Abstract
Phospholipase C (PLC) enzymes are key virulence factors in several pathogenic bacteria. Burkholderia pseudomallei, the causative agent of melioidosis, possesses at least three plc genes (plc1, plc2 and plc3). We found that in culture medium plc1 gene expression increased with increasing pH, whilst expression of the plc3 gene was pH (4.5 to 9.0) independent. Expression of the plc2 gene was not detected in culture medium. All three plc genes were expressed during macrophage infection by B. pseudomallei K96243. Comparing B. pseudomallei wild-type with plc mutants revealed that plc2, plc12 or plc123 mutants showed reduced intracellular survival in macrophages and reduced plaque formation in HeLa cells. However, plc1 or plc3 mutants showed no significant differences in plaque formation compared to wild-type bacteria. These findings suggest that Plc2, but not Plc1 or Plc3 are required for infection of host cells. In Galleria mellonella, plc1, plc2 or plc3 mutants were not attenuated compared to the wild-type strain, but multiple plc mutants showed reduced virulence. These findings indicate functional redundancy of the B. pseudomallei phospholipases in virulence.
Subject terms: Immunology, Microbiology
Introduction
Burkholderia pseudomallei, a Gram-negative facultative intracellular bacterium, is the etiological agent of melioidosis in humans and in animals. Melioidosis in humans was at one time thought to be largely restricted to Southeast Asia and Northern Australia, but it is now thought to occur in many tropical and sub-tropical regions of the world1. The annual global burden of meliodosis is estimated to be 165,000 cases with 89,000 deaths from the disease1. A feature of B. pseudomallei is its ability to modulate a range of host-cell responses and to evade phagocyte killing activity2,3. B. pseudomallei has evolved mechanisms to evade phagocyte activities, including escape from phagosomes and entry into host cell cytosol where it multiplies and forms actin tails allowing cell-to-cell spreading4. This complex intracellular lifestyle is contributed by several bacterial virulence factors including type three secretion systems (T3SS), type six secretion systems (T6SS), polysaccharide capsule, lipopolysaccharide (LPS), and various secreted effector proteins5. Additionally, B. pseudomallei can produce many enzymes which play roles in virulence including proteases, catalase, peroxidase, superoxide dismutase1, and phospholipase C (Plc) enzymes6. Plc enzymes play roles in the pathogenesis of several Gram-positive and Gram-negative bacterial infections including those caused by Mycobacterium tuberculosis7, Pseudomonas aeruginosa8, Clostridium perfringens9, Listeria monocytogenes10, and Legionella pneumophila11. Also, different of Plcs play different roles in virulence12, including tissue colonization, evasion of host defense mechanisms, escape from host cell phagosomes and/or the induction of mediators of inflammation13.
Analysis of the B. pseudomallei K96243 genome reveals genes encoding three Plc enzymes (Plc1, Plc2 and Plc3). The genes encoding Plc1 (bpsl2403) and Plc2 (bpsl0338) are located on chromosome 1. These encoded proteins are predicted to be acidic, have the ability to hydrolyze phospholipids including phosphatidylcholine and sphingomyelin and are non-hemolytic14. The gene encoding Plc3 (bpss0067) is located on chromosome 25. At present, the conditions under which the plc genes are expressed, and their roles in virulence are poorly understood. We have previously characterized the B. pseudomallei Plc1 and Plc2 and found that Plc2 was cytotoxic14. Additionally, these Plc enzymes appear to play a role in nutrient acquisition14. Subsequently, Burtnick et al.15 showed that B. pseudomallei Plc1 and Plc2 are secreted from the bacterial cell via the type II secretion system (T2SS) in a GspD-dependent manner15.
Little is known about the B. pseudomallei Plc3 enzyme. Whole-genome microarrays have revealed that plc3 is up-regulated in vivo, and in hamsters a plc3 mutant shows reduced virulence compared to the wild-type, suggesting that it is required for virulence16. However, the mechanisms underlying attenuation are unknown. Dowling et al.17 reported that Plc3 might be a potential candidate vaccine requiring further study.
In this study, the expression of the plc genes in culture medium and in J774A.1 macrophage-like cells was analyzed using RT-PCR. B. pseudomallei plc1, plc2 or plc3 single mutants and plc12 or plc123 mutants were assessed for virulence in macrophages and in Galleria mellonella larvae. Our work provides new insights into the role of Plc enzymes in the pathogenesis of disease caused by B. pseudomallei.
Results and discussion
Culture pH differentially affects plc1 gene expression
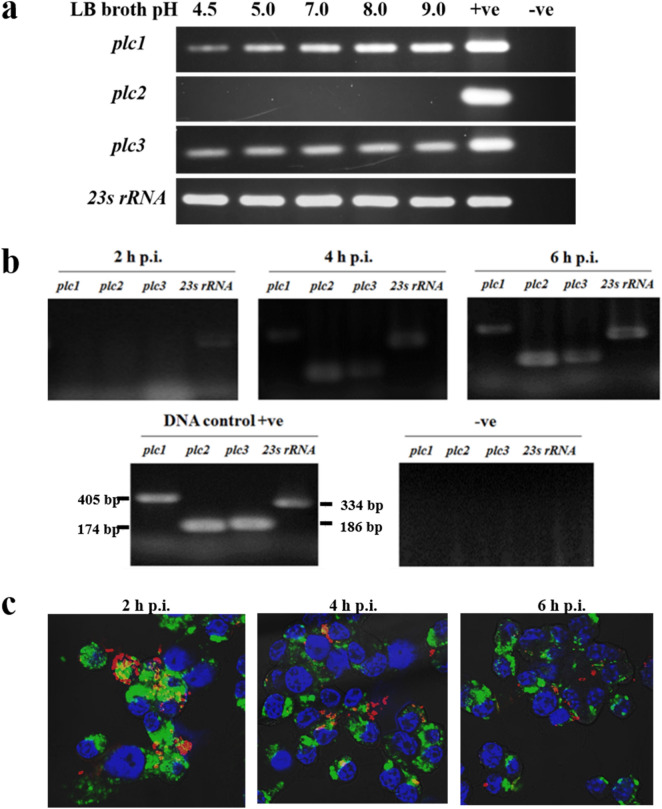
B. pseudomallei is an intracellular bacterium. After phagocytosis by phagocytes, the bacteria encounter acidic condition within the phagosome, before escaping to survive in the cytosol4. To determine the effect of pH on the expression of the plc1-3 genes, B. pseudomallei K96243 was incubated in LB broth which had been adjusted to pH 4.5, 5.0, 7.0, 8.0 or 9.0 before bacterial mRNA was extracted and tested. Using RT-PCR we first showed that expression of B. pseudomallei 23S rRNA was similar at all pH conditions tested. RT-PCR revealed that the level of transcription of the plc1 gene increased between pH 4.5 and 9.0. This result suggests that the pH of bacterial cultures can differentially affect B. pseudomallei plc1 gene expression. In contrast, the level of expression of plc3 was similar at all pH values tested. Expression of the plc2 gene was not detected under any of the conditions tested (Fig. 1a) (full-length gels are presented in Supplementary Fig. 1). Our finding that plc2 was not expressed in strain K96243 is similar to the results reported by Ooi et al.18. However, plc2 is expressed in B. pseudomallei strain 2218, and the Plc2 protein was detected in B. pseudomallei strain MSHR66815 culture supernatant. These findings indicate that plc2 gene expression is strain-dependent.
