Abstract
Background
Trichostrongylus is one of the most important zoonotic trichostrongylid nematodes, infecting mostly livestock. Data on its genetic characteristics are lacking in Iran.
Methods
We determined the phylogenetic relationships of Trichostrongylus species in three counties of Kohgiloyeh and Boyerahmad (K-B) province, southwest Iran. Small intestine and abomasum of 70 sheep and goats were investigated.
Results
A total of 35 isolates of Trichostrongylus worms were detected and all were genetically identified as Trichostrongylus vitrinus. Analysis of 321 bp of the internal transcribed spacer 2 (ITS2) of ribosomal DNA revealed 16 genotypes. All genotypes were single nucleotide polymorphisms, including some hypervariable points. All sequences were trimmed to 170 bp, compared with sequences on GenBank including short sequences from other endemic foci of Iran and other countries and all isolates were used to generate a maximum likelihood phylogenetic tree, which consisted of two clades A and B. Clade A included isolates from Iran, Russia, New Zealand, Australia and the UK; clade B only contained South African isolates. Most clade A isolates (north, southwest and west Iran, Russia, New Zealand, Australia and UK) were in a similar phylogenetic position. One subclade was detected in clade A (isolates from Southwest Iran, New Zealand and UK).
Conclusions
We hypothesize that drug resistant T. vitrinus may account for its exclusive detection in our samples. The high similarity of genotypes from Iran, New Zealand and UK may be due to their close political relationships during the colonial era. More research is needed to understand better the phylogeny of T. vitrinus and its relationship with drug resistance and human transmission. 
Keywords: Trichostrongylus vitrinus, Phylogenetic analysis, ITS2-rDNA
Background
Parasitic infections affect populations all over the world, especially in developing countries. Among the gastrointestinal nematodes, Trichostrongylus (order Strongylida, family Trichostrongylidae), has a substantial economic, medical and veterinary impact. Trichostrongylus spp. comprise more than ten principal zoonotic species, that affect human and livestock such as goats, sheep and cattle and cause gastroenteritis, diarrhea, weight loss and losses of production in livestock [1] and a spectrum of manifestations from subclinical symptoms to rash, abdominal pain, diarrhea, mild anemia, leukocytosis and eosinophilia in humans [2]. Moreover, anthelmintic resistance of nematodes in this genus is common to drugs like pyrantel pamoate, levamizole and albendazole and results in treatment failure and increased livestock morbidity [3, 4].
Trichostrongylosis in livestock has been reported from diverse geographical settings, including Southeast Asia [5], the Middle East [6], Africa [7], Europe [8, 9] and Australian continent [4]. Similarly, human infection has been reported from Italy [10], France [11], Morocco and Egypt [12, 13], Brazil [14], the Caribbean [15] and Australia [16]. Iran is one of the main foci of livestock and human infections and trichostrongylosis has been reported frequently [17, 18].
The life-cycle of Trichostrongylus spp. is direct, simple and without an intermediate host. Host infection occurs upon ingestion of filariform larvae found in contaminated vegetables or water. These larvae migrate to the small intestine or abomasum, grow to adults which mate and produce eggs that are excreted in the feces into the external environment. Under optimal temperature and humidity, rhabditiform larvae hatch from eggs within a few days and, after two moltings (L1 and L2) over 5 to 10 days, become infective filariform larvae (third-stage) [3, 19].
Clinical diagnosis of infection is based on the finding of eggs in the feces of human and herbivores but the majority of eggs of species of the family Trichostrongylidae, except for the genus Nematodirus, are indistinguishable from each other. Moreover, Trichostrongylus spp. eggs may be confused with hookworm eggs. Although traditional morphological and morphometric methods can identify the adult male worms from different Trichostrongylus species, they are time-consuming and laborious and cannot be used for the species identification of ova and female worms [20].
With the advent of molecular techniques in recent years, many problems in identification have been resolved. Several DNA based techniques can discriminate different species of Trichostrongylus, including all life-cycle stages and sexes, e.g. PCR-restriction fragment length polymorphism (PCR-RFLP) [21], randomized amplified polymorphism DNA (RAPD) [22], PCR-single-strand conformational polymorphism (PCR-SSCP) [23], PCR-denaturing gradient gel electrophoresis (PCR-DGGE) [24], multiplex-PCR [25], quantitative PCR [26] and sequence analysis [27].
RAPD-PCR, PCR-SSCP and PCR-sequence analysis have been used to study the phylogenetic relationships and population structure of strongylid nematodes [28] and gene sequencing has led to the development of genetic databases for inter- and intraspecific identification [29].
The internal transcribed spacer 2 region of the ribosomal RNA gene (ITS2-rDNA) is a useful marker to detect interspecific [30–32] and intraspecific differences [32–34] and establish the phylogenetic relationships within the family Trichostrongylidae.
There are few data on the genetics of Trichostrongylus species in Iran. We applied sequence analysis of ITS2 to conduct a study with the aim of investigation of phylogenetic relationships of Trichostrongylus isolates in southwest Iran.
Methods
Study area
This study was conducted in the three main counties of Kohgiloyeh and Boyerahmad (K-B) Province: Boyerahmad, Gachsaran and Kohgiloyeh. This southwestern province is mostly mountainous with 20% of the region comprising plains. The highest peak and the plains are 4283 and 115 m above sea level, respectively. The difference in elevation in the northeast (Boyerahmad county) and the south-southwest (Gachsaran and Kohgiloyeh counties) results in two different types of weather patterns; Boyerahmad county is cold and wet and covered with large oak forests while Gachsaran and Kohgiloyeh are warm and semi-arid regions.
