMap, a survey of the impact of CRISPR-induced null alleles in a large collection of cell lines (https://depmap.org), showing that loss of K17 results in reduced proliferation in 324 out of 739 cell lines (note loss of K14, a similar type I keratin, is neutral in this setting).
Next, we explored whether the proliferation deficit observed in KRT17-null cells can be correlated to reduced GLI1 activity. First, we measured the mRNA levels for GLI1 target genes with an known role in cell proliferation. By RT-qPCR, we determined that the PTCH1, CCND1, CCND2, MYC and GLI1 mRNAs are each markedly decreased in KRT17-null relative to parental HeLa cells (Fig. 4I). Second, we conducted luciferase assays using a reporter of GLI1 transcriptional activity in KRT17 KO HeLa cells (see Materials and Methods). Expression of GFP–K17 WT, but not GFP–K17ΔNLS, significantly stimulated GLI1 reporter activity in KRT17 KO HeLa cells (Fig. S4D). These findings support the possibility that mislocalization of LAP2β in cells lacking K17 leads to reductions in GLI1-dependent gene expression and cell proliferation. Whether this is related to the impact of K17 on nuclear shape and chromatin organization remains unclear.
In conclusion, our findings establish a novel role for nuclear-localized K17 as a regulator of nuclear architecture, chromatin organization and epigenetic modification of gene expression in keratinocytes. We report on >80 novel nuclear proteins (confirmed and candidates) in the ‘K17 interactome’ with defined roles in chromatin dynamics, gene expression, RNA processing, nucleo-cytoplasmic trafficking, or DNA repair. These findings expand the realm of potential functions for nuclear keratins and point to exciting lines of future investigations.
MATERIALS AND METHODS
Cell culture
HeLa (cervical, epithelial) and A431 (vulva, epidermoid) keratinocyte cell lines (sourced from the ATCC) were routinely cultured in normal growth medium (NGM), which consists of Dulbecco's modified Eagle's medium (DMEM; Life Technologies, ref# 11995-065), 10% fetal bovine serum (FBS; Atlanta Biologicals ref# S11150), 100 units/ml Penicillin and 100 μg/ml Streptomycin (P/S; Life Technologies ref# 15140-122), at 37°C and 5% CO2. Cells were routinely passaged with 0.05% trypsin (Corning, ref# 25-052-Cl) when they were ∼75–80% confluent. The protocols for generating HeLa cells null for KRT17 and confirming the anticipated genome recombination are identical to those previously used to generate A431 cells null for KRT17 (Hobbs et al., 2015). Prior to use in these studies, all cell lines were treated with a Plasmocin treatment regimen for 2 weeks and confirmed to be mycoplasma free (data not shown). Established cell lines are authenticated on a regular basis using short tandem repeat (STR) profiling.
Newborn mouse skin keratinocytes were initiated in primary culture as detailed in Sections 3.2–3.3 in Wang et al. (2016a).
For all microscopy experiments, cells were seeded on coverslips (Globe Scientific Inc., ref# 1404-15) that had been previously coated with rat collagen type I (Corning ref# 354236). At the time of harvest, cells were fixed with 4% paraformaldehyde, rinsed with PBS and mounted on microscopy slides using mounting medium containing DAPI (Duolink ref# 82040-0005). For indirect immunofluorence detection of K17, cells were permeabilized with ice-cold 0.2% Triton X-100 diluted in PBS for 5 min on ice prior to antibody incubation as previously described (McGowan and Coulombe, 1998). For the co-culture cell competition assay, KRT17 WT and KRT17 KO cells (10,000 each per cover slip) were cultured for a defined number of days prior to fixation and mounting. For transient transfection assays, GFP, GFP–K17 WT or GFP–K17ΔNLS cDNA (pEGFP-C3 vector backbone; Clontech ref# 6082-1) were transfected into cells using FuGENE HD Transfection Reagent (Promega ref# E2311) following the manufacturer's recommendations. Cells were cultured on coverslips, as described above, for 1 day prior to transfection and for 2 days following transfection. Microscopy-based analyses and western immunoblotting assays established that the expression levels of the DNA constructs being compared were highly similar if not the same.
