Abstract
Radiotherapy has been the major treatment strategy for nasopharyngeal carcinoma (NPC), while the occurrence of radioresistance may lead to cancer recurrence or progression. This study aimed to identify the key microRNAs (miRNAs) and their target genes in the development of NPC radioresistance. Public microarray data were searched and analyzed to screen the differentially expressed miRNAs (DEMs) and genes (DEGs) between radioresistant and radiosensitive NPC samples. MiRNA-mRNA networks were constructed. As a result, 5 DEMs and 195 DEGs were screened out. The DEGs were enriched in various signaling pathways, such as Cytokine-cytokine receptor interaction, Jak-STAT signaling pathway, and Toll-like receptor signaling pathway. Several hub genes, such as IGF2, OLA1, BBS10, MMP9, and BBS7 were identified. A regulatory miRNA-mRNA network containing 87 miRNA-mRNA pairs was constructed. Then, 14 key miRNA-mRNA pairs that contained the hub genes were further filtered out. In the networks, miR-203a-3p had the largest number of target genes. Afterwards, the candidate pairs (miR-203a-3p/BTK and miR-484/OLA1) have been verified by a qRT-PCR assay. In summary, we identified several miRNAs and hub genes via big data screening. A total of 87 miRNA-mRNA pairs (including 14 key pairs) were predicted to play a crucial role in the development of NPC radioresistance. These data provide a bioinformatics basis for further exploring the molecular mechanism of radiotherapy resistance in NPC. Future studies are needed to validate the results.
Keywords: Nasopharyngeal carcinoma, Radioresistance, MicroRNA, Microarray, qRT-PCR
Introduction
Nasopharyngeal carcinoma (NPC) is a kind of malignant tumor originated from the nasopharynx, which is characterized by low differentiation squamous cell carcinoma. Although NPC is a rare malignancy in the world, it is endemic in Southeast Asia (Gioacchini et al. 2017). Given its high radiosensitivity, the primary and standard treatment for NPC is radiotherapy. Nevertheless, in the course of treatment, the emergence of radioresistance significantly reduced the efficacy of radiotherapy. Thus, the survival rate of a proportion of NPC patients remains low after radiotherapy. Evidence indicated that the occurrence of radioresistance may result in local recurrence or distant metastasis within 1.5 years in NPC patients (Zhao et al. 2017). To date, there have been few clinical measures to overcome the resistance of radiotherapy in NPC patients (Perri et al. 2019).
A number of studies have been devoted to the molecular mechanisms underlying the development of NPC radioresistance. Evidence indicates that LAPTM4B was up-regulated in radioresistant NPC cells, and LAPTM4B knockdown decreases radioresistance by inhibiting autophagy (Chu et al. 2019). Likewise, SALL4 may act as an oncogene and play a role in the radioresistance of NPC cells. Thus, its silencing increased radiation-induced DNA damage and cell apoptosis in radioresistant NPC cells (Nie et al. 2019). Conversely, as a tumor suppressor, FOXO3a silencing may promote tumor radioresistance of NPC by activating Wnt/β-catenin signal pathway (Luo et al. 2019). Nevertheless, little is known about the precise molecular mechanisms involved in the development of NPC radioresistance.
In recent decades, microRNAs (miRNAs) have attracted much attention. miRNAs are a class of short non-coding RNA (ncRNA) sequences that bind to mRNAs and repress their expression, thus modulating gene expressions via post-transcriptional regulation (Lekka and Hall 2018). On the basis of this feature, miRNAs have been implicated in the onset and development of various disorders, including carcinomas (Dragomir et al. 2018). A number of miRNAs have been studied in the development of NPC. For example, miR-155 can inhibit cell proliferation and increase cell apoptosis of NPC by targeting PI3K/AKT-FOXO3a signaling (Wu et al. 2019). Overexpression of miR-543 promotes cell proliferation and cell invasion of NPC by targeting JAM-A (Jiang et al. 2019a). A miRNA can target a number of genes, whereas a gene can be targeted by various miRNAs. Hence, a regulatory network involving different miRNAs and their target genes might exist. To study the miRNA-mRNA network may help us better understand the molecular mechanisms in the development of radioresistance in NPC.
A recently published study (Guo et al. 2019) has been devoted to the screening of the key miRNAs and genes in the development of NPC radioresistance. However, a comprehensive miRNA-mRNA network regarding this issue had not been established. To our knowledge, few studies have been devoted to this subject. Thus, in the present study, we aimed to construct a miRNA-mRNA interaction network and screen the key miRNA-mRNA pairs that might play a critical role in the radioresistance of NPC by conducting a bioinformatics analysis.
Material and methods
Data source
To obtain differentially expressed miRNAs (DEMs) and genes (DEGs) between radioresistant and radiosensitive NPC samples, microarray-based datasets were retrieved from the Gene Expression Omnibus database (ncbi.nlm.nih.gov/geo). Datasets that met the following criteria were selected: (1) Datasets were restricted to Homo sapiens; (2) Datasets should contain radioresistant and radiosensitive NPC samples; (3) Datasets that contained the largest number of samples were chosen.
As a result, GSE48502 (miRNA) and GSE32389 (mRNA) were selected. The information of these datasets was downloaded.
The dataset GSE48502 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48502) was deposited by Xiao et al. in 2013 (Li et al. 2014), containing 3 samples of NPC radioresistant cell line and 3 samples of their parent cell line. The experiment type was an ncRNA profiling based on Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray 030840 (miRBase release 14.0 miRNA ID version). The dataset GSE32389 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32389) was deposited by Yang et al. in 2013 (Yang et al. 2012), comprising 12 samples of radioresistant NPC tissues and 8 samples of radiosensitive NPC ones. The experiment type was Expression profiling by array, and the platform was UGI Human 14112 V1.0. The samples in these datasets were from China.
