Abstract
The global shrimp industry has suffered bacterial diseases caused mainly by Vibrio species. The typical vibriosis, acute hepatopancreatic necrosis disease (AHPND), has resulted in mass mortality and devastating economic losses. Thus, therapeutic strategies are highly needed to decrease the risk of vibriosis outbreaks. Herein, we initially identified that the growth of the causative agent of AHPND, Vibrio parahaemolyticus (VPAHPND) and other vibrios in Pacific white shrimp (Litopenaeus vannamei) was inhibited by a Bacillus subtilis strain BSXE-1601. The natural products amicoumacins A, B, and C were purified from the cell-free supernatant from the strain BSXE-1601, but only amicoumacin A was demonstrated to be responsible for this anti-Vibrio activity. Our discovery provided the first evidence that amicoumacin A was highly active against shrimp pathogens, including the representative strain VPAHPND. Furthermore, we elucidated the amicoumacin A biosynthetic gene cluster by whole genome sequencing of the B. subtilis strain BSXE-1601. In addition to amicoumacin A, the strain BSXE-1601 genome harbored other genes encoding bacillibactin, fengycin, surfactin, bacilysin, and subtilosin A, all of which have previously reported antagonistic activities against pathogenic strains. The whole-genome analysis provided unequivocal evidence in support of the huge potential of the strain BSXE-1601 to produce diverse biologically antagonistic natural products, which may facilitate further studies on the effective therapeutics for detrimental diseases in shrimp.
Keywords: amicoumacin A, genome sequence, Bacillus subtilis, AHPND, vibriosis, Litopenaeus vannamei
Introduction
The greatest challenge in the global shrimp industry is a disease particularly that of bacterial origin and chief among these is Vibrio species (Ajadi et al., 2016; Hoseinifar et al., 2018). The acute hepatopancreatic necrosis disease (AHPND), formerly known as shrimp “early mortality syndrome” (EMS), is a newly emerging vibriosis that has caused severe mortality (up to 100%) and devastating economic effect in the global shrimp industry (Lightner et al., 2012; Joshi et al., 2014; Kongrueng et al., 2015; Boonsri et al., 2017). Infected shrimp show slow growth, an empty stomach and midgut, and severe atrophy of hepatopancreas (Joshi et al., 2014; Kongrueng et al., 2015; Sirikharin et al., 2015; Han et al., 2020). Originally, the etiological agent of AHPND has been reported to be Vibrio parahaemolyticus (VPAHPND) (Tran et al., 2013). Recent studies have shown that other Vibrio spp., such as Vibrio punensis (Restrepo et al., 2018), Vibrio owensii (Liu et al., 2015), Vibrio harveyi-like (Kondo et al., 2015), and Vibrio campbellii (Dong et al., 2017a,b) are also capable of inducing AHPND in shrimp. Besides AHPND, other vibrioses were frequently reported in farmed shrimp caused by Vibrio alginolyticus, Vibrio anguillarum, V. harveyi, Vibrio vulnificus, Vibrio splendidus, V. campbellii and Vibrio fischeri (Lavilla-Pitogo et al., 1990; Lightner, 1996; Lavilla-Pitogo et al., 1998; Chen et al., 2000; Jayasree et al., 2006; Longyant et al., 2008; Zheng et al., 2016; Chandrakala and Priya, 2017; Karnjana et al., 2019). Furthermore, non-Vibrio species such as Aeromonas spp. (Dierckens et al., 1998; Zhou et al., 2019), Streptococcosis spp. (Hasson et al., 2009), Shewanella spp. (Wang et al., 2000), Flavobacterium spp. (Chandrakala and Priya, 2017) and Pseudoalteromonas spp. (Zheng et al., 2016) were also documented to cause severe diseases in fish and shrimp aquaculture. Hence, strategies that focus on restraining the growth or activity of pathogenic bacteria are highly needed.
Recently, Bacillus spp. have been suggested to present a high interest as a promising therapeutic agent against vibriosis and other diseases in aquaculture, because of their antagonistic properties against aquatic pathogenic microorganisms, and their non-pathogenic and non-toxic characteristics (Rengpipat et al., 2003; Aly et al., 2008; Ran et al., 2012; Hoseinifar et al., 2018; Kuebutornye et al., 2019; Wang et al., 2019). The bacteriostatic properties of Bacillus spp. are linked primarily to the bioactive natural products produced by themselves (Kuebutornye et al., 2019). These natural products are not necessary for the survival of their producers but offer advantages under particular environmental conditions (Hug et al., 2020). They are highly optimized in the course of evolution for interactions with biological targets and, thus, exhibit various biological functions, such as antibacterial, antifungal and immunosuppressive activities (Hug et al., 2020). Genus Bacillus can produce a large variety of biologically active natural products, including non-ribosomal peptides (NRPs) (e.g., bacillibactin, bacilysin, fengycin, surfactin), polyketides (PKs) (e.g., bacillaene, difficidin, and macrolactin), ribosomally synthesized and post-translationally modified peptides (RiPPs) (e.g., lanthipeptides, lasso peptides, sactipeptides), polyketide-peptide hybrids (e.g., amicoumacin and ieodoglucomide), terpenes and alkaloids (Reimer and Bode, 2014; Li et al., 2015; Bartholomae et al., 2017; Greule et al., 2017; Kaspar et al., 2019; Hug et al., 2020). Nonetheless, the Bacillus spp. with broad-spectrum bacteriostatic activity utilized to control shrimp diseases especially AHPND are poorly documented and it is less clear by what mode of action these bacteria mitigate infections and facilitate shrimp culture.
Therefore, the present study aimed to explore the biologically active natural products produced by Bacillus spp. against AHPND and other vibriosis in Pacific white shrimp (Litopenaeus vannamei). To fulfill this goal, we chose a bacterial strain named BSXE-1601 as a therapeutic agent candidate. This strain was isolated from both the intestine of shrimp and the sediments of shrimp culture ponds during August 2013 at Lianyungang City, Jiangsu Province, China. In the present study, an assessment of the antagonistic activity in vitro was conducted to identify the therapeutic potential of the strain BSXE-1601 against vibriosis. Moreover, a bioactive metabolite responsible for this anti-Vibrio activity was isolated, structure analyzed, and bioactivity determined. To interrogate the potential biosynthetic pathways of the bioactive metabolites and the biosynthetic capability of the strain BSXE-1601, the whole genome sequencing of the strain BSXE-1601 was conducted. Our whole-genome analysis of BSXE-1601 provides a better understanding of the mechanisms involved in the anti-Vibrio properties and its high potential to be used as a multifunctional biological agent in aquaculture by suppressing pathogenic microbes of shrimp.
Materials and Methods
Bacterial Strain Identification
BSXE-1601 was obtained from the Microbial Culture Collection Center, Laboratory of Aquaculture Ecology, Ocean University of China (also preserved in China General Microbiological Culture Collection Center, CGMCC NO. 15949). The morphological, physiological, and biochemical characteristics of the strain BSXE-1601 were tested according to Bergey’s Manual of Systematic Bacteriology (Krieg and Holt, 1984). The genetic identification was performed using 16S rRNA gene sequencing as described earlier with some modifications (Gomez-Gil et al., 2012). Briefly, the DNA of the strain BSXE-1601 was extracted by simple boiling (Ribeiro et al., 2016). Then polymerase chain reaction (PCR) amplification of the 16S rRNA gene was undertaken using universal primers (27F, AGAGTTTGATCMTGGCTCAG; 1491R, TACGGYTACCTTGTTACGACTT) with the following amplification program: one cycle at 94°C for 2 min followed by 35 cycles at 94°C for 1 min, 55°C for 1 min 30 s, 72°C for 2 min and a final extension step at 72°C for 5 min. After visualizing the PCR products using 1% gel electrophoresis at a constant voltage of 110 V, fragments were sequenced by Sangon (Shanghai, China). Nucleotide sequence was subjected to homology search using the BLAST software of the NCBI. The phylogenetic tree was constructed by the neighbor-joining (NJ) method using MEGA 6.0 software according to Hall (2013).
Safety Assessment of the Strain BSXE-1601
To identify the toxicity of the strain BSXE-1601 to shrimp, its safety in shrimp was identified in the laboratory via immersion, dietary, and injection administrations. The strain BSXE-1601 was applied at a concentration of 104, 106, and 108 CFU ml–1 in the immersion assay, 104, 106, and 108 CFU g–1 in the dietary assay, and 104, 106, and 108 CFU shrimp–1 in the injection test. The detailed procedure is specified in “SI toxicity assay” in Supplementary Material.
Aquatic Pathogenic Strains
The selected indicator bacteria were 10 common aquatic pathogenic strains (Table 1). V. harveyi SRTT9, V. alginolyticus AR-1, and V. vulnificus S01P2 were isolated from diseased L. vannamei. V. splendidus BSD11 and Pseudoalteromonas sp. LPE40 were isolated from diseased sea cucumber Apostichopus japonicus. These five strains were stored in the Microbial Culture Collection Center, Laboratory of Aquaculture Ecology, KLMME, Ocean University of China, Qingdao, China. V. parahaemolyticus 20130629002S01 isolated from the AHPND-infected L. vannamei were kindly provided by the Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, PR China. Moreover, V. parahaemolyticus 20130629002S01 was identified as an AHPND-causing strain (VPAHPND 2S01 for short) (Dong et al., 2017a,b; Chen et al., 2018). Edwardsiella tarda HC01090721 and Streptococcus iniae NUF849 were kindly provided by the Laboratory of Pathology and Immunology of Aquatic Animals, KLMME, Ocean University of China, Qingdao, PR China and proved to be pathogenic strains for Japanese flounder Paralichthys olivaceus (Tang et al., 2010; Sheng et al., 2018). Aeromonas hydrophila AP40301 described as a pathogen of turbot Scophthalmus maximus (Xu et al., 2018) and Shewanella marisflavi AP629 considered as a novel pathogen of sea cucumber A. japonicus (Li et al., 2010) were both kindly provided by the Key Laboratory of Mariculture and Stock Enhancement in North China’s Sea, Agriculture Ministry, PRC, Dalian Ocean University, Dalian, China.
