Abstract
A bioassay-guided phytochemical investigation of propolis samples from Tanzania and Zambia that screened for activity against Trypanosoma brucei has led to the isolation of two novel flavanones with promising antitrypanosomal activity. The compounds were characterized based on their spectral and physical data and identified as 6-(1,1-dimethylallyl) pinocembrin and 5-hydroxy-4″,4″-dimethyl-5″-methyl-5″-H-dihydrofuranol [2″,3″,6,7] flavanone. The two compounds, together with the propolis extracts and fractions, were assayed against a standard drug-sensitive strain of T. b. brucei (s427 wild-type), multi-drug resistant-resistant T. b. brucei (B48), drug-sensitive T. congolense (1L300) and a derived diminazene-resistant T. congolense strain (6C3), and for toxicity against U947 human cells and RAW 246.7 murine cells. Activity against T. b. brucei was higher than against T. congolense. Interestingly, the Tanzanian propolis extract was found to be more active than its fractions and purified compounds in these assays, with an IC50 of 1.20 μg/mL against T. b. brucei. The results of a cytotoxicity assay showed that the propolis extracts were less toxic than the purified compounds with mean IC50 values > 165.0 μg/mL.
Keywords: Trypanosomiasis, Flavanones, Tanzania, Zambia
Graphical abstract
Highlights
-
•
Two samples of propolis from East Africa display good activity against Trypanosoma brucei and T. congolense.
-
•
Activity against both wild type and pentamidine and diminazene resistant forms.
-
•
Two novel flavonoids and one known flavonoid were isolated from Tanzanian and Zambian propolis samples and characterized.
-
•
Pure isolated compounds not much more active than crude extracts.
-
•
Repeated observation of anti-protozoal activity shows the importance of propolis indefending the hive against infections.
1. Introduction
Propolis or ‘bee glue’ is a sticky resinous substance, produced by honeybees to seal holes in their hives. This maintains a healthy environment in the hive and protects them from microbes and infections (Ghisalberti 1979). The biological activities of propolis have been well-studied and reported in the literature. Previously, 22 propolis samples from nine sub-Saharan African countries including Zambia and Tanzania, were profiled using high resolution LC-MS, GC-MS and HPLC and were observed to have a high degree of diversity in chemical composition. However, there was no clear geographic discrimination between the samples (Zhang et al., 2014). It has also been reported that a Tanzanian propolis extract showed greater antibacterial activity against a range of gram positive bacteria than a Zambian propolis extract (Seidel et al., 2008). However, there is still a lack of information on propolis from Zambia and Tanzania, particularly the bioactivity of isolated compounds from these samples. In this study, a bioassay guided technique has been used to identify some active metabolites from these samples using in vitro assays against Trypanosoma brucei, drug-resistant T. b. brucei, T. congolense, and drug-resistant T. congolense. The subspecies of T. b. brucei, T. b. gambiense and T. b. rhodesiense, cause sleeping sickness, or Human African Trypanosomiasis (HAT) in humans (De Koning 2020) and T. b. brucei and T. congolense cause a severe disease in animals, called nagana, or African Animal Trypanosomiasis (AAT)) (Giordani et al., 2016). There is increasing evidence that bees produce propolis to protect their hives against protozoal infections (Alotaibi et al., 2019). The in vitro cytotoxicity of the samples was also evaluated against human U937 cells and murine RAW 246.7 cells.
2. Materials and methods
2.1. General
The Zambian propolis sample was provided by Forest Fruits Co Lusaka and originated from North West Zambia, while the Tanzanian propolis sample was obtained from the area around the city of Moshi in Northern Tanzania. Solvents, reagents and other consumables were purchased from Sigma-Aldrich, Fisher Scientific, Bio-Wittaker or Merck UK. Column chromatography was carried out using silica gel 60 of particle size 0.063–0.200 mm. A UNICAM UV 300 spectrophotometer was used to determine the UV–Vis absorption spectra of the compounds in the range from 190 to 750 nm. The IR spectra of isolated compounds were recorded on a PerkinElmer Fourier transform infrared (FT-IR, model 1725X) with an attenuated total reflection (ATR) probe in the region 400 to 4000 cm−1. Melting points were determined using a Stuart Scientific melting point apparatus (Bibby, UK). NMR spectra were obtained on a Bruker AV3HD500 spectrophotometer using CDCl3 or DMSO‑d6 as solvents. NMR spectra were processed using Mnova (Mestrelab Research, Santiago de Compostela, Spain).
2.2. Extraction and fractionation
About 8.2 g of the Zambian propolis sample was extracted three times for 1 h, with 150 mL of ethanol at room temperature, under sonication (Clifton ultrasonic bath, Fisher Scientific, Loughborough, UK) yielding 6.48 g of extract. The extracts were combined, the solvent was evaporated using a rotary evaporator (Buchi, VWR, Leicestershire, UK), and the residue was weighed. 7.7 g of the Tanzanian propolis sample was extracted consecutively with solvents of increasing polarity starting with n-hexane, followed by ethyl acetate and methanol and the extracts were finally combined to yield a total extract weight of 3.4 g. It was hoped that the consecutive solvent extraction method might give better recovery but this was apparently not the case. Recovery of residues and re-extraction with the solvent was carried out using sonication. The extracts were filtered and concentrated under vacuum using a Buchi rotary evaporator. Fractionation of the extracts was carried out as described in Section S1 (Supplementary information).
2.3. LC and LC-MS analysis
All profiled samples and fractions were dissolved in methanol to give a concentration of 1 mg/mL. LC-MS analysis was carried out on an Accela 600 HPLC system combined with an Exactive (Orbitrap) mass spectrometer from (ThermoFisher, Hemel Hempstead, UK). MS detection range was from 100 to 1200 m/z and the scanning was performed under electrospray ionization polarity switching mode. The needle voltages were set at −4.0 kV (negative) and 4.5 kV (positive); and sheath and auxiliary gases were at 50 and 17 arbitrary units respectively. Separation was performed on an ACE C18 column (Hichrom Reading UK, 150 × 3 mm, 3 μm) with 0.1% v/v formic acid in water as mobile phase A and 0.1% v/v formic acid in acetonitrile as B at a flow rate of 0.3 mL/min, using the following gradient: 25% B for 30 min, 5 min 100% B, and 5 min 25% B, injecting 10 μL of sample solution. The MS2 spectra were obtained on an LTQ Orbitrap Fourier Transform Mass Spectrometer (FTMS) under the same conditions described for the Exactive instrument, with a collision energy of 35 V, and data were processed using Xcalibur software.