Identification of DEMs and DEGs
The matrixes of the datasets were downloaded, in which lines with blanks or insufficient data were firstly deleted. Then, the data were analyzed using the Limma package of R software (version 3.6.1). A threshold of P value or FDR < 0.05 was considered significant. Cut-off criteria for the fold-change were set as |fold-change| = 2.
Functional annotation of the DEGs
To annotate the functions of the screened DEGs, Gene Ontology (GO) and KEGG pathway enrichment analysis were conducted through the use of the GATHER tool (Chang and Nevins 2006). A P value of less than 0.05 was considered statistically significant.
Protein–protein interaction network construction
To screen the possible hub genes/proteins that might play a crucial role in the development of NPC radioresistance, the DEGs were submitted to the STRING tool (Szklarczyk et al. 2019) to predict their interaction relationships. A combined score of not less than 0.40 (median confidence score) was set as the cut-off criterion. The hub proteins were sorted by the degree or betweenness value in the network. Cytoscape (version 3.6.0) was used to help visualize the networks.
MiRNA target gene prediction and miRNA-mRNA network construction
A number of computational microRNA-target prediction tools, such as Targetscan, miRanda, and miRDB, have been developed. Among these tools, mirDIP database was selected because it included the integration of different tools and thus can minimize the shortcomings of individual databases (Tokar et al. 2018). Candidate mRNAs with a score class of more than ‘high’ were selected as the target genes of the DEMs.
To limit the target genes to the genes related to radioresistance of NPC, The intersection of the target genes and the DEGs that had converse expression levels with the relevant DEMs were obtained by Venn analysis.
MiRNA-mRNA regulatory network was constructed by using Cytoscape software (version 3.6.0). Based on the network, the key miRNA-mRNA pairs were identified according to the hub genes.
Cell culture
The human nasopharyngeal cell line, HONE1 (ATCC, Manassas, VA, USA) was conserved in our laboratory. The radioresistant HONE1-IR cells were successfully established according to the method as described in the literature (Wang et al. 2017; Zhuo et al. 2015). Cells were cultured in RPMI‑1640 media (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum in a humidified atmosphere containing 5% CO2 at 37 °C.
MiR-203a-3p mimic and miR-484 inhibitor were purchased from Chongqing Aozhe Biotech Company. HONE1-IR was cultured. MiR-203a-3p mimic or miR-484 inhibitor was transfected at a final concentration of 100 nM, by using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacture’s protocol. After incubation for 48 h, the cells were lysed for RNA isolation.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cultured cells with Trizol reagent (Invitrogen) according to the manufacturer’s protocol. qRT-PCR was conducted using an ABI PRISM 7900 sequence detection system (Applied Biosystems). All reactions were conducted in triplicate. The relative expression of the amplified RNA samples was calculated using the 2−ΔΔCt method. Gene level was normalized to GAPDH, and miRNA level was normalized to U6 expression. The results are presented as the fold changes of the mRNAs/miRNAs in the HONE1-IR (radioresistant) relative to the HONE1 (radiosensitive).
Statistical analysis
The comparisons of the mRNA/miRNA expression levels between two groups were analyzed by the T test. A P value of less than 0.05 was considered as statistically significant differences. Statistical analysis was performed using Medcalc software (version 15.2.2, Belgium).
Results
Identification of DEMs and DEGs associated with NPC radioresistance
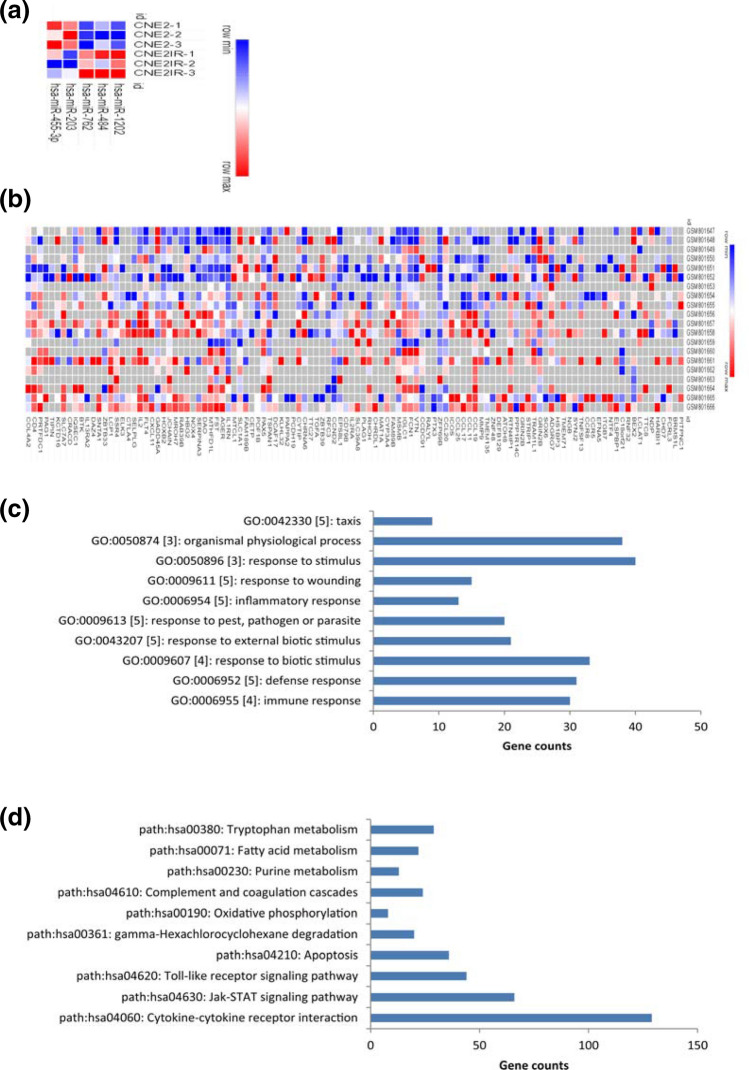
To get the DGMs, the dataset GSE48502 was analyzed, in which the radioresistant and the radiosensitive samples were compared. FDR was not used because no DEMs could be screened out when this index was used. Thus, P < 0.05 was considered significant statistically. As a result, 5 miRNAs met the criteria (Fig. 1a). Of these DEMs, 3 were up-regulated and the remaining 2 were down-regulated in radioresistant cells.