Generation of Krt17ΔNLS/ΔNLS mice
A mouse strain, Krt17ΔNLS/ΔNLS, carrying K399A and K401A point mutations at the Krt17 locus, was generated using the RNA-guided CRISPR-Cas9 system as described (Wang et al., 2013, 2016b). A guide RNA (gRNA) (5′-GTACAAGCCAAAAGAACGTAAGG-3′; PAM motif is underlined) was selected based on having a cut site proximal to codon K399 and low predicted off-target recognition (http://crispr.technology) (Jaskula-Ranga et al., 2016 preprint), cloned into the pT7gRNA vector using oligonucleotides (Fwd, 5′-CCATCGATAATACGACTCACTATAGTACAAGCCAAAAGAACGTAG-3′; Rev, 5′-CATGTTCGGTTTTCTTGCATCAAAATCTCGATCTTTATCGTTCAA-3′), and in vitro transcribed and purified (NEB HiScribe T7 High Yield RNA Synthesis Kit) prior to injection. The homology-directed repair (HDR) template was purchased as a 160-nt single stranded Ultramer (IDT), and encoded a AAG (Lys) to GCG (Ala) mutation at codon 399 and a AAA (Lys) to GCA (Ala) mutation at codon 401 of the mouse K17 coding sequence (5′-GCATTAGGGGTGAGGAAGGTACTCTCGGATTTCATCTAATCT CTCCAACTTTTTTTTTCCAGCCTGACTCAGTACGCGCCAGCAGAACGTAAGGATATTGGTAACTGAGGGCTGGGGTAGAAGGATGCATGTGGCAGGAATCGCCTAGCAGATTGCTAGG-3′; mutation sites are underlined). The gRNA, Cas9 mRNA and HDR template were co-injected into C57Bl/6 zygotes by the JHU Transgenic Core Facility. Potential transgenic founders were screened using restriction digestion of PCR product extending beyond the repair template oligonucleotide and findings were confirmed by direct DNA sequencing (data not shown). One male homozygote and one male heterozygote founder were selected and independently backcrossed by mating to wild-type C57Bl/6 females for three generations to eliminate potential off-target effects. The Krt17ΔNLS/ΔNLS homozygotes used in this study resulted from heterozygous×heterozygous breedings. All animal experiments were performed according to approved institutional guidelines.
Analysis of shape, size and surface area of cell nuclei
Cells on coverslips were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, ref# 15710) for 10 min at room temperature with gentle agitation on a plate rocker and mounted on microscope slides (VWR ref# 48311-703) with mounting medium containing DAPI. Images of nuclei were acquired with a Zeiss LSM700 confocal laser scanning microscope (40× and 63× objectives). Confocal micrographs were processed to maximum intensity projections (MIPs) in order to quantify two-dimensional area measurements of cell nuclei, using the freehand selections tool in the ImageJ processing software (Schneider et al., 2012). Three-dimensional measurements (e.g. volume, sphericity and surface area) of confocal micrographs were acquired with the Imaris version 8 microscopy image analysis software (Bitplane; http://bitplane.com). Statistical analyses were conducted using the two-tailed unpaired t-test function on the GraphPad Prism 8 software to calculate P-values.
Whole-cell lysis, SDS-PAGE and immunoblotting
Whole-cell lysates were prepared in urea sample buffer [8 M urea, 1% SDS, 10% glycerol, 60 mM Tris-HCl pH 6.8, 0.005% Pyronin Y, and 5% β-mercaptoethanol (β-ME)], homogenized using QIAshredders (QIAGEN ref# 79656), and subjected to Bradford assay (Bio-Rad ref# 500-0006) prior to SDS-PAGE electrophoresis on a 4-15% gradient gel (Bio-Rad ref# 456-1086). 10 µg of protein lysate was loaded per well. Immunoblotting was performed using nitrocellulose membranes (Bio-Rad ref# 162-0115) and a blocking buffer of 5% milk dissolved in either PBS or Tris-buffered saline (TBS).
