Abstract
Orofacial clefts and their management impose a substantial burden on patients, on their families, and on the health system. Under the current standard of care, affected patients are subjected to a lifelong journey of corrective surgeries and multidisciplinary management to replace bone and soft tissues, as well as restore esthetics and physiologic functions while restoring self-esteem and psychological health. Hence, a better understanding of the dynamic interplay of molecular signaling pathways at critical phases of palate development is necessary to pioneer novel prenatal interventions. Such pathways include transforming growth factor–β (Tgfβ), sonic hedgehog (Shh), wingless-integrated site (Wnt)/β-catenin, bone morphogenetic protein (Bmp), and fibroblast growth factor (Fgf) and its associated receptors, among others. Here, we summarize commonly used surgical methods used to correct cleft defects postnatally. We also review the advances made in prenatal diagnostics of clefts through imaging and genomics and the various in utero surgical corrections that have been attempted thus far. An overview of how key mediators of signaling that drive palatogenesis are emphasized in the context of the framework and rationale for the development and testing of therapeutics in animal model systems and in humans is provided. The pros and cons of in utero therapies that can potentially restore molecular homeostasis needed for the proper growth and fusion of palatal shelves are presented. The theme advanced throughout this review is the need to develop preclinical molecular therapies that could ultimately be translated into human trials that can correct orofacial clefts at earlier stages of development.
Keywords: cleft palate, craniofacial, signaling molecules, replacement therapies, prenatal drug delivery, translational medicine
Introduction
Orofacial clefts (OFCs) are the most common craniofacial birth defect in humans, arising early in pregnancy after a disruption or failure in the growth and fusion of craniofacial tissues. The most common OFCs are clefts of the lip alone (CL), cleft lip with palate (CLP), or cleft palate (CP) (Leslie and Marazita 2013). Multiple genes and environmental factors contribute to this group of complex disorders that present as isolated/nonsyndromic defects or those that involve other organ systems as part of a syndrome. Nonsyndromic clefts of the lip and/or palate (CL/P) show a distribution of prevalence across ethnic and geographic groups, with rates ranging from 1:500 to 1:2,500 live human births. Since embryological origins are different for the upper lip and secondary palate, CL and CLP are viewed as variants of the same defect that differ in severity (Marazita 2012).
One-third of OFCs are part of syndromes, including DiGeorge, Van der Woude, and Treacher-Collins syndromes, as well as the Pierre Robin sequence (Pereira et al. 2018). Among the known environmental associations are maternal smoking or passive smoke exposure (Sabbagh et al. 2015), viral infection (James et al. 2014), phenytoin and other antiepileptic medication use (Veroniki et al. 2017), isotretinoin (Lammer et al. 1985), and deficiencies of zinc, folic acid, and other micronutrients (Wehby and Murray 2010). A maternal history of diabetes mellitus, overweight and obesity, advanced maternal and/or paternal age, and parental consanguinity have also been identified as factors that contribute to cleft development (Sabbagh et al. 2014). However, current evidence does not support the speculation that fetal exposure to alcohol early in development may be implicated in OFCs (Bell et al. 2014). The genetic origin of OFCs is supported by familial studies that show a 32-fold higher risk in the proband if first-degree relatives are affected by CL/P (Sivertsen et al. 2008). Furthermore, genome-wide association studies (GWASs) have identified a range of DNA variants influencing the risk of OFCs (Leslie and Marazita 2013). Next-generation sequencing has allowed for the rapid discovery of cleft-associated genes for cases that are extremely rare, are clinically heterogeneous, or lack a strong family history (Weinberg et al. 2018). The recent use of a “cleft map” allowed the dissection and visualization of the contribution of shared genetic effects to phenotypic heterogeneity of OFCs and revealed that CL and CLP share GWAS loci (Carlson et al. 2019).
Postnatal Surgical Correction of Cleft Palate Defects
The current standard of care for patients affected by craniofacial cleft anomalies includes postnatal surgical interventions that are based upon a timeline that coincides with midfacial growth and development. A variety of well-established CL and palatoplasty techniques are available and are dictated by the severity of clefting in individuals. CP repair, in general, is dependent upon the principles of a tension-free and multilayered closure with repositioning of the velar muscle sling (Dao and Goudy 2016). The consensus for the optimum timing of intervention falls between 10 and 12 mo of age (Liao and Mars 2006) but most customarily within 18 mo. While closure of the palatal defect will facilitate feeding and speech, the benefits of early palate repair must be carefully weighed against concerns of negatively affecting the patient’s midfacial growth (Liao and Mars 2006). A successful repair involves 1) complete closure of the oral and nasal layers without fistula formation, 2) velopharyngeal competence with feeding and speech, 3) minimal impact on midfacial growth, and 4) improved eustachian tube function.