Fig. 1.
a The DEMs from the microarray-based miRNA expression profile GSE48502. b The DEGs from the microarray-based gene expression profile GSE32389. The horizontal axis stands for the name of the DEMs or DEGs; the right vertical stands for the name of the samples. Red stands for up-regulated miRNAs/ mRNAs, while blue stands for down-regulated miRNAs/mRNAs. c–d The top terms of the Go analysis (c) and KEGG enrichment analysis (d) of the DEGs. The terms were generated from the GATHER tool
The DEGs were generated from the analysis of GSE32389. Likewise, P < 0.05 and |fold-change| = 2 was set as a cut-off. Consequently, 121 up-regulated and 74 down-regulated genes were screened out (Fig. 1b and Table 1).
Table 1.
The DEMs and DEGs in the radioresistant samples of NPC
| Row names (tT) | log FC | t | P value | adj. P value | B |
|---|---|---|---|---|---|
| Upregulated DEMs | |||||
| hsa-miR-762 | − 1.327 | − 6.296 | 0.000 | 0.048 | 0.410 |
| hsa-miR-484 | − 1.250 | − 2.794 | 0.027 | 0.289 | − 3.932 |
| hsa-miR-1202 | − 1.196 | − 7.151 | 0.000 | 0.045 | 1.213 |
| Downregulated DEMs | |||||
| hsa-miR-455-3p | 1.121 | 2.757 | 0.028 | 0.289 | − 3.987 |
| hsa-miR-203 | 1.738 | 4.788 | 0.002 | 0.093 | − 1.240 |
| Upregulated DEGs (Top5) | |||||
| RNF32 | − 2.902 | − 3.961 | 0.003 | 0.34 | − 3.088 |
| ITGAM | − 2.703 | − 3.690 | 0.005 | 0.372 | − 3.203 |
| CTLA4 | − 2.583 | − 4.064 | 0.001 | 0.32 | − 2.274 |
| CCL19 | − 2.242 | − 4.325 | 0.000 | 0.242 | 0.683 |
| CHIT1 | − 2.176 | − 3.592 | 0.005 | 0.37 | − 2.934 |
| Downregulated DEGs (Top5) | |||||
| PCDH19 | 3.1267 | 5.199 | 0.000 | 0.254 | − 2.143 |
| ZNF502 | 3.100 | 4.801 | 0.000 | 0.26 | − 2.306 |
| CTTNBP2 | 2.413 | 3.424 | 0.005 | 0.372 | − 2.763 |
| PAPPA2 | 2.404 | 3.281 | 0.009 | 0.429 | − 3.394 |
| CHD7 | 2.261 | 3.408 | 0.006 | 0.405 | − 3.048 |
Functional annotation of the DEGs
To annotate the functions of the DEGs, we submitted these genes to the GATHER tool for analysis. As shown in Fig. 1c, the significant GO terms were mainly related to stress responses such as immune response, defense response, response to biotic stimulus, response to external biotic stimulus, and inflammatory response.
The pathway enrichment analysis showed that the most significant dysfunctional pathways were varied, such as Cytokine-cytokine receptor interaction, Jak-STAT signaling pathway, Toll-like receptor signaling pathway, Apoptosis, and gamma-Hexachlorocyclohexane degradation (Fig. 1d).
Thus, multiple signaling pathways associated with stress response and metabolisms may be involved in the development of NPC radioresistance.
Protein–protein interaction network construction
To screen the hub genes/proteins of the DEGs, they were submitted to the STRING tool for analysis. The genes with a median confidence (0.40) were chosen.
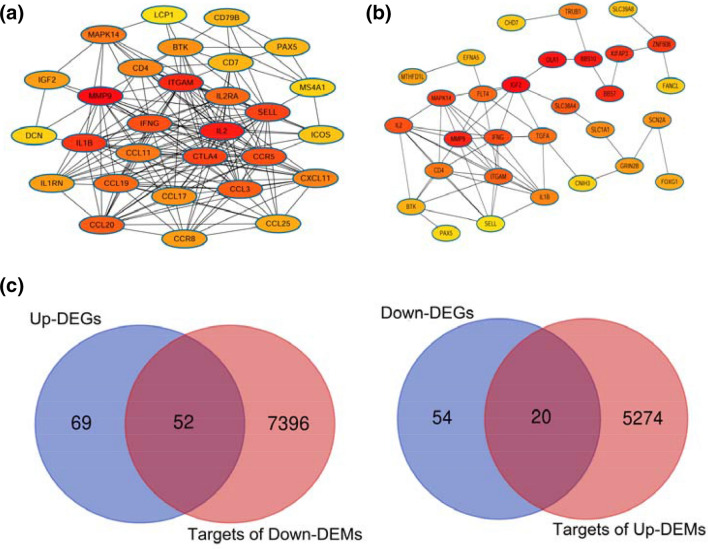
The plug-in unit, CytoHubba, in Cytoscape software (version 3.6.0) was used to calculate the degree and betweenness values, respectively, of the genes. Then, the interaction networks based on these two parameters were established, respectively (Table 2). In the network based on degree, the top 5 node proteins were MMP9, IL2, ITGAM, IL1B, and CTLA4 (Fig. 2a). Nevertheless, in the network based on betweenness, the top 5 node proteins were IGF2, OLA1, BBS10, MMP9, and BBS7 (Fig. 2b).