Primary antibodies utilized for immunoblotting include: rabbit polyclonal anti-K17 (McGowan and Coulombe, 1998), mouse monoclonal anti-lamin A/C (Santa Cruz Biotechnology, 1:500, ref# sc-376248), mouse anti-lamin B1/B2 (‘2B2’; 1:2000, generously provided by Dr Robert Goldman, Department of Cell and Developmental Biology, Northwestern University, USA), mouse anti-pan isoform LAP2 (‘LAP12’; 1:5, generously provided by Dr Roland Foisner, Max F. Perutz Laboratories, Medical University of Vienna, Austria), rabbit polyclonal anti-emerin (Santa Cruz Biotechnology, 1:2000, ref# sc-15378), rabbit monoclonal anti-GAPDH (Cell Signaling Technology, 1:1000, ref# 5174), mouse monoclonal anti-acetyl-histone H3 (H3K9Ac+H3K14Ac; Santa Cruz Biotechnology, 1:500, ref# sc-518011), mouse monoclonal anti-acetyl-histone H4 (H4S1Ac+H4K5Ac+H4K8Ac+H4K12Ac; Santa Cruz Biotechnology, 1:500, ref# sc-377520) and mouse monoclonal anti-monomethyl-histone H4 (H4K20me1; Santa Cruz Biotechnology, 1:1000, ref# sc-134221), mouse monoclonal anti-β-actin (Santa Cruz Biotechnology, 1:1000, ref# sc-47778), rabbit polyclonal anti-histone H3 (Cell Signaling Technology, 1:1000, ref# 9715S), mouse monoclonal anti-RNA polymerase II (Santa Cruz Biotechnology, 1:500, ref# sc-55492), mouse monoclonal anti-karyopherin β1 (GeneTex, 1:5000, ref# GTX22811), goat polyclonal anti-exportin 1 (Santa Cruz Biotechnology, 1:500, ref# sc-7825), mouse monoclonal anti-histone H2B (Santa Cruz Biotechnology, 1:500, ref# sc515808), mouse monoclonal anti-histone H2A.X (Santa Cruz Biotechnology, 1:500, ref# sc-517336), and mouse monoclonal anti-histone γH2A.X (Santa Cruz Biotechnology, 1:500, ref# sc-517348). Secondary antibodies for immunoblotting include donkey anti-mouse IgG and goat anti-rabbit IgG (LI-COR, ref# 925-32212 and 926-32211). Immunodetection and image capture occurred by infrared imaging (LI-COR Biosciences). Statistical analyses were conducted using the two-tailed unpaired t-test function on the GraphPad Prism 8 software to calculate P-values.
Indirect immunofluorescence staining for microscopy and quantitative analysis
Cells were fixed and mounted as described above. Primary antibodies utilized for indirect immunofluorescence include: mouse monoclonal anti-lamin A/C (Santa Cruz Biotechnology, 1:1000, ref# sc-376248), mouse anti-lamin B1/B2 (2B2; 1:500, generously provided by Dr Robert Goldman), mouse monoclonal anti-pan isoform LAP2 (‘LAP12’; 1:2), rabbit polyclonal anti-LAP2α (‘245-3’; 1:500), mouse monoclonal anti-LAP2β (‘LAP17’; 1:2) (all generously provided by Dr Roland Foisner), mouse monoclonal anti-emerin (Santa Cruz Biotechnology, 1:2000, ref# sc-25284), mouse monoclonal anti-acetyl-histone H3 (H3K9Ac+H3K14Ac; Santa Cruz Biotechnology, 1:50, ref# sc-518011), mouse monoclonal anti-acetyl-histone H4 (H4S1Ac+H4K5Ac+H4K8Ac+H4K12Ac; Santa Cruz Biotechnology, 1:50, ref# sc-377520), mouse monoclonal anti-monomethyl-histone H4 (H4K20me1; Santa Cruz Biotechnology, 1:50, ref# sc-134221), and rabbit polyclonal anti-K17 (1:500; McGowan and Coulombe, 1998). Species-appropriate Alexa Fluor-conjugated secondary antibodies (488 or 568) were utilized prior to mounting on slides as described above.