Figure 1.
Reverse transcription (RT)-PCR analysis of B. pseudomallei plc expression. (a) Expression of plc genes in LB broth adjusted to pH 4.5, 5, 7, 8 or 9. B. pseudomallei wild-type K96243 was incubated for 15 min in LB broth at the pH indicated before RNA extraction. Positive control (+ ve) was B. pseudomallei genomic DNA. DNase-treated bacterial RNA was used as a negative control (− ve) to confirm the absence of DNA contamination in RNA samples (full-length gels are presented in Supplementary Fig. 1). (b) Expression of plc genes in J774A.1 macrophage-like cells infected with B. pseudomallei. RNA of B. pseudomallei K96243 was harvested from infected macrophages at 2, 4 or 6 h p.i. and converted to cDNA for PCR analysis with primers specific to each plc gene. Positive control (+ ve) was B. pseudomallei genomic DNA. DNase-treated bacterial RNA was used as a negative control (−ve). Unfortunately we are not able to reproduce the full length image of this gel because this image file was inadvertently deleted after this project was completed and before we submitted this manuscript. (c) Confocal micrographs of J774A.1 macrophages infected with B. pseudomallei (MOI 10) showed that majority of B. pseudomallei were within phagosomes at 2 h post-infection (p.i.). Escape of bacteria from the vacuoles was first observed at 4 h p.i., and most bacteria were within the cytosol at 6 h p.i.
The B. pseudomallei plc genes are induced in infected macrophages
We next investigated expression of the plc genes in macrophages. J774A.1 macrophage-like cells were infected with B. pseudomallei K96243. At 2, 4, and 6 h post-infection (p.i.), the cells were lysed, and mRNAs corresponding to the plc gene detected using RT-PCR. The results showed no expression of the plc1, plc2 or plc3 genes at 2 h p.i. The expression of all of the plc genes was detected at 4 h p.i., but the level of expression of plc2 was relatively higher than the levels of expression of plc1 and plc3. At 6 h p.i., the levels of expression of all plc genes increased compared with 4 p.i. (Fig. 1b). Our finding that the expression of plc2 gene was induced in macrophages, even though we could not detect expression in culture medium (Fig. 1a), suggests a role of Plc2 in the infection of macrophages.
We stained infected macrophages for LAMP-1 and B. pseudomallei using probes labelled with Alexa Fluor 488 or Alexa Fluor 568 and nuclei were stained with DAPI (Fig. 1c). This revealed that at 2 h p.i. most B. pseudomallei K96243 were trapped within the phagosome. However, at 4 h p.i, co-localization of B. pseudomallei with lysosomes was rare indicating bacterial escape from phagosomes, and almost all of the B. pseudomallei cells were not associated with the phagosome at 6 h p.i. It is possible that the intracellular environment induces expression of plc1, plc2 and plc3 genes.
Several previous reports show the expression of plc genes of other species of bacteria in host tissues12,13. For example, the Mycobacterium tuberculosis plc genes are up-regulated in macrophages, and the Plcs are cytotoxic to mouse macrophages7,19. The Clostridium perfringens PLC (α-toxin) is produced in host tissues and can activate the arachidonic acid cascade in cells, with consequent modulation of host immune responses20, and the induction of ERK1/2 pathway, resulting in cytotoxicity21.
B. pseudomallei plc2, but not plc1 or plc3, is required for bacterial survival and replication in macrophages
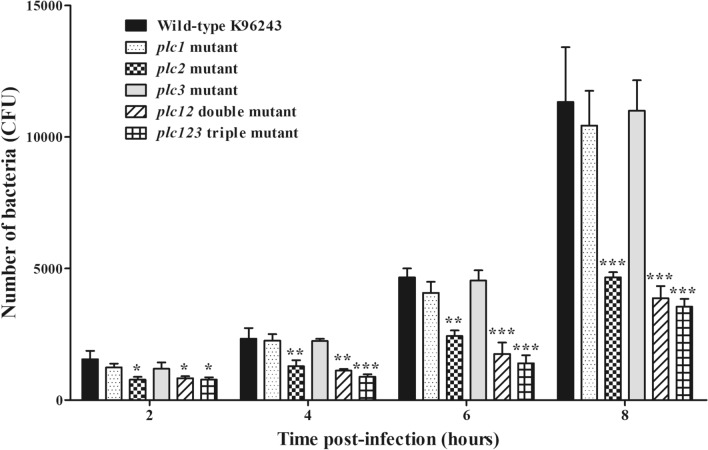
To provide insight into the role of plc1, plc2 and plc3 genes in virulence, we tested a range of single and multiple mutants. We have previously constructed plc1, plc2 and plc12 mutants14 and these were included in our study. Additionally, for this study we constructed plc3 single and plc123 mutants by insertion mutagenesis14. Mutagenesis of the plc genes was confirmed by Southern blotting (data not shown). J774A.1 macrophage-like cells were infected with the mutants. At 2, 4, 6 and 8 h p.i., the numbers of recoverable plc2, plc12 or plc123 mutants were significantly lower (*P < 0.05, **P < 0.01, ***P < 0.001) than the number of viable wild-type bacteria (Fig. 2). In contrast, there was no significant difference (P > 0.05) in the numbers of viable single plc1 or plc3 mutants compared with the number of viable wild-type bacteria at all tested time points (2, 4, 6 or 8 h p.i.). These findings indicate that plc2 gene was required for B. pseudomallei survival and replication inside the macrophage. Our results also indicate that plc1 and plc3 play no role in survival and replication in macrophages, but we cannot discount the possibility of functional redundancy between these enzymes, which would mask the phenotype associated with the single plc1 and plc3 mutants.