TABLE 1.
Aquatic animal pathogenic strains used in this study.
| Pathogenic strain | Aquatic animal species (source of the strain) | References |
| Vibrio alginolyticus AR-1 | Pacific white shrimp (Litopenaeus vannamei) | – |
| Vibrio harveyi SRTT9 | – | |
| Vibrio vulnificus S01P2 | – | |
| Vibrio parahaemolyticus 20130629002S01 | Dong et al., 2017a,b | |
| Vibrio splendidus BSD11 | Sea cucumber (Apostichopus japonicas) | – |
| Pseudoalteromonas sp. LPE40 | – | |
| Shewanella marisflavi AP629 | Li et al., 2010 | |
| Aeromonas hydrophila AP40301 | Turbot (Scophthalmus maximus) | Xu et al., 2018 |
| Edwardsiella tarda HC01090721 | Japanese flounder (Paralichthys olivaceus) | Tang et al., 2010 |
| Streptococcus iniae NUF849 | Sheng et al., 2018 |
The references of the pathogenic strains isolated by our lab are not shown here.
Microbial Fermentation
A single colony of the strain BSXE-1601 grown on nutrient agar (NA) at 28°C overnight was inoculated into a 500 ml Erlenmeyer flask containing 100 ml of nutrient broth medium. After incubating at 28°C overnight in an aerobic atmosphere on a rotary shaker with 180 rpm, 1 ml of cultural liquid was transferred as a seed into 100 ml of nutrient broth (NB) medium. The flasks were incubated at 28°C with 180 rpm overnight. Similarly, all of the aquatic pathogenic strains used for bacteriostatic experiments were grown in LB35 medium at 28°C overnight under aerobic conditions.
Assessment of Antagonistic Activity in vitro Based on Agar Diffusion Assay
The antagonistic activities of culture broth and step purification fractions were evaluated by an agar well diffusion method (Lertcanawanichakul and Sawangnop, 2008). The inoculum was prepared using the 10 selected pathogenic strains grown overnight in LB broth medium and adjusted to a concentration of 106 CFU ml–1. Aliquots of 100 μl of the culture broths were inoculated onto LB nutrient agar medium. The wells (diameter, 6 mm) were then punched in each plate and a 100 μl aliquot of strain BSXE-1601 culture broth or step purification fractions was filled into each well. The plates were aerobically incubated at 28°C for 24–48 h. The inhibition zones were then measured. Well diffusion tests were performed in triplicate for each strain.
Assessment of Antagonistic Activity in vivo Based on a Feeding Assay
Experimental Diets
To check the resistant activity of the strain BSXE-1601 against vibrios in vivo in shrimps, a feeding test was conducted in the lab.
Basal diet (control diet) was formulated with commercial shrimp food (No. 2 feed for L. vannamei, from Nantong Chia Tai Co., Ltd, Nantong, China), covered with a layer of sodium alginate and fish oil (3.15 g 500 g–1 and 4.2 ml 500 g–1, respectively) (Sun et al., 2012). Strain BSXE-1601 was cultured overnight in NB medium as described above, and then harvested by centrifugation (2800 × g for 10 min). The fresh cells were resuspended in sterile normal saline, and then sprayed to the surface of commercial shrimp food, and the mixture was covered with a layer of sodium alginate and fish oil (the same ratio with control diet). On basis of basal diet, two treatment diets were supplemented with 3.5 × 107 and 3.5 × 108 CFU g–1 of strain BSXE-1601 (named as BSXE07 and BSXE08). All the diets were kept at 4°C for using later.
Experimental Animals and Feeding Experiment
L. vannamei, obtained from hatchery of Baorong Aquatic Science and Technology Development Co., Ltd. (Qingdao, China), were fed with basal diet and acclimated simultaneously for 10 days by increasing salinity 2‰ per two days from 21 to 30‰. Then 108 similar-sized individuals (4.00 ± 0.07 g) which had been starved for 24 h were randomly distributed into 9 aquariums (53 cm × 28 cm × 34 cm, 50 l), with a density of 12 shrimps per aquarium. The shrimps were fed daily at 8 am and 5 pm for 7 days, with the feeding amount of 4–5% of shrimp weight. Each experimental group (BSXE07 and BSXE08) was repeated at three times. Uneaten feed and feces in the tank were collected by pipetting before the next feeding. During the feeding trial, the environmental conditions were suitable for shrimps (temperature, 23 ± 1°C; salinity, 30‰; pH, 8.0 ± 0.1; dissolved oxygen, >5 mg l–1). The food consumption and mortality were recorded daily.
Sample Collection
At day 4 and day 7, three shrimps were selected from each aquarium. Hepatopancreas from surface-disinfected shrimp were aseptically removed and homogenized. A series of 10-fold dilutions was prepared using sterile normal saline, and 0.1 ml from each dilution was plated on TCBS agar using spread plating. Vibrio species were enumerated after a 48-h-incubation at 28°C.
Characterization of Molecules Responsible for the Antagonistic Activity
Preliminary Isolation
(i) Preparation of cell-free supernatant (CFS). The overnight culturing broth of the strain BSXE-1601 was centrifuged at 448 × g for 10 min. The supernatant was treated with filtration using a 0.22 μm Millipore filter to obtain CFS, and the bacteria were resuspended in normal saline. The inhibitory activities of both the CFS and cell resuspension solution were determined by the agar well diffusion method as described before, using the strain VPAHPND 2S01 as the indicator bacterium.
(ii) Preliminary purification of CFS by ammonium sulfate precipitation. The strain BSXE-1601 CFS was subjected to ammonium sulfate precipitation to obtain protein precipitation and deproteinized supernatant. Protein precipitate was solubilized in 0.01 M PBS (pH 7.2) and then dialyzed to remove salt to obtain dialysate. The inhibitory activities of both the dialysate and deproteinized supernatant were determined by the agar well diffusion method, using the strain VPAHPND 2S01 as the indicator bacterium, contrasted with untreated CFS.
Physicochemical and Enzymatic Treatments
For thermostability assay, the CFS sample from the strain BSXE-1601 was subjected to heat treatment for 15, 30, 45, and 60 min at 60, 80, and 100°C, respectively. For pH stability, the strain BSXE-1601 CFS was incubated for 6 h at 28°C at different pH values, adjusted with NaOH and HCl. Sensitivity to proteinase K was tested by incubation for 30, 60, and 120 min at 37°C. The proteinase K was used at a final concentration of 1 mg ml–1. The antagonistic activity was determined before and after each treatment by the agar well diffusion method, using the strain VPAHPND 2S01 as the indicator bacterium.
Isolation and Characterization
The cell-free supernatant from the strain BSXE-1601 (5 l) was extracted with ethyl acetate (2 × 5 l) to give a yellow oily residue (6.3 g) by evaporation of menstruum in vacuum, which was subjected to chromatography over a silica gel column eluting with CH2Cl2/MeOH mixtures of a stably growing polarity (100:0, 50:1, 30:1, 20:1, 10:1, 1:1, v/v) to afford six parts (F1, 820 mg; F2, 3.60 g; F3, 187 mg; F4, 225 mg; F5, 126 mg and F6, 367 mg). Bioassay results indicated that F5 and F6 (10:1 and 1:1 CH2Cl2/MeOH) had a potent antagonistic effect against the selected pathogens. The mixture of Fraction 5 (126 mg) and Fraction 6 (367 mg) was subjected to Sephadex LH-20 chromatography (20 mm × 800 mm) and further purified by HPLC over ODS (40% MeCN-H2O, 0.05% TFA, v/v) to give compounds 1 (tR 8.37 min; 5 mg), 2 (tR 8.84 min) and 3 (tR 9.05 min). Due to the fast interconversion between 2 and 3 in methanol under room temperature, the structures of 2 and 3 were determined from the spectroscopic data of their mixture (3 mg), through careful comparison of their ESI-MS and NMR with previously reported data (Iron et al., 1982; Itoh et al., 1982).
Determination of MICs
The minimal inhibitory concentrations (MICs) of the purified antagonistic compounds for the 10 selected aquatic pathogenic strains were determined by the broth microdilution method according to Wiegand with a few modifications (Wiegand et al., 2008). Briefly, broth dilution used LB35 growth medium containing geometrically increasing concentrations (a twofold dilution series) of the antimicrobial substances, which was inoculated with 106 CFU ml–1 of pathogenic cells. The MICs were considered to be the lowest concentration of antagonistic substances that caused total inhibition of bacterial growth. MICs were also determined in triplicate for each strain.