For proximity ligation assays (PLAs), fixed cells were double immunolabeled with rabbit polyclonal anti-K17 (1:500; McGowan and Coulombe, 1998) and mouse anti-K17 (Santa Cruz Biotechnology, 1:500, ref# sc-393002) prior to incubation with PLA probes (Sigma-Aldrich ref# DUO92002 and DUO92004) for 60 min. Before mounting, cells were gently rinsed multiple times with Wash Buffer A (Sigma-Aldrich ref# DUO82049) prior to DNA oligonucleotide incubation for 30 min and amplification for for 100 min at 37°C (Sigma-Aldrich ref# DUO92007).
Subcellular fractionation, K17 co-IP and MS-based proteomics
When cells cultured on 15-cm tissue culture dishes reached 80% confluence, they were rinsed with PBS at room temperature, scraped into ice-cold low-salt lysis buffer [10 mM Tris-HCl pH 7.5, 1 mM MgCl2, 10 mM KCl, plus protease inhibitor cocktail (PIC), which includes 1 mM PMSF, 1 mM Na3VO4, 1 mM Na4P2O7, 50 mM NaF, 2 µg/ml antipain, 10 µg/ml aprotinin, 10 µg/ml benzamidine, 1 µg/ml leupeptin, 1 µg/ml chymostatin and 1 µg/ml pepstatin-A], incubated on ice for 30 min, and sheared with a 22-gauge needle to release cell nuclei. After pelleting by centrifugation (800 g, 5 min, 4°C), supernatant was removed and pelleted nuclei were reconstituted in a high-salt lysis buffer (40 mM HEPES pH 7.5, 450 mM NaCl, 1 mM EDTA, plus PIC) and twice incubated on ice for 10 min followed by 1 min vortex prior to ice water bath sonication (8 cycles total; 1 cycle, 30 s on, 30 s off) and centrifugation (17,000 g, 10 min, 4°C) to pellet insoluble nuclear debris. The resulting supernatant was subjected to pre-clearing of non-specific-binding proteins via incubation with TrueBlot Anti-Rabbit Ig IP Agarose Beads followed by K17 immunoprecipitation (IP), as described previously (Wang et al., 2016a).
The concentrated eluate was then analyzed by immunoblotting or submitted to the Mass Spectrometry and Proteomics Core Facility at the Johns Hopkins School of Medicine for identification of co-immunoprecipitated proteins.
Mass spectrometry and bioinformatics analysis
For MS, the immunoprecipitation eluates were each adjusted to pH 8.0 using 500 mM TEAB buffer, reduced with 16 mg/ml DTT for 1 h at 60°C, and alkylated with 36 mg/ml iodoacetomide for 15 min at room temperature under low-light conditions. Proteins were then precipitated by TCA/acetone extraction overnight at −20°C, reconstituted in 100 mM TEAB, and proteolyzed into peptide fragments with 1 µg of lyophilized trypsin overnight at 37°C. Tryptic peptides were then labeled with TMT 10pplex isobaric tags (ThermoFisher ref# 90110; TMT10-127N and TMT10-130N reagents were used to label the eluates from the KRT17 WT and KRT17 KO cell lysates, respectively) for 1 h at room temperature. TMT-labeled peptides were then mixed together and step-fractionated by basic reverse-phase (bRP) chromatography (Waters Oasis u-HLB plates) using 10 mM TEAB containing 5%, 15%, 20%, 30% and 75% acetonitrile. The resulting 5 bRP fractions were evaporated until dry and then reconstituted in 2% acetonitrile/0.1% formic acid. Finally, samples were analyzed by liquid chromatography/tandem mass spectrometry (LC-MS/MS). Labeled peptides were first separated by a nano-Acquity LC system (Waters) using reverse-phase chromatography on a 75-µm×150 mm ProntoSIL-120-5-C18 H column (2%-90% acetonitrile/0.1% formic acid gradient over 90 min at 300 nl/min) and then detected using a Q Exactive HF in FTNT Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Tandem MS/MS spectra were processed by Proteome Discoverer software (ThermoFisher Scientific, v1.4) and analyzed with Mascot v.2.5.1 Matrix Science (www.matrixscience.com) using 2015RefSeq_72r_human_plus (human peptide database).