2.4. Determination of cytotoxic effect on U937 and RAW 246.7 mammalian cell lines
The U937 cells (a human cell line from the European Collection of Cell Cultures Cat. No. 85011440, supplied by Sigma-Aldrich, Dorset, UK) were cultured as described in RPMI 1640 medium (Lonza) supplemented with 5% FBS, 1% L-Glutamine and 1% Penicillin/Streptomycin at 37 °C in a humidified atmosphere of 5% CO2 as previously described (Passmore et al., 2001). RAW 264.7 (ATCC-TIB-71), a murine macrophage cell line, was cultured in Dulbecco's Modified Eagle's Medium (DMEM; ATCC, Catalogue No. 30–2002) supplemented with the following additions: 10% fetal bovine serum (FBS) (Biosera), 1% (v/v) each of 2 mM L-glutamine (Life Technologies, Paisley, UK) and 100 IU/100 μg/mL penicillin/streptomycin (Life Technologies, Paisley, UK). The cytotoxicity assay method with RAW246.7 and the cell density determinations were carried out as described by Han et al. (2002) (Han et al., 2002). U937 cells were grown to log phase at 37 °C and harvested at a density of 1 × 105 cells/ml in a 96-well plate (TPP, Switzerland). 100 μl of the cell suspension was added to each well and the plate was incubated for 24 h at 37 °C, 5% CO2, 100% humidity. In order to determine the EC50 value for each sample, a 2-fold serial dilution of the test compound was carried out in a separate 96-well plate, using the appropriate culture medium, and 100 μl of each dilution was then transferred to the cultured cells using a multichannel pipette, followed by another incubation for 24 h. To each well 20 μL of 5 mM resazurin sodium salt (Sigma-Aldrich) was added and the plates were incubated for a further 24 h, after which fluorescence was measured using a Wallac Victor 2 microplate reader (λ Ex/Em: 560/590 nm). The compound was tested in triplicate and cell viability was expressed as a percentage of the drug-free control. The resulting data were analyzed using GraphPad Prism 8 to obtain dose-response curves and corresponding 50% inhibitory concentration (IC50) values.
2.5. Trypanosoma strains and cultures, and anti-trypanosomal assay
Pairs of standard drug-sensitive laboratory strains and derived clonal lines made resistant to first-line drugs by in vitro exposure were used to assess general efficacy of propolis samples and isolated compounds, and potential cross-resistance issues, respectively. For T. b. brucei we used Lister s427 WT (De Koning et al., 2000) and clone B48 (Bridges et al., 2007), which was generated by knockout of the TbAT1 drug transporter (Matovu et al., 2003) followed by adaptation to high concentrations of pentamidine which resulted in loss of the TbAQP2 gene and high levels of resistance to the diamidine and melaminophenyl arsenical drugs (Munday et al., 2014). Culturing of T. brucei bloodstream forms was performed in complete HMI-9 medium with 10% fetal bovine serum at 37 °C/5% CO2, as described (Gudin et al., 2006). The T. congolense bloodstream forms of strain IL3000 (Savannah-type), and the derived diminazene-resistant clone 6C3, were cultured in Minimal Essential Medium (MEM) base with 10% goat serum, supplemented with 14 μL/L β-mercapto-ethanol, glutamine and antibiotics at 34 °C/5% CO2 as described (Cerone et al., 2019). The extract and the purified compounds were tested against T. brucei and T. congolense using the resazurin viability indicator exactly as described previously (Alotaibi et al., 2019; Omar et al., 2017). Briefly, T. brucei (2 × 104/well) or T. congolense (5 × 104/well) were incubated with a series of 23 doubling dilutions and no-drug controls in 96-well plates for 48 h prior to the addition of resazurin and a further incubation of 24 h. Stock solutions for each compound or extract concentration were prepared in DMSO, ensuring that there was a constant percentage of 1% (v/v) DMSO per well.
3. Results and discussion
3.1. In-vitro antitrypanosomal activity and cross-resistance studies of Zambian and Tanzanian propolis samples
In order to determine the in vitro anti-Trypanosoma activity of the Zambian and Tanzanian propolis samples, the crude extracts and fractions were tested against T. brucei S427WT and the multi-drug resistant strain B48 (Table 1). The result revealed that fraction ZP-F3 and ZP-F4 from the Zambian propolis sample gave the best activity against T. brucei s427 WT in vitro (EC50 values of 0.94 and 1.10 μg/mL) respectively, while the most active fraction from the Tanzanian sample was TP-F4 (EC50 = 2.36 μg/mL). There were no significant differences in the EC50 values for the standard drug-sensitive strain Lister T. brucei 427 WT and its derived resistant cell line B48 apart from ZP-F3, which showed a just-significant increase in its EC50 value with a resistance factor (RF) of 1.59, compared to an RF of 158 for pentamidine (P < 0.0001; Student's unpaired t-test).
Table 1.
EC50 values of ZP and TP propolis samples and its fractions on T. brucei s427 wild-type and B48 (n = 3).
| Samples |
T. brucei S427WT |
T. brucei B48 |
|||
|---|---|---|---|---|---|
| EC50, AVG ± SEM (μg/mL) | EC50, AVG ± SEM (μg/mL) | R. F | t-test | ||