Genome Sequencing, Assembly, and Annotation
The genomic DNA of the strain BSXE-1601 was extracted using the TIANamp bacteria DNA kit (Tiangen, China), and then sequenced with a 10-kb SMRTbellTM template library using Pacific Biosciences (PacBio) Sequel Single-Molecule Real-Time (SMRT) sequencing. Pre-assembly error correction was performed with the hierarchical genome assembly process (HGAP) of SMRT Analysis software using default parameters. Then all of the obtained data were assembled into a single circular chromosome without gaps. The genome circle was prepared using Circos ver. 0.641. Bacterial genes were predicted by Glimmer 3.02 software (Delcher et al., 2007) and gene functions were annotated by BLASTP using NR, KEGG, GO, COG, Swiss-Prot, and CAZy databases. The rRNAs and tRNAs were identified using RNAmmer 1.22 and tRNAscan-SE 1.3.13, respectively. The strain BSXE-1601 genome was analyzed using AntiSMASH (Blin et al., 2019) to predict the gene clusters coding for secondary metabolites. The Comprehensive Antibiotic Resistance Database (CARD) (Jia et al., 2016) was used to identify the genes relating to antimicrobial resistance.
Statistics
The Vibrio counts in hepatopancreas of shrimp were subjected to two-way repeated measures ANOVA, followed by Bonferroni post-test, with BSXE-1601 adding concentration and time as factors. Statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS) version 25.0 using a significance level of 5%.
Results
Identification of the Strain BSXE-1601
The strain BSXE-1601 was identified as a Gram-positive and spore-producing bacterium by Gram-staining and spore staining. The fully developed single colony of the strain BSXE-1601 grown on NA at 28°C for 24 h was moist, round or irregular in shape, and opaque and off-white in color. Further, this strain showed positive results for catalase, V-P test, and nitrate reduction test (Supplementary Table 1). The minimal inhibitory concentration of NaCl was 7% (Supplementary Table 1). With these features, the strain BSXE-1601 matched the physiological and biochemical properties of Bacillus subtilis (Krieg and Holt, 1984).
A 1,463 base pairs (bp) fragment of the 16S rRNA gene was amplified from the strain BSXE-1601. The BLAST matching analysis showed that the 16S rRNA gene sequence of the strain had a high similarity (99%) to that of B. subtilis. The phylogenetic tree was constructed by the neighbor-joining (NJ) method using MEGA 6.0 software, and it clearly showed that the strain BSXE-1601 clustered with members of B. subtilis strains (Figure 1).
FIGURE 1.
Phylogenetic tree of Bacillus subtilis BSXE-1601 and its closest relatives based on 16S rRNA sequence. The phylogenetic tree was constructed by the neighbor-joining (NJ) method using MEGA 6.0 software. The bootstrap values are shown at the branch points.
Safety Assessment of the Strain BSXE-1601
The safety assessment of the strain BSXE-1601 in shrimp was identified via the immersion, dietary, and injection administrations. None of these three detection methods caused diseases or mortality at the end of the experiment.
In vitro Antagonistic Activities of the Strain BSXE-1601
According to the agar well diffusion method of antagonistic testing, of all the ten tested pathogenic strains, B. subtilis BSXE-1601 inhibited the activities of seven strains, including VPAHPND 2S01, V. alginolyticus AR-1, V. harveyi SRTT9, V. vulnificus S01P2, Pseudoalteromonas sp. LPE40 and S. marisflavi AP629. The result shown in Table 2 indicated that BSXE-1601 certainly had potential antagonistic activity against aquatic pathogenic strains.
TABLE 2.
The inhibition activity of Bacillus subtilis BSXE-1601 to different aquatic pathogens.
| Test organisms | The diameter of the inhibition zones (mm) | Antimicrobial susceptibility |
| V. vulnificus S01P2 | 22.33 | ++ |
| V. harveyi SRTT9 | 21.43 | ++ |
| V. alginolyticus AR-1 | 19.99 | ++ |
| V. parahaemolyticus 20130629002S01 | 19.03 | ++ |
| Pseudoalteromonas sp. LPE40 | 11.08 | + |
| S. marisflavi AP629 | 14.09 | + |
| V. splendidus BSD11 | <10 | − |
| A. hydrophila AP40301 | <10 | − |
| E. tarda HC01090721 | <10 | − |
| S. iniae NUF849 | <10 | − |
“++”: susceptible; “+”: intermediate; “-”: non-susceptible or resistant.
In vivo Antagonistic Activities of the Strain BSXE-1601
The resistant activities of BSXE-1601 against vibrios in the hepatopancreas of shrimp were checked on day 4 and day 7 with two concentrations of BSXE-1601 (3.5 × 107 and 3.5 × 108 CFU g–1). The results showed that both of the concentrations of BSXE-1601 (BSXE07 and BSXE08 treatments) had significant antagonistic effect on the vibrios in the hepatopancreas of shrimp as compared with the control at each time point (P < 0.05) (Figure 2). Comparing the vibrio counts on day 7 with that on day 4, there were no significant increase in BSXE08 but the significant increase in BSXE07 and the control group (P < 0.05), which demonstrated that the high concentration of the tested B. subtilis strain exhibited a promising antagonistic effect against vibrios.
FIGURE 2.
The resistant activity of BSXE-1601 against Vibrio spp. in the hepatopancreas of shrimp on day 4 and day 7 with different concentrations of BSXE-1601. Results are expressed as mean ± standard error of three replicates. Values marked with a different letter are significantly different (P < 0.05).
Antagonistic Compounds Extraction
Preliminary Identification of the Metabolites
The antagonistic testing of the preliminary purified metabolites showed that the inhibition zone of the cell-free supernatant (21.35 mm) was bigger than that of the resuspending solution (15.25 mm). Only deproteinized supernatant had the inhibition activity (19.54 mm) while the protein in the dialysate had no activity, which indicated that the inhibitory substances were extracellular and non-proteinaceous.
Effects of Heat, pH, and Proteinase K on Antagonistic Activity
The antagonistic activity against the strain VPAHPND 2S01 was heat stable and remained unchanged after 60 min of incubation at 80°C. According to the inhibition zone, the antagonistic activity decreased slightly (14%) at 100°C after 60 min of incubation but kept active over a wide range of pH values, from 3 to 9. However, it was not resistant to strong alkaline pH, and the activity was noted to disappear at pH 12. Treatment with proteinase K did not affect the antimicrobial activity of the CFS.
Structure Elucidation of the Compounds
Careful analysis of the bioactive fractions obtained from B. subtilis BSXE-1601 using LC-MS revealed three isocoumarins like components from the chromatogram, which were further tested to be bioactive against several aquatic pathogens (Figure 3A). Large-scale fermentation, as well as chromatographic purification, allow further characterization of these three compounds as amicoumacins A-C (compounds 1-3) (Figure 3B), based on a careful comparison of their 1D NMR and ESI-MS with the published data (Itoh et al., 1982; Table 3). We observed the fast interconversion between 2 and 3 in methanol under room temperature, which was consistent with the previous description (Itoh et al., 1982), partly due to the spontaneous intramolecular condensation between the -COOH and its γ-OH in 2, as well as the hydrolysis of 5-membered lactone ring in 3. The product ratio of 1-3 was calculated to be 4.8: 1.5: 1 based on their peak area.
FIGURE 3.
LC-MS profile of ethyl acetate (EA) extract from B. subtilis BSXE-1601 cell-free supernatant. (A) Amicoumacins A-C (1–3) were revealed from the chromatogram (DAD, λ = 314 nm) based on their specific m/z: 424 [M + H]+ for 1, 425 [M + H]+ for 2 and 407 [M + H]+ for 3; (B) Structures of amicoumacins.
TABLE 3.
13C NMR (150 MHz) data of amicoumacins A-C (1–3) in DMSO-d6.
| Pos. | 1 | 2 | 3 |
| δC, type | δC, type | δC, type | |
| 1 | 169.0, C | 169.0, C | 169.1, C |
| 2 | |||
| 3 | 81.0, CH | 80.8, CH | 81.1, CH |
| 4 | 38.6, CH2 | 38.7, CH2 | 38.7, CH2 |
| 5 | 118.6, CH | 118.6, CH | 118.6, CH |
| 6 | 136.4, CH | 136.5, CH | 136.5, CH |
| 7 | 115.3, CH | 115.4, CH | 115.5, CH |
| 8 | 160.9, C | 160.9, C | 160.9, C |
| 9 | 108.4, C | 108.5, C | 108.5, C |
| 10 | 140.6, C | 140.6, C | 140.4 C |
| 1′ | 21.5, CH3 | 21.5, CH3 | 21.6, CH3 |
| 2′ | 23.4, CH3 | 23.4, CH3 | 23.4, CH3 |
| 3′ | 25.3, CH | 24.1, CH | 24.1, CH |
| 4′ | 31.5, CH2 | 29.2, CH2 | 29.2, CH2 |
| 5′ | 49.8, CH | 47.7, CH | 48.3, CH |
| 6′ | |||
| 7′ | 173.9, C | 172.6, C | 170.3, C |
| 8′ | 70.9, CH | 70.9, CH | 72.0, CH |
| 9′ | 73.8, CH | 71.3, CH | 84.0, CH |
| 10′ | 51.2, CH | 48.7, CH | 48.6, CH |
| 11′ | 33.3, CH2 | 32.3, CH2 | 34.2, CH2 |
| 12′ | 174.6, C | 174.3, C | 174.3, C |
MICs of Amicoumacins for Pathogenic Strains
Be consistent with the report of Iron et al. (1982), among the three amicoumacins, only amicoumacin A exhibited antibacterial activity (Table 4), and amicoumacins B and C were inactive against the pathogens (data didn’t show). The results revealed that amicoumacin A had good resistant activity against the tested bacterial pathogens of shrimp, especially VPAHPND 2S01, V. alginolyticus AR-1, V. harveyi SRTT9, V. vulnificus S01P2, even at a low concentration of 1.25 μg ml–1.