Figure 2.
Intracellular growth of B. pseudomallei wild-type or plc mutants in J774A.1 macrophage-like cells. B. pseudomallei wild-type, plc1, plc2, plc3, plc12 or plc123 mutants were used to infect J774A.1 cells at an MOI of 0.5. At 2, 4, 6 or 8 h p.i., the infected cells were lysed and bacteria enumerated after plating onto agar. Error bars represent standard error of mean for data collected from 3 independent experiments. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001, one-way ANOVA followed by Dunnett's post hoc test) between B. pseudomallei wild-type K96243 and mutant strains. The figure was prepared using GraphPad Prism version 7.05 for Windows (www.graphpad.com).
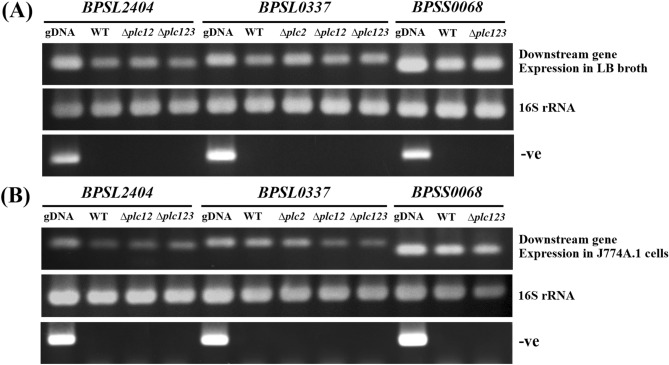
To investigate whether the phenotypes we observed with the plc mutants was due to polar effects on downstream genes we measured expression of the genes downstream of plc1, plc2 or plc3 (bpsl2404, bpsl0337 or bpss0068 respectively) using RT-PCR. B. pseudomallei wild-type and the plc mutants were cultured in LB broth, or extracted from intracellular bacteria after macrophage infection. We demonstrated similar bpsl2404, bpsl0337 and bpss0068 amplicons with mRNA from wild-type or plc1, plc2, or plc3 mutants cultured in LB broth (Fig. 3a) and extracted from intracellular bacteria (Fig. 3b) (full-length gels are presented in Supplementary Fig. 2). These results indicate that the insertional mutation in the plc1, plc2, or plc3 genes did not abolish the expression of downstream genes. However, we cannot discount the possibility that expression of downstream genes was affected.
Figure 3.
Expression of genes downstream of plc1, plc2 or plc3, assessed using RT-PCR. The mRNA from wild-type, plc1, plc2, plc3, plc12, or plc123 mutants cultured in LB broth (a), or isolated from infected J774A.1 macrophages (b) was extracted before converting to cDNA as outlined in material and methods (full-length gels are presented in Supplementary Fig. 2). The cDNA was amplified using PCR primers specific to the bpsl2404, bpsl0337 or bpss0068 genes which are downstream of plc1, plc2 or plc3, respectively (upper panel). The 16S rRNA (middle panel) and DNase-treated mRNA (lower panel) were included as a normalization control and negative control, respectively.
The defect in intracellular survival of the plc2 mutant is not due to delayed escape from the phagolysosome
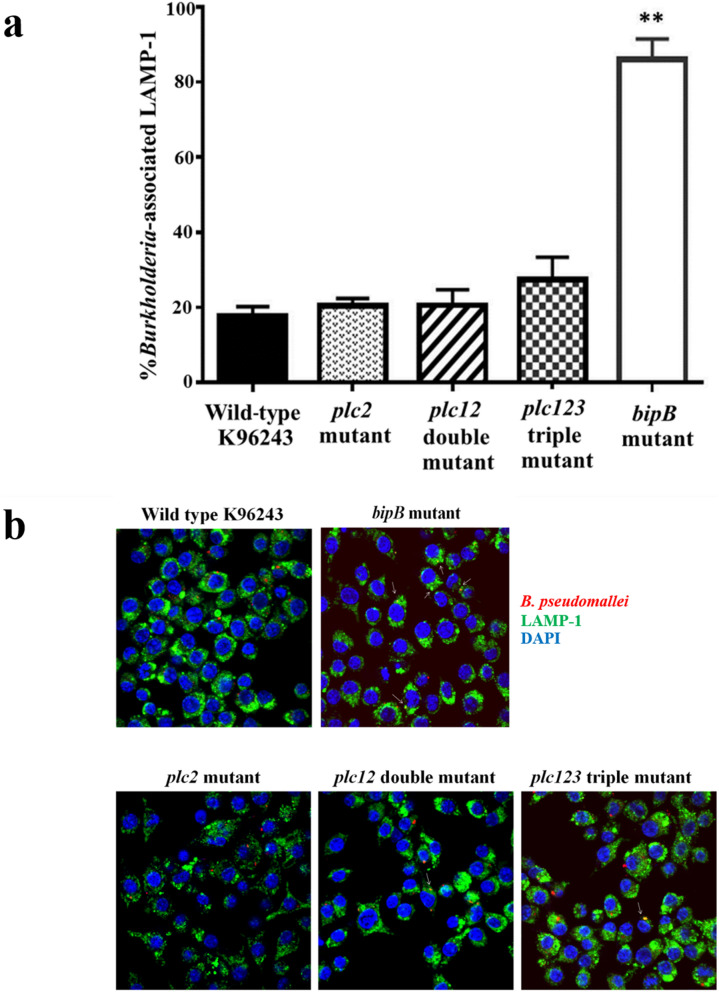
The ability of B. pseudomallei to escape from the phagosome thought to be considered to be a mechanism by which the bacteria evade phagocyte killing. We had already showed that the plc1, plc2 and plc3 genes are expressed in macrophages (Fig. 1c) and that the B. pseudomallei plc2 mutant was defective in survival in macrophages (Fig. 2). Here we investigated whether Plc2 enzyme played a role in escape from the phagolyosome. J774A.1 macrophage-like cells were infected with either B. pseudomallei wild-type, plc2, plc12, or plc123 mutants, and co-localization with lysosomes at 3 h p.i. was investigated by immunostaining with antibodies specific to LAMP-1. A B. pseudomallei bipB mutant, which is known to be delayed in phagosome escape22,23, was included as a control in our experiments. As expected, we found that the majority of bipB mutant cells showed delayed escape from phagosome as evidence the increased association with LAMP-1 (86 ± 3.3% association) when compared with the wild-type bacteria (Fig. 4a). The B. pseudomallei plc2, plc12 and plc123 mutants rarely co-localized with LAMP-1 (20.3 ± 1.2%, 20.7 ± 2.3%, and 27.3 ± 3.5% co-localization, respectively; Fig. 4b), similar to the degree of co-localization of the wild-type bacteria with LAMP-1 (17.7 ± 1.5% co-localization). This finding suggests that mutation of the plc genes did not affect escape of B. pseudomallei from phagosome.