| Zambian propolis (ZP) and its fractions | |||||
| ZP Crudea | 4.14 ± 0.12 | 7.39 ± 0.50 | 1.78 | 0.0032 | |
| ZP-F1a | 17.05 ± 0.75 | 16.66 ± 1.90 | 0.98 | 0.8557 | |
| ZP-F2a | 1.76 ± 0.06 | 1.95 ± 0.15 | 1.11 | 0.2976 | |
| ZP-F3a | 0.94 ± 0.06 | 1.50 ± 0.13 | 1.59 | 0.0165 | |
| ZP-F4a | 1.10 ± 0.03 | 0.97 ± 0.05 | 0.89 | 0.0821 | |
| ZP-F5a | 1.23 ± 0.03 | 1.07 ± 0.07 | 0.87 | 0.0986 | |
| ZP-F6a | 2.26 ± 0.03 | 2.30 ± 0.20 | 1.02 | 0.8813 | |
| ZP-F7a | 4.31 ± 0.17 | 4.36 ± 0.28 | 1.01 | 0.8733 | |
| ZP-F8a | 5.41 ± 0.15 | 6.10 ± 0.27 | 1.13 | 0.1761 | |
| ZP-F9a | 7.75 ± 0.35 | 7.46 ± 0.48 | 0.96 | 0.6509 | |
| Tanzanian propolis (ZP) and its fractions | |||||
| TP crudea | 1.20 ± 0.04 | 1.02 ± 0.07 | 0.85 | 0.0843 | |
| TP-F1a | 5.78 ± 0.44 | 6.07 ± 0.67 | 1.05 | 0.7392 | |
| TP-F2a | 8.76 ± 0.44 | 8.54 ± 0.65 | 0.98 | 0.8001 | |
| TP-F3a | 4.87 ± 0.21 | 5.78 ± 0.61 | 1.19 | 0.2272 | |
| TP-F4a | 2.36 ± 0.18 | 2.58 ± 0.33 | 1.10 | 0.5751 | |
| TP-F5a | 4.23 ± 0.21 | 5.45 ± 0.41 | 1.29 | 0.0565 | |
| TP-F6a | 15.78 ± 0.74 | 16.84 ± 1.26 | 1.07 | 0.5110 | |
| TP-F7a | 4.45 ± 0.36 | 6.04 ± 0.66 | 1.36 | 0.1021 | |
| TP-F8a | 8.23 ± 0.48 | 7.82 ± 1.38 | 0.95 | 0.7936 | |
| TP-F9a | 138.73 ± 8.18 | 152.80 ± 12.07 | 1.10 | 0.3893 | |
| Control | |||||
| Pentamidineb | 0.0031 ± 0.0003 | 0.4816 ± 0.02 | 157.84 | <0.0001 | |
The EC50 values are the average ± SEM of at least 3 independent determinations. a EC50 in μg/mL; b EC50 in μM. RF = Resistance factor, being EC50(WT)/EC50(B48). Statistical significance was determined using an unpaired two-tailed Student's t-test comparing the EC50 values of the resistant strain with those obtained for the control strain s427. Pentamidine is a known trypanocide and used as positive control and strain control for B48
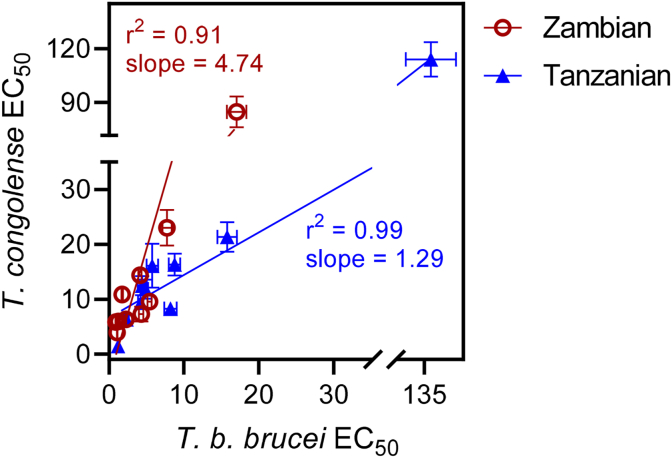
The same fractions were also tested against the main AAT pathogen, T. congolense, and its resistant strain, finding no significant differences between the two strains (P > 0.05), showing that these propolis fractions were not cross-resistant with first-line trypanosomiasis drugs such as diminazene aceturate (Table 2). Interestingly, the same order of potency for these fractions applied for T. brucei and T. congolense, with ZP-F3 and ZP-F4 being the most active fractions from the Zambian propolis sample against T. congolense IL3000 as well (EC50 values of 5.85 and 3.92 μg/mL, respectively), and TP-F4 exhibiting the most activity among Tanzanian fractions (EC50 = 6.34 μg/mL). Fig. 1 shows strong correlation between the effects against T. brucei and T. congolense, for both Zambian and Tanzanian fractions, but while the Zambian fractions were much more potent against T. brucei than against T. congolense (slope of 4.74 in the correlation plot; AVG of 3.85-fold), the Tanzanian propolis differentiated much less between the two Trypanosoma species, indicating broader trypanocidal activity (slope of 1.29 in the correlation plot; AVG of 1.92-fold).
Table 2.
EC50 values of ZP and TP propolis fractions on T. congolense IL3000 and the diminazene-resistant T. congolense clone 6C3 (n = 3).
| Samples |
Trypanosoma congolense IL3000 |
Trypanosoma congolense 6C3 |
||
|---|---|---|---|---|
| EC50, AVG ± SEM (μg/mL) | EC50, AVG ± SEM (μg/mL) | R. F | t-test | |
| Zambian propolis (ZP) and its fractions | ||||
| ZP Crudea | 14.39 ± 0.37 | 13.48 ± 1.12 | 0.94 | 0.4821 |
| ZP-F1a | 84.65 ± 4.98 | 76.45 ± 2.68 | 0.90 | 0.2205 |
| ZP-F2a | 10.89 ± 0.72 | 14.22 ± 1.47 | 1.31 | 0.1117 |
| ZP-F3a | 5.85 ± 0.54 | 5.50 ± 0.29 | 0.94 | 0.5943 |
| ZP-F4a | 3.92 ± 0.39 | 4.25 ± 0.23 | 1.08 | 0.5081 |
| ZP-F5a | 5.97 ± 0.74 | 8.92 ± 0.82 | 1.50 | 0.0557 |
| ZP-F6a | 6.30 ± 0.74 | 6.70 ± 1.01 | 1.06 | 0.7715 |
| ZP-F7a | 7.28 ± 0.78 | 8.84 ± 0.67 | 1.21 | 0.1656 |
| ZP-F8a | 9.57 ± 0.64 | 9.35 ± 0.36 | 0.98 | 0.7578 |
| ZP-F9a | 23.00 ± 1.90 | 23.15 ± 1.97 | 1.01 | 0.9607 |
| Tanzanian propolis (ZP) and its fraction | ||||
| TP crudea | 1.46 ± 0.29 | 1.62 ± 0.27 | 1.11 | 0.7031 |
| TP-F1a | 16.09 ± 2.32 | 17.55 ± 1.16 | 1.09 | 0.6045 |
| TP-F2a | 16.32 ± 1.16 | 19.17 ± 0.88 | 1.18 | 0.1214 |
| TP-F3a | 11.83 ± 1.00 | 12.40 ± 1.49 | 1.05 | 0.7652 |
| TP-F4a | 6.34 ± 0.67 | 5.91 ± 0.60 | 0.93 | 0.6596 |
| TP-F5a | 9.34 ± 0.58 | 9.63 ± 0.67 | 1.03 | 0.7613 |
| TP-F6a | 21.34 ± 1.58 | 21.69 ± 0.86 | 1.02 | 0.8540 |
| TP-F7a | 12.49 ± 1.02 | 15.48 ± 1.31 | 1.24 | 0.1464 |
| TP-F8a | 8.29 ± 0.61 | 9.66 ± 0.89 | 1.17 | 0.2734 |
| TP-F9a | 113.97 ± 5.55 | 103.03 ± 2.84 | 0.90 | 0.1544 |
| Control | ||||
| Diminazeneb | 0.20 ± 0.03 | 1.51 ± 0.09 | 7.75 | 0.0002 |
The EC50 values are the average ± SEM of at least 3 independent determinations. a EC50 in μg/mL; b EC50 in μM. RF = Resistance factor, being EC50(WT)/EC50(6C3). Statistical significance was determined using an unpaired two-tailed Student's t-test comparing the EC50 values of the resistant strain with those obtained for the control strain IL3000. Diminazene is the most used trypanocide against AAT and used as positive control and strain control for 6C3.
Fig. 1.