The raw MS data was subject to appropriate filtering prior to analysis, as follows. Entries with a 127N/130N isobaric tag labeling ratio equal to or greater than 1.20 (n=140), which reflects enrichment over negative control K17 IP from KRT17 KO cells, along with a 127N/130N isobaric tag count equal to or greater than 4 (n=77), were selected for further analysis. Of the top 77 proteins that persisted through this filtering, 58 (75%) had at least one defined nuclear role according to Gene Ontology terms in the Universal Protein Resource (UniProt) database (https://www.uniprot.org/). These 77 proteins were then subjected to association network analysis using the online STRING-db freeware (Jensen et al., 2009; Szklarczyk et al., 2015).
Luciferase assays
Renilla luciferase control plasmid pRL-TK (Promega, E2241), GLi activity responsive Firefly luciferase plasmid 8xGLiBs-luc (Bianchi et al., 2005), pEGFP-C3 K17 WT (Pan et al., 2013), the pCIG empty vector (Holtz et al., 2013), the GLi1-pCIG overexpression vector (Carpenter et al., 2015) and the GFP-K17ΔNLS vector (Hobbs et al., 2015) were nucleofected into HeLa cells using SE Cell Line 4D X nucleofector Kit S (Lonza #V4XC-1032) with setting DS-138, plated in a 96-well plate, and subjected to the Promega Dual Luciferase Reporter Assay System (Promega, PR-E1910) to measure firefly and Renilla luciferase activities 24 h post nucleofection. The firefly relative light unit (RLU) value was normalized to the internal Renilla RLU per well after measurements were recorded. Five biological replicates of normalized Firefly RLUs were combined for each parameter, and the means of each parameter were compared using a Mann–Whitney test. Data presented were transformed by dividing individual RLUs of each parameter by the mean of pRL-TK control parameter alone which was then subjected to statistical analysis.
Supplementary Material
Acknowledgements
We are grateful to Amar Mirza, Anthony Oro, Beau Su, Daniela Drummond-Barbosa, Hoku West-Foyle, Loza Lee, Robert Cole, Robert Goldman, Roland Foisner, Scot C. Kuo, Tatiana N. Boronina and Catherine Redmond for support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.T.J., R.R.N., B.G.P., R.P.H., M.J.M., P.A.C.; Methodology: J.T.J., R.R.N., B.G.P., C.M.P., P.A.C.; Validation: J.T.J., R.R.N., B.G.P., C.M.P., R.P.H., M.J.M., P.A.C.; Formal analysis: J.T.J., R.R.N., B.G.P., C.M.P., P.A.C.; Investigation: J.T.J., R.R.N., B.G.P., C.M.P.; Resources: M.J.M., P.A.C.; Data curation: J.T.J., R.R.N., B.G.P., C.M.P., P.A.C.; Writing - original draft: J.T.J., P.A.C.; Writing - review & editing: J.T.J., R.R.N., R.P.H., M.J.M., P.A.C.; Visualization: J.T.J., P.A.C.; Supervision: R.P.H., M.J.M., P.A.C.; Project administration: J.T.J., P.A.C.; Funding acquisition: P.A.C.