Figure 4.
Quantitative analysis of bacterial co-localization with LAMP-1 in J774A.1 macrophage-like cells. (a) J774A.1 cells were infected with B. pseudomallei wild-type K96243, plc2, plc12, plc123 or bipB mutants for 3 h. Cells were fixed, permeabilized and immunostained with rat anti-LAMP1 and mouse anti-Burkholderia monoclonal antibodies. The y-axis shows the number of bacteria co-localizing with LAMP-1 × 100/total number of intracellular bacteria. Error bars represent standard error of the mean for data collected from 3 independent experiments. Asterisks indicate significant differences (P < 0.01, students t-test) between B. pseudomallei wild-type K96243 and mutants. The figure was prepared using GraphPad Prism version 7.05 for Windows (www.graphpad.com). (b) Representative confocal micrographs of the association of B. pseudomallei with endocytic vesicles in J774A.1 cells. B. pseudomallei wild-type K96243, plc2, plc12 double, plc123 triple or bipB mutants were used to infect J774A.1 cells. At 3 h p.i., the infected macrophages and the bacteria were stained and visualised using confocal microscopy. Macrophage LAMP-1 was stained green with rat monoclonal antibody (1D4B) and Alexa Fluor 488 goat anti-rat IgG antibody, and nuclei were stained blue with DAPI. Bacteria were stained red with mouse anti-B. pseudomallei monoclonal antibody (9D5) and Alexa Fluor 568 goat anti-mouse IgG antibody. White arrows indicated the co-localization of B. pseudomallei and LAMP-1.
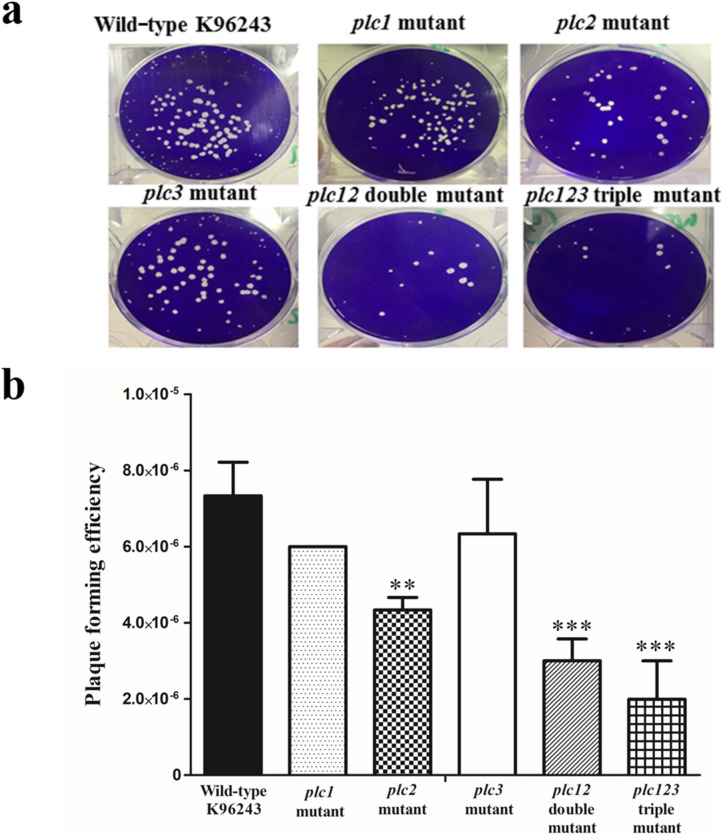
B. pseudomallei plc2 mutant shows deficiency in plaque formation
We next measured plaque formation in monolayers of HeLa cells infected with either B. pseudomallei wild-type or the plc1 plc2, plc3, plc12 or plc123 mutants. Plaque-formation reflects the ability of bacteria to invade, survive within and then spread from cell to cell. As shown in Fig. 5, plaque-formation in HeLa cells was significantly reduced in cells infected with the plc2, plc12 or plc123 mutants, compared to the wild-type strain (**P < 0.01, ***P < 0.001, ***P < 0.001, respectively). In contrast, there was no significant reduction in plaque formation after infection with the plc1 or plc3 mutants (P > 0 0.05) (Fig. 5). This result shows correlation with our previous study14 which showed that plaque-formation efficiency in HeLa cells was significantly reduced after infection with plc2 or plc12 double mutants compared to the wild-type strain. Plaque-formation was restored in a plc2 complemented strain. This finding suggested that the defective phenotype was due to the plc2 gene mutation14. However, complementation of plc12 double and plc123 triple mutants was not possible because of restrictions on the use of multiple antibiotic resistance markers in B. pseudomallei.
Figure 5.