Correlation plot for EC50 values of T. b. brucei and T. congolense for extracts of Zambian and Tanzanian propolis. The trend lines were generated by linear regression using Prism 8.4. Values were abstracted from Table 1, Table 2 Error bars are SD; when not shown, fall within the symbol.
Having successfully identified the most active fractions from each propolis sample we proceeded to purify and identify some of the active compounds from the selected fractions. Three new trypanocides were isolated. The fractionation methods are described in section S1. General profiling of the composition of the most active fractions from the two propolis samples was carried out by high resolution mass spectrometry and the results are presented in section S2. The elemental compositions of the peaks in both active fractions were consistent with most of the components being modified flavonoids.
3.2. Characterization of 6-(1,1-dimethylallyl)pinocembrin (1)
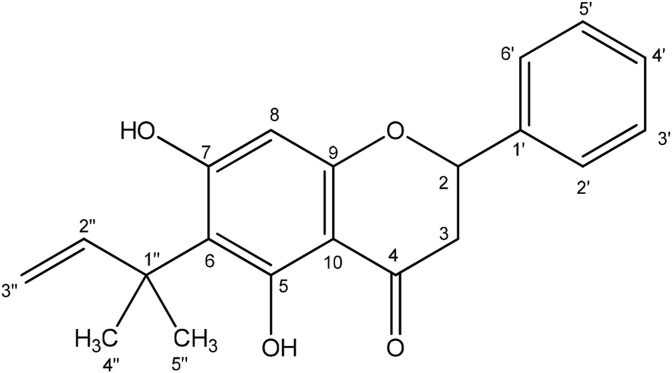
Compound 1 was obtained from the combined fractions 3 and 4 from open column chromatography of the ethanol extract of Zambian propolis, with further purification as detailed in section S1, as a white crystalline solid with a melting point of 158–160 °C while the UV spectrum gave absorption maxima at λ 296 nm. The IR spectrum showed absorption bands at 3225 (-OH) 1720, 1700 (C O), 1450 (C C) cm−1, and 1650, 1471 cm−1 for the presence of aromatic rings. The mass spectrum of the compound gave an [M-H]- ion at m/z = 323.1291 (Calc for C20H19O4, 323.1283). The NMR spectra are shown in section S3 and summarized in Table 3. The most deshielded proton in its proton spectrum was at δH 13.13 ppm for the H-bonded 5-hydroxyl group in flavonoids. The spectrum also showed a multiplet integrated for five protons of a mono-substituted benzene ring. The aliphatic protons at 5.41, 3.08 and 2.84 indicated the compound is a flavanone as these signals are typical of the H-2, H-3a and H-3b of a flavanone moiety. The other aliphatic proton signals observed must be from a side chain in the compound. An aromatic proton singlet at 5.99 ppm indicates the side chain must be on ring A of the flavanone and probably at position C-6 or C-8. A vinyl group on the side chain was revealed by the proton signals at 6.47 (dd, J = 17.8, 10.5 Hz), 5.38 (dd, J = 10.5, 0.9 Hz) and 5.48 (dd, J = 17.8, 0.9 Hz) in addition to a two methyl singlet, hence the side chain must be isopentyl. The 13C spectrum showed signals for a carbonyl carbon at δC 196.1 ppm expected for the saturated ketone of a flavanone. There were 12 aromatic carbon signals, five aliphatic carbons and two olefinic carbons at 113.4 and 149.7 ppm. The structure of the compound was further deduced from correlations in its 2D NMR. From the HMBC spectrum, the 5-OH proton showed 2J/3J correlations to C-5, C-6 and C-10 and the methyl protons of the side chain also showed 3J correlations to C-6 hence the side chain must be at position C-6. Similarly, the 7-OH proton showed correlations to C-6, C-7 and C-8. From the HSQC spectrum C-8 has the proton singlet at 5.99 ppm attached to it, thus confirming the substitution of the side chain at C-6 and not at C-8. The 2J/3J correlations, in the HMBC spectrum, from H-2 and H-3 to the carbonyl carbon confirm the flavanone moiety and other correlations from these protons to aromatic ring carbons indicate the phenyl ring is at C-2. Using these correlations, the chemical shifts for the protons and carbons were fully assigned as given in Table 3. This novel compound was characterized to be 6-(1,1-dimethyl allyl)pinocembrin (Fig. 2). Fig. S3-6 shows MS2 spectrum of compound 1 at 35 V collision energy, the fragments at m/z 219.07, and 245.08 are consistent with a fragment containing ring A, while the fragment at m/z 267.10 consists of both rings A and B. A proposed fragmentation scheme is shown in Fig. S3-7.
Table 3.
NMR Spectroscopic Data (500 MHz, CDCl3) for compound 1.
| Position | Proton δ ppm (mult, J Hz) | Carbon δppm (mult) | COSY (NOESY) | HMBC | ||
|---|---|---|---|---|---|---|
| 2 | 5.37 (1H, dd, 5.8, 1.0) | 78.7 (CH) | H-3 | C-1′, C-2′, C-6′ | ||
| 3 | 2.85 (1H, dd, 17.1, 3.1) 3.10 (1H, dd, 17.2, 12.9) | 43.5 (CH2) | H-2, H-3 | C-2, C-4 | ||
| 4 | – | 196.0 (C) | – | – | ||
| 5 | – | 163.4 (C) | – | – | ||
| 6 | – | 111.7 (C) | – | – | ||
| 7 | – | 164.6 (C) | – | – | ||
| 8 | 5.99 (1H, s) | 96.8 (CH) | – | C-6, C-7, C-9, C-10 | ||
| 9 | – | 160.8 (C) | – | – | ||
| 10 | – | 103.1 (C) | – | – | ||
| 1′ | – | 138.5 (C) | – | – | ||
| 2′ | 7.46 (1H, d, 1.1) | 126.1 (CH) | H-3′ | C-2, C-4′, C-6′ | ||
| 3′ | 7.45 (1H, m) | 128.8 (CH) | H-2′ | C-1′ | ||
| 4′ | 7.42 (1H, m) | 128.8 (CH) | C-2′, C-6′ | |||
| 5′ | 7.45 (1H, m) | 128.8 (CH) | C-1′ | |||
| 6′ | 7.47 (1H, d, 2.7) | 126.1 (CH) | C-2, C-2′, C-4′ | |||
| 1″ | – | 40.7 (C) | – | |||
| 2″ | 6.47 (1H, dd, 17.8, 10.5) | 149.7 (CH) | H-3″ | C-1″, C-4″, C-5″ | ||
| 3″ | 5.39 (1H, d, 1.1) 5.48 (1H, dd, 17.8, 0.9) | 113.4 (CH2) | H-2″ | C-1″, C-2″ | ||
| 4″ | 1.62 (s) | 27.3 (CH3) | – | C-1″, C-2″, C-6 | ||
| 5″ | 1.60 (s) | 26.6 (CH3) | – | C-1″, C-2″, C-6 | ||
| 5-OH | 13.13 (s) | – | – | C-5, C-6, C-10 | ||
| 7-OH | 7.50 (s) | – | – | C-6, C-7, C-8 | ||
Fig. 2.