TABLE 4.
The minimal inhibitory concentrations (MICs) of amicoumacin A for pathogenic strains.
| Test organisms | M.I.C (μg ml–1) |
| V. vulnificus S01P2 | 1.25 |
| V. harveyi SRTT9 | 1.25 |
| V. alginolyticus AR-1 | 1.25 |
| V. parahaemolyticus 20130629002S01 | 1.25 |
| Pseudoalteromonas sp. LPE40 | 1.25 |
| S. marisflavi AP629 | 1.25 |
| S. iniae NUF849 | 10 |
| A. hydrophila AP40301 | >10 |
| E. tarda HC01090721 | >10 |
| V. splendidus BSD11 | >10 |
Genome Sequencing, Assembly, and Annotation
The genome of B. subtilis strain BSXE-1601 consisted of a circular chromosome 4,242,126 bp in size with an average GC content of 44.03%. The further analysis predicted the chromosome contains 4,325 coding DNA sequences (CDSs), 30 rRNAs, and 86 tRNAs (Table 5 and Figure 4).
TABLE 5.
General genome features of B. subtilis BSXE-1601.
| Features | Chromosome |
| Genome topology | Circular |
| Assembly size (bp) | 4,242,126 |
| G + C content (%) | 44.03 |
| Protein coding sequences (CDS) | 4,325 |
| tRNA genes | 86 |
| rRNA genes | 30 |
| Secondary metabolite gene clusters | 10 |
| GenBank accession | CP028812 |
FIGURE 4.
Circular representation of B. subtilis BSXE-1601 genome. The outer scale is in mega bases (Mb). Circle 1 (from outside to inside), the marker of genome size. Cycles 2 and 3, CDS with positive and negative chain, different colors represent different functional classifications; Circle 4, rRNA and tRNA; Circle 5, GC content, the higher value makes redder, the lower makes bluer. Circle 6, the GC-skew value, the algorithm is (G-C)/(G + C). Generally, when GC-skew > 0, the histogram is outward and expressed in red, GC-skew < 0, and the histogram is inward and expressed in blue.
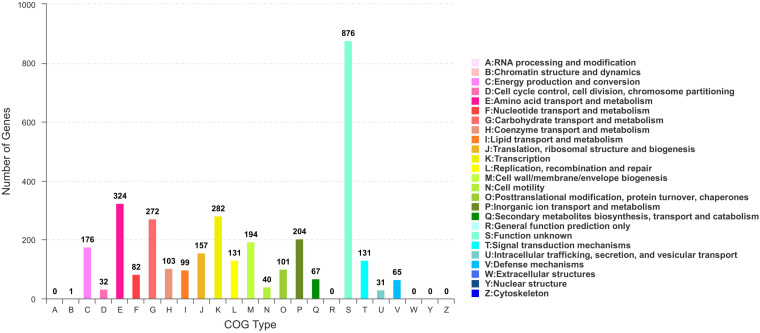
Based on COG analysis (Tatusov et al., 2003), 3315 proteins were classified into 21 functional categories. Among them, 1,327 proteins were associated with metabolism (40.03%): “amino acid transport and metabolism” (COG initial E, 9.77%), “carbohydrate transport and metabolism” (G, 8.21%), “inorganic ion transport and metabolism” (P, 6.15%), “energy production and conversion” (C, 5.31%), “coenzyme transport and metabolism” (H, 3.11%), “lipid transport and metabolism” (I, 2.99%), “nucleotide transport and metabolism” (F, 2.47%) and “secondary metabolites biosynthesis, transport, and catabolism” (Q, 2.02%) (Figure 5).
FIGURE 5.
COG functional classification in B. subtilis BSXE-1601 coding sequences.
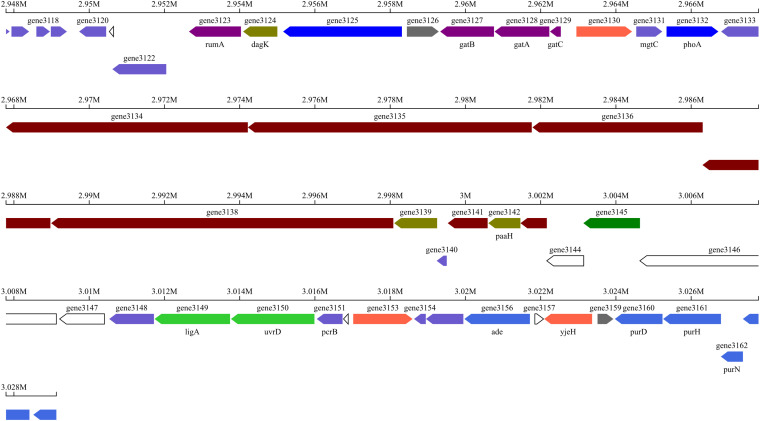
10 putative gene clusters were identified to be responsible for secondary metabolite biosynthesis using antiSMASH v.5.0 (Table 6). Of these, cluster 7 displayed high similarity to the known amicoumacin biosynthetic gene cluster. Cluster 7 included 17 genes involved in the biosynthesis of amicoumacin A, which are 3 known genes mgtC, phoA, paaH and 14 unknown genes (Table 7). The gene structure of cluster 7 was shown in Figure 6. Moreover, the secondary metabolic gene cluster 1, cluster 4, cluster 8, cluster 9 and cluster 10 had high similarity to the biosynthetic gene clusters of bacillibactin, fengycin, surfactin, bacilysin and subtilosin A, respectively.
TABLE 6.
Secondary metabolic gene clusters of B. subtilis BSXE-1601.
| Cluster | Type | Location | Similar cluster | Gene number |
| 1 | NRPS | 432092-481833 | Bacillibactin biosynthetic gene cluster, NRPS | 50 |
| 2 | Type III PKS | 1415819-1456916 | 48 | |
| 3 | Terpene | 1506043-1527939 | 19 | |
| 4 | TransATPKS-NRPS | 1625563-1757058 | Fengycin biosynthetic gene cluster, hybrid | 65 |
| 5 | Terpene | 2442478-2463282 | 25 | |
| 6 | TransATPKS-otherKS | 2651018-2751176 | Elansolid biosynthetic gene cluster, polyketide | 42 |
| 7 | Type I PKS-NRPS | 2947790-3029128 | Zwittermycin A biosynthetic gene cluster, hybrid | 49 |
| 8 | NRPS | 3305557-3370946 | Surfactin biosynthetic gene cluster, NRPS | 48 |
| 9 | NRPS | 4064957-4106375 | Bacilysin biosynthetic gene cluster, NRPS | 48 |
| 10 | Sactipeptide | 4110050-4131660 | Subtilosin A biosynthetic gene cluster, RiPP | 21 |
TABLE 7.
Secondary metabolic function genes related to the biosynthesis of amicoumacin A in B. subtilis BSXE-1601.
| Protein | Gene name | Size (aa) | Proposed function | Identity/similarity (%) | Accession No |
| Orf 1 | gene3147 | 397 | MFS transporter | 99.7 | WP_003240131.1 |
| AmiA | gene3146 | 1497 | Non-ribosomal peptide synthetase | 98.5 | WP_003240128.1 |
| AmiB | gene3145 | 501 | Serine hydrolase | 98.2 | WP_003240126.1 |
| AmiC | gene3144 | 327 | Hypothetical protein | 97.6 | WP_003240124.1 |
| AmiD | gene3143 | 233 | Thioesterase | 99.1 | WP_003240123.1 |
| AmiEb | gene3142 (paaH) | 284 | 3-Hydroxybutyryl-CoA dehydrogenase | 99.6 | WP_003240121.1 |
| AmiFb | gene3141 | 353 | Hypothetical protein | 100 | WP_003240119.1 |
| AmiGb | gene3140 | 90 | WP_003240119.1 | ||
| AmiHb | gene3139 | 380 | Acyl-CoA dehydrogenase (NADP(+)) | 98.9 | WP_003240115.1 |
| AmiI | gene3138 | 3031 | Hybrid non-ribosomal peptide synthetase/Type 1 polyketide synthase | 98.9 | WP_003240114.1 |
| AmiJ | gene3137 | 889 | Non-ribosomal peptide synthetase | 99.2 | WP_003240112.1 |
| AmiK | gene3136 | 1508 | Polyketide synthase | 99.2 | WP_003240111.1 |
| AmiL | gene3135 | 2514 | Polyketide synthase | 98.8 | WP_003240108.1 |
| AmiM | gene3134 | 2142 | Polyketide synthase | 99.3 | WP_003240106.1 |
| AmiN | gene3133 | 333 | Kinase | 99.7 | WP_003240104.1 |
| AmiO | gene3132 (phoA) | 459 | Alkaline phosphatase | 99.3 | WP_003240102.1 |
| Orf 2 | gene3131 (mgtC) | 230 | Membrane component | 99.6 | WP_003240098.1 |
FIGURE 6.
Specific gene structures in cluster 7.