Plaque-forming efficiencies of B. pseudomallei wild-type or plc mutants in HeLa cells. (a) Representative images of plaques. (b) Plaque-forming efficiency of either B. pseudomallei wild-type K96243, plc1, plc2, plc3, plc12 or plc123 mutants in HeLa cells infected at an MOI of 10. Plaques were visualized by crystal violet staining of the monolayers at 24 h p.i. Error bars represent standard error of the mean for data collected in 3 independent experiments. Asterisks indicate significant differences (**P < 0.01, and ***P < 0.001, one-way ANOVA followed by Dunnett's post hoc test) between B. pseudomallei wild-type K96243 and mutant strains. The figure was prepared using GraphPad Prism version 7.05 for Windows (www.graphpad.com).
To assess whether the reduction in plaque formation reflects a reduced ability to adhere to or to invade HeLa cells, we assessed invasion efficiency. There was no significant difference (P > 0 0.05) in the number of culturable intracellular bacteria at 2 h p.i. between wild-type and either plc2, plc12 double, or plc123 triple mutants (Fig. 6). This finding indicates that the absence of the plc 2 gene had no effect of the bacteria to adhere to or invade HeLa cells. Overall, our findings indicate that Plc2 is required for survival and replication of B. pseudomallei in non-phagocytic cells.
Figure 6.
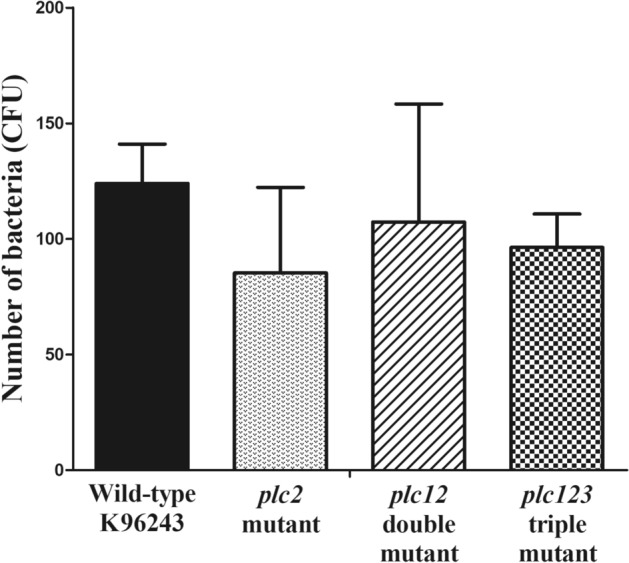
Invasion efficiency of B. pseudomallei wild-type and plc mutants in HeLa cells. The B. pseudomallei wild-type, plc2, plc12 or plc123 mutants were used to infect HeLa cells at an MOI of 50. At 2 h p.i., the infected cells were lysed and the numbers of intracellular bacteria were enumerated by colony count. Error bars represent standard error of mean for data collected from 3 independent experiments. No significant difference between B. pseudomallei wild-type K96243 and mutant strains was detected (P > 0.05, one-way ANOVA followed by Dunnett's post hoc test). The figure was prepared using GraphPad Prism version 7.05 for Windows (www.graphpad.com).
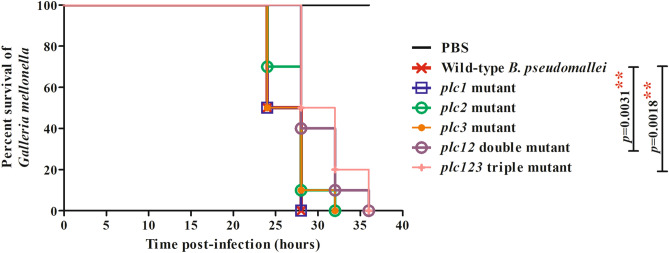
The plc12 and plc123 mutants are attenuated in G. mellonella
Our results above showed that Plc2 was required for intracellular survival and replication in host cells. To investigate the roles of Plc1, Plc2 and Plc3 in virulence of B. pseudomallei, a G. mellonella larvae infection model was used24.
There was no significant difference (P > 0 0.05) in the survival of larvae infected with the plc1, plc2 or plc3 mutants compared with the wild-type strain (Fig. 7). However, larvae infected with the plc12 or plc123 mutants showed significantly (P = 0.0031, P = 0.0018. respectively) increased survival, compared to larvae infected with the wild-type strain. This finding suggests redundancy of the functions of the phospholipases in virulence of B. pseudomallei. Our finding that there was no significant difference (P > 0 0.05) in the survival of larvae infected with the plc12 and plc123 mutants suggests that plc3 does not contribute to virulence in G. mellonella larvae. Because of the restrictions on the number of antibiotic markers we could introduce into B. pseudomallei, we could not generate plc12 or plc123 complemented mutants further validate our findings.
Figure 7.
Virulence of B. pseudomallei strains in G. mellonella larvae. Groups of 10 larvae were challenged with 102 CFU of B. pseudomallei wild-type, plc1, plc2, plc3, plc12 or plc123 mutants. The number of dead larvae was scored at 24, 28, 32 and 36 h p.i.. GraphPad Prism version 7.05 for Windows (www.graphpad.com) was used to graph and analyze the data using a Log-rank (Mantel-Cox) test. Asterisks indicate significant differences (P < 0.05), between B. pseudomallei wild-type K96243 and mutant strains. The figure was prepared using GraphPad Prism version 7.05 for Windows (www.graphpad.com).
Our finding contrasts with a previous study where a plc3 mutant was found to 104-fold attenuated Syrian hamsters compared to the wild-type bacterium16. This might reflect differences in the immune system between G. mellonella and mammals. G. mellonella possess an innate immune system which involves a cellular immune response mediated by hemocytes, and a humoral immune response orchestrated by antimicrobial peptides25, However, they lack the complement system found in mammals and G. mellonella also lacks an adaptive immune system26. Additionally, it is known that B. pseudomallei infection of hamsters is not similar to infection of other mammals such as mice. Hamsters are highly susceptible to infection with B. pseudomallei whereas mice are relatively resistant27. There are also reports of the different behaviour of B. pseudomallei mutants in hamsters and in mice. For example fliC28 and fliD16 mutants are not attenuated in hamsters but a fliC mutant is highly attenuated in mice29.