Compound 1, 6-(1,1-dimethyl allyl)pinocembrin.
3.3. Characterization of 6-(1,1-Dimethylallyl)naringenin (2)
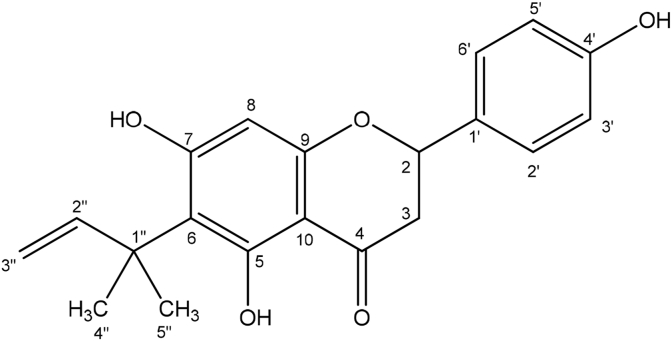
A second compound was obtained from fractions 2 and 3 of the ethanol extract of the Zambian propolis as an amorphous yellow solid (38.8 mg) from the ethanol extract. The mass spectrum gave an [M-H]- ion at 339.1240 (Calc for C20H19O5, 339.1233). The NMR spectra and a summary table are given in section S4 of the Supplementary Information. Its proton spectrum was similar to that of compound 1 except for two aromatic doublets at δH 6.87 (2H, d, J = 8.6 Hz) and 7.31 (2H, d, J = 8.6 Hz) replacing the 5H aromatic multiplets in compound 1. The carbon signals were also identical except for the signals at 115.8 (2 × CH), 128.0 (2 × CH) and 156.5 (C) replacing the carbon signals of the phenyl group in 1. Therefore, C-4′ must be substituted with a hydroxyl group. This was further confirmed by the difference of 16 mass units between compound 2 and compound 1. The correlations in its 2D NMR spectra were similar to those of compound 1 and correlations from the protons in ring B confirmed the –OH attachment to C-4′. The full chemical shift assignments (Table S6-1) were thus made from the 2D spectra and were confirmed by comparison with literature reports (Seo et al., 1997) and the structure was assigned as 6-(1,1-dimethylallyl)naringenin (Fig. 3).
Fig. 3.
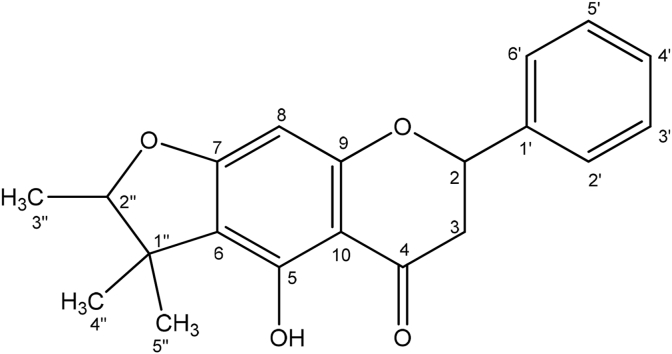
Structure of compound 2, 6-(1,1-dimethylallynaringenin. Characterization of 5-hydroxy-4″,4″-dimethyl-5″-methyl-5″-H-dihydrofurano [2″,3″,6,7]flavanone (3).
Compound 3 was obtained as a mixture, which, by comparing ring B protons in the NMR, can be estimated to be composed of 52.4% compound 3 with 47.6% compound 2. The LC-MS analysis of TP-F4-(7–9) exhibited many pseudo-molecular ion peaks, with a major peak at m/z 339.1246 (C20H19O5, 2.368 ppm). For the compound isolated the IR spectrum showed the presence of an OH group at 3225 cm−1, carbonyl group (1700, 1650 cm−1) and 1650, 1500 cm−1 for the presence of aromatic rings. The proton spectrum of the fraction was similar to the one for compounds 1 and 2, the major difference between the spectra was the absence of the isopentyl side chain. The NMR spectra are shown in section S5 and summarized in Table 4. This was replaced by a saturated furan ring fused to ring A of the flavanone at positions C-6 and C-7. The oxymethine furan proton was observed at δH 4.46 ppm while the three methyl groups on the furan ring were at 1.18, 1.43 and 1.36 ppm. The carbon spectrum (Dept-q) showed 20 carbon signals with three methyls, two oxygenated methines, one methylene, one carbonyl and 12 aromatic carbons. The structure was confirmed using correlations in its 2D spectra as follows; the furan proton (H-2″) showed 3J correlations to C-6 and C-7 in the HMBC spectrum thus confirming the furan ring being fused at those positions. Similar correlations for the 5-OH indicated C-6 to be a quaternary carbon, hence further confirming the furan ring being fused at C-6 and C-7. The signals for the H-3 protons were observed at δ 2.73 (1H, dt, J = 16.8, 4.2, H-3a) and 3.19 (1H, dd, J = 17.0, 13.1, H-3b) and were coupled to each other in COSY. Furthermore, the signal of the vicinal methyl protons was observed at δH 1.43 (3H, d, J = 7.0, H-3″) and was coupled to H-2” in the COSY spectrum (Fig. S8-4). Its chemical shift values were fully assigned as given in Table 4. The structure was an assigned as the novel compound 5-Hydroxy-4″,4″-dimethyl-5″-methyl-5″-H-dihydrofurano [2″,3″,6,7]flavanone (Fig. 4). The proton and carbon spectra were similar to those obtained by Seo et al. where the compound they isolated differs from compound 3 in that the B ring is substituted with hydroxyl at position C-4 (Seo et al., 1997). Thus the carbon shifts obtained for compound 3 in rings A, C and the furan ring were similar to those in their compound but the carbon signals in ring B were generally at lower shift values. In the proton spectrum there was an obvious envelope for the aromatic protons at δH 6.90 ppm. The MS2 spectrum was consistent with the proposed structure and is shown in figure S5-6 along with a proposed fragmentation scheme in Fig. S5-7.
Table 4.