Discussion
The growth of AHPND causative agent, VPAHPND 2S01, as well as V. alginolyticus AR-1, V. harveyi SRTT9, V. vulnificus S01P2, Pseudoalteromonas sp. LPE40 and S. marisflavi AP629 was inhibited by the bioactive strain BSXE-1601 isolated from both shrimp intestine and shrimp culture ponds in this study. These bacterial pathogens are not only the most important causative agents of bacterial infections in shrimp but also responsible for fatal diseases in fish, shellfish and sea cucumber (Wang et al., 1999, 2000; Li et al., 2010, 2011; Austin et al., 2012; Petton et al., 2015; Zheng et al., 2016; Boonsri et al., 2017; Chandrakala and Priya, 2017; Karnjana et al., 2019; Nguyen et al., 2019). The isolated strain BSXE-1601 was then identified as B. subtilis by sequencing the 16S rRNA gene. Recently, Bacillus species have been frequently reported to have the antagonistic properties against aquatic pathogenic bacteria, such as V. parahaemolyticus (Vinoj et al., 2013; Xu et al., 2013), V. harveyi (Liu et al., 2014; Zokaeifar et al., 2014), V. splendidus (Zhao et al., 2012), V. alginolyticus (Zhang et al., 2011), A. hydrophila and E. tarda (Ran et al., 2012). Hence, Bacillus species presented a high interest as a promising therapeutic agent against vibriosis and other diseases in aquaculture (Aly et al., 2008; Ran et al., 2012; Hoseinifar et al., 2018).
The biologically active natural products are mainly responsible for the bacteriostatic properties of Bacillus spp. (Kaspar et al., 2019; Kuebutornye et al., 2019). In the past decade, a new complex of the isocoumarins group has been extracted from the cell-free supernatant of Bacillus spp. (Saeed, 2016). Amicoumacins were the most studied and well-characterized isocoumarins which are composed of the low molecular weight phenylpropanol derivative substances. Amicoumacin A was first discovered from B. pumilus by Itoh et al. (1981) and then from different Bacillus strains (Pinchuk et al., 2002; Zidour et al., 2017; Terekhov et al., 2018) and Xenorhabdus bovienii (Park et al., 2016). The strong anti-inflammatory and antiulcer properties of amicoumacin A were reported in the 1980s (Itoh et al., 1981; Iron et al., 1982), which was subsequently associated with its anticancer activity (Li et al., 2012; Prokhorova et al., 2016). Furthermore, amicoumacin A was documented to display a pronounced bactericidal activity against clinically relevant bacterial pathogens, such as Helicobacter pylori (Pinchuk et al., 2001) and the “superbug” methicillin-resistant Staphylococcus aureus (MRSA) (Berrue et al., 2009; Lama et al., 2012; Han et al., 2013; Terekhov et al., 2020). Noteworthily, besides the clinical applications, amicoumacin A produced by B. pumilus H2 also exhibited antagonistic effect toward the fish pathogen V. vulnificus CZ-A2 in vitro (Gao et al., 2017). The discovery gave a new insight into the mechanism of the anti-Vibrio activity of Bacillus spp. However, there was limited information about the mitigation infection of amicoumacin A on aquatic animal pathogens, and its activity against the representative strain VPAHPND and other shrimp pathogens was not previously described.
In our research, the biologically active metabolite which was responsible for the antagonistic activity of B. subtilis BSXE-1601 was identified as amicoumacin A, according to its physicochemical characterization and NMR spectral data analysis. Furthermore, the MICs determination for familiar aquatic animal pathogens suggested that amicoumacin A was very effective on VPAHPND 2S01, V. alginolyticus AR-1, V. harveyi SRTT9, V. vulnificus S01P2, Pseudoalteromonas sp. LPE40 and S. marisflavi AP629, which was matched by the antagonistic properties of the strain BSXE-1601 culture broth. Our discovery provided the first evidence that amicoumacin A was highly active against the especially important Gram-negative shrimp pathogenic bacteria. These pathogens are also responsible for fatal diseases in fish, shellfish and sea cucumber (Li et al., 2010; Austin et al., 2012; Nguyen et al., 2019). Hence, our work may facilitate the application of the B. subtilis strain BSXE-1601 as the effective therapeutic for detrimental diseases in aquaculture.
To elucidate the mechanism of the action of amicoumacin A, many functional studies were conducted. Lama et al. (2012) found that when amicoumacin A was exposed to MRSA, it reduced the activity of murein hydrolase, called autolysin, which was highly needed in the regulation of continued wall extension and cellular division (Higgins et al., 1970; Gilpin et al., 1972). The recent research demonstrated that the mode of action of penicillin-induced explosive lysis of Streptococcus pneumonia, a decades-long puzzle, was also because of autolysin disorder (Flores-Kim et al., 2019). Interestingly, with the assistance of confocal microscopy and scanning electron microscopy, Gao et al. (2017) found that amicoumacin A, secreted by a marine probiotic strain, can damage the surface structure of V. vulnificus cells, as reflected by the formation of cell cavities, the presence of membrane holes, and consequent cell lysis. As a further step, Polikanov et al. (2014) suggested that amicoumacin A, as a potent protein synthesis inhibitor, could interfere with translation by locking the mRNA in the mRNA-binding channel of the 30S subunit. It indicated that amicoumacin A could interfere with bacterial and eukaryotic translation, which could be the reason for its antibacterial, antifungal, anticancer, and anti-inflammatory properties (Itoh et al., 1981; Canedo et al., 1997; Li et al., 2012; Polikanov et al., 2014; Prokhorova et al., 2016). Noteworthy, according to proteomics and metabolomics, Terekhov et al. (2018) revealed a distinct mechanism of Bacillus self-resistance to amicoumacin A, based on a subtle equilibrium of its activation and deactivation by phosphatase AmiO and kinase AmiN, respectively. Understanding the target mechanism of amicoumacin A and the on/off mode of activity switching has the potential to find novel ways of developing therapeutic treatments against aquaculture diseases.
To interrogate the potential biosynthetic pathway of amicoumacin A, the whole genome sequencing of the strain BSXE-1601 was conducted in our study. Gene clusters related to the biosynthesis of secondary metabolites were predicted based on the analysis with antiSMASH. It revealed that the secondary metabolite biosynthesis gene cluster 7 in the strain BSXE-1601 encoded a hybrid modular polyketide synthase-non-ribosomal peptide synthetase (PKS-NRPS), and 17 genes (gene3131-3147) in this cluster were proposed to be responsible for amicoumacin A biosynthesis. Inspection of this genetic locus identified a PKS-NRPS hybrid protein encoded by gene3138, three PKS encoded by gene3134-3136, and two NRPSs encoded by gene3137 and gene3146. This was consistent with the results of Li et al. (2015), who succeeded in the heterologous expression of amicoumacin A biosynthesis genes amiA-O and provided unequivocal evidence that the ami locus encodes amicoumacin biosynthesis. Moreover, with the successful heterologous expression of the amicoumacin biosynthetic gene cluster and the ability to readily inactivate individual ami genes, they identified that inactive preamicoumacin precursors were first synthesized and then hydrolyzed by the asparagine-specific peptidase (encoded by amiB) into the active component amicoumacin A.
Additionally, in the present study, with the antiSMASH analysis, another 9 secondary metabolic gene clusters were found in B. subtilis BSXE-1601 genomes: three encoding NRPSs, two terpene synthases (TSs), one Type III PKS, one trans-Acyl transferase polyketide synthetase (TransATPKS)-otherKS, one TransATPKS-NRPS hybrid, one sactipeptide synthase. The diverse secondary metabolic pathways indicated the high biosynthetic capability of the strain BSXE-1601. Cluster 1, 4, 8, 9, and 10 were highly similar to the known clusters encoding bacillibactin, fengycin, surfactin, bacilysin, and subtilosin A, all of which have antimicrobial/antibacterial activities (Arguelles-Arias et al., 2009; Chen et al., 2009). For instance, bacilysin is one of the simplest non-ribosomal peptides produced by B. subtilis, with antagonistic properties against several bacteria and fungi (Kenig and Abraham, 1976). Surfactin, the most powerful biosurfactant known to date, is another NRP which inhibits the biofilm formation of Escherichia coli, Salmonella enterica, Proteus mirabilis, and Staphylococcus aureus (Mireles et al., 2001; Liu et al., 2019). Subtilosin A is one of the ribosomally synthesized and post-translationally modified peptides from B. subtilis, which displayed activity against gram-positive and gram-negative bacteria (Shelburne et al., 2007; Huang et al., 2009). A large number of beneficial genes BSXE-1601 possessing means that the strain BSXE-1601 may have a high potential to be used as a multifunctional biological agent in aquaculture by suppressing pathogenic microbes of aquatic animals. However, the related functional genes still need to be verified. Since the yields of secondary metabolites differ in different species and even under different culture conditions, the isolation and identification of the biologically active metabolites from B. subtilis BSXE-1601 still need to be investigated for a better understanding of BSXE-1601. Furthermore, because of ecologically adaptive changes, members of B. subtilis exhibit considerable genomic diversity (Earl et al., 2008). Hence, to testify if the biosynthetic capabilities of amicoumacin A, bacillibactin, fengycin, surfactin, bacilysin and subtilosin A are BSXE-1601 specific, a further comparative genome study should be performed between BSXE-1601 and other B. subtilis strains.