While these initial goals can be achieved through velar muscle repositioning on first intervention, patients with a deficient or absent bone of the primary palate typically undergo a second surgical intervention (alveolar bone graft) during or prior to the mixed dentition stage, essential for the development of proper anatomical relationships of midface structures (Dao and Goudy 2016). Within the United States, pre- and postsurgical consultations involve a multidisciplinary panel of care providers, including otorhinolaryngologists, oral and maxillofacial surgeons, speech therapists, audiologists, orthodontists, pediatric dental specialists, and social workers.
Prenatal Approaches
Diagnostic Screening
At present, both invasive and noninvasive methods exist to collect fetal material for prenatal screening with a range of diagnostic approaches. For pregnancies arising from in vitro fertilization, it is possible to undertake preimplantation genetic screening (Vermeesch et al. 2016). For natural pregnancies, routine invasive approaches involve harvesting fetal cells by real-time ultrasound-guided transabdominal aspiration of amniotic fluid (i.e., amniocentesis), typically at 16 wk of gestation. Alternatively, placental cells (i.e., chorionic villus sampling [CVS]), retrieved between 10 and 13 wk of gestation, can be used (Ghidini 2018). Since CVS results can be initially reported within 48 h of testing and final results from long-term culture available in 7 to 10 d, fetal anomalies may be detected as early as the 11th gestational week. In contrast, amniocentesis usually only provides an answer after 17 wk of gestation (Alfirevic et al. 2017).
Harvesting cell-free nucleic acids (cfDNA and cfRNA) from maternal blood is a more recent noninvasive approach to prenatal screening initially introduced to detect common fetal autosomal and sex chromosome aneuploidies, now with the capacity to identify DNA microdeletions, copy number variants (CNVs), and monogenic disorders (Ghidini et al. 2019). This approach has already been used to diagnose cleft-associated syndromes, such as DiGeorge syndrome (Wapner et al. 2015). Sampling maternal biofluids is advantageous as it obviates the risk of fetal loss that accompanies invasive sampling of fetal tissue directly (Bianchi 2012). Since a negative result in cfRNA/DNA analysis has upward of 99% negative predictive value, there has been a 70% decrease worldwide in invasive testing, with an associated reduction in costs to the health care system (Ghidini et al. 2019). However, the mixture of fetal and maternal material does confer additional downstream analytic complexity.
While karyotyping and microarray-based cytogenic techniques can detect aneuploidy, microarray is preferred, as it avails additional genetic information (Vermeesch et al. 2016), providing higher sensitivity and shorter turnaround times than conventional metaphase karyotyping (Bianchi 2012). Recently, next-generation technologies have surpassed chromosomal microarray techniques for the diagnosis of genetic disease (Clark et al. 2018). Prenatal whole-exome sequencing (WES) assists in the diagnosis of dysmorphic fetuses by identification of single-nucleotide polymorphisms (SNPs) as well as insertion or deletion events (Thevenon et al. 2016). Cleft detection with fetal-parental-trio WES has recently been linked to enhanced detection of fetal structural anomalies relative to cytogenetic or microarray techniques (Lord et al. 2019). However, in recently published protocols, this technique follows ultrasound at approximately 11 wk of gestation, as it relies on the triangulation of data to predict the pathogenicity of each molecular variant based on fetal phenotype. Notwithstanding, prenatal molecular diagnoses derived from similar protocols have arisen almost twice as frequently in fetuses with craniofacial morphological abnormalities (46%) compared to those without (24%) (Normand et al. 2018). Thus, first-trimester molecular diagnoses may be made possible by advances in imaging technology that allow morphologic assessment earlier in development. Pertinent also is that some (Ghidini et al. 2019), but not all, published studies provided real-time molecular diagnoses during pregnancy. The clinical utility of sequencing modalities will be enhanced by the advent of more efficient techniques and the refinement of pathology workflows to accelerate the turnaround time of molecular testing. While studies that have implemented WES have derived fetal tissue via invasive methods that involve CVS, amniotic fluid, or fetal blood, it remains to be investigated whether less invasive strategies for obtaining fetal tissue could also be employed to promptly detect dysmorphology in offspring via analysis of genetic aberrations in fetal cfDNA/RNA. Cell-free fetal transcriptomics may be a useful diagnostic tool in the future, but its implementation would necessitate the acquisition of reference data for the expression of key genes across a range of developmental stages and accurate determination of the gestational age.