Table 2.
Screening of the hub genes by constructing protein–protein interaction networks
| Node_name | Degree | Closeness | Betweenness | Clustering coefficient |
|---|---|---|---|---|
| Sorted by degree (top 10) | ||||
| MMP9 | 29 | 57.16825 | 2680.223 | 0.27833 |
| IL2 | 26 | 53.91349 | 1035.989 | 0.44615 |
| ITGAM | 25 | 53.56349 | 870.1263 | 0.42667 |
| IL1B | 24 | 52.61349 | 619.9493 | 0.42754 |
| SELL | 22 | 51.23016 | 372.9203 | 0.52381 |
| CTLA4 | 22 | 50.98016 | 258.224 | 0.55411 |
| IFNG | 21 | 52.55159 | 1285.687 | 0.55238 |
| CCR5 | 21 | 50.14683 | 131.4213 | 0.63333 |
| CCL3 | 17 | 48.24683 | 147.8977 | 0.66912 |
| CCL20 | 17 | 47.39683 | 55.42792 | 0.77206 |
| Sorted by betweenness (top 10) | ||||
| IGF2 | 12 | 48.47976 | 5251.501 | 0.15152 |
| OLA1 | 4 | 37.37619 | 4008.284 | 0.16667 |
| BBS10 | 8 | 33.84524 | 3962 | 0.10714 |
| MMP9 | 29 | 57.16825 | 2680.223 | 0.27833 |
| BBS7 | 3 | 27.33214 | 2352 | 0.33333 |
| KIFAP3 | 2 | 23.96667 | 2178 | 0 |
| ZNF608 | 6 | 23.725 | 2064 | 0.06667 |
| MAPK14 | 15 | 49.68492 | 1416.393 | 0.26667 |
| IFNG | 21 | 52.55159 | 1285.687 | 0.55238 |
| IL2 | 26 | 53.91349 | 1035.989 | 0.44615 |
Fig. 2.
The DEGs were submitted to the STRING tool to screen the hub genes. The hub genes were sorted by degree (a) or betweenness (b) from protein–protein interaction analysis, respectively. c Venn analysis on the intersection between the predicted target genes of the DEMs and the reversely expressed DEGs
Prediction of miRNA target genes
To predict the target genes of the DEMs, mirDIP database was used. The class score was set as ‘high’. As a result, a total of 5294 genes were predicted to be the potential target genes of the up-regulated DEMs (hsa-miR-484, hsa-miR-1202, and hsa-miR-762), and 7448 genes might be those of the down-regulated DEMs (hsa-miR-203a-3p and hsa-miR-455-3p).
Given that not all target genes may be associated with NPC radioresistance, Venn analysis was conducted to obtain the intersection between the target genes and the reversely expressed DEGs. As shown in Fig. 2a, a total of 20 DEGs were chosen as the targets of the up-regulated DEMs, while 52 DEGs were selected as the targets of the down-regulated DEMs.
Regulatory miRNA-mRNA network analysis
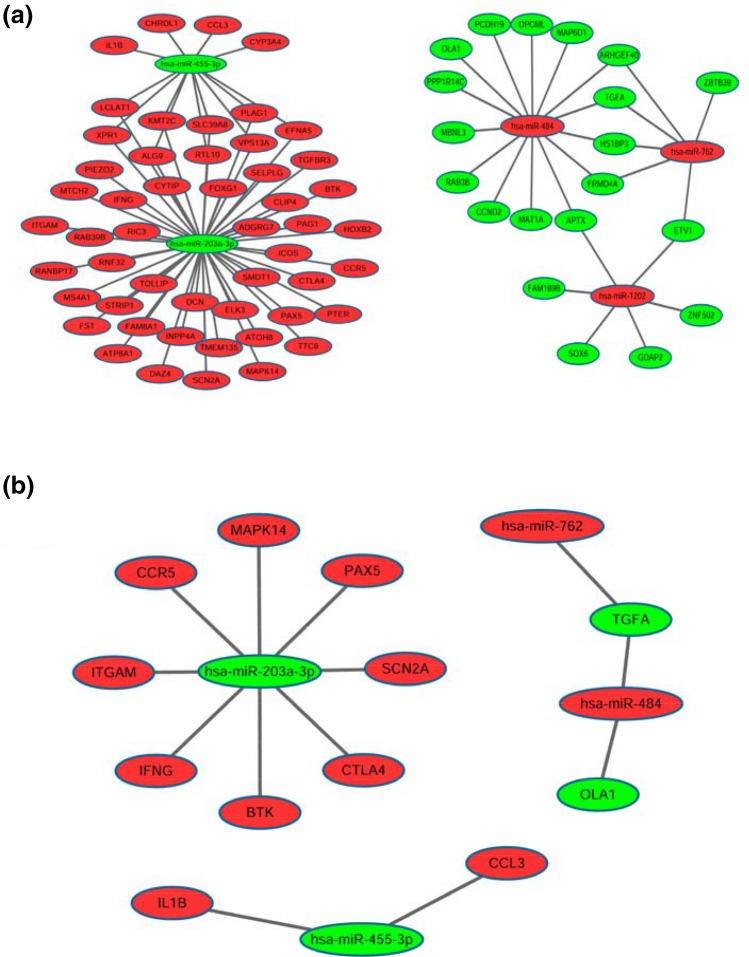
A miRNA-mRNA network was established by using Cytoscape software (version 3.6.0). In this network, there were a total of 87 miRNA-target gene pairs of reverse associations (Fig. 3a). Of the down-regulated miRNAs, miR-203a-3p targeted the largest number of genes in this network. Among the up-regulated miRNAs, miR-484 targeted the largest number of genes.