NMR Spectroscopic Data (500 MHz, Acetone-d6) for compound 3.
| Position | Proton δppm (mult, J Hz) | Carbon δppm (mult) | COSY (NOESY) | HMBC |
|---|---|---|---|---|
| 2 | 5.43 (1H, dd, J = 13.0,3.0) | 79.8 (CH) | H-3b | C-3 |
| 3 | 2.73(1H, dt, J = 16.8, 4.2) 3.19 (1H, dd, J = 17.0,13.1) | 43.3 (CH2) | H-3a- H-3b H-2 |
C-2, C-4 |
| 4 | – | 197.1 (C) | – | – |
| 5 | – | 103.3 (C) | – | – |
| 6 | – | 114.3 (C) | – | – |
| 7 | – | 163.6 (C) | – | – |
| 8 | 5.89 (1H, s) | 90.9 (CH) | – | C-6, C-10 |
| 1′ | – | 129.7 (C) | – | – |
| 2′ | 7.39 (1H, dd, J = 8.7, 2.8) | 128.8 (CH) | H-3′ | C-2, C-6′ |
| 3′ | 6.90 (1H, m) | 115.9 (CH) | H-2′ | C-1′, C-5′ |
| 4′ | 6.90 (1H, m) | 115.9 (CH) | C-2′, C-3′, C-5, C-6′ | |
| 5′ | 6.90 (1H, m) | 115.9 (CH) | H-6′ | C-1′ C-3′ |
| 6′ | 7.39 (1H, dd, J = 8.7, 2.8) | 128.8 (CH) | H-5′ | C-2, C-2′ |
| 1″ | – | 42.5 (C) | – | – |
| 2″ | 4.46 (1H, q, J = 6.5) | 91.4 (CH) | H-3″ | |
| 3″ | 1.36 (3H, d, J = 7.0) | 25.3 (CH3) | H-2″ | C-1″, C-2″ |
| 4″ | 1.43 (3H, s) | 14.2 (CH3) | C-1″, C-2″, C-6 | |
| 5″ | 1.18 (3H, s) | 20.7 (CH3) | C-1″, C-2″ C-4″, C-6 | |
| 5-OH | 12.45 (s) | 96.5 (C) | C-6, C-9, C-10 |
Fig. 4.
The structure of 5-hydroxy-4″,4″-dimethyl-5″-methyl-5″-H-dihydrofurano [2″,3″,6,7] flavanone (3).
3.4. Activity of the isolated compounds against Trypanosoma species
The isolated compounds were also tested on the same drug sensitive and multi-drug resistant clones of T. b. brucei and T. congolense. The results are presented in Table 5. Compound 2 from Zambian propolis displayed the highest activity against T. b. brucei, with an EC50 value of 2.24 μg/mL. Compounds 3 + 2 and 1 also displayed activity below 5 μg/mL, with EC50 values of 3.02 and 4.01 μg/mL, respectively. The EC50 values for the multidrug resistant strain B48 were within 1–1.2-fold (P > 0.05), thus the compounds were not cross-resistant with first-line HAT treatments pentamidine and melarsoprol (Bridges et al., 2007). The RF for pentamidine, included as control, was 144 (P = 0.0003; Table 5). Like the fractions tested above, the purified compounds displayed somewhat less potent activity against T. congolense than against T. brucei under the assay conditions, with EC50 values of 7.35, 10.47 and 13.77 μg/mL for compounds 2, 1, and 3 + 2, respectively. Again, no cross-resistance was observed with the main AAT treatment, diminazene aceturate (RF~1; P > 0.05; compare RF = 9.8 for diminazene) (see Table 6).
Table 5.
EC50 values of purified compounds isolated from ZP and TP propolis samples against T. brucei (n = 3).
| Purified Compounds | T. b. brucei s427 WT | T. b. brucei B48 | ||||
|---|---|---|---|---|---|---|
| EC50, AVG ± SEM (μg/mL) | EC50, AVG ± SEM (μM) | EC50, AVG ± SEM (μg/mL) | EC50, AVG ± SEM (μM) | RF | P | |
| (1) | 4.01 ± 0.26 | 12.4 ± 0.78 | 4.94 ± 0.22 | 15.3 ± 0.69 | 1.23 | 0.051 |
| (2) | 2.24 ± 0.13 | 6.58 ± 0.39 | 2.28 ± 0.16 | 6.69 ± 0.48 | 1.02 | 0.87 |
| (3 + 2) | 3.02 ± 0.27 | – | 3.61 ± 0.23 | – | 1.20 | 0.17 |
| Pentamidine | 0.0034 ± 0.0004 | 0.4929 ± 0.04 | 144.08 | 0.0003 | ||
The EC50 values are the average and SEM of at least 3 independent determinations. RF = Resistance factor, being EC50(WT)/EC50(B48). Statistical significance was determined using an unpaired two-tailed Student's t-test comparing the EC50 values of the resistant strain with those obtained for the control strain s427.
Table 6.
EC50 values of purified compounds isolated from ZP and TP propolis samples on T. congolense IL300, and a T. congolense cell line resistant to diminazene (n = 3).
| Purified Compounds | Trypanosoma congolense IL3000 | Trypanosoma congolense 6C3 | ||||
|---|---|---|---|---|---|---|
| EC50, AVG ± SEM (μg/mL) | EC50, AVG ± SEM (μM) | EC50, AVG ± SEM (μg/mL) | EC50, AVG ± SEM (μM) | RF | P | |
| (1) | 10.47 ± 0.96 | 32.30 ± 2.96 | 9.03 ± 0.43 | 27.85 ± 1.33 | 0.86 | 0.2430 |
| (2) | 7.35 ± 0.71 | 21.61 ± 2.08 | 5.69 ± 0.61 | 16.74 ± 1.79 | 0.77 | 0.1506 |
| (3 + 2) | 13.77 ± 1.26 | – | 12.94 ± 1.04 | – | 0.94 | 0.6383 |
| Diminazene | 0.15 ± 0.03 | 1.45 ± 0.04 | 9.81 | <0.0001 | ||
The EC50 values are the average and SEM of at least 3 independent determinations. RF= Resistance factor, being EC50(WT)/EC50(6C3). Statistical significance was determined using an unpaired two-tailed Student’s t-test comparing the EC50 values of the resistant strain with those obtained for the control strain Il3000.
The purified compounds and the most active fractions that were isolated from the Tanzanian and Zambian propolis samples were tested in vitro on the human cell line U937 and a murine cell line, RAW 246.7, using the resazurin assay. The results showed very low levels of toxicity, with similar EC50 values for both cell lines (Table 7). As the fractions and isolated compounds were more active against T. b. brucei than against T. congolense, the Selectivity Index (SI), being the ratio of the mammalian cell line EC50 and the parasite cell line EC50, was higher for T. b. brucei, with values for the isolated compounds, between 15 and 70, compared to 7 up to 17 for T. congolense. Interestingly, as the crude fractions displayed both higher trypanocidal activity and lower toxicity, the SI values for the fractions were much higher, e.g. >100 for Tanzanian crude propolis relative to both Trypanosoma species (Table 7).
Table 7.