In addition to exploration of DNA- and RNA-based methodologies, discernment of variation at the protein level may aid in prenatal diagnosis. For decades, α-fetoprotein in maternal blood samples has been routinely used to assist in the detection of developmental anomalies, including Down syndrome (Cheng et al. 1993) and neural tube defects (Milunsky et al. 1980). Proteomic approaches have since arisen to better diagnose a range of chromosomal aberrations (Narasimhan et al. 2013). More recent advances in prenatal diagnostics have involved putative protein biomarkers to discriminate normal pregnancies from those with congenital heart defects, with changes in protein levels having been identified from as early as 8 wk of gestation (Chen et al. 2016). Interestingly, studied cardiac structural anomalies were polygenic in nature, which foreshadows that the diagnosis of clefts with diverse genetic diagnoses may be possible by appraisal of a handful of biomarkers from known cleft-associated pathways. Conversely, analysis of these prenatal samples may enable us to glean a deeper understanding of the molecular events underpinning craniofacial dysmorphogenesis.
In Utero Surgical Repair
Surgical interventions are currently undertaken for a subset of fetuses diagnosed with congenital anomalies linked to high mortality or severe morbidity if treatment is delayed. However, there is considerable debate about the practicality and efficacy of these high-risk approaches. The appeal of prenatal interventions for OFC repair stems from their promise to definitively restore form and function prior to birth, thereby mitigating the need for complex postnatal surgical manipulations and multidisciplinary dental/maxillofacial treatments that often commit patients to ongoing care into their adult years of life. Significantly, prenatal surgery can circumvent scarring sequelae associated with postnatal interventions, namely, secondary dentoalveolar and midfacial growth deformities (Moore et al. 2018). The distinct benefit to intervening surgically in utero is the nearly scarless mechanism of wound healing prenatally (Longaker et al. 1991; Ozturk et al. 2001). This is particularly advantageous in the case of OFCs, as the resultant therapy is heavily reliant on the reconstructive, functional, and aesthetic outcome, given the physical and psychological impact of midfacial developmental anomalies (Wójcicki and Drozdowski 2011). Furthermore, prominent scars form within integumentary tissues, with underlying fibrous banding and contraction in facial morphology postsurgically. Some postulate that the less-intense inflammatory response during fetal development is responsible for scarless mechanisms of fetal wound repair in utero; there are also theories related to the impact of amniotic fluid sterility and richness in growth factors (e.g., hyaluronic acid) in promoting wound healing (Longaker et al. 1994; Ozturk et al. 2001).
Mucosal wounds and minor bone defects heal during the course of fetal development without the formation of tissue callus, the equivalent of scarring in integument (Stelnicki et al. 1999). Furthermore, maxillary growth restriction was not identified in early cleft treatment in utero in animal models. This is of particular clinical significance, as earlier repair of cleft anomalies during fetal development may allow for postrepair growth of midface morphology beyond that which can be achieved postnatally. Thus, earlier cleft repair in utero has a potential for CLP healing with minimal scar formation, lower or no restriction of mandibular/maxillary bone growth, and potential elimination for need of postnatal corrective procedures. However, important barriers must be overcome to consider in utero surgical correction of OFCs clinically feasible. As false-positive results during prenatal morphological screening can lead to unnecessary, risk-provoking intervention for mother and fetus, it is critical that prenatal diagnostic techniques are first developed and optimized in larger animal models.
Targeted Molecular Therapeutics
Evidence of the efficacy of molecular agents to restore palatal morphogenesis in the prenatal period is also emerging. Beyond the environmental and stochastic factors known to influence palatogenesis, precision therapies can target genetic defects of craniofacial development. Precision therapies for Mendelian diseases, such as those that replace deficient proteins, directly target disease-associated pathways, or influence expression of disease-relevant genes, are clinically available for a number of conditions, such as lysosomal storage disorders, cystic fibrosis, tuberous sclerosis, and spinal muscular atrophy (Dugger et al. 2018). While these approaches have historically been applied to monogenic conditions, there is scope for their extrapolation to conditions for which there is genetic heterogeneity but pathway homogeneity. Hence, while a range of causative mutations are implicated in CLP, targeting interrelated pathways may be of therapeutic benefit to cleft patients with a range of genetic etiologies. The most significant among the barriers that currently exist for the effective translation and application of biologically driven therapies to human OFCs is the underlying complexity of cleft genetics itself. While the molecular basis of an individual cleft condition is genetically heterogenous, known variants account for a minority of the estimated heritability. Hence, new gene discoveries along with the knowledge of interacting signaling pathways are critical for the advancement of the field.
In contrast to inherited metabolic diseases, which require chronic treatments with repeated infusion of replacement proteins or other bioactive molecules (Hughes 2018), therapies able to affect nonreversible developmental fate decisions can produce permanent effects in response to short-term treatments. Such transient therapies have potential benefits over longer-acting gene therapy approaches that risk incorporation of transgenic products into maternal or filial DNA. Known cleft-associated genetic aberrations represent potential targets for appropriately timed precision gene product replacement therapies to restore the delicate molecular equilibrium required for normal embryonic development. The efficacy of such therapies at various timepoints in animal models has advanced contemporary knowledge of when the activity of key proteins peaks during palatogenesis. Trialed preclinical interventions and their outcomes are discussed below.