Conclusion
The genes encoding three B. pseudomallei Plc enzymes are expressed within macrophage-like cells, but at different expression levels. The plc2 gene was expressed in infected macrophages but not in culture medium, suggesting a role in virulence. Our findings suggest that plc2 either alone, or in combination with plc1 and plc3, contributes to growth in host cells and our finding that virulence in G. mellonella was dependent on the inactivation of genes encoding combinations of Plc enzymes, indicates functional redundancy. The data reported in this study provide important new insight into the roles of Plcs in virulence of B. pseudomallei and open new opportunities for further research into the roles on these enzymes in virulence.
Materials and methods
Primers, bacterial strains and cell lines
Primers used in this study are shown in Table 1. Escherichia coli, B. pseudomallei K96243 and the mutant strains were routinely cultured in Luria–Bertani (LB) or trypticase soy medium. B. pseudomallei K96243 plc1, plc2 single and plc12 double mutants were constructed in the previous study14. All cultures were typically grown for 24–48 h at 37 °C. Appropriate antibiotics (Sigma-Aldrich) i.e. chloramphenicol 50 µg/mL, kanamycin 400 µg/mL and tetracycline 50 µg/mL were added into the medium if required. All manipulations of B. pseudomallei were approved by the Technical Biosafety Committee (TBC), National Center for Genetic Engineering and Biotechnology (BIOTEC).
Table 1.
Oligonucleotide primers used in this study.
| Primers | Oligonucleotide sequences (5′–3′) | Purposes | Sources |
|---|---|---|---|
| PLC88 | AGACCGTGCTGCTCGTGAA | Forward primer for construction of plc3 mutant | This study |
| PLC89 | GGCTCGTTGTTCGGTCGCA | Reverse primer for construction of plc3 mutant | This study |
| Plc1F | TGATGCAGGAAAACCGCTC | Forward primer for internal fragment of plc1 gene | This study |
| Plc1R | AGCCCGTCCACATGTAGTAG | Reverse primer for internal fragment of plc1 gene | This study |
| Plc2F | GCTCGACAACAGCGATTACG | Forward primer for internal fragment of plc2 gene | This study |
| Plc2R | TTCTGCAGGATGTTCGTCCC | Reverse primer for internal fragment of plc2 gene | This study |
| Plc3F | TCAAGGAAGACATCCGTGCG | Forward primer for internal fragment of plc3 gene | This study |
| Plc3R | CGTCGAAATTCACGAGCAGC | Reverse primer for internal fragment of plc3 gene | This study |
| 23s F | TTTCCCGCTTAGATGCTTT | Forward primer for internal fragment of 23s RNA gene | 33 |
| 23s R | AAAGGTACTCTGGGGATAA | Reverse primer for internal fragment of 23s RNA gene | 33 |
| 16s F | AGACACGGCCCAGACTCCTAC | Forward primer for internal fragment of 16s RNA gene | 33 |
| 16s R | CAGTCACCAATGCAGTTCCCA | Reverse primer for internal fragment of 16s RNA gene | 33 |
| bpsl2404F-173 | GGCAAGGATCTGCAAAACGG | Forward primer for amplification of bpsl2404 gene | This study |
| bpsl2404R-173 | ACGACCGACACCTTCTTGTC | Reverse primer for amplification of bpsl2404 gene | This study |
| bpsl0337F-184 | TCCCCAGTTCCTCCTCGATT | Forward primer for amplification of bpsl0337 gene | This study |
| bpsl0337R-184 | ATGCAACACACCGAACAACC | Reverse primer for amplification of bpsl0337 gene | This study |
| bpss0068F-158 | CTGCCGATGCCGGATTATCA | Forward primer for amplification of bpss0068 gene | This study |
| bpss0068R-158 | AACGAATTTGCTTGCTCGGG | Reverse primer for amplification of bpss0068 gene | This study |
J774A.1 murine macrophage-like and human epithelial HeLa cells were obtained from the American Type Culture Collection (ATCC) and were cultured in Dulbecco’s Modified Eagle medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Invitrogen) under a 5% CO2 atmosphere at 37 °C in a humidified incubator.
RNA preparation and reverse transcription (RT)-PCR analysis
B. pseudomallei K96243 was grown in LB broth for 6 h before incubation at 37 °C for 15 min in LB broth pH 4.5, 5.0, 6.0, 7.0, 8.0 or 9.0. Total RNA was extracted from 108 CFUs B. pseudomallei cultured in each condition using TRIZOL (Invitrogen) according to manufacturer’s instructions. The isolated bacterial RNA was then treated with DNase I (Ambion) to remove any genomic DNA contamination before use.
To detect B. pseudomallei genes expression within macrophages, monolayers of J774A.1 murine macrophage-like cells were infected with the bacteria. At the indicated time points, the infected cell monolayers were washed and subsequently lysed with 500 μL of 0.1% Triton X-100 (Sigma-Aldrich) to allow intracellular bacteria released from infected cells. Then, 500 μL of 1 × PBS was added and the intracellular bacterial RNA were extracted using TRIZOL (Invitrogen) according to manufacturer’s instructions.
To convert the extracted total RNA to cDNA, SuperScript III First-Strand Synthesis System (Invitrogen) was used. The cDNA was quantified and adjusted so that similar quantities were included in the PCR reactions. The cDNA was amplified using the PCR with primers (Table 1), GoTaq DNA polymerase (Promega) and cycling conditions of 94 °C, 3 min and 30 cycles of 94 °C for 30 s, 50 °C for 1 min, and 72 °C for 45 s, followed by incubation at 72 °C for 5 min. In each PCR experiment, the amplification of 23S rRNA was used as a normalization control.
Construction of B. pseudomallei plc3 single and plc123 triple mutants
A B. pseudomallei plc3 mutant was constructed by insertion mutagenesis30. A 406-bp (nucleotide positions 1052–1457) internal region of the plc3 gene was amplified from B. pseudomallei K96243 genomic DNA with primers PLC88 and PLC89 (Table 1). The amplified DNA fragment was ligated into EcoRV digested pKNOCK-Cm, a suicide vector30 to generate recombinant plasmid pVSK3 for insertion mutagenesis. The constructed plc3 mutant was selected on Pseudomonas agar base supplemented with SR103E (Oxoid) and chloramphenicol.