Fig. 3.
a miRNA-mRNA interaction network regarding the radioresistance of NPC; there were a total of 87 miRNA-mRNA pairs. b Key miRNA-mRNA network containing the hub genes; there were 14 key miRNA-mRNA pairs. Red stands for up-regulated miRNAs/mRNAs, while green stands for down-regulated miRNAs/mRNAs
Screening the key miRNA-mRNA pairs in the network
Because of the different functions among genes, different miRNA-mRNA pairs have different weights in the biological process. Thus, miRNA-mRNA pairs that included hub genes may be regarded as the key miRNA-mRNA pairs.
To select the hub genes, the gene list based on the STRING analysis was sorted by ‘degree’ and ‘betweenness’, respectively. The genes that had a degree of not less than 10 or had a betweenness value of not less than 500 were selected as the hub genes. Then, the intersection of the target genes and the hub genes was obtained by Venn analysis. Next, the key miRNA-mRNA network was constructed (Fig. 3b).
In this network, a total of 14 pairs were shown. Interestingly, miR-203a-3p still had the largest number of target genes in this network.
qRT-PCR validation of candidate miRNA-mRNA pairs in radioresistant and radiosensitive NPC cells
To validate the expression levels of the DEMs and DEGs in radioresistant and radiosensitive NPC cells. Two candidate miRNA-mRNA pairs (miR-203a-3p-BTK, and miR-484-OAL1) were tested in the cultured HONE1-IR and HONE1 cells. The data showed that the expression trends of the candidate miRNA-mRNA pairs were in line with those in the networks (Fig. 4a).
Fig. 4.
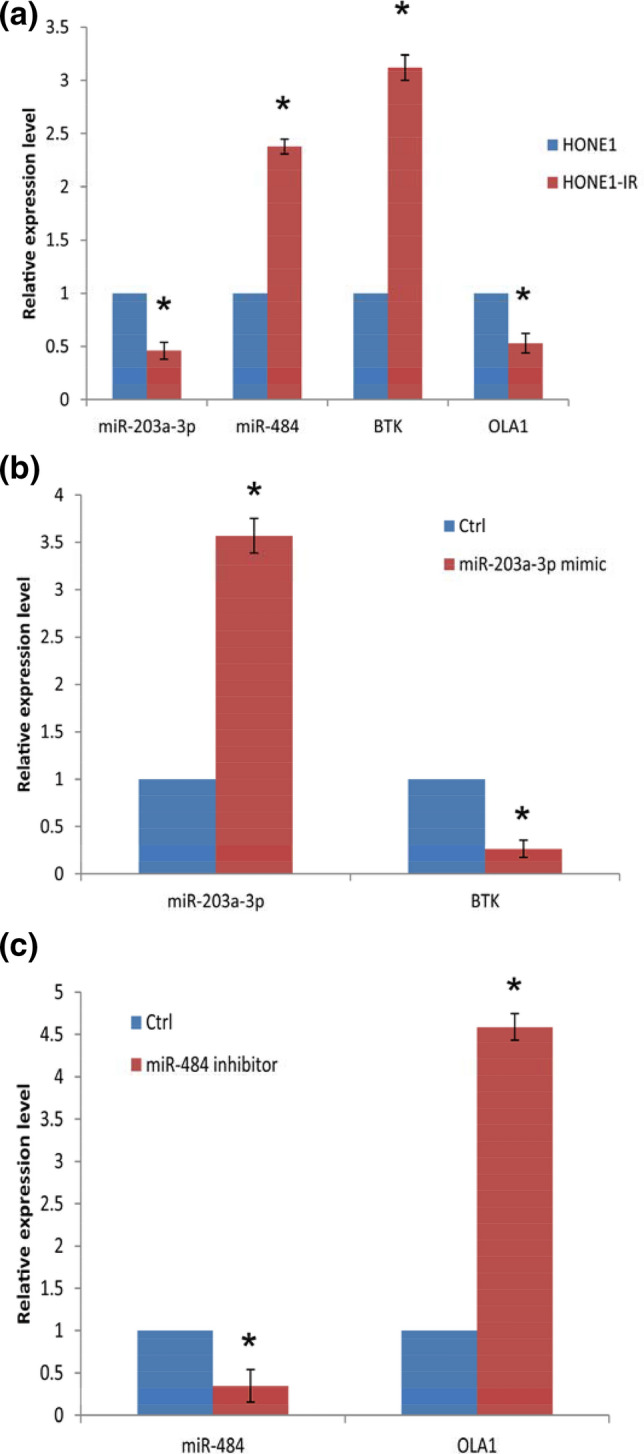
a The expression levels of the candidate miRNAs and mRNAs in HONE1 and HONE1-IR cells, as determined by qRT-PCR (P < 0.05, HONE1-IR vs HONE1 in the four comparisons, respectively). b The expression levels of miR-203a-3p/BTK mRNA in HONE1-IR cells with or without miR-203a-3p mimic administration (qRT-PCR; P < 0.05, miR-203a-3p mimic vs controls). c The expression levels of miR-484/OLA1 mRNA in HONE1-IR cells with or without miR-484 inhibitor administration (qRT-PCR; P < 0.05, miR-484 inhibitor vs controls)
To validate this, further experiments were conducted. HONE1-IR was cultured and miR-203a-3p mimic or miR-484 inhibitor was transfected, respectively. The data showed that transfection of miR-203a-3p mimic significantly elevated the expression of miR-203a-3p, and led to a decrease in BTK expression in HONE1-IR cells (Fig. 4b). Transfection of miR-484 inhibitor markedly downregulated miR-484 expression levels and resulted in an increase in OAL1 expression in HONE1-IR cells (Fig. 4c).