Although most of people live in rural and urban areas in K-B, this province also has a significant nomadic population. Every year, the nomads herd their sheep and goats to and from their summer (Yailaq) and winter quarters (Qishlag). Data from the Veterinary Bureau consensus report that the number of sheep, goats and cattle in K-B province is 1,543,300.
Sample collection
Samples were obtained from a total of 70 slaughtered sheep and goats from the three counties. Their small intestines and abomasa were separated from the carcasses in the slaughterhouses, separately packed in plastic bags, and sent to the Parasitology laboratory of Yasuj University of Medical Science. Mucosa were scraped and the contents of the small intestines and abomasa were washed separately onto a 100-mesh sieve (aperture size 0.149 mm). Washed contents and mucosal scrapings were examined under a stereo microscope to facilitate the recovery of worms. Samples were stored in 70% ethanol until molecular analysis.
The identification of Trichostrongylus spp. was carried out based on morphological characteristics. Each adult male worm was identified by its morphological and morphometric characteristics and then subjected to sequence analysis of the ITS2 region for further identification [3].
DNA extraction
300 μl of lysis buffer (NaCl 0.1M, EDTA 0.01M, Tris-HCL 0.1M, SDS 1%) was added to the microtube containing the Trichostrongylus worm and DNA extraction performed in two steps. Step one used the freezing-thawing technique to disrupt the worm tegument and cells. Samples were frozen in liquid nitrogen (1–2 min) and thawed at room temperature with extra crushing using a steel bullet that was added to each microtube (20–30 s). In the second step, 30 µg/ml of proteinase K was added and the samples were incubated at 56 °C for 1 h; then the DNA was extracted once with phenol/chloroform (25:24 v/v) and again with chloroform. DNA was precipitated with equal volumes of isopropanol and also one tenth volume of 3 M NaAc. The DNA precipitate was washed with 70% ethanol, dried, and dissolved in 50 µl deionized water, and stored at – 20 °C.
ITS2 PCR and sequencing
Primers NC1 (5'-ACG TCT GGT TCA GGG TTG TT-3') and NC2 (5'-TTA GTT TCT TTT CCT CCG CT-3') [35] were used to amplify the ITS2 region of all samples and confirm the genus Trichostrongylus. The PCR reactions were performed in a final volume of 25 μl containing 12.5 μl of 2× premix (Ampliqon, Skovlunde, Denmark), 20 pmol of each primer, 5 μl of template DNA, and enough water made up to 25 μl. Optimal conditions for PCR were initial denaturation at 95 °C (7 min) followed by 35 cycles of 95 °C (45 s), 56 °C (45 s), and 72 °C (50 s), and a final extension step at 72 °C (5 min). The PCR products were visualized after electrophoresis on a 1.5% agarose gel with 0.5 μg/ml ethidium bromide. A 100 bp DNA marker was used for sizing the bands in each run.
The PCR products were also sent to Bioneer Company (Daejeon, South Korea) and sequenced using the Applied Biosystems automated DNA sequencer (3730 XL; Applied Biosystems, Foster City, USA). Sequencing was performed in both directions using the same PCR primers. The sequences were deposited in the GenBank database under accession numbers MK271662-MK271677.
Phylogenetic analysis
BLAST software (https://www.ncbi.nlm.nih.gov) was used to compare the newly generated sequences with published ITS2 sequences in GenBank to identify and reconfirm species identification based on morphological and morphometric data. The sequences were analyzed using Geneious Pro 5.5.6 software. Two phylogenetic trees were generated using: (i) all our isolates; and (ii) including our genotypes (STs) and all other T. vitrinus ITS2 sequences from other regions of Iran and the other countries that were deposited in GenBank (Fig. 1). Alignment was performed using ClustalW and nucleotide distances were calculated by the BioEdit software (version 7.0.5.3) [36]. Maximum likelihood trees were inferred by using MEGA 6 software [37] after trimming all sequences at both ends. Bootstrap values for the ML method were based on 1000 replicates.
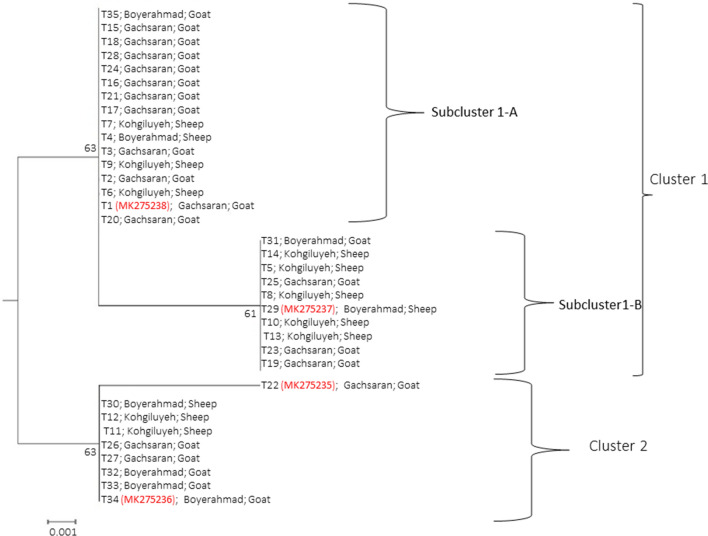
Fig. 1.