Conclusion
In conclusion, the results may provide new insights into the possible mechanism of antagonistic activity of B. subtilis strain BSXE-1601 against the bacterial pathogens of shrimp. Amicoumacins A, B, and C belonging to isocoumarins, were purified from the cell-free supernatant from the strain BSXE-1601 but only amicoumacin A was demonstrated to be responsible for this anti-Vibrio activity. Our reports gave the first description in which the resistances of amicoumacin A against shrimp pathogens, including the AHPND-causing strain V. parahaemolyticus VPAHPND 2S01. However, whether amicoumacin A specifically targets the primary virulence factor mediating AHPND etiology and mortality in shrimp or not still needs to be clarified. The complete genome sequence of BSXE-1601 aided to elucidate amicoumacin A biosynthetic gene cluster and its potential metabolic pathway. This work may further facilitate the heterologous expression of the natural product amicoumacin A biosynthetic pathway to increase the yield of amicoumacin A. The predicted diverse secondary metabolic pathways indicated the ability of BSXE-1601 to produce a variety of effective antibacterial products, and its high potential to be applied as a multifunctional biological agent in aquaculture by suppressing pathogenic microbes of aquatic animals. Sustainable aquaculture may be assured with the exploration of this modest microbicidal technique.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP028812.
Ethics Statement
All animal experiments were carried out in accordance with the U.K. Animals (Scientific Procedures) Act, 1986, and associated guidelines, EU Directive 2010/63/EU for animal experiments.
Author Contributions
XT and WZ provided the experimental ideas and the design of this study. DW did the experiment and wrote the manuscript with assistance of JL, GZ, and KZ. WW, WJ, HL, and VK helped to analyze the data and revise the manuscript. SD helped to revise the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Laboratory of Aquaculture Ecology and Key Laboratory of Marine Drugs, Ocean University of China, for the support.
Funding. This work was supported by National Key Research and Development Program of China (Grant No. 2019YFD0900403 & 2017YFE0122100), National Natural Science Foundation of China (Grant No. 41976095), Natural Science Foundation of Shandong (Grant No. ZR2017MC027), Specialized Project of City Demonstration for the Innovation and Development of Marine Economy of Qingdao (Grant No. 2016-476), and the Specialized Project of Regional Demonstration for the Innovation and Development of Marine Economy of Guangdong Province (Grant No. GD2013-B003-005).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.581802/full#supplementary-material
References
- Ajadi A., Sabri M. Y., Dauda A. B., Ina-Salwany M. Y., Hasliza A. H. (2016). Immunoprophylaxis: A Better Alternative Protective Measure against Shrimp Vibriosis -A Review. PJSRR 2 2028–2462. [Google Scholar]
- Aly S. M., Ahmed Y. A.-G., Ghareeb A. A.-A., Mohamed M. F. (2008). Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreochromis niloticus) to challenge infections. Fish Shellf. Immunol. 25 128–136. 10.1016/j.fsi.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Arguelles-Arias A., Ongena M., Halimi B., Lara Y., Brans A., Joris B., et al. (2009). Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microbial. Cell Fact. 8:63. 10.1186/1475-2859-8-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin B., Austin D. A., Austin B., Austin D. A. (2012). Bacterial fish pathogens The Netherlands: Springer, (Vol. 481 pp 482). [Google Scholar]
- Bartholomae M., Buivydas A., Viel J. H., Montalbán-López M., Kuipers O. P. (2017). Major gene-regulatory mechanisms operating in ribosomally synthesized and post-translationally modified peptide (RiPP) biosynthesis. Mole. Microbiol. 106 186–206. 10.1111/mmi.13764 [DOI] [PubMed] [Google Scholar]
- Berrue F., Ibrahim A., Boland P., Kerr R. G. (2009). Newly isolated marine Bacillus pumilus (SP21): A source of novel lipoamides and other antimicrobial agents. Pure Appl. Chem. 81 1027–1031. 10.1351/pac-con-08-09-25 [DOI] [Google Scholar]
- Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucl. Acids Res. 47 W81–W87. 10.1093/nar/gkz310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsri N., Rudtanatip T., Withyachumnarnkul B., Wongprasert K. (2017). Protein extract from red seaweed Gracilaria fisheri prevents acute hepatopancreatic necrosis disease (AHPND) infection in shrimp. J. Appl. Phycol. 29 1597–1608. 10.1007/s10811-016-0969-2 [DOI] [Google Scholar]
- Canedo L. M., Puentes J. L. F., Baz J. P., Acebal C., Calle F. D. E. L., Grávalos D. G., et al. (1997). PM-94128, a new isocoumarin antitumor agent produced by a marine bacterium. J. Antibiot. 50 175–176. 10.7164/antibiotics.50.175 [DOI] [PubMed] [Google Scholar]
- Chandrakala N., Priya S. (2017). Vibriosis in Shrimp Aquaculture A Review. Int. J. Scientif. Res. Sci. 3 27–33. [Google Scholar]
- Chen F.-R., Liu P.-C., Lee K.-K. (2000). Lethal attribute of serine protease secreted by Vibrio alginolyticus strains in kuruma prawn Penaeus japonicus. Z. Naturforsch. C 55 94–99. 10.1515/znc-2000-1-218 [DOI] [PubMed] [Google Scholar]
- Chen M., Dong X., Qiu L., Wan X., Xie G., Huang J. (2018). Quantitative Analysis of Acute Hepatopancreatic Necrosis Disease Causing Vibrio parahaemolyticus (VPAHPND) in Infected Litopenaeus vannamei. Progr. Fish. Sci. 39 93–100. 10.19663/j.issn2095-9869.20170414001 [DOI] [Google Scholar]
- Chen X. H., Koumoutsi A., Scholz R., Schneider K., Vater J., Süssmuth R., et al. (2009). Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 140 27–37. 10.1016/j.jbiotec.2008.10.011 [DOI] [PubMed] [Google Scholar]
- Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23 673–679. 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckens K. R., Vandenberghe J., Beladjal L., Huys G., Mertens J., Swings J. (1998). Aeromonas hydrophila causes “black disease” in fairy shrimps (Anostraca; Crustacea). J. Fish Dis. 21 113–119. [DOI] [PubMed] [Google Scholar]
- Dong X., Bi D., Wang H., Zou P., Xie G., Wan X., et al. (2017a). pirABvp-Bearing Vibrio parahaemolyticus and Vibrio campbellii pathogens isolated from the Same AHPND-affected pond possess highly similar pathogenic plasmids. Front. Microbiol. 8 1–9. 10.3389/fmicb.2017.01859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Wang H., Xie G., Zou P., Guo C., Liang Y., et al. (2017b). An isolate of Vibrio campbellii carrying the pir VP gene causes acute hepatopancreatic necrosis disease. Emerg. Microbes. Infect. 6:e2. 10.1038/emi.2016.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl A. M., Losick R., Kolter R. (2008). Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16 269–275. 10.1016/j.tim.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Kim J., Dobihal G. S., Fenton A., Runder D. Z., Bernhardt T. G. (2019). A switch in surface polymer biogenesis triggers growth-phase-dependent and antibiotic-induced bacteriolysis. ELife 8 1–23. 10.7554/eLife.44912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. Y., Liu Y., Miao L. L., Li E. W., Hou T. T., Liu Z. P. (2017). Mechanism of anti-Vibrio activity of marine probiotic strain Bacillus pumilus H2, and characterization of the active substance. AMB Expr. 7:23 10.1186/s13568-017-0323-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin R. W., Chatterjee A. N., Young F. E. (1972). Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of Staphylococcus aureus. J. Bacteriol. 111 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gil B., Roque A., Chimetto L., Moreira A. P. B., Lang E., Thompson F. L. (2012). Vibrio alfacsensis sp. nov., isolated from marine organisms. Int. J. System. Evol. Microbiol. 62 2955–2961. 10.1099/ijs.0.033191-0 [DOI] [PubMed] [Google Scholar]
- Greule A., Zhang S., Paululat T., Bechthold A. (2017). From a natural product to its biosynthetic gene cluster: A demonstration using polyketomycin from Streptomyces diastatochromogenes tü6028. J. Visual. Exper. 2017 1–9. 10.3791/54952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mole. Biol. Evol. 30 1229–1235. 10.1093/molbev/mst012 [DOI] [PubMed] [Google Scholar]
- Han J. E., Choi S.-K., Han S.-H., Lee S. C., Jeon H. J., Lee C., et al. (2020). Genomic and histopathological characteristics of Vibrio parahaemolyticus isolated from an acute hepatopancreatic necrosis disease outbreak in Pacific white shrimp (Penaeus vannamei) cultured in Korea. Aquaculture 524:735284 10.1016/j.aquaculture.2020.735284 [DOI] [Google Scholar]
- Han X., Liu S., Wang F., Liu J., Hu X., You X., et al. (2013). Amicoumacin Research Progress of Amicoumacin Group Antibiotic. World Notes Antibio. 34 106–115. 10.13461/j.cnki.wna.004659 [DOI] [Google Scholar]