To construct the B. pseudomallei plc123 triple mutant, the amplified internal plc3 fragment (nucleotide positions 1052-1457) was ligated into EcoRV digested pKNOCK-Km30. This constructed plasmid, designated pVSK4, was introduced into B. pseudomallei plc12 double mutant14. The mutants were selected on Pseudomonas agar base (Oxoid) supplemented with SR103E (Oxoid) containing chloramphenicol, kanamycin and tetracycline (Sigma-Aldrich). The plc123 mutant was verified by PCR and Southern blotting.
Intracellular survival and plaque assays
Intracellular replication of B. pseudomallei in macrophage-like cells was assessed as described previously31 with some modifications. Briefly, J774A.1 murine macrophage-like cells were seeded at a density of 2.5 × 105 cells per well of a 24-well tissue culture plate and infected approximately 24 h later with B. pseudomallei wild-type K96243 or plc mutant at a multiplicity of infection (MOI) of approximately 0.5, for 2 h. Then infected cells were overlaid with DMEM medium (Invitrogen) containing gentamicin 128 μg/mL and spectinomycin 256 μg/mL (Sigma-Aldrich) to kill extracellular bacteria. The infected cell monolayers were subsequently lysed at 2, 4, 6 and 8 h p.i. with 0.1% Triton X-100 (Sigma-Aldrich). The numbers of intracellular bacteria were quantified by serial dilution and plating on tryptic soy agar. Bacterial colony forming units (CFU) were counted after 36–48 h of incubation at 37 °C. Plaque forming assays were performed as described previously32. The plaque-forming efficiency was calculated as the number of plaques/bacterial CFU added per well.
Confocal analysis of bacterial co-localization with LAMP-1
The intracellular localizations of B. pseudomallei wild-type K96243 and the plc mutants in J774A.1 macrophages cells relative to LAMP-1 containing vesicles were investigated according to previously described33. Briefly, Macrophages were infected for 2 h at a multiplicity of infection (MOI) of 2 and incubated at 37 °C, 5% CO2. At different time points, B. pseudomallei infected J774A.1 cells were fixed in 4% paraformaldehyde, the monolayers were permeabilized with 0.5% (v/v) Triton X-100, and blocked with 1% (w/v) bovine serum albumin. Bacteria were detected with a 1:10 dilution of mouse anti-Burkholderia monoclonal antibody and detected with a 1:200 dilution of Alexa Fluor 568-goat anti-mouse IgG (Invitrogen, USA). LAMP1 was stained green with a 1:100 dilution of rat monoclonal antibody (1D4B; Abcam, USA) and 1:500 Alexa Fluor 488 goat anti-rat IgG antibody (Molecular Probes, USA), and nuclei were stained blue with a 1:500 dilution of 4,6-diamidino-2-phenylindole (DAPI). Cells were examined by a laser-scanning confocal microscope equipped with LSM5 Image Browser (LSM 510 META, Carl Zeiss, Germany). The association of Burkholderia with LAMP1 was considered when the red fluorescent bacteria co-localized with the green fluorescence of LAMP1-positive vacuoles, represented as an area of yellow staining.
The percentage of intracellular Burkholderia associated with LAMP-1 was determined as the number of bacteria co-localized with LAMP-1/total number of intracellular bacteria × 100. For the quantitative analysis of the association of intracellular B. pseudomallei strains with LAMP-1 containing vesicles, at least 200 individual bacteria associated LAMP-1 containing vesicles were monitored.
Virulence in G. mellonella
Virulence in G. mellonella larvae was tested as described previously24 with some modifications. Larvae between 2–2.5 cm and free of melanization or injury were used in the experiments. To prepare the bacterial culture for infection, the overnight cultures of B. pseudomallei wild-type and the mutants were adjusted to a concentration of 104 CFUs per ml in PBS. A 701 N fixed needle syringe (Hamilton, Nevada) was used to inject 10 µl aliquots of the bacterial suspension into the G. mellonella larvae to get the final concentration of 102 CFUs. Injections were performed directly into the larval body cavity and groups of 10 larvae were injected with each bacterial strain. Control larvae were injected with PBS. Following injection, larvae were incubated in the dark at 37 °C and the number of dead larvae were recorded at a variety of times post injection. The Galleria mellonella study was approved by the Mahidol University-Institute Animal Care and Use Committee (U1-05763-2559).
Statistical analysis
For in vivo mutant characterization, a log-rank (Mantel-Cox) test was used to compare survival curves and the experiments for comparison between groups were performed using the one-way ANOVA followed by Dunnett's post hoc test within the GraphPad Prism version 7.05 for Windows (www.graphpad.com, GraphPad Software, CA, USA). P-values less than 0.05 were considered statistically significant (*P < 0.05, **P < 0.01, and ***P < 0.001).
Supplementary information
Acknowledgements
The authors are grateful to Songprakhon P (Department of Research and Development, Siriraj Hospital) and Janesomboon S (Department of Immunology, Siriraj Hospital) for their kind technical assistances. Dr. Narisara Chantratita (Faculty of Tropical Medicine, Mahidol University, Thailand) for kindly provided mouse anti-Burkholderia monoclonal antibody. We used the laser scanning confocal microscope (LSM 800, Zeiss, Jena, Germany) at the Division of Molecular Medicine, Faculty of Medicine Siriraj Hospital, Mahidol University.
Author contributions
Conceived and designed the experiments: V.S., S.K. Performed the experiments: V.S., P.W., S.C., M.T., C.M. Analyzed the data: V.S., P.W., S.K. Wrote the paper: V.S., P.W., J.M.S., R.W.T., S.K.
Funding
Korbsrisate S is supported by the Siriraj Grant for Research and Development, Faculty of Medicine Siriraj Hospital, Mahidol University. Meethai C is supported by the Royal Golden Jubilee Ph.D programme, Thailand Research Fund (PHD/0070/2559).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Varintip Srinon and Patoo Withatanung.
Supplementary information
is available for this paper at 10.1038/s41598-020-76186-z.