Maps showing Kohgiloyeh and Boyerahmad Province counties (a) and the Iranian provinces (b) from which T. vitrinus isolates were used for generating the phylogenetic trees. The geographical origins of T. vitrinus are shown in dark blue
Results
Overall, 35 Trichostrongylus isolates were obtained from more than 2000 nematodes in the abomasum and small intestines: (i) 8 from Boyerahmad county (sample ID: T4 and T29 to T35); (ii) 10 from Kohgiloyeh county (T5 to T14); and (iii) 17 from Gachsaran county (T1 to T3 and T15 to T28) (Table 1). Trichostrongylus vitrinus was the only species identified based on the morphological and morphometric characteristics. Amplification of the ITS2 fragment resulted in bands of about 321 bp for all 35 isolates and identified 16 different genotypes. The highest number of isolates belonged to genotypes 1 (n = 7), 6 (n = 4), 7 (n = 4), 5 (n = 3) and 10 (n = 3). Genotypes 1, 2 and 6 were found in all three counties, while genotypes 7 and 10 were observed only in Kohgiluyeh/Gachsaran and Boyerahmad/Gachsaran, respectively. Genotypes 3 and 5 were each seen in two isolates and genotypes 9 to 16 each contained only one isolate (Table 2).
Table 1.
The sex, host type, isolation site and geographical origin of T. vitrinus isolates in this study
| Sample ID | Sex | Host | Isolation site | Geographical origin (county) |
|---|---|---|---|---|
| T1 | Male | Goat | Small intestine | Gachsaran |
| T2 | Female | Goat | Small intestine | Gachsaran |
| T3 | Male | Goat | Small intestine | Gachsaran |
| T4 | Female | Sheep | Abomasum | Boyerahmad |
| T5 | Male | Sheep | Abomasum | Kohgiluyeh |
| T6 | Male | Sheep | Abomasum | Kohgiluyeh |
| T7 | Male | Sheep | Abomasum | Kohgiluyeh |
| T8 | Male | Sheep | Abomasum | Kohgiluyeh |
| T9 | Male | Sheep | Abomasum | Kohgiluyeh |
| T10 | Male | Sheep | Abomasum | Kohgiluyeh |
| T11 | Male | Sheep | Abomasum | Kohgiluyeh |
| T12 | Male | Sheep | Abomasum | Kohgiluyeh |
| T13 | Male | Sheep | Small intestine | Kohgiluyeh |
| T14 | Male | Sheep | Abomasum | Kohgiluyeh |
| T15 | Female | Goat | Small intestine | Gachsaran |
| T16 | Male | Goat | Small intestine | Gachsaran |
| T17 | Male | Goat | Small intestine | Gachsaran |
| T18 | Male | Goat | Small intestine | Gachsaran |
| T19 | Male | Goat | Small intestine | Gachsaran |
| T20 | Male | Goat | Small intestine | Gachsaran |
| T21 | Male | Goat | Small intestine | Gachsaran |
| T22 | Male | Goat | Small intestine | Gachsaran |
| T23 | Male | Goat | Small intestine | Gachsaran |
| T24 | Male | Goat | Small intestine | Gachsaran |
| T25 | Male | Goat | Small intestine | Gachsaran |
| T26 | Male | Goat | Small intestine | Gachsaran |
| T27 | Male | Goat | Small intestine | Gachsaran |
| T28 | Male | Goat | Small intestine | Gachsaran |
| T29 | Male | Sheep | Abomasum | Boyerahmad |
| T30 | Male | Sheep | Abomasum | Boyerahmad |
| T31 | Male | Goat | Abomasum | Boyerahmad |
| T32 | Male | Goat | Abomasum | Boyerahmad |
| T33 | Male | Goat | Abomasum | Boyerahmad |
| T34 | Male | Goat | Abomasum | Boyerahmad |
| T35 | Male | Goat | Abomasum | Boyerahmad |
Table 2.
Single nucleotide polymorphism positions of 16 genotypes and 4 haplotypes among long (321 bp) and short (170 bp) ITS2 sequences, respectively, from T. vitrinus isolates in southwest Iran
| Haplotype (170 bp) | SNPs | Sample ID | SNPs | Genotype and GenBank ID (321 bp) |
|---|---|---|---|---|
| Haplotype A | 85T > C | T1; T2; T3; T4; T17; T18; T28 | 147T > C | Genotype 1 (MK271667) |
| T9; T24; T35 | 37C > A; 147T > C | Genotype 2 (MK271664) | ||
| T6; T16 | 62C > T; 147T > C | Genotype 3 (MK271663) | ||
| T7; T21 | 37C > A; 62C > T; 147T > C | Genotype 4 (MK271666) | ||
| T15; T20 | 51T > C; 147T > C | Genotype 5 (MK271674) | ||
| Haplotype B | T8; T25; T29; T31 | Genotype 6 (MK271662) | ||
| T10; T14; T19; T23 | 51T > C | Genotype 7 (MK271665) | ||
| T13 | 47_48insT; 51T > C | Genotype 8 (MK271673) | ||
| T5 | 37C > A | Genotype 9 (MK271669) | ||
| Haplotype C | 34T > A; 85T > C | T26; T33; T34 | 62C > T; 96T > A; 147T > C | Genotype 10 (MK271675) |
| T11 | 47_48insT; 51T > C; 96T > A; 147T > C | Genotype 11 (MK271672) | ||
| T27 | 51T > C; 96T > A; 147T > C | Genotype 12 (MK271676) | ||
| T32 | 96T > A; 147T > C | Genotype 13 (MK271677) | ||
| T12 | 37C > A; 96T > A; 147T > C; 246T > C | Genotype 14 (MK271670) | ||
| T30 | 37C > A; 62C > T; 96T > A; 147T > C | Genotype 15 (MK271668) | ||
| Haplotype D | 34T > A; 85T > C; 147A > G | T22 | 37C > A; 62C > T; 96T > A; 147T > C; 209A > G; 246T > C | Genotype 16 (MK271671) |
Most of the nucleotide polymorphisms were observed in nucleotides that were close to the 5'-end of the sequences and all were single nucleotide polymorphisms (SNP). T to C transition at nucleotide 147 was the most frequent SNP observed in all genotypes except genotypes 6–9. Additional transitions found were T to C at nucleotides 51 (genotypes 5, 7, 8, 11–12) and 246 (genotypes 14 and 16), C to T at nucleotides 62 (genotypes 3–4, 10, 15–16), A to G at nucleotide 209 (genotype 16). T to A transversion at nucleotide 96 (genotypes 10–16) and C to A transversion at nucleotide 37 (genotypes 2, 4, 9 and 14–16) were also observed as well as insertion of A at nucleotide 48 (genotypes 8 and 11) (Table 2).