TGF-β3 Rescue
Given its role in medial edge epithelial fusion of the left and right palatal processes, transient, high-level expression of Tgfβ3 is critical to murine palatogenesis (Funato et al. 2015). To remedy the cleft phenotype of Tgfβ3-null mice, Spivak et al. (2007) pioneered a virally mediated intra-amniotic Tgfβ3 gene transfer therapy and showed palatal fusion without adverse effects in 100% of pups following injections at E12.5 and E13.5. Another group tested delayed timing of therapy at E14.5 and E15.5, resulting in 82% and 75% of palatal closures, respectively (Wu et al. 2012). Since TGFβ3 polymorphisms are also associated with nonsyndromic clefts in humans (Zhu et al. 2010), successful translation into human fetuses was hypothesized to yield successful outcomes between gestational weeks 8 and 10 (Spivak et al. 2007). Promising results from murine palatal cultures foreshadow the potential versatility of TGFβ3-based therapies for CP attributed to environmental causes, including dioxin exposure (Thomae et al. 2005). In 2019, a group of researchers were able to rescue a core binding factor β (Cbfb) (a cofactor of the Runx1 family of transcription factors) deficiency-induced anterior CP phenotype through the in vitro administration of folic acid (Sarper et al. 2019). In these mutants, TGFβ3 expression is disrupted in the area of failed anterior palatal fusion and affects the phosphorylation of Stat3, a downstream effector molecule of cellular proliferation, migration, and apoptosis. This reversal of CP using folic acid highlights the obligatory function of the Runx1/Cbfb-Stat3-TGFβ3 signaling axis in anterior palatal fusion.
Activation of Shh Signaling
The intestinal cell kinase (ICK) gene encoding for a mitogen-activated protein (MAP) kinase involved in activating Shh signaling during development is downregulated during endocrine-cerebro-osteodysplasia (ECO), a syndrome that includes CP. As the first study to attempt prenatal pharmaceutical activation of Shh signaling, Shin et al. (2019) injected a small-molecule agonist for Smoothened (SAG) intraperitoneally into pregnant Icktm1a/+ mice at various stages. Although success in palatal closures showed variable efficacy, the highest efficiency was achieved at E11.25, indicating that developmental windows are critical for restoring molecular homeostasis in Shh-mediated actions.
Modulation of the Wnt Signaling Pathway
The importance of Wnt signaling and related pathway genes in palate formation is elegantly reviewed by Reynolds et al. (2019). The first Wnt pathway intervention study was performed on mice deficient in glycogen synthase kinase–3β (GSK-3β), a key mediator of canonical Wnt signaling. Subcutaneous rapamycin was administered every 12 h to pregnant dams at E13.5 to E15 to stabilize the recombinant FRB-tagged Gsk-3β protein. Complete and partial alleviation of the cleft phenotype was subsequently reported in 5 of 9 and 1 of 9 treated pups, respectively (Liu et al. 2007). Rescue was not observed with later injection regimens, which attests to the developmental window during which Gsk-3β functions in palatogenesis. While an elegant proof of concept of targeted Wnt pathway manipulation, the clinical utility of this study is limited by its development in a conveniently inducible transgenic system that cannot be clinically reproduced in humans.
Another set of approaches targets the activities of dickkopf-related (Dkk) proteins that are extracellular antagonists of Wnt signal transduction and function by high-affinity binding to Wnt coreceptors Lrp5/6. In situ application of dickkopf 1 (Dkk1) to the maxillary processes of chick embryos via protein-impregnated beads elicits downregulation of osteochondrogenic targets of Wnt signaling, including Bmp4, Tbx22, Sox9, and Barx1, thereby hindering growth of the maxillary and palatine bones (Shimomura et al. 2019).
Small-molecule inhibitors of Dkk1 and Dkk2 are now known to share a functional molecular relationship with the paired box domain-containing transcription factor, Pax9. Pax9 hemizygosity in humans led to a decreased level of Wnt signaling and concurrent increased level of Dkks, suggesting that Pax9 deficiency-related phenotypes could be mitigated with direct Dkk antagonism (Schuffenhauer et al. 1999). Researchers have subsequently revealed the potential linkages between Pax9 and CP in cases from the United States, Japan, Korea, and China (Song et al. 2013). The controlled delivery of a small-molecule WAY-262611 that has been shown to potentiate downstream Wnt/β-catenin signaling via Dkk1 antagonism (Pelletier et al. 2009) faithfully reversed secondary CPs in Pax9-deficient mice in utero (Jia et al. 2017) (see Fig.). However, WAY-262611 could not rescue the expression of Msx1 and Bmp4, which appear restricted to the anterior palatal mesenchyme. Jia et al. (2017) also trialed another Dkk inhibitor, IIIc3a (Li et al. 2012), via tail vein injections and showed complete closure of secondary palate in 80% (12/15) of embryos. Maternal intraperitoneal injections at E12.5, E13.5, and E14.5 also resulted in resolution of the cleft defect in middle and posterior regions of 7/11 Pax9-deficient palates, as shown in independent studies by Li et al. (2017).