Funding
This work was supported by grants R01AR044232 (P.A.C.) and T32GM007315 (C.M.P.) from the National Institutes of Health. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.254094.supplemental
References
- Akoumianaki T., Kardassis D., Polioudaki H., Georgatos S. D. and Theodoropoulos P. A. (2009). Nucleocytoplasmic shuttling of soluble tubulin in mammalian cells. J. Cell Sci. 122, 1111-1118. 10.1242/jcs.043034 [DOI] [PubMed] [Google Scholar]
- Bettinger B. T., Gilbert D. M. and Amberg D. C. (2004). Actin up in the nucleus. Nat. Rev. Mol. Cell Biol. 5, 410-415. 10.1038/nrm1370 [DOI] [PubMed] [Google Scholar]
- Bianchi N., Depianto D., McGowan K., Gu C. and Coulombe P. A. (2005). Exploiting the keratin 17 gene promoter to visualize live cells in epithelial appendages of mice. Mol. Cell Biol.25, 7249-7259. 10.1128/MCB.25.16.7249-7259.2005 [DOI] [PMC free article] [PubMed]
- Blake D. A., Sahiner N., John V. T., Clinton A. D., Galler K. E., Walsh M., Arosemena A., Johnson P. Y. and Ayyala R. S. (2006). Inhibition of cell proliferation by mitomycin C incorporated into P(HEMA) hydrogels. J. Glaucoma 15, 291-298. 10.1097/01.ijg.0000212236.96039.9c [DOI] [PubMed] [Google Scholar]
- Bouameur J.-E. and Magin T. M. (2017). Lessons from animal models of cytoplasmic intermediate filament proteins. Subcell. Biochem. 82, 171-230. 10.1007/978-3-319-49674-0_7 [DOI] [PubMed] [Google Scholar]
- Castano E., Philimonenko V. V., Kahle M., Fukalová J., Kalendová A., Yildirim S., Dzijak R., Dingová-Krásna H. and Hozak P. (2010). Actin complexes in the cell nucleus: new stones in an old field. Histochem. Cell Biol. 133, 607-626. 10.1007/s00418-010-0701-2 [DOI] [PubMed] [Google Scholar]
- Carpenter B. S., Barry R. L., Verhey K. J. and Allen B. L. (2015). The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. J. Cell Sci.128, 1034-1050. 10.1242/jcs.162552 [DOI] [PMC free article] [PubMed]
- Depianto D., Kerns M. L., Dlugosz A. A. and Coulombe P. A. (2010). Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat. Genet. 42, 910-914. 10.1038/ng.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erber A., Riemer D., Bovenschulte M. and Weber K. (1998). Molecular phylogeny of metazoan intermediate filament proteins. J. Mol. Evol. 47, 751-762. 10.1007/PL00006434 [DOI] [PubMed] [Google Scholar]
- Escobar-Hoyos L. F., Shah R., Roa-Pena L., Vanner E. A., Najafian N., Banach A., Nielsen E., Al-Khalil R., Akalin A., Talmage D. et al. (2015). Keratin-17 promotes p27KIP1 nuclear export and degradation and offers potential prognostic utility. Cancer Res. 75, 3650-3662. 10.1158/0008-5472.CAN-15-0293 [DOI] [PubMed] [Google Scholar]
- Hobbs R. P., Depianto D. J., Jacob J. T., Han M. C., Chung B. M., Batazzi A. S., Poll B. G., Guo Y., Han J., Ong S. et al. (2015). Keratin-dependent regulation of Aire and gene expression in skin tumor keratinocytes. Nat. Genet. 47, 933-938. 10.1038/ng.3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs R. P., Jacob J. T. and Coulombe P. A. (2016). Keratins are going nuclear. Dev. Cell 38, 227-233. 10.1016/j.devcel.2016.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz A. M., Peterson K. A., Nishi Y., Morin S., Song J. Y., Charron F., McMahon A. P. and Allen B. L. (2013). Essential role for ligand-dependent feedback antagonism of vertebrate hedgehog signaling by PTCH1, PTCH2 and HHIP1 during neural patterning. Development140, 3423-3434. 10.1242/dev.095083 [DOI] [PMC free article] [PubMed]
- Jaskula-Ranga V. and Zack D. J. (2016). grID: A CRISPR-Cas9 guide RNA database and resource for genome-editing. bioRxiv 10.1101/097352 [DOI]
- Jacob J. T., Coulombe P. A., Kwan R. and Omary M. B. (2018). Types I and II keratin intermediate filaments. Cold Spring Harb. Perspect Biol. 10, a018275 10.1101/cshperspect.a018275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M. et al. (2009). STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412-D416. 10.1093/nar/gkn760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor P., Chen M., Winkler D. D., Luger K. and Shen X. (2013). Evidence for monomeric actin function in INO80 chromatin remodeling. Nat. Struct. Mol. Biol. 20, 426-432. 10.1038/nsmb.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumeta M., Hirai Y., Yoshimura S. H., Horigome T. and Takeyasu K. (2013). Antibody-based analysis reveals “filamentous vs. non-filamentous” and “cytoplasmic vs. nuclear” crosstalk of cytoskeletal proteins. Exp. Cell Res. 319, 3226-3237. 10.1016/j.yexcr.2013.07.021 [DOI] [PubMed] [Google Scholar]