Because there was a number of short STs in the GenBank, all STs from our study, other parts of Iran and other countries were trimmed and shortened after alignment and eventually a 170 nucleotide fragment of the sequences was obtained for the phylogenetic analysis (Fig. 2). Based on the polymorphisms in this short fragment, only 4 haplotypes were defined for our K-B isolates (named A to D). Haplotypes A, B, and C were observed in Boyerahmad, Gachsaran and Kohgiloyeh but Haplotype D was reported only from Gachsaran. Haplotypes B and D had the greatest genetic distance (0.018%) (Tables 2, 3).
Fig. 2.
Maximum likelihood phylogenetic tree based on the Tamura-Nei model constructed from ITS2 sequences of T. vitrinus from Kohgiloyeh and Boyerahmad Province in southwest Iran. Geographical origin (county) and host for each isolate is shown. Representative sequences generated in the present study are indicated in red
Table 3.
Genetic distances as calculated by BioEdit (7.05.8) between haplotypes A, B, C and D of isolates in this study
| Haplotype | A | B | C | D |
|---|---|---|---|---|
| A | ||||
| B | 0.006 | |||
| C | 0.006 | 0.012 | ||
| D | 0.012 | 0.018 | 0.006 |
The constructed phylogenetic tree of our 35 T. vitrinus isolates (Fig. 2) revealed two main clades, 1 and 2. Clade 1 contains two subclades 1-A and 1-B. Subclade 1-A consisted of all isolates of haplotype A, including: 11 isolates from Gachsaran; 3 from Kohgiluyeh; and 2 from Boyerahmad. Subclade 1-B included 4 isolates from Gachsaran, 5 from Kohgiluyeh and 1 isolate from Boyerahmad; all from haplotype B. Clade 2 consisted of 9 isolates from haplotypes C and D. Eight isolates (4 from Boyerahmad, 2 each from Gachsaran and Kohgiluyeh) showed haplotype C. The haplotype D isolate from Gachsaran was in a more distant phylogenetic position. Both main clades included isolates from both hosts and both small intestine and abomasum. The second phylogenetic tree consisted of representatives of haplotypes of K-B province (A to D haplotypes) and all reliable ITS2 STs from other regions of Iran and other countries that retrieved from GenBank. Haplotypes distribution by region is shown is Table 4. Based on the tree topology, there were two clades, A and B (Fig. 3). Clade A showed little resolution and contained more genotypes from a broader geographical reach encompassing north (KY355024, KY355027 and KF872228), northwest (KJ755061 and KF880745) west (JF276024, JF276025 and JF276026) and K-B province in southwest (MK271662, MK271667, MK271671 and MK271675) Iran, the Ryazan region from Russia (KR020010, KR020011 and KR020012), the UK (AY439027 and JF680986), Australia (X78064) and New Zealand (NZ) (KC998731, KC998732 and KC998733), while Clade B isolates were all from one national park in South Africa (KP688062 and KP688063). Most clade A genotypes had similar phylogenetic positions but some NZ (KC998732 and KC998733) and the UK (JF680986) isolates and haplotypes C and D from Kohgiloyeh and Boyerahmad Province (MK271671 and MK271675) consisted one subclade in this clade, though it was not well supported by bootstrap analysis. Also, one isolate from west Iran (JF276026) was placed in the main part of clade A.
Table 4.
Trichostrongylus vitrinus ITS2 genotypes (STs) from the present study, other parts of Iran and other countries retrieved from GenBank used to generate the phylogenetic tree
| Host | Location | GenBank ID | Reference |
|---|---|---|---|
| Soay sheep | UK (Scotland) | AY439027 | Wimmer et al. [49] |
| Sheep | New Zealand | KC998731-KC998733 | Bisset et al. [48] |
| African buffalo | South Africa: Kruger National Park | KP688062; KP688063 | Budischak et al. [52] |
| Sheep | Australia: Victoria | X78064 | Hoste et al. [47] |
| Sheep | UK | JF680986 | Unpublished |
| Roe deer | Russia: Ryazan region | KR020010-KR020012 | Unpublished |
| Goat; cattle | Iran: Khuzestan Province | JF276024-JF276026 | Ghasemikhah et al. [27] |
| Sheep | Iran: Ardebil Province, Meshkin Shahr district | KJ755061 | Unpublished |
| Sheep | Iran: Ardebil Province, Meshkin Shahr district | KF880745 | Unpublished |
| Human | Iran: Guilan Province, Langroud district | KF872228 | Unpublished |
| Human | Iran: Guilan Province, Fouman district | KY355024; KY355027 | Sharifdini et al. [32] |
| Goat; sheep | Iran: Kohgiluyeh and Boyerahmad Province | MK275235-MK275238 | Present study |
Fig. 3.
Maximum likelihood phylogenetic tree of T. vitrinus from Iran and other countries
Discussion
In the present study, the only Trichostrongylus species isolated from different parts of one of the southwestern Iranian provinces was T. vitrinus. Sixteen ITS2 genotypes were found among the studied isolates, all of the nucleotide changes were SNPs and most in the 5'-end of the fragment. Comparison of the present isolates with those from other parts of Iran and other countries revealed two main clades. Isolates from Iran, the UK, New Zealand, Australia and Russia were distributed in one main clade and South African samples comprised another clade.