- Hasson K. W., Wyld E. M., Fan Y., Lingsweiller S. W., Weaver S. J., Cheng J., et al. (2009). Streptococcosis in farmed Litopenaeus vannamei: a new emerging bacterial disease of penaeid shrimp. Dis. Aqua. Organ. 86 93–106. 10.3354/dao02132 [DOI] [PubMed] [Google Scholar]
- Higgins M. L., Pooley H. M., Shockman G. D. (1970). Site of initiation of cellular autolysis in Streptococcus faecalis as seen by electron microscopy. J. Bacteriol. 103 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseinifar S. H., Sun Y. Z., Wang A., Zhou Z. (2018). Probiotics as means of diseases control in aquaculture, a review of current knowledge and future perspectives. Front. Microbiol. 9:1–18. 10.3389/fmicb.2018.02429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Geng H., Miyyapuram V. R., Sit C. S., Vederas J. C., Nakano M. M. (2009). Isolation of a variant of subtilosin A with hemolytic activity. J. Bacteriol. 191 5690–5696. 10.1128/JB.00541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug J. J., Krug D., Müller R. (2020). Bacteria as genetically programmable producers of bioactive natural products. Nat. Rev. Chem. 4 172–193. 10.1038/s41570-020-0176-1 [DOI] [PubMed] [Google Scholar]
- Iron J., Shomura T., Omoto S., Miyado S., Yuda Y., Shibata U., et al. (1982). Isolation, physicochemical properties and biological activities of amicoumacins produced by Bacillus pumilus. Agricul. Biol. Chem. 46 1255–1259. 10.1080/00021369.1982.10865227 [DOI] [Google Scholar]
- Itoh J., Omoto S., Nishizawa N., Kodama Y., Inouye S. (1982). Chemical structures of amicoumacins produced by Bacillus pumilus. Agricul. Biol. Chem. 46 2659–2665. 10.1080/00021369.1982.10865491 [DOI] [Google Scholar]
- Itoh J., Omoto S., Shomura T., Nishizawa N., Miyado S., Yuda Y., et al. (1981). Amicoumacin-A, a new antibiotic with strong antiinflammatory and antiulcer activity. J. Antibio. 34 611–613. 10.7164/antibiotics.34.611 [DOI] [PubMed] [Google Scholar]
- Jayasree L., Janakiram P., Madhavi R. (2006). Characterization of Vibrio spp. associated with diseased shrimp from culture ponds of Andhra Pradesh (India). J. World Aquac. Soc. 37 523–532. 10.1111/j.1749-7345.2006.00066.x [DOI] [Google Scholar]
- Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., Tsang K. K., et al. (2016). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucl. Acids Res. 45 D566–D573. 10.1093/nar/gkw1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi J., Srisala J., Truong V. H., Chen I. T., Nuangsaeng B., Suthienkul O., et al. (2014). Variation in Vibrio parahaemolyticus isolates from a single Thai shrimp farm experiencing an outbreak of acute hepatopancreatic necrosis disease (AHPND). Aquaculture 42 297–302. 10.1016/j.aquaculture.2014.03.030 [DOI] [Google Scholar]
- Karnjana K., Soowannayan C., Wongprasert K. (2019). Ethanolic extract of red seaweed Gracilaria fisheri and furanone eradicate Vibrio harveyi and Vibrio parahaemolyticus biofilms and ameliorate the bacterial infection in shrimp. Fish Shellf. Immunol. 88 91–101. 10.1016/j.fsi.2019.01.058 [DOI] [PubMed] [Google Scholar]
- Kaspar F., Neubauer P., Gimpel M. (2019). Bioactive Secondary Metabolites from Bacillus subtilis: A Comprehensive Review. J. Nat. Prod. 82 2038–2053. 10.1021/acs.jnatprod.9b00110 [DOI] [PubMed] [Google Scholar]
- Kenig M., Abraham E. P. (1976). Antimicrobial activities and antagonists of bacilysin and anticapsin. Microbiology 94 37–45. 10.1099/00221287-94-1-37 [DOI] [PubMed] [Google Scholar]
- Kondo H., Van P. T., Dang L. T., Hirono I. (2015). Draft genome sequence of non-Vibrio parahaemolyticus acute hepatopancreatic necrosis disease strain KC13. 17.5, isolated from diseased shrimp in Vietnam. Gen. Announc. 3 915–978e. 10.1128/genomeA.00978-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongrueng J., Yingkajorn M., Bunpa S., Sermwittayawong N., Singkhamanan K., Vuddhakul V. (2015). Characterization of Vibrio parahaemolyticus causing acute hepatopancreatic necrosis disease in southern Thailand. J. Fish Dis. 38 957–966. 10.1111/jfd.12308 [DOI] [PubMed] [Google Scholar]
- Krieg N. R., Holt J. G. (1984). Bergey’s manual of systematic bacteriology. Taipe: Yi Hsien Publishing Co. [Google Scholar]
- Kuebutornye F. K. A., Abarike E. D., Lu Y. (2019). A review on the application of Bacillus as probiotics in aquaculture. Fish Shellf. Immunol. 87 820–828. 10.1016/j.fsi.2019.02.010 [DOI] [PubMed] [Google Scholar]
- Lama A., Pané-Farré J., Chon T., Wiersma A. M., Sit C. S., Vederas J. C., et al. (2012). Response of methicillin-resistant Staphylococcus aureus to amicoumacin a. PLoS One 7:e34037. 10.1371/journal.pone.0034037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavilla-Pitogo C. R., Baticados M. C. L., Cruz-Lacierda E. R., Leobert D. (1990). Occurrence of luminous bacterial disease of Penaeus monodon larvae in the Philippines. Aquaculture 91 1–13. 10.1016/0044-8486(90)90173-K [DOI] [Google Scholar]
- Lavilla-Pitogo C. R., Leaño E. M., Paner M. G. (1998). Mortalities of pond-cultured juvenile shrimp, Penaeus monodon, associated with dominance of luminescent vibrios in the rearing environment. Aquaculture 164 337–349. 10.1016/S0044-8486(98)00198-7 [DOI] [Google Scholar]
- Lertcanawanichakul M., Sawangnop S. (2008). A comparison of two methods used for measuring the antagonistic activity of Bacillus species. Wala. J. Sci. Technol. 5 161–171. [Google Scholar]
- Li H., Qiao G., Li Q., Zhou W., Won K. M., Xu D., et al. (2010). Biological characteristics and pathogenicity of a highly pathogenic Shewanella marisflavi infecting sea cucumber, Apostichopus japonicus. J. Fish Dis. 33 865–877. 10.1111/j.1365-2761.2010.01189.x [DOI] [PubMed] [Google Scholar]
- Li Q., Qi N., Yang L., Li H., Wang Y. (2011). Partial characterization of polyclonal antibody against bacterium Shewanella marisflavi. J. Dalian Ocean Univ. 4. [Google Scholar]
- Li Y., Li Z., Yamanaka K., Xu Y., Zhang W., Vlamakis H., et al. (2015). Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Scientif. Rep. 5 1–7. 10.1038/srep09383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Xu Y., Liu L., Han Z., Lai P. Y., Guo X., et al. (2012). Five new amicoumacins isolated from a marine-derived Bacterium Bacillus subtilis. Mar. Drugs 10 319–328. 10.3390/md10020319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner D. V. (1996). A Handbook of Shrimp Pathology and Diagnostic Procedures for Diseases of Cultured Penaeid Shrimp, Baton Rouge, LA: World Aquaculture Society. [Google Scholar]
- Lightner D. V., Redman R. M., Pantoja C. R., Noble B. L., Tran L. (2012). Early mortality syndrome affects shrimp in Asia. Global Aquac. Adv. 15:40. [Google Scholar]
- Liu H., Li Z., Tan B., Lao Y., Duan Z., Sun W., et al. (2014). Isolation of a putative probiotic strain S12 and its effect on growth performance, non-specific immunity and disease-resistance of white shrimp, Litopenaeus vannamei. Fish Shellf. Immunol. 41 300–307. 10.1016/j.fsi.2014.08.028 [DOI] [PubMed] [Google Scholar]
- Liu J., Li W., Zhu X., Zhao H., Lu Y., Zhang C., et al. (2019). Surfactin effectively inhibits Staphylococcus aureus adhesion and biofilm formation on surfaces. Appl. Microbiol. Biotechnol. 103 4565–4574. 10.1007/s00253-019-09808-w [DOI] [PubMed] [Google Scholar]
- Liu L., Xiao J., Xia X., Pan Y., Yan S., Wang Y. (2015). Draft genome sequence of Vibrio owensii strain SH-14, which causes shrimp acute hepatopancreatic necrosis disease. Genom. Announc. 3 1315–1395e. 10.1128/genomeA.01395-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longyant S., Rukpratanporn S., Chaivisuthangkura P., Suksawad P., Srisuk C., Sithigorngul W., et al. (2008). Identification of Vibrio spp. in vibriosis Penaeus vannamei using developed monoclonal antibodies. J. Invertebr. Pathol. 98 63–68. 10.1016/j.jip.2007.10.013 [DOI] [PubMed] [Google Scholar]
- Mireles J. R., Toguchi A., Harshey R. M. (2001). Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183 5848–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. V., Alfaro A. C., Merien F. (2019). Omics approaches to investigate host–pathogen interactions in mass mortality outbreaks of Crassostrea gigas. Rev. Aquacul. 11 1308–1324. 10.1111/raq.12294 [DOI] [Google Scholar]
- Park H. B., Perez C. E., Perry E. K., Crawford J. M. (2016). Activating and attenuating the amicoumacin antibiotics. Molecules 21 1–17. 10.3390/molecules21070824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petton B., Bruto M., James A., Labreuche Y., Alunno-Bruscia M., Le Roux F., et al. (2015). Crassostrea gigas mortality in France: the usual suspect, a herpes virus, may not be the killer in this polymicrobial opportunistic disease. Front. Microbiol. 6:686. 10.3389/fmicb.2015.00686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinchuk I. V., Bressollier P., Sorokulova I. B., Verneuil B., Urdaci M. C. (2002). Amicoumacin antibiotic production and genetic diversity of Bacillus subtilis strains isolated from different habitats. Res. Microbiol. 153 269–276. 10.1016/S0923-2508(02)01320-7 [DOI] [PubMed] [Google Scholar]