Discussion
In the present study, we screened out several miRNAs and genes that might play a key role in the development of radioresistance in NPC patients, and further constructed a regulatory miRNA-mRNA network by using bioinformatics approaches. As a result, a total of 5 significant DEMs with high fold-changes (> 2) between radioresistant and radiosensitive samples were identified. A total of 195 DEGs were screened out, which were enriched in various signaling pathways, such as Cytokine-cytokine receptor interaction, Jak-STAT signaling pathway, and Toll-like receptor signaling pathway. Then, a number of genes were regarded as the hub genes through PPI analysis. A regulatory miRNA-mRNA network containing 87 pairs were constructed, miR-203a-3p targeted the largest number of genes in this network. Afterwards, 14 key miRNA-mRNA pairs that contained the hub genes were further filtered out. In the key miRNA-mRNA network, miR-203a-3p still had the largest number of target genes.
Previously, two published studies (Guo et al. 2019; Li et al. 2014) have focused on the construction of miRNA-mRNA network regarding NPC radioresistance. However, the authors only used microarray data based on cell lines to screen the DEGs. In the present study, we have chosen a dataset involving NPC tissues with a relatively large sample size. In addition, we also filtered out the key miRNA-mRNA pairs on the basis of the PPI results.
To screen the hub genes from the DEGs, PPI networks were constructed. In the networks, two parameters, namely, ‘Degree’ and ‘Betweenness’, have been accepted to be used to measure the interactions among genes. In the present study, PPI networks were constructed based on these parameters, respectively. In these two network diagrams, one part of the hub genes overlapped, while the other parts did not. Hence, in the following miRNA-mRNA network construction, the hub genes obtained by both algorithms are considered.
When the miRNA-mRNA network was constructed, 87 miRNA-target gene pairs were involved. The data indicated that multiple miRNA-mRNA pairs may participate in the process of radioresistance in NPC cells. However, given that not all pairs have the same weight in the complex regulatory network, we screened out the key miRNA-mRNA pairs that contained the hub genes. As a result, a simple network containing 14 miRNA-mRNA pairs was created. In this network, all the target genes belonged to the hub genes, suggesting that this network may be crucial in the development of NPC radioresistance. It is worthy of note that in the above two miRNA-mRNA networks, the node ‘miR-203a-3p’ and its target genes formed the largest subnetwork. miR-203a-3p was shown to be down-regulated in radioresistant NPC samples, while its target genes were up-regulated in the same samples, indicating that the oncogenes might play important roles in the development of NPC radioresistance.
miR-203a-3p is a miRNA of great concern, which plays diverse roles in different cancers. For example, in hepatocellular carcinoma and colorectal carcinoma, miR-203a-3p acts as an onco-miRNA by targeting IL-24 (Huo et al. 2017) and PDE4D (Chen et al. 2018), respectively. In contrast, miR-203a-3p was down-regulated and may act as a tumor suppressor in NPC (Jiang et al. 2017). In the present study, miR-203a-3p was shown to be down-regulated in radioresistant NPC samples, indicating that elevated expression of miR-203a-3p may increase the radiosensitivity of NPC cells. This speculation is in line with a recent study on gastric cancer, in which miR-203 (miR-203a-3p) promoted the radiosensitivity of gastric cancer cells by targeting ZEB1 (Jiang et al. 2019b). Another down-regulated DEM, miRNA-455-3p, has been shown to act as a tumor suppressor in several cancers. For instance, miR-455-3p acts as a tumor suppressor by targeting hTERT in melanoma cells (Chai et al. 2018). Likewise, miR-455-3p also functions as a tumor suppressor by directly targeting HOXB5 in non-small cell lung cancer progression (Gao et al. 2018). The evidence was in accordance with our results that miR-455-3p was down-regulated in radioresistant NPC cells.
MiR-1202 was downregulated in cervical cancer cells, and it thus might be used as a potential biomarker distinguishing cervical cases from healthy controls (Yang et al. 2019). Likewise, downregulated miR-1202 was also detected in glioma (Quan et al. 2017) and liver cancer cells (Du et al. 2017). Thus, it might act as a tumor suppressor in the development of these cancers. By contrast, miR-1202 was indicated to regulate Th-1/Th-2 status, T-reg cell status, and immune checkpoints, and was also thought to be one of the reliable prognostic indicators (Takashima et al. 2020). Hence, miR-1202 might also play different roles in various cancers. MiR-762 was shown to target MEN1, a tumor suppressor gene, in ovarian carcinoma. Thus, it has been suggested to be a potential treatment target for this cancer (Hou et al. 2017). In breast cancer, miR-762 may promote cancer invasion and metastasis by targeting IRF7 (Li et al. 2015). Moreover, upregulation of miR-762 in lung cancer may not only have a correlation with the development of tyrosine kinase inhibitor resistance but also predict a poor post-chemotherapy prognosis in patients (Ge et al. 2019).