Figure.
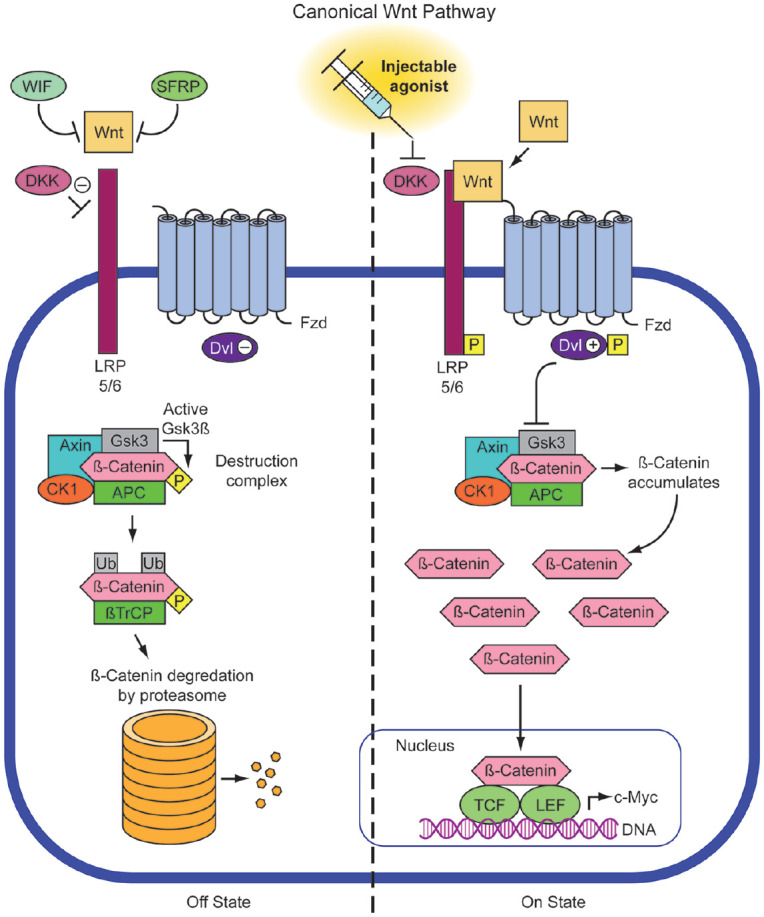
Small-molecule inhibitors of dickkopf (Dkk) 1 and Dkk2 (inhibitors of LRP5/6) allow for the activation of Wnt/β-catenin signaling transduction. Several different molecules have been described (e.g., WAY262611; IIIc3a) and trialed to rescue the phenotype of murine cleft models in which Dkk1/2 are upregulated in the posterior palate to the detriment of Wnt signaling.
Since Wnt signaling pathway is critical for organogenesis and is also implicated in tumorigenesis (particularly bone tumors), it is crucial to perform thorough toxicology analyses. Jia et al. (2017) performed magnetic resonance imaging (MRI) on mothers injected intraperitoneally with Wnt agonist small-molecule therapies, along with surviving wild-type and heterozygous progeny, showing no toxic effects or tumor development up to 18 mo following the last injection.
Challenges and Considerations for a Way Forward
Prior to the translation of prenatal intervention for the correction of human OFCs, a number of barriers remain to be overcome. The implementation of in utero therapies is constrained by the inability of contemporary approaches to identify structural anomalies in the first trimester of pregnancy. Since the primary and secondary palates form between 6 to 7 wk and 8 to 10 wk of gestation, respectively (Yu et al. 2017), cleft detection prior to or during this window would be essential for maximum therapeutic effect. Clearly, more research is needed on whether therapies can indeed operate retroactively and if intervention times can be targeted later in palatogenesis.
Whereas the maxillary lip and alveolar ridge can be confidently evaluated with conventional ultrasonography, isolated clefts of the secondary palate have historically gone undetected. While conventional screening protocols subject to initial ultrasound can identify cleft phenotypes at week 20, high-frequency ultrasonic technology can accurately visualize structures as early as 13 wk of gestation (Maarse et al. 2010). Novel 2- and 3-dimensional ultrasound techniques have ameliorated detection rates of secondary CP but are largely implemented in the latter stage of pregnancy. Despite reports of first-trimester diagnoses with select ultrasound techniques, these novel approaches have yet to be widely adopted in routine screening. Similarly, while MRI has higher positive predictive value than ultrasound for posterior CPs, it is not typically employed until weeks 20 to 39 (Tian et al. 2019).