- Pinchuk I. V., Bressollier P., Verneuil B., Fenet B., Sorokulova I. B., Mégraud F., et al. (2001). In vitro anti-Helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicr. Agents Chemother. 45 3156–3161. 10.1128/AAC.45.11.3156-3161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov Y. S., Osterman I. A., Szal T., Tashlitsky V. N., Serebryakova M. V., Kusochek P., et al. (2014). Amicoumacin A Inhibits Translation by Stabilizing mRNA Interaction with the Ribosome. Mole. Cell 56 531–540. 10.1016/j.molcel.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova I. V., Akulich K. A., Makeeva D. S., Osterman I. A., Skvortsov D. A., Sergiev P. V., et al. (2016). Amicoumacin A induces cancer cell death by targeting the eukaryotic ribosome. Scientif. Rep. 6 1–10. 10.1038/srep27720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran C., Carrias A., Williams M. A., Capps N., Dan B. C. T., Newton J. C., et al. (2012). Identification of Bacillus strains for biological control of catfish pathogens. PLoS One 7:e45793. 10.1371/journal.pone.0045793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer D., Bode H. B. (2014). A natural prodrug activation mechanism in the biosynthesis of nonribosomal peptides. Nat. Prod. Rep. 31 154–159. [DOI] [PubMed] [Google Scholar]
- Rengpipat S., Tunyanun A., Fast A. W., Piyatiratitivorakul S., Menasveta P. (2003). Enhanced growth and resistance to Vibrio challenge in pond-reared black tiger shrimp Penaeus monodon fed a Bacillus probiotic. Dis. Aquat. Organ. 55 169–173. 10.3354/dao055169 [DOI] [PubMed] [Google Scholar]
- Restrepo L., Bayot B., Arciniegas S., Bajaña L., Betancourt I., Panchana F., et al. (2018). PirVP genes causing AHPND identified in a new Vibrio species (Vibrio punensis) within the commensal Orientalis clade. Scientif. Rep. 8:13080. 10.1038/s41598-018-30903-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro J. C., Tamanini R., Soares B. F., De Oliveira A. M., De Godoi Silva F., Da Silva F. F., et al. (2016). Efficiency of boiling and four other methods for genomic DNA extraction of deteriorating spore-forming bacteria from milk. Sem.:Cienc. Agrar. 37 3069–3078. 10.5433/1679-0359.2016v37n5p3069 [DOI] [Google Scholar]
- Saeed A. (2016). Isocoumarins, miraculous natural products blessed with diverse pharmacological activities. Eur. J. Med. Chem. 116 290–317. 10.1016/j.ejmech.2016.03.025 [DOI] [PubMed] [Google Scholar]
- Shelburne C. E., An F. Y., Dholpe V., Ramamoorthy A., Lopatin D. E., Lantz M. S. (2007). The spectrum of antimicrobial activity of the bacteriocin subtilosin A. J. Antimicr. Chemother. 59 297–300. 10.1093/jac/dkl495 [DOI] [PubMed] [Google Scholar]
- Sheng X., Gao J., Liu H., Tang X., Xing J., Zhan W. (2018). Recombinant phosphoglucomutase and CAMP factor as potential subunit vaccine antigens induced high protection against Streptococcus iniae infection in flounder (Paralichthys olivaceus). J. Appl. Microbiol. 125 997–1007. 10.1111/jam.13948 [DOI] [PubMed] [Google Scholar]
- Sirikharin R., Taengchaiyaphum S., Sanguanrut P., Chi T. D., Mavichak R., Proespraiwong P., et al. (2015). Characterization and PCR detection of binary, Pir-like toxins from Vibrio parahaemolyticus isolates that cause acute hepatopancreatic necrosis disease (AHPND) in shrimp. PloS One 10:e0126987. 10.1371/journal.pone.0126987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Liu F., Song X.-L., Mai K.-S., Li Y.-H., Huang J. (2012). Effects of adding probiotics in the feed on non-specific immune gene expression and disease resistance of Litopenaeus vannamei. Oceanologia et Limnologia Sinica 43 845–851. [Google Scholar]
- Tang X., Zhan W., Sheng X., Chi H. (2010). Immune response of Japanese flounder Paralichthys olivaceus to outer membrane protein of Edwardsiella tarda. Fish Shellf. Immunol. 28 333–343. 10.1016/j.fsi.2009.11.015 [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinform. 4 1–41. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terekhov S. S., Nazarov A. S., Mokrushina Y. A., Baranova M. N., Potapova N. A., Malakhova M. V., et al. (2020). Deep functional profiling facilitates the evaluation of the antibacterial potential of the antibiotic amicoumacin. Antibiotics 9 1–12. 10.3390/antibiotics9040157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terekhov S. S., Smirnov I. V., Malakhova M. V., Samoilov A. E., Manolo A. I., Nazarov A. S., et al. (2018). Ultrahigh-throughput functional profiling of microbiota communities. Proc. Nat. Acad. Sci. U S A 115 9551–9556. 10.1073/pnas.1811250115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L., Nunan L., Redman R. M., Mohney L. L., Pantoja C. R., Fitzsimmons K., et al. (2013). Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aqua. Organ. 105 45–55. 10.3354/dao02621 [DOI] [PubMed] [Google Scholar]
- Vinoj G., Vaseeharan B., DavidJayaseelan B., Rajakumaran P., Ravi C. (2013). Inhibitory effects of Bacillus licheniformis (DAB1) and Pseudomonas aeruginosa (DAP1) against Vibrio parahaemolyticus isolated from Fenneropenaeus indicus. Aquacult. Int. 21 1121–1135. 10.1007/s10499-012-9617-2 [DOI] [Google Scholar]
- Wang C., Yang B., Song X., Huang J. (1999). Detecting vibriosis (Vibro vulnificus) of the abalone fester disease by double antibody sandwich elisa. Mar. Fish. Res. 1. [Google Scholar]
- Wang A., Ran C., Wang Y., Zhang Z., Ding Q., Yang Y., et al. (2019). Use of probiotics in aquaculture of China—a review of the past decade. Fish Shellf. Immunol. 86 734–755. 10.1016/j.fsi.2018.12.026 [DOI] [PubMed] [Google Scholar]
- Wang Y. G., Lee K. L., Najiah M., Shariff M., Hassan M. D. (2000). A new bacterial white spot syndrome (BWSS) in cultured tiger shrimp Penaeus monodon and its comparison with white spot syndrome (WSS) caused by virus. Dis. Aqua. Organ. 41 9–18. 10.3354/dao041009 [DOI] [PubMed] [Google Scholar]
- Wiegand I., Hilpert K., Hancock R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Proto. 3 163–175. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- Xu D., Wang Y., Sun L., Liu H., Li J. (2013). Inhibitory activity of a novel antibacterial peptide AMPNT-6 from Bacillus subtilis against Vibrio parahaemolyticus in shrimp. Food Control 30 58–61. 10.1016/j.foodcont.2012.07.025 [DOI] [Google Scholar]
- Xu Y., Yao H., Guo Q., Li H., Ye S., Li R., et al. (2018). Isolation, virulence and ERIC-PCR genotyping of Aeromonas hydrophila in farmed turbot Scophthalmus maximus (L.). J. Dalian Ocean Unive. 33 430–434. 10.16535/j.cnki.dlhyxb.2018.04.003 [DOI] [Google Scholar]
- Zhang Q., Tan B., Mai K., Zhang W., Ma H., Ai Q., et al. (2011). Dietary administration of Bacillus (B. licheniformis and B. subtilis) and isomaltooligosaccharide influences the intestinal microflora, immunological parameters and resistance against Vibrio alginolyticus in shrimp, Penaeus japonicus (Decapoda: Penaeidae). Aquacul. Res. 42 943–952. 10.1111/j.1365-2109.2010.02677.x [DOI] [Google Scholar]
- Zhao Y., Zhang W., Xu W., Mai K., Zhang Y., Liufu Z. (2012). Effects of potential probiotic Bacillus subtilis T13 on growth, immunity and disease resistance against Vibrio splendidus infection in juvenile sea cucumber Apostichopus japonicus. Fish Shellf. Immunol. 32 750–755. 10.1016/j.fsi.2012.01.027 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Yu M., Liu Y., Su Y., Xu T., Yu M., et al. (2016). Comparison of cultivable bacterial communities associated with Pacific white shrimp (Litopenaeus vannamei) larvae at different health statuses and growth stages. Aquaculture 451 163–169. 10.1016/j.aquaculture.2015.09.020 [DOI] [Google Scholar]
- Zhou H., Gai C., Ye G., An J., Liu K., Xu L., et al. (2019). Aeromonas hydrophila, an Emerging Causative Agent of Freshwater-Farmed Whiteleg shrimp Litopenaeus vannamei. Microorganisms 7:450. 10.3390/microorganisms7100450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidour M., Chevalier M., Belguesmia Y., Cudennec B., Grard T., Drider D., et al. (2017). Isolation and characterization of bacteria colonizing Acartia tonsa copepod eggs and displaying antagonist effects against Vibrio anguillarum, Vibrio alginolyticus and other pathogenic strains. Front. Microbiol. 8:1919. 10.3389/fmicb.2017.01919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zokaeifar H., Babaei N., Saad C. R., Kamarudin M. S., Sijam K., Balcazar J. L. (2014). Administration of Bacillus subtilis strains in the rearing water enhances the water quality, growth performance, immune response, and resistance against Vibrio harveyi infection in juvenile white shrimp, Litopenaeus vannamei. Fish Shellf. Immunol. 36 68–74. 10.1016/j.fsi.2013.10.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP028812.