Most of the target genes in the simple miRNA-mRNA network were up-regulated in radioresistant NPC samples, indicating that oncogenes might play a predominant role in this process. For example, the hub gene, BTK (Bruton's tyrosine kinase), is a non-receptor intracellular kinase that belongs to the TEC-family tyrosine kinases, which plays a key role in the intracellular signaling of both B and T lymphocytes and thus contributes to cancer progression. Hence, its inhibitor has been suggested to be a potential agent for cancer therapy (Molina-Cerrillo et al. 2017). Overexpression of BTK in ovarian cancer may have a correlation with cisplatin resistance and predict poor survival (Zucha et al. 2015). PAX5 (Paired-box gene 5) is a B-cell essential transcription factor. Overexpression of PAX5 was detected in cisplatin-resistant bladder cancer cells (Wang et al. 2018). PAX5 promotes tumor growth in prostate cancer by regulating ATG5-mediated autophagy (Zhang et al. 2019). Moreover, PAX acts as a biomarker for L-cell type rectal carcinoids detection (Fu et al. 2019). Hence, the screened hub genes may play critical roles in the development of radioresistance in NPC patients.
Several limitations need to be addressed in the present study. First, in the process of screening the DEMs and DEGs, P values instead of FDRs were used. If we used FDRs as a cut-off, only a few miRNAs/genes met the inclusion criteria. Thus, P values were used in the present study. Second, in the validation process, only one cell line, HONE1, was used. Future studies using more cell lines are needed to confirm the results.
In summary, five miRNAs (miR-203a-3p, miR-455-3p, miR-1202, miR-484 and miR-762) and several hub genes, such as IGF2, OLA1, BBS10, MMP9, and BBS7 were identified as the key miRNAs and hub genes, respectively. A total of 87 miRNA-mRNA pairs (including 14 key pairs) were predicted to play a crucial role in the development of NPC radioresistance. The data provide a basis for further investigating the molecular mechanism of radioresistance in NPC. Future studies using both cell and animal models are needed to validate the results.
Author contributions
XZ and HZ designed the study; AC, JL, and WZ, performed the analysis; XZ, and HZ prepared the manuscript; AC, and HY prepared the figures. All authors have reviewed the paper and agreed with the submission.
Funding
This study was partly supported by the National Natural Science Foundation of China (51963006) and the Special Funding from China Science Foundation for Postdoctors (2015T80962).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Chai L, et al. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res. 2018;10:989–1003. doi: 10.2147/CMAR.S163335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- Chen L, et al. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am J Cancer Res. 2018;8:2387–2401. [PMC free article] [PubMed] [Google Scholar]
- Chu C, Niu X, Ou X, Hu C. LAPTM4B knockdown increases the radiosensitivity of EGFR-overexpressing radioresistant nasopharyngeal cancer cells by inhibiting autophagy. Onco Targets Ther. 2019;12:5661–5677. doi: 10.2147/OTT.S207810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragomir M, Mafra ACP, Dias SMG, Vasilescu C, Calin GA. Using microRNA networks to understand cancer. Int J Mol Sci. 2018 doi: 10.3390/ijms19071871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du B, Zhang P, Tan Z, Xu J. MiR-1202 suppresses hepatocellular carcinoma cells migration and invasion by targeting cyclin dependent kinase 14. Biomed Pharmacother. 2017;96:1246–1252. doi: 10.1016/j.biopha.2017.11.090. [DOI] [PubMed] [Google Scholar]
- Fu Z, Zuo C, Sheehan CE, Patil DT, Lin J, Yang Z, Lee H. Novel finding of paired box 5 (PAX5) cytoplasmic staining in well-differentiated rectal neuroendocrine tumors (Carcinoids) and its diagnostic and potentially prognostic utility. Appl Immunohistochem Mol Morphol. 2019;27:454–460. doi: 10.1097/PAI.0000000000000635. [DOI] [PubMed] [Google Scholar]
- Gao X, et al. miR-455-3p serves as prognostic factor and regulates the proliferation and migration of non-small cell lung cancer through targeting HOXB5. Biochem Biophys Res Commun. 2018;495:1074–1080. doi: 10.1016/j.bbrc.2017.11.123. [DOI] [PubMed] [Google Scholar]
- Ge P, Cao L, Chen X, Jing R, Yue W. miR-762 activation confers acquired resistance to gefitinib in non-small cell lung cancer. BMC cancer. 2019;19:1203. doi: 10.1186/s12885-019-6416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioacchini FM, Tulli M, Kaleci S, Magliulo G, Re M. Prognostic aspects in the treatment of juvenile nasopharyngeal carcinoma: a systematic review. Eur Arch Otorhinolaryngol. 2017;274:1205–1214. doi: 10.1007/s00405-016-4154-7. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zhang Y, Zhang SJ, Ma YN, He Y. Comprehensive analysis of key genes and microRNAs in radioresistant nasopharyngeal carcinoma. BMC Med Genomics. 2019;12:73. doi: 10.1186/s12920-019-0507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou R, Yang Z, Wang S, Chu D, Liu Q, Liu J, Jiang L. miR-762 can negatively regulate menin in ovarian cancer. OncoTargets Ther. 2017;10:2127–2137. doi: 10.2147/OTT.S127872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo W, Du M, Pan X, Zhu X, Gao Y, Li Z. miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma cell growth and metastasis. FEBS Open Bio. 2017;7:1085–1091. doi: 10.1002/2211-5463.