Previous studies in Iran has detected other Trichostrongylus spp. in livestock. In Isfahan Province (central Iran), T. axei, T. vitrinus and T. colubriformis were isolated from sheep [38] and T. vitrinus, T. capricola, T. probolorus, T. skrjabini, T. axei and T. colubriformis from sheep and goats [39]. In Khuzestan Province (West Iran), T. vitrinus, T. colubriformis [40] and in Golestan Province in North Iran, T. axei were isolated from goats and sheep [41].
Human infections with several Trichostrongylus spp. have also been reported in Iran, including T. lerouxi from northern, central, and southwestern regions [42], T. orientalis, T. colubriformis, and T. axei, T. vitrinus, T. capricola, T. probolurus and T. skrjabini in Isfahan (central), T. orientalis, T. colubriformis, T. axei, and T. vitrines in Khuzestan Province (West) and also along the Caspian Sea in North Iran [43]. Recent studies from northern provinces showed T. colubriformis, T. vitrinus, T. longispicularis and T. axei in Guilan Province [32] and T. colubriformis, and T. axei in Mazandaran Province [44]. Also, in East Azerbaijan in northwest Iran, T. probolurus, T. colubriformis and T. vitrinus, have been reported.
Trichostrongylus vitrinus has broad distribution within Iran and is the dominant species isolated from 95% of sheep breeding farms in England, Scotland, Wales and Northern Ireland [8]. The widespread distribution of T. vitrinus may be related to its ability to withstand a wide range of environmental conditions; moreover, Blackburn et al. [45] showed that T. vitrinus larvae can grow in temperatures < 8 °C and this is one distinguishing feature compared to other Trichostrongylus species.
Although, we sampled in three parts of Kohgiloyeh and Boyerahmad Province, finding only T. vitrinus in our study remains unclear. This contrasts with multiple species found in other Iranian studies with samples usually obtained from a single region or slaughterhouse in a province. Amongst the myriad environmental- (temperature, humidity, microclimate, season and type of vegetation), host (age, host movement, anthelmintic treatment, immunity and genetic resistance), and parasite-related factors (larval survival) [3], drug resistance is a possible explanation. There is only one Iranian, drug resistance study from Khuzestan Province the neighboring region of Kohgiloyeh and Boyerahmad Province and this showed a higher levamisole drug resistance of T. vitrinus compared to other Trichostrongylus species in sheep flocks [40]. A recent study in the UK also showed T. vitrinus as the sole Trichostrongylus species with monepantel resistance in a sheep/cattle farm [46]. Another study revealed higher anti-helminthic resistance and higher prevalence of T. vitrinus in treated livestock in comparison to other species including T. colibriformis and T. axei in Scotland [9]. These results contrast to the findings in New Zealand that demonstrated greater resistance to albendazole/levamizole in T. colubriformis compared to T. vitrinus and T. axei [4]. Veterinary authorities in Kohgiloyeh and Boyerahmad Province report an increasing use of anti-parasitic drugs by nomadic, rural and industrial herders (M. Sadghi, personal communication) that may reinforce the hypothesis of drug resistance in T. vitrinus isolates in studied areas.
Our study included a relatively high number of T. vitrinus isolates in comparison to other studies, resulting to detecting several haplotypes in a number of different phylogenetic positions; 16 (1 to 16) and 4 (A to D) haplotypes were found in the long and short size fragments of ITS2 sequences of T. vitrinus, respectively. The long nucleotide fragment of the ITS2 gene was associated with eight nucleotide changes between haplotypes and five of eight polymorphic points (37, 51, 62, 96 and 147 nucleotides) were observed more frequently in several haplotypes. The DNA chromatograph showed > 1 peak (concomitant existence of ≥ two nucleotides) at these points in some isolates in the forward and reverse sequences, although the nucleotide chromatograph was dominant for one nucleotide in each position. The alignment of our sequences with those from GenBank showed that these nucleotide positions were also highly polymorphic in a number of isolates. These highly variable points seem not to be the same in the different ITS copies even in one isolate. These findings are consistent with those of Hoste et al. [47] who reported these SNPs in the ITS2 copies of T. vitrinus, which account for intra-individual variation.
The topology of phylogenetic tree of all isolates showed two distinct clades, A and B, with clade A containing more haplotypes and countries than clade B (South Africa only). All north-northwest and some southwest (haplotypes A and B) and western isolates of Iran as well as isolates from Ryazan region of Russia, Australia [47], New Zealand [48] and the UK [49] had identical phylogenetic position in clade A. Haplotypes C and D from southwest Iran and two haplotypes from New Zealand [48] and one from the UK formed a subclade within clade A. Similarly, Ghasemikhah et al. [27] showed by Bayesian inference that T. vitrinus from Khuzestan Province (JF2760224) was in a sub cluster with T. vitrinus from the UK (AY439027). The link between the UK, Australia, New Zealand and Iran could be the export of livestock from the UK during colonial times. In the 19th and 20th centuries, British forces were stationed in some parts of Iran and the British Oil Company, established in 1909, was headquartered in Khuzestan [50]. Moreover, distribution of haplotypes of Kohgiloyeh and Boyerahmad Province in two positions in the clade A is consistent with higher genetic flow due to the movement of livestock, notably sheep and goats, by nomadic tribes who travel to and from cold to warmer regions during different seasons of the year [51] and exposure to strains of Trichostrongylus from various foci in a confined time interval. The similarity of isolates from Russian and the northern provinces of Iran is due to close geographic proximity and presence of Russian troops before and after both World Wars in North Iran.