References
- 1.Limmathurotsakul D, et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 2016;1:15008. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 2.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White NJ. Melioidosis. Lancet. 2003;361:1715–1722. doi: 10.1016/S0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- 4.Willcocks SJ, Denman CC, Atkins HS, Wren BW. Intracellular replication of the well-armed pathogen Burkholderia pseudomallei. Curr. Opin. Microbiol. 2016;29:94–103. doi: 10.1016/j.mib.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Holden MT, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeShazer D, Brett PJ, Burtnick MN, Woods DE. Molecular characterization of genetic loci required for secretion of exoproducts in Burkholderia pseudomallei. J. Bacteriol. 1999;181:4661–4664. doi: 10.1128/JB.181.15.4661-4664.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raynaud C, et al. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2002;45:203–217. doi: 10.1046/j.1365-2958.2002.03009.x. [DOI] [PubMed] [Google Scholar]
- 8.Ostroff RM, Wretlind B, Vasil ML. Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect. Immun. 1989;57:1369–1373. doi: 10.1128/IAI.57.5.1369-1373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Awad MM, Bryant AE, Stevens DL, Rood JI. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 1995;15:191–202. doi: 10.1111/j.1365-2958.1995.tb02234.x. [DOI] [PubMed] [Google Scholar]
- 10.Camilli A, Tilney LG, Portnoy DA. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aurass P, et al. The Legionella pneumophila Dot/Icm-secreted effector PlcC/CegC1 together with PlcA and PlcB promotes virulence and belongs to a novel zinc metallophospholipase C family present in bacteria and fungi. J. Biol. Chem. 2013;288:11080–11092. doi: 10.1074/jbc.M112.426049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores-Diaz M, Monturiol-Gross L, Naylor C, Alape-Giron A, Flieger A. Bacterial sphingomyelinases and phospholipases as virulence factors. Microbiol. Mol. Biol. Rev. 2016;80:597–628. doi: 10.1128/MMBR.00082-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumari Bandana KJ. Kaur Jagdeep Phospholipases in bacterial virulence and pathogenesis. Adv. Biotech. Micro. 2018;10(5):555798. [Google Scholar]
- 14.Korbsrisate S, et al. Characterization of two distinct phospholipase C enzymes from Burkholderia pseudomallei. Microbiology. 2007;153:1907–1915. doi: 10.1099/mic.0.2006/003004-0. [DOI] [PubMed] [Google Scholar]
- 15.Burtnick MN, Brett PJ, DeShazer D. Proteomic analysis of the Burkholderia pseudomallei type II secretome reveals hydrolytic enzymes, novel proteins, and the deubiquitinase TssM. Infect. Immun. 2014;82:3214–3226. doi: 10.1128/IAI.01739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuanyok A, Tom M, Dunbar J, Woods DE. Genome-wide expression analysis of Burkholderia pseudomallei infection in a hamster model of acute melioidosis. Infect. Immun. 2006;74:5465–5476. doi: 10.1128/IAI.00737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowling AJ, et al. Genome-wide analysis reveals loci encoding anti-macrophage factors in the human pathogen Burkholderia pseudomallei K96243. PLoS ONE. 2010;5:e15693. doi: 10.1371/journal.pone.0015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ooi WF, et al. The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 2013;9:e1003795. doi: 10.1371/journal.pgen.1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakala N’goma JC, Schue M, Carriere F, Geerlof A, Canaan S. Evidence for the cytotoxic effects of Mycobacterium tuberculosis phospholipase C towards macrophages. Biochim. Biophys. Acta. 2010;1801:1305–1313. doi: 10.1016/j.bbalip.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Oda M, et al. Signal transduction mechanism involved in Clostridium perfringens alpha-toxin-induced superoxide anion generation in rabbit neutrophils. Infect. Immun. 2006;74:2876–2886. doi: 10.1128/IAI.74.5.2876-2886.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monturiol-Gross L, et al. Internalization of Clostridium perfringens alpha-toxin leads to ERK activation and is involved on its cytotoxic effect. Cell Microbiol. 2014;16:535–547. doi: 10.1111/cmi.12237. [DOI] [PubMed] [Google Scholar]
- 22.Suparak S, et al. Multinucleated giant cell formation and apoptosis in infected host cells is mediated by Burkholderia pseudomallei type III secretion protein BipB. J. Bacteriol. 2005;187:6556–6560. doi: 10.1128/JB.187.18.6556-6560.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinon V, et al. Comparative assessment of the intracellular survival of the Burkholderia pseudomallei bopC mutant. J. Microbiol. 2013;51:522–526. doi: 10.1007/s12275-013-2557-3. [DOI] [PubMed] [Google Scholar]
- 24.Wand ME, Muller CM, Titball RW, Michell SL. Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis. BMC Microbiol. 2011;11:11. doi: 10.1186/1471-2180-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai CJ, Loh JM, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7:214–229. doi: 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao J, Chard LS, Wang Z, Wang Y. Syrian hamster as an animal model for the study on infectious diseases. Front. Immunol. 2019;10:2329. doi: 10.3389/fimmu.2019.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Titball RW, et al. Burkholderia pseudomallei: animal models of infection. Trans. R. Soc. Trop. Med. Hyg. 2008;102(Suppl 1):S111–116. doi: 10.1016/S0035-9203(08)70026-9. [DOI] [PubMed] [Google Scholar]
- 28.DeShazer D, Brett PJ, Carlyon R, Woods DE. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 1997;179:2116–2125. doi: 10.1128/JB.179.7.2116-2125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chua KL, Chan YY, Gan YH. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 2003;71:1622–1629. doi: 10.1128/IAI.71.4.1622-1629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexeyev MF. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques. 1999;26:824–826. doi: 10.2144/99265bm05. [DOI] [PubMed] [Google Scholar]
- 31.Muangsombut V, et al. Inactivation of Burkholderia pseudomallei bsaQ results in decreased invasion efficiency and delayed escape of bacteria from endocytic vesicles. Arch. Microbiol. 2008;190:623–631. doi: 10.1007/s00203-008-0413-3. [DOI] [PubMed] [Google Scholar]
- 32.Pinweha P, et al. Inactivation of bpsl1039-1040 ATP-binding cassette transporter reduces intracellular survival in macrophages, biofilm formation and virulence in the murine model of Burkholderia pseudomallei infection. PLoS ONE. 2018;13:e0196202. doi: 10.1371/journal.pone.0196202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muangsombut V, et al. Burkholderia pseudomallei evades Nramp1 (Slc11a1)- and NADPH oxidase-mediated killing in macrophages and exhibits Nramp1-dependent virulence gene expression. Front. Cell Infect. Microbiol. 2017;7:350. doi: 10.3389/fcimb.2017.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.