The potential for earlier OFC identification through screening of prospective parents for pathogenic mutations that could induce palatal dysmorphogenesis in their offspring is an exciting forefront in precision medicine. Current diagnostic panels only include genes known to cause lethal and/or debilitating diseases. Nonsyndromic OFCs and those that are part of syndromes (i.e., WNT7A and MKS1) are not captured in routine screening panels and will be overlooked prior to pregnancy. Even as panels are updated with more genes definitively linked to birth defects, preconception screening is limited by the inability to detect de novo mutations. The latter are causative in 3.5-fold more cases of fetal structural anomalies detected in utero than inherited genetic abnormalities (Lord et al. 2019). Importantly, these methods of screening would not be effective in detecting environmentally induced OFCs (i.e., those due to factors such as vitamin deficiencies and teratogenic exposures), and they depend upon a priori knowledge of specific single SNPs, insertion or deletion events, and CNVs. This necessitates prenatal screening to adequately determine whether a developing fetus harbors a molecular aberration that can lead to disordered morphogenesis and be considered for targeted putative therapies.
Current strategies for prenatal correction of OFC phenotypes in the animal models discussed have focused solely on single-gene mutations but offer valuable proof of concepts for replacement therapy approaches that restore molecular homeostasis during critical stages of development. It is also important to consider the potentially undesirable developmental implications or delayed pathological outcomes of therapies, given that many palate-specific molecular targets function within signaling cascades with diverse molecular outcomes. For example, beyond its developmental roles, Wnt signaling also orchestrates tumorigenesis in several organ systems (Zhan et al. 2017). Notwithstanding, the controlled dosage of agonist or antagonist drugs that target discrete ligand-receptor complexes can be effective as pathologic levels are likely modulated by cellular regulatory mechanisms (Komiya and Habas 2008).
Targeted drug delivery systems must be optimized to drive translation of preclinical methodologies. Due to variability in the transplacental passage of therapeutic agents administered systemically to pregnant women, this route of delivery is considered suboptimal (Hermes et al. 2014). In primates, less than 1% of maternal serum concentrations of a protein tagged with the Fc portion of an IgG1 reached the fetus following maternal systemic delivery (Schneider et al. 2018). Thus, to compensate, mothers would be exposed to large quantities of exogenous molecules with potentially detrimental effects. However, no signs of maternal or fetal drug-related toxicity have been demonstrated in primates at the maximum dose administered in humans to remedy ectodermal dysplasia.
Although it is believed that lower quantities of molecules administered intra-amniotically are needed (as amniotic fluid may serve as a reservoir promoting targeted drug uptake), drug kinetics must be measured to rule out toxicity and side effects to both mother and fetus (Hermes et al. 2014). While fetal and maternal outcomes may be compromised by invasive injection protocols, less invasive ultrasound-guided injections have been employed in mice and humans (Schneider et al. 2018). In murine models, even fetuses in high-dose cohorts survived intra-amniotic injection and were born without complications (Hermes et al. 2014). Of the 3 human babies born following intra-amniotic ectodysplasin therapy, the twins were born prematurely, as is common of multiple pregnancies, but the single pregnancy was carried for a normal gestational term. Consequently, more studies are required to determine whether an association exists between premature births and in utero therapy and whether this has clinically significant implications.
With the dawn of direct-to-consumer genomic analysis assays, a high propensity for misinterpretation of test results will persist, further reinforcing the need for effective communication between knowledgeable provider and responsive patient. The International Fetal Transplantation and Immunology Society’s In Utero Gene Therapy Consensus Statement dictates prenatal intervention only should be considered when both a reliable molecular diagnosis and a strong genotype-phenotype correlation exist (Almeida-Porada et al. 2019). Multiple barriers presently preclude the widespread adoption of genomics into prenatal clinical practice, compromising shared decision-making processes in prenatal diagnostics and management. The advent of CRISPR/Cas9 technology to readily generate craniofacial disease models with specific human mutations should aid in the translation of new technologies (Neben et al. 2016). The 1998 UK Gene Therapy Advisory Committee recommends that in utero treatment should be reserved for cases that confer a clear advantage over postnatal intervention (Kingdom, 1998). While precision medicine gives rise to a range of other ethical conundrums, we believe that the promise of precision-driven approaches for early diagnosis and therapy for the treatment of orofacial clefts is an essential step forward.
Author Contributions
J.D. Oliver, L.R. Halpern, P. Schneider, R.N. D’Souza, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; E.C. Turner, contributed to conception, design, data acquisition, analysis, and interpretation, drafted the manuscript; S. Jia, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Acknowledgments
We also thank Russell Hendrycks for his assistance in generating the Figure.