- Lawrence M., Daujat S. and Schneider R. (2016). Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 32, 42-56. 10.1016/j.tig.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Mastrangelo E. and Milani M. (2018). Role and inhibition of GLI1 protein in cancer. Lung Cancer (Auckl) 9, 35-43. 10.2147/LCTT.S124483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza D., Infante P., Colicchia V., Greco A., Alfonsi R., Siler M., Antonucci L., Po A., De Smaele E., Ferretti E. et al. (2013). PCAF ubiquitin ligase activity inhibits Hedgehog/Gli1 signaling in p53-dependent response to genotoxic stress. Cell Death Differ. 20, 1688-1697. 10.1038/cdd.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan K. M. and Coulombe P. A. (1998). Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143, 469-486. 10.1083/jcb.143.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza A. N., Mckellar S. A., Urman N. M., Brown A. S., Hollmig T., Aasi S. Z. and Oro A. E. (2019). LAP2 proteins chaperone GLI1 movement between the lamina and chromatin to regulate transcription. Cell 176, 198-212.e15. 10.1016/j.cell.2018.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee R. N., Chen P. and Levy D. L. (2016). Recent advances in understanding nuclear size and shape. Nucleus 7, 167-186. 10.1080/19491034.2016.1162933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Hobbs R. P. and Coulombe P. A. (2013). The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr. Opin. Cell Biol. 25, 47-56. 10.1016/j.ceb.2012.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. (2008). As functional nuclear actin comes into view. is it globular, filamentous, or both? J. Cell Biol. 180, 1061-1064. 10.1083/jcb.200709082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Knop M. and Schiebel E. (1998). Spc98p directs the yeast γ-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol. Biol. Cell 9, 775-793. 10.1091/mbc.9.4.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. and Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenenberger C.-A., Buchmeier S., Boerries M., Sütterlin R., Aebi U. and Jockusch B. M. (2005). Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J. Struct. Biol. 152, 157-168. 10.1016/j.jsb.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Schweizer J., Bowden P. E., Coulombe P. A., Langbein L., Lane E. B., Magin T. M., Maltais L., Omary M. B., Parry D. A., Rogers M. A. et al. (2006). New consensus nomenclature for mammalian keratins. J. Cell Biol. 174, 169-174. 10.1083/jcb.200603161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P. et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447-D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506-d515. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N. and Percipalle P. (2010). Nuclear functions of actin. Cold Spring Harb. Perspect Biol. 2, a000620 10.1101/cshperspect.a000620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., Zhang F. and Jaenisch R. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell153: 910-918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed]
- Wang F., Zieman A. and Coulombe P. A. (2016a). Skin Keratins. Methods Enzymol. 568, 303-350. 10.1016/bs.mie.2015.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Huang X., Fang X., Zhang Y. and Wang W. (2016b). CRISPR-Cas9 system as a versatile tool for genome engineering in human cells. Mol. Ther. Nucleic Acids5, e388. 10.1038/mtna.2016.95 [DOI]
- Yeh T.-S., Hsieh R.-H., Shen S.-C., Wang S.-H., Tseng M.-J., Shih C.-M. and Lin J.-J. (2004). Nuclear βII-tubulin associates with the activated notch receptor to modulate notch signaling. Cancer Res. 64, 8334-8340. 10.1158/0008-5472.CAN-04-2197 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang J., Huang W., Cai J., Ba J., Wang Y., Ke Q., Huang Y., Liu X., Qiu Y. et al. (2018). Nuclear Nestin deficiency drives tumor senescence via lamin A/C-dependent nuclear deformation. Nat. Commun. 9, 3613 10.1038/s41467-018-05808-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieman A. G., Poll B. G., Ma J. and Coulombe P. A. (2019). Altered keratinocyte differentiation is an early driver of keratin mutation-based palmoplantar keratoderma. Hum. Mol. Genet. 28, 2255-2270. 10.1093/hmg/ddz050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.