EC50 of cytotoxicity of Zambian and Tanzanian propolis samples and the purified compounds against U937 cell line and RAW 246.7 cell line.
| Sample/Compound | U937 |
RAW 246.7 |
||||||
|---|---|---|---|---|---|---|---|---|
| EC50, AVG ± SEM (μg/mL) | EC50, AVG ± SEM (μM) | Selectivity Index (SI)1 |
EC50, AVG ± SEM (μg/mL) | EC50, AVG ± SEM (μM) | Selectivity Index (SI)1 |
|||
| Tbb427 WT | TcoIL3000 | Tbb427 WT | TcoIL3000 | |||||
| ZP crude | 166.2 ± 3.3 | – | 40.1 | 11.5 | 199.5 ± 1.7 | – | 48.2 | 13.9 |
| ZP-F2+F3 | 71.6 ± 2.8 | – | 70.2 | 13.0 | 74.9 ± 2.8 | – | 73.4 | 13.6 |
| (1) | 59.9 ± 5.2 | 184.7 ± 16.2 | 14.9 | 6,6 | 60.7 ± 1.4 | 187.3 ± 4.4 | 15.1 | 6.7 |
| (2) | 51.2 ± 3.1 | 150.5 ± 9.2 | 22.8 | 9.0 | 79.9 ± 5.6 | 235.0 ± 16.4 | 35.7 | 14.0 |
| TP crude | 148.0 ± 2.4 | – | 123.4 | 101.4 | 91.3 ± 3.2 | – | 76.1 | 62.6 |
| TP-F4 | 98.2 ± 4.8 | – | 41.6 | 16.6 | 114.4 ± 5.9 | – | 48.5 | 19.3 |
| (3 + 2) | 90.5 ± 4.2 | – | 30.0 | 7.0 | 97.5 ± 7.9 | – | 3 2.3 | 7.5 |
All EC50 values are the average and SEM of at least 3 independent experiments. SI = EC50 (mammalian cell line)/EC50 (Trypanosoma species). 1Note: the SI was based on EC50 values of the mammalian cells after 24 h incubation with the test compounds/extracts, whereas the EC50 values for the protozoan species were determined using 48 h of incubation prior to the addition of resazurin (see Methods section).
4. Conclusion
This study identified and characterized two novel flavanones isolated from Tanzanian and Zambian propolis samples. Both flavanones demonstrated antitrypanosomal activity against drug-susceptible and drug-resistant T. b. brucei and T. congolense. Interestingly, the Tanzanian propolis extract was found to have greater antitrypanosomal activity than its fractions and isolated compounds. EC50 values for this crude extract vs. T. b. brucei and T. congolense were 1.2 μg/mL and 1.46 μg/mL, respectively, with similar values against pentamidine-resistant T. b. brucei (EC50 1.02 μg/mL) and diminazene-resistant T. congolense (EC50 1.62 μg/mL). These findings are in line with previous reports of greater antitrypanosomal activity of crude propolis extracts from Nigeria and Libya compared with their isolated phytochemicals (Siheri et al., 2014, 2016); Omar et al., 2017). Taken together, these results consistently point to the possibility of a synergistic effect of the isolated compounds, although additive effects of multiple active compounds in the same extract cannot be excluded. These observations make the prospect of a more complex propolis extract, which would be decidedly cheaper to produce if it can be standardized, seem quite appealing, especially as the most active fractions displayed a very good Selectivity Index.
Propolis, whatever its origin, appears to always exhibit high levels of anti-trypanosomatid activity, particularly exhibited by flavonoids (Alotaibi et al., 2019; Omar et al., 2016; Siheri et al., 2016). This is believed to be a self-selecting feature due to the bee's vulnerability to certain trypanosomatid parasites. The Scottish honey bee microbiome contains high levels of genetic material derived from the trypanosomatid Lotmarim passim (Regan et al., 2018) and this organism has been found to be widespread in other bee populations (Castelli et al., 2019; Ravoet et al., 2015; Schwarz et al., 2015). Thus far there is no evidence that bees ingest propolis but since the spread of the protozoal infection occurs via insect feces (Ruiz-González and Brown, 2006), coating the surfaces in the hive with propolis that is active against trypanosomatids could prevent transmission. Interestingly, it has recently been reported that feeding propolis to bees also reduces infection with the microsporidium bee parasite Nosema ceranae (Mura et al., 2020). In addition, it has been observed that propolis has beneficial effects in stabilising the honey bee microbiome (Saelao et al., 2020). We conclude that East African propolis, like propolis from other African locations, has promising anti-kinetoplastid properties.
Funding
The work was funded through a PhD studentship from the Ministry of Health State of Kuwait. The support of the Saudi Arabian Ministry of Health for support of MJN is gratefully acknowledged.
Acknowledgements
We are grateful to the technical staff in the Strathclyde Institute of Pharmacy and Biomedical Sciences, Strathclyde University, and the Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and the Life Sciences, University of Glasgow, Glasgow, UK, for their technical assistance. Special thanks also go to the previous Minister of Health in Kuwait, Dr Ali Al-Obaidi, for his support of this research.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2020.10.011.
Contributor Information
Harry P. de Koning, Email: harry.de-koning@glasgow.ac.uk.