Footnotes
This work was supported by the National Institute of Health/National Institute of Dental and Craniofacial Research grants DE019471 and DE027255 (R.N. D’Souza) and supported in part by the American Cleft Palate-Craniofacial Association (S. Jia) and a PhD training stipend from the Department of Biomedical Engineering, University of Utah to J.D. Oliver. P. Schneider is supported by the Swiss National Science Foundation (grant 310030A-176356).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iDs: J.D. Oliver  https://orcid.org/0000-0003-3387-5770
https://orcid.org/0000-0003-3387-5770
P. Schneider  https://orcid.org/0000-0003-0677-9409
https://orcid.org/0000-0003-0677-9409
R.N. D’Souza  https://orcid.org/0000-0002-1505-5173
https://orcid.org/0000-0002-1505-5173
References
- Alfirevic Z, Navaratnam K, Mujezinovic F. 2017. Amniocentesis and chorionic villus sampling for prenatal diagnosis. Cochrane Database Syst Rev. 9(9):CD003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-Porada G, Waddington SN, Chan JKY, Peranteau WH, MacKenzie T, Porada CD. 2019. In utero gene therapy consensus statement from the IFeTIS. Mol Ther. 27(4):705–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JC, Raynes-Greenow C, Turner RM, Bower C, Nassar N, O’Leary CM. 2014. Maternal alcohol consumption during pregnancy and the risk of orofacial clefts in infants: a systematic review and meta-analysis. Paediatr Perinat Epidemiol. 28(4):322–332. [DOI] [PubMed] [Google Scholar]
- Bianchi DW. 2012. From prenatal genomic diagnosis to fetal personalized medicine: progress and challenges. Nat Med. 18(7):1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JC, Anand D, Butali A, Buxo CJ, Christensen K, Deleyiannis F, Hecht JT, Moreno LM, Orioli IM, Padilla C, et al. 2019. A systematic genetic analysis and visualization of phenotypic heterogeneity among orofacial cleft GWAS signals. Genet Epidemiol. 43(6):704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gu H, Li J, Yang Z-Y, Sun X, Zhang L, Shan L, Wu L, Wei X, Zhao Y, et al. 2016. Comprehensive maternal serum proteomics identifies the cytoskeletal proteins as non-invasive biomarkers in prenatal diagnosis of congenital heart defects. Sci Rep. 6(1):19248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EY, Luthy DA, Zebelman AM, Williams MA, Lieppman RE, Hickok DE. 1993. A prospective evaluation of a second-trimester screening test for fetal down syndrome using maternal serum alpha-fetoprotein, hCG, and unconjugated estriol. Obstet Gynecol. 81(1):72–77. [PubMed] [Google Scholar]
- Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, Kingsmore SF. 2018. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med. 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao AM, Goudy SL. 2016. Cleft palate repair, gingivoperiosteoplasty, and alveolar bone grafting. Facial Plast Surg Clin North Am. 24(4):467–476. [DOI] [PubMed] [Google Scholar]
- Dugger SA, Platt A, Goldstein DB. 2018. Drug development in the era of precision medicine. Nat Rev Drug Discov. 17(3):183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato N, Nakamura M, Yanagisawa H. 2015. Molecular basis of cleft palates in mice. World J Biol Chem. 6(3):121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidini A, Bianchi DW, Levy B, Van Mieghem T, Deprest J, Chitty LS. 2019. In case you missed it: the prenatal diagnosis editors bring you the most significant advances of 2018. Prenat Diagn. 39(2):61–69. [DOI] [PubMed] [Google Scholar]
- Health Departments of the United Kingdom. 1998. Gene therapy advisory committee: report on the potential use of gene therapy in utero. Dis Markers. 14(3):151–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes K, Schneider P, Krieg P, Dang A, Huttner K, Schneider H. 2014. Prenatal therapy in developmental disorders: drug targeting via intra-amniotic injection to treat X-linked hypohidrotic ectodermal dysplasia. J Invest Dermatol. 134(12):2985–2987. [DOI] [PubMed] [Google Scholar]
- Hughes JH, Liu K, Plagge A, Wilson PJM, Sutherland H, Norman BP, et al. 2019. Conditional targeting in mice reveals that hepatic homogentisate 1,2-dioxygenase activity is essential in reducing circulating homogentisic acid and for effective therapy in the genetic disease alkaptonuria. Hum Mol Genet. 28(23):3928–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A, Oluwatosin B, Njideka G, Babafemi, Benjamin OG, Olufemi D, Leo R, Folorunso I, Phylis Olusina O. 2014. Cleft palate in HIV-exposed newborns of mothers on highly active antiretroviral therapy. Oral Surg. 7(Suppl 1):102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Zhou J, Fanelli C, Wee Y, Bonds J, Schneider P, Mues G, D’Souza RN. 2017. Small-molecule Wnt agonists correct cleft palates in Pax9 mutant mice in utero. Development. 144(20):3819–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya Y, Habas R. 2008. Wnt signal transduction pathways. Organogenesis. 4(2):68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer EJ, Chen DT, Hoar RM, Agnish ND, Benke PJ, Braun JT, Curry CJ, Fernhoff PM, Grix AW, Jr, Lott IT, et al. 1985. Retinoic acid embryopathy. N Engl J Med. 313(14):837–841. [DOI] [PubMed] [Google Scholar]