Conclusions
Trichostrongylus vitrinus was the only species isolated from sheep and goat in Kohgiloyeh and Boyerahmad Province southwest Iran where higher drug resistance may contribute in the dominance of this species. The similarity of Iranian, UK, New Zealand and Australian isolates may be explained by colonial activity of the UK and notable relationship with these countries in previous centuries. High genetic variations of southwest Iran may be due to annual movement of livestock by nomad’s activities and exposure of livestock with different strains of T. vitrinus in different regions and higher genetic flow between these isolates. To our knowledge, this is the first comprehensive study to evaluate the intraspecific population structure of T. vitrinus isolates from different foci of Iran and other countries. More research is needed to characterize further the molecular epidemiology of Trichostrongylus in Iran and examine important factors like treatment practice and the possible role of drug pressure and drug resistance on the predominance of certain isolates in different regions. Moreover, understanding better the transmission between livestock and man could lead to better control strategies.
Acknowledgments
The authors appreciate Mr Mehrdad Arabi for his help in sampling.
Abbreviations
- ITS2
Internal transcribed spacer 2
- ITS2-rDNA
Internal transcribed spacer 2 of ribosomal DNA
- PCR-RFLP
PCR-restriction fragment length polymorphism
- RAPD
Randomized amplified polymorphism DNA
- PCR-SSCP
PCR-single-strand conformational polymorphism
- PCR-DGGE
PCR-denaturing gradient gel electrophoresis
- K-B
Kohgiloyeh and Boyerahmad
- SNPs
Single nucleotide polymorphisms
- STs
Genotypes
Authors’ contributions
MAG conceived and designed the study. SAAM, MM, HM and AJ collected the data. MAG and IM analysed and interpreted the data. MAG, WRT, SAAM, MM and HM wrote the first draft of the manuscript. MAG, MK and WRT reviewed and edited the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by the award grant for youth assistant professors of Iran Ministry of Health and Medical Education (MOHME) and the Vice- Chancellor of Yasuj University of Medical Sciences (Project no. P-23-2-263).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Yasuj University of Medical Sciences (93.12.25.11).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Amin Ghatee, Email: ghateea1980@gmail.com.
Seyed Ali Asghar Malek Hosseini, Email: malekhosseini91@gmail.com.
Masoud Marashifard, Email: masoud.marashifard@gmail.com.
Mehdi Karamian, Email: karamianm@yahoo.com.
Walter Robert Taylor, Email: bob@tropmedres.ac.
Ali Jamshidi, Email: dani1364@gmail.com.
Iraj Mobedi, Email: mobedii@yahoo.com.
Hasan Azarmehr, Email: hasanazarmehr811@gmail.com.
References
- 1.Holmes PH. Pathogenesis of trichostrongylosis. Vet Parasitol. 1985;18:89–101. doi: 10.1016/0304-4017(85)90059-7. [DOI] [PubMed] [Google Scholar]
- 2.Watthanakulpanich D, Pongvongsa T, Sanguankiat S, Nuamtanong S, Maipanich W, Yoonuan T, et al. Prevalence and clinical aspects of human Trichostrongylus colubriformis infection in Lao PDR. Acta Trop. 2013;126:37–42. doi: 10.1016/j.actatropica.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Roeber F, Jex AR, Gasser RB. Impact of gastrointestinal parasitic nematodes of sheep, and the role of advanced molecular tools for exploring epidemiology and drug resistance-an Australian perspective. Parasit Vectors. 2013;6:153. doi: 10.1186/1756-3305-6-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waghorn TS, Knight JS, Leathwick DM. The distribution and anthelmintic resistance status of Trichostrongylus colubriformis, T. vitrinus and T. axei in lambs in New Zealand. N Z Vet J. 2014;62:152–159. doi: 10.1080/00480169.2013.871193. [DOI] [PubMed] [Google Scholar]
- 5.Tan TK, Panchadcharam C, Low VL, Lee SC, Ngui R, Sharma RS, et al. Co-infection of Haemonchus contortus and Trichostrongylus spp. among livestock in Malaysia as revealed by amplification and sequencing of the internal transcribed spacer II DNA region. BMC Vet Res. 2014;10:38. doi: 10.1186/1746-6148-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter AG, Heath PJ. Ovine internal parasitism in the Yemen Arab Republic. Trop Anim Health Prod. 1984;16:95–106. doi: 10.1007/BF02239853. [DOI] [PubMed] [Google Scholar]
- 7.Sissay MM, Uggla A, Waller PJ. Prevalence and seasonal incidence of nematode parasites and fluke infections of sheep and goats in eastern Ethiopia. Trop Anim Health Prod. 2007;39:521–531. doi: 10.1007/s11250-007-9035-z. [DOI] [PubMed] [Google Scholar]
- 8.Burgess CG, Bartley Y, Redman E, Skuce PJ, Nath M, Whitelaw F, et al. A survey of the trichostrongylid nematode species present on UK sheep farms and associated anthelmintic control practices. Vet Parasitol. 2012;189:299–307. doi: 10.1016/j.vetpar.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Melville LA, McBean D, Fyfe A, Campbell SJ, Palarea-Albaladejo J, Kenyon F. Effect of anthelmintic treatment strategy on strongylid nematode species composition in grazing lambs in Scotland. Parasit Vectors. 2016;9:199. doi: 10.1186/s13071-016-1493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancrini G, Boemi G, Iori A, Corselli A. Human infestations by Trichostrongylus axei, T. capricola and T. vitrinus: 1st report in Italy. Parassitologia. 1982;24:145–149. [PubMed] [Google Scholar]