David G. Watson, Email: d.g.watson@strath.ac.uk.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Alotaibi A., Ebiloma G.U., Williams R., Alenezi S., Donachie A.M., Guillaume S., Igoli J.O., Fearnley J., De Koning H.P., Watson D.G. European propolis is highly active against trypanosomatids including Crithidia fasciculata. Sci. Rep. 2019;9:11364. doi: 10.1038/s41598-019-47840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D.J., Gould M.K., Nerima B., Mäser P., Burchmore R.J., De Koning H.P. Loss of the high-affinity pentamidine transporter is responsible for high levels of cross-resistance between arsenical and diamidine drugs in African trypanosomes. Mol. Pharmacol. 2007;71:1098–1108. doi: 10.1124/mol.106.031351. [DOI] [PubMed] [Google Scholar]
- Castelli L., Branchiccella B., Invernizzi C., Tomasco I., Basualdo M., Rodriguez M., Zunino P., Antúnez K. Detection of Lotmaria passim in Africanized and European honey bees from Uruguay, Argentina and Chile. J. Invertebr. Pathol. 2019;160:95–97. doi: 10.1016/j.jip.2018.11.004. [DOI] [PubMed] [Google Scholar]
- Cerone M., Uliassi E., Prati F., Ebiloma G.U., Lemgruber L., Bergamini C., Watson D.G., de A M Ferreira T., Roth Cardoso G.S.H., Soares Romeiro L.A., De Koning H.P., Bolognesi M.L. Discovery of sustainable drugs for neglected tropical diseases: cashew nut shell liquid (CNSL)-based hybrids target mitochondrial function and ATP production in Trypanosoma brucei. ChemMedChem. 2019;14:621–635. doi: 10.1002/cmdc.201800790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning H.P. The drugs of sleeping sickness: their mechanisms of action and resistance, and a brief history. Trav. Med. Infect. Dis. 2020;5:14. doi: 10.3390/tropicalmed5010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning H.P., MacLeod A., Barrett M.P., Cover B., Jarvis S.M. Further evidence for a link between melarsoprol resistance and P2 transporter function in African trypanosomes. Mol. Biochem. Parasitol. 2000;106:181–185. doi: 10.1016/s0166-6851(99)00206-6. [DOI] [PubMed] [Google Scholar]
- Ghisalberti E. Propolis: a review. Bee World. 1979;60:59–84. [Google Scholar]
- Giordani F., Morrison L.J., Rowan T.G., De Koning H.P., Barrett M.P. The animal trypanosomiases and their chemotherapy: a review. Parasitology. 2016;143:1862–1889. doi: 10.1017/S0031182016001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudin S., Quashie N.B., Candlish D., Al-Salabi M.I., Jarvis S.M., Ranford-Cartwright L.C., De Koning H.P. Trypanosoma brucei: a survey of pyrimidine transport activities. Exp. Parasitol. 2006;114:118–125. doi: 10.1016/j.exppara.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Han S., Sung K.-H., Lee S., Cho K., Lee C.-K., Ha N.-J., Kim K. Activation of murine macrophage cell line RAW 264.7 by Korean propolis. Arch Pharm. Res. (Seoul) 2002;25:895–902. doi: 10.1007/BF02977011. [DOI] [PubMed] [Google Scholar]
- Matovu E., Stewart M., Geiser F., Brun R., Mäser P., Wallace L.J.M., Burchmore R.J., Enyaru J.C.K., Barrett M.P., Kaminsky R., Seebeck T., De Koning H.P. Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryot. Cell. 2003;2:1003–1008. doi: 10.1128/EC.2.5.1003-1008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday J.C., Eze A.A., Baker N., Glover L., Clucas C., Aguinaga Andrés D., Natto M.J., Teka I.A., McDonald J., Lee R.S., Graf F.E., Ludin P., Burchmore R.J.S., Turner C.M.R., Tait A., MacLeod A., Mäser P., Barrett M.P., Horn D., De Koning H.P. Trypanosoma brucei aquaglyceroporin 2 is a high-affinity transporter for pentamidine and melaminophenyl arsenic drugs and the main genetic determinant of resistance to these drugs. J. Antimicrob. Chemother. 2014;69:651–663. doi: 10.1093/jac/dkt442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura A., Pusceddu M., Theodorou P., Angioni A., Floris I., Paxton R.J., Satta A. Propolis consumption reduces Nosema ceranae infection of European honey bees (Apis mellifera) Insects. 2020;11:124. doi: 10.3390/insects11020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar R.M., Igoli J., Gray A.I., Ebiloma G.U., Clements C., Fearnley J., Ebel R.A., Zhang T., De Koning H.P., Watson D.G. Chemical characterisation of Nigerian red propolis and its biological activity against Trypanosoma brucei Phytochem. Anal. 2016;27:107–115. doi: 10.1002/pca.2605. [DOI] [PubMed] [Google Scholar]
- Omar R., Igoli J.O., Zhang T., Gray A.I., Ebiloma G.U., Clements C.J., Fearnley J., Edrada Ebel R., Paget T., De Koning H.P., Watson D.G. The chemical characterization of Nigerian propolis samples and their activity against Trypanosoma brucei. Sci. Rep. 2017;7:923. doi: 10.1038/s41598-017-01038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore J., Lukey P., Ress S. The human macrophage cell line U937 as an in vitro model for selective evaluation of mycobacterial antigen-specific cytotoxic T-cell function. Immunology. 2001;102:146–156. doi: 10.1046/j.1365-2567.2001.01164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravoet J., Schwarz R.S., Descamps T., Yañez O., Tozkar C.O., Martin-Hernandez R., Bartolomé C., De Smet L., Higes M., Wenseleers T., Schmid-Hempel R., Neumann P., Kadowaki T., Evans J.D., De Graaf D.C. Differential diagnosis of the honey bee trypanosomatids Crithidia mellificae and Lotmaria passim. J. Invertebr. Pathol. 2015;130:21–27. doi: 10.1016/j.jip.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Regan T., Barnett M.W., Laetsch D.R., Bush S.J., Wragg D., Budge G.E., Highet F., Dainat B., de Miranda J.R., Watson M., Blaxter M., Freeman T.C. Characterisation of the British honey bee metagenome. Nat. Commun. 2018;9:4995. doi: 10.1038/s41467-018-07426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-González M.X., Brown M.J.F. Honey bee and bumble bee trypanosomatids: specificity and potential for transmission. Ecol. Entomol. 2006;31:616–622. [Google Scholar]
- Saelao P., Borba R.S., Ricigliano V., Spivak M., Simone-Finstrom M. Honeybee microbiome is stabilized in the presence of propolis. Biol. Lett. 2020;16:20200003. doi: 10.1098/rsbl.2020.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R.S., Bauchan G.R., Murphy C.A., Ravoet J., De Graaf D.C., Evans J.D. Characterization of two species of trypanosomatidae from the honey bee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. gen., n. sp. J. Eukaryot. Microbiol. 2015;62:567–583. doi: 10.1111/jeu.12209. [DOI] [PubMed] [Google Scholar]
- Seidel V., Peyfoon E., Watson D.G., Fearnley J. Comparative study of the antibacterial activity of propolis from different geographical and climatic zones. Phytother Res. 2008;22:1256–1263. doi: 10.1002/ptr.2480. [DOI] [PubMed] [Google Scholar]
- Seo E.-K., Silva G.L., Chai H.-B., Chagwedera T.E., Farnsworth N.R., Cordell G.A., Pezzuto J.M., Kinghorn A.D. Cytotoxic prenylated flavanones from Monotes engleri. Phytochemistry. 1997;45:509–515. doi: 10.1016/s0031-9422(96)00871-0. [DOI] [PubMed] [Google Scholar]
- Siheri W., Igoli J.O., Gray A.I., Nasciemento T.G., Zhang T., Fearnley J., Clements C.J., Carter K.C., Carruthers J., Edrada-Ebel R., Watson D.G. The isolation of antiprotozoal compounds from Libyan propolis. Phytother Res. 2014;28:1756–1760. doi: 10.1002/ptr.5194. [DOI] [PubMed] [Google Scholar]
- Siheri W., Zhang T., Ebiloma G.U., Biddau M., Woods N., Hussain M.Y., Clements C.J., Fearnley J., Ebel R.E., Paget T., Muller S., Carter K.C., Ferro V.A., De Koning H.P., Watson D.G. Chemical and antimicrobial profiling of propolis from different regions within Libya. PloS One. 2016;11 doi: 10.1371/journal.pone.0155355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Omar R., Siheri W., Al Mutairi S., Clements C., Fearnley J., Edrada-Ebel R., Watson D.G. Chromatographic analysis with different detectors in the chemical characterisation and dereplication of African propolis. Talanta. 2014;120:181–190. doi: 10.1016/j.talanta.2013.11.094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.