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, et al. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2017;36:138. doi: 10.1186/s13046-017-0604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Dai B, Feng L. miR-543 promoted the cell proliferation and invasion of nasopharyngeal carcinoma by targeting the JAM-A. Hum Cell. 2019;32:477–486. doi: 10.1007/s13577-019-00274-0. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Jin S, Tan S, Shen Q, Xue Y. MiR-203 acts as a radiosensitizer of gastric cancer cells by directly targeting ZEB1. Onco Targets Ther. 2019;12:6093–6104. doi: 10.2147/OTT.S197539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018 doi: 10.1002/1873-3468.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XH, et al. Integrated analysis of differential miRNA and mRNA expression profiles in human radioresistant and radiosensitive nasopharyngeal carcinoma cells. PLoS ONE. 2014;9:e87767. doi: 10.1371/journal.pone.0087767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang R, Wang L, Hao J, Zhang Q, Ling R, Yun J. microRNA-762 promotes breast cancer cell proliferation and invasion by targeting IRF7 expression. Cell Prolif. 2015;48:643–649. doi: 10.1111/cpr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, et al. FOXO3a knockdown promotes radioresistance in nasopharyngeal carcinoma by inducing epithelial-mesenchymal transition and the Wnt/beta-catenin signaling pathway. Cancer Lett. 2019;455:26–35. doi: 10.1016/j.canlet.2019.04.019. [DOI] [PubMed] [Google Scholar]
- Molina-Cerrillo J, Alonso-Gordoa T, Gajate P, Grande E. Bruton’s tyrosine kinase (BTK) as a promising target in solid tumors. Cancer Treat Rev. 2017;58:41–50. doi: 10.1016/j.ctrv.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Nie X, et al. SALL4 induces radioresistance in nasopharyngeal carcinoma via the ATM/Chk2/p53 pathway. Cancer Med. 2019;8:1779–1792. doi: 10.1002/cam4.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri F, et al. Management of recurrent nasopharyngeal carcinoma: current perspectives. OncoTargetsTher. 2019;12:1583–1591. doi: 10.2147/ott.S188148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Y, Song Q, Wang J, Zhao L, Lv J, Gong S. MiR-1202 functions as a tumor suppressor in glioma cells by targeting Rab1A. Tumour Biol. 2017;39:1010428317697565. doi: 10.1177/1010428317697565. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, et al. miR-101, miR-548b, miR-554, and miR-1202 are reliable prognosis predictors of the miRNAs associated with cancer immunity in primary central nervous system lymphoma. PLoS ONE. 2020;15:e0229577. doi: 10.1371/journal.pone.0229577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokar T, et al. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46:D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Dong XM, Zhang FL, Zhang JR. miR-206 enhances nasopharyngeal carcinoma radiosensitivity by targeting IGF1. Kaohsiung J Med Sci. 2017;33:427–432. doi: 10.1016/j.kjms.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Wang G, Dong J, Deng L. Overview of cantharidin and its analogues. Curr Med Chem. 2018;25:2034–2044. doi: 10.2174/0929867324666170414165253. [DOI] [PubMed] [Google Scholar]
- Wu S, Xie DL, Dai XY. Down-regulation of miR-155 promotes apoptosis of nasopharyngeal carcinoma CNE-1 cells by targeting PI3K/AKT-FOXO3a signaling. Eur Rev Med Pharmacol Sci. 2019;23:7391–7398. doi: 10.26355/eurrev_201909_18847. [DOI] [PubMed] [Google Scholar]
- Yang S, et al. Identification of prognostic biomarkers for response to radiotherapy by DNA microarray in nasopharyngeal carcinoma patients. Int J Oncol. 2012;40:1590–1600. doi: 10.3892/ijo.2012.1341. [DOI] [PubMed] [Google Scholar]
- Yang X, Yan Z, Yang H, Ni H, Zhang L, Wang Y. Clinical value of combined detection of miR-1202 and miR-195 in early diagnosis of cervical cancer. Oncology Lett. 2019;17:3387–3391. doi: 10.3892/ol.2019.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Li Z, Bai F, Zhang S. PAX5-induced upregulation of IDH1-AS1 promotes tumor growth in prostate cancer by regulating ATG5-mediated autophagy. Cell Death Dis. 2019;10:734. doi: 10.1038/s41419-019-1932-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. High expression of Ki-67 acts a poor prognosis indicator in locally advanced nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2017;494:390–396. doi: 10.1016/j.bbrc.2017.09.118. [DOI] [PubMed] [Google Scholar]
- Zhuo X, Chang A, Huang C, Yang L, Zhao H, Wu Y, Zhou Q. Nanoparticle-mediated down-regulation of TWIST increases radiosensitivity of nasopharyngeal carcinoma cells via ERK pathway. Am J Cancer Res. 2015;5:1571–1579. [PMC free article] [PubMed] [Google Scholar]
- Zucha MA, Wu AT, Lee WH, Wang LS, Lin WW, Yuan CC, Yeh CT. Bruton’s tyrosine kinase (Btk) inhibitor ibrutinib suppresses stem-like traits in ovarian cancer. Oncotarget. 2015;6:13255–13268. doi: 10.18632/oncotarget.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]