- 11.Lattès S, Ferté H, Delaunay P, Depaquit J, Vassallo M, Vittier M, et al. Trichostrongylus colubriformis nematode infections in humans, France. Emerg Infect Dis. 2011;17:1301–1302. doi: 10.3201/eid1707.101519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawless DK, Kuntz RE, Strome CP. Intestinal parasites in an Egyptian village of the Nile Valley with emphasis on the protozoa1. Am J Trop Med Hyg. 1956;5:1010–1014. doi: 10.4269/ajtmh.1956.5.1010. [DOI] [PubMed] [Google Scholar]
- 13.Poirriez J, Dei-Cas E, Guevart E, Abdellatifi M, Giard P, Vernes A. Human infestation by Trichostrongylus vitrinus in Morocco. Ann Parasitol Hum Comp. 1984;59:636–638. doi: 10.1051/parasite/1984596636. [DOI] [PubMed] [Google Scholar]
- 14.Souza RP, Souza JN, Menezes JF, Alcântara LM, Soares NM, Aquino Teixeira MC. Human infection by Trichostrongylus spp. in residents of urban areas of Salvador city, Bahia Brazil. Biomédica. 2013;33:439–445. doi: 10.7705/biomedica.v33i3.770. [DOI] [PubMed] [Google Scholar]
- 15.Bundy DA, Terry SI, Murphy CP, Harris EA. First record of Trichostrongylus axei infection of man in the Caribbean region. Trans R Soc Trop Med Hyg. 1985;79:562–563. doi: 10.1016/0035-9203(85)90100-2. [DOI] [PubMed] [Google Scholar]
- 16.Boreham RE, McCowan MJ, Ryan AE, Allworth AM, Robson JM. Human trichostrongyliasis in Queensland. Pathology. 1995;27:182–185. doi: 10.1080/00313029500169842. [DOI] [PubMed] [Google Scholar]
- 17.Sahba GH, Arfaa F, Bijan H. Intestinal helminthiasis in the rural area of Khuzestan, south-west Iran. Ann Trop Med Parasitol. 1967;61:352–357. doi: 10.1080/00034983.1967.11686498. [DOI] [PubMed] [Google Scholar]
- 18.Daryani A, Sharif M, Nasrolahei M, Khalilian A, Mohammadi A, Barzegar G. Epidemiological survey of the prevalence of intestinal parasites among schoolchildren in Sari, northern Iran. Trans R Soc Trop Med Hyg. 2012;106:455–459. doi: 10.1016/j.trstmh.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Gasser RB, Bott NJ, Chilton NB, Hunt P, Beveridge I. Toward practical, DNA-based diagnostic methods for parasitic nematodes of livestock - bionomic and biotechnological implications. Biotechnol Adv. 2008;26:325–334. doi: 10.1016/j.biotechadv.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Van Wyk JA, Cabaret J, Michael LM. Morphological identification of nematode larvae of small ruminants and cattle simplified. Vet Parasitol. 2004;119:277–306. doi: 10.1016/j.vetpar.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Gasser RB, Chilton NB, Hoste H, Stevenson LA. Species identification of trichostrongyle nematodes by PCR-linked RFLP. Int J Parasitol. 1994;24:291–293. doi: 10.1016/0020-7519(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 22.Humbert JF, Cabaret J. Use of random amplified polymorphic DNA for identification of ruminant trichostrongylid nematodes. Parasitol Res. 1995;81:1–5. doi: 10.1007/BF00932409. [DOI] [PubMed] [Google Scholar]
- 23.Gasser RB, Chilton NB. Applications of single-strand conformation polymorphism (SSCP) to taxonomy, diagnosis, population genetics and molecular evolution of parasitic nematodes. Vet Parasitol. 2001;101:201–213. doi: 10.1016/S0304-4017(01)00567-2. [DOI] [PubMed] [Google Scholar]
- 24.Gasser R, Nansen P, Guldberg P. Fingerprinting sequence variation in ribosomal DNA of parasites by DGGE. Mol Cell Probes. 1996;10:99–105. doi: 10.1006/mcpr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 25.Zarlenga DS, Chute MB, Gasbarre LC, Boyd PC. A multiplex PCR assay for differentiating economically important gastrointestinal nematodes of cattle. Vet Parasitol. 2001;97:199–209. doi: 10.1016/S0304-4017(01)00410-1. [DOI] [PubMed] [Google Scholar]
- 26.von Samson-Himmelstjerna G, Harder A, Schnieder T. Quantitative analysis of ITS2 sequences in trichostrongyle parasites. Int J Parasitol. 2002;32:1529–1535. doi: 10.1016/S0020-7519(02)00163-7. [DOI] [PubMed] [Google Scholar]
- 27.Ghasemikhah R, Sharbatkhori M, Mobedi I, Kia EB, Harandi MF, Mirhendi H. Sequence analysis of the second internal transcribed spacer (ITS2) region of rDNA for species identification of Trichostrongylus nematodes isolated from domestic livestock in Iran. Iran J Parasitol. 2012;7:40–46. [PMC free article] [PubMed] [Google Scholar]
- 28.Seesao Y, Gay M, Merlin S, Viscogliosi E, Aliouat-Denis CM, Audebert C. A review of methods for nematode identification. J Microbiol Methods. 2017;138:37–49. doi: 10.1016/j.mimet.2016.05.030. [DOI] [PubMed] [Google Scholar]
- 29.Archie EA, Ezenwa VO. Population genetic structure and history of a generalist parasite infecting multiple sympatric host species. Int J Parasitol. 2011;41:89–98. doi: 10.1016/j.ijpara.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Gasser RB, Hoste H. Genetic markers for closely-related parasitic nematodes. Mol Cell Probes. 1995;9:315–320. doi: 10.1016/S0890-8508(95)91588-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhao GH, Jia YQ, Cheng WY, Zhao W, Bian QQ, Liu GH. Characterization of the complete mitochondrial genomes of Nematodirus oiratianus and Nematodirus spathiger of small ruminants. Parasit Vectors. 2014;7:319. doi: 10.1186/1756-3305-7-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharifdini M, Derakhshani S, Alizadeh SA, Ghanbarzadeh L, Mirjalali H, Mobedi I, et al. Molecular identification and phylogenetic analysis of human Trichostrongylus species from an endemic area of Iran. Acta Trop. 2017;176:293–299. doi: 10.1016/j.actatropica.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Newton LA, Chilton NB, Beveridge I, Gasser RB. Differences in the second internal transcribed spacer of four species of Nematodirus (Nematoda: Molineidae) Int J Parasitol. 1998;28:337–341. doi: 10.1016/S0020-7519(97)00150-1. [DOI] [PubMed] [Google Scholar]
- 34.Akkari H, Jebali J, Gharbi M, Mhadhbi M, Awadi S, Darghouth MA. Epidemiological study of sympatric Haemonchus species and genetic characterization of Haemonchus contortus in domestic ruminants in Tunisia. Vet Parasitol. 2013;193:118–125. doi: 10.1016/j.vetpar.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 35.Gasser RB, Chilton NB, Hoste H, Beveridge I. Rapid sequencing of rDNA from single worms and eggs of parasitic helminths. Nucleic Acids Res. 1993;21:2525–2526. doi: 10.1093/nar/21.10.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestechian N, Baghaei M, Yosefi H. Study on Trichostrongylus eggs isolated from sheep by PCR-RFLP in Isfahan. J Feyz. 2005;35:23–29. [Google Scholar]
- 39.Talari SA, Arbabi M. Prevalence of gastrointestinal tract Trichostrogylus in sheep and goats in slaughterhous of Kashan. J Feyz. 2005;9:34–38. [Google Scholar]
- 40.Gholamian A, Eslami A, Nabavi L, Rasekh A. A field survey on resistance of gasterointestinal nematodes to levamisole in sheep in Khuzestan Province of Iran. J Vet Res. 2006;61:7–13. [Google Scholar]
- 41.Ranjbar-Bahadori SH, Eslami A, Samani RA. Study on the helminth infection of native ruminants of Golestan province. Iranian J Vet Res. 2007;63:303–305. [Google Scholar]
- 42.Ghadirian E. Human infection with Trichostrongylus lerouxi (Biocca, Chabaud, and Ghadirian, 1974) in Iran. Am J Trop Med Hyg. 1977;26:1212–1213. doi: 10.4269/ajtmh.1977.26.1212. [DOI] [PubMed] [Google Scholar]
- 43.Ghadirian E, Arfaa F. Present status of trichostrongyliasis in Iran. Am J Trop Med Hyg. 1975;24:935–941. doi: 10.4269/ajtmh.1975.24.935. [DOI] [PubMed] [Google Scholar]
- 44.Sharifdini M, Heidari Z, Hesari Z, Vatandoost S, Kia EB. Molecular phylogenetics of Trichostrongylus species (Nematoda: Trichostrongylidae) from humans of Mazandaran province Iran. Korean J Parasitol. 2017;55:279–285. doi: 10.3347/kjp.2017.55.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blackburn PJ, Carmichael IH, Walkden-Brown SW, Greenslade S. Use of developmental temperature and gastrointestinal tract location to isolate pure Trichostrongylus vitrinus from mixed, naturally acquired trichostrongylid infections in sheep. Aust Vet J. 2015;93:221–224. doi: 10.1111/avj.12325. [DOI] [PubMed] [Google Scholar]
- 46.Hamer K, Bartley D, Jennings A, Morrison A, Sargison N. Lack of efficacy of monepantel against trichostrongyle nematodes in a UK sheep flock. Vet Parasitol. 2018;257:48–53. doi: 10.1016/j.vetpar.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Hoste H, Chilton NB, Gasser RB, Beveridge I. Differences in the second internal transcribed spacer (ribosomal DNA) between five species of Trichostrongylus (Nematoda: Trichostrongylidae) Int J Parasitol. 1995;25:75–80. doi: 10.1016/0020-7519(94)00085-3. [DOI] [PubMed] [Google Scholar]
- 48.Bisset SA, Knight JS, Bouchet CL. A multiplex PCR-based method to identify strongylid parasite larvae recovered from ovine faecal cultures and/or pasture samples. Vet Parasitol. 2014;200:117–127. doi: 10.1016/j.vetpar.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Wimmer B, Craig BH, Pilkington JG, Pemberton JM. Non-invasive assessment of parasitic nematode species diversity in wild Soay sheep using molecular markers. Int J Parasitol. 2004;34:625–631. doi: 10.1016/j.ijpara.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 50.Lotfi E, Dehqhannejad M. Food shortage and Iranians riots during the Second World War (1941–1945) J Historical R. 2014;6:1–16. [Google Scholar]
- 51.Ghatee MA, Haghdoost AA, Kooreshnia F, Kanannejad Z, Parisaie Z, Karamian M, et al. Role of environmental, climatic risk factors and livestock animals on the occurrence of cutaneous leishmaniasis in newly emerging focus in Iran. J Infect Public Health. 2018;11:425–433. doi: 10.1016/j.jiph.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Budischak SA, Hoberg EP, Abrams A, Jolles AE, Ezenwa VO. A combined parasitological molecular approach for noninvasive characterization of parasitic nematode communities in wild hosts. Mol Ecol Resour. 2015;15:1112–1119. doi: 10.1111/1755-0998.12382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.