- Leslie EJ, Marazita ML. 2013. Genetics of cleft lip and cleft palate. Am J Med Genet C Semin Med Genet. 163C(4):246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lan Y, Krumlauf R, Jiang R. 2017. Modulating Wnt signaling rescues palate, morphogenesis in Pax9 mutant mice. J Dent Res. 2017;96(11):1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shan J, Chang W, Kim I, Bao J, Lee HJ, Zhang X, Samuel VT, Shulman GI, Liu D, et al. 2012. Chemical and genetic evidence for the involvement of Wnt antagonist dickkopf2 in regulation of glucose metabolism. Proc Natl Acad Sci USA. 109(28):11402–11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YF, Mars M. 2006. Hard palate repair timing and facial growth in cleft lip and palate: a systematic review. Cleft Palate Craniofac J. 43(5):563–570. [DOI] [PubMed] [Google Scholar]
- Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT. 2007. Chemical rescue of cleft palate and midline defects in conditional GSK-3beta mice. Nature. 446(7131):79–82. [DOI] [PubMed] [Google Scholar]
- Longaker MT, Chiu ES, Adzick NS, Stern M, Harrison MR, Stern R. 1991. Studies in fetal wound healing. V. A prolonged presence of hyaluronic acid characterizes fetal wound fluid. Ann Surg. 213(4):292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longaker MT, Whitby DJ, Ferguson MW, Lorenz HP, Harrison MR, Adzick NS. 1994. Adult skin wounds in the fetal environment heal with scar formation. Ann Surg. 219(1):65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord J, McMullan DJ, Eberhardt RY, Rinck G, Hamilton SJ, Quinlan-Jones E, Prigmore E, Keelagher R, Best SK, Carey GK, et al. 2019. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet. 393(10173):747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maarse W, Berge SJ, Pistorius L, van Barneveld T, Kon M, Breugem C, Mink van der Molen AB. 2010. Diagnostic accuracy of transabdominal ultrasound in detecting prenatal cleft lip and palate: a systematic review. Ultrasound Obstet Gynecol. 35(4):495–502. [DOI] [PubMed] [Google Scholar]
- Marazita ML. 2012. The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet. 13:263–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunsky A, Alpert E, Neff RK, Frigoletto FD. 1980. Prenatal diagnosis of neural tube defects: IV. Maternal serum alpha-fetoprotein screening. Obstet Gynecol. 55(1):60–66. [PubMed] [Google Scholar]
- Moore AL, Marshall CD, Barnes LA, Murphy MP, Ransom RC, Longaker MT. 2018. Scarless wound healing: transitioning from fetal research to regenerative healing. Wiley Interdiscip Rev Dev Biol. 7(2):10.1002/wdev.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K, Lin SL, Tong T, Baig S, Ho S, Sukumar P, Biswas A, Hahn S, Bajic VB, Choolani M. 2013. Maternal serum protein profile and immune response protein subunits as markers for non-invasive prenatal diagnosis of trisomy 21, 18, and 13. Prenat Diagn. 33(3):223–231. [DOI] [PubMed] [Google Scholar]
- Neben CL, Roberts RR, Dipple KM, Merrill AE, Klein OD. 2016. Modeling craniofacial and skeletal congenital birth defects to advance therapies. Hum Mol Genet. 25(R2):R86–R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand EA, Braxton A, Nassef S, Ward PA, Vetrini F, He W, Patel V, Qu C, Westerfield LE, Stover S, et al. 2018. Clinical exome sequencing for fetuses with ultrasound abnormalities and a suspected mendelian disorder. Genome Med. 10(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk S, Deveci M, Sengezer M, Gunhan O. 2001. Results of artificial inflammation in scarless foetal wound healing: an experimental study in foetal lambs. Br J Plast Surg. 54(1):47–52. [DOI] [PubMed] [Google Scholar]
- Pelletier JC, Lundquist JT, Gilbert AM, Alon N, Bex FJ, Bhat BM, Bursavich MG, Coleburn VE, Felix LA, Green DM, et al. 2009. (1-(4-(Naphthalen-2-yl)pyrimidin-2-yl)piperidin-4-yl)methanamine: a wingless β-catenin agonist that increases bone formation rate. J Med Chem. 52(22):6962–6965. [DOI] [PubMed] [Google Scholar]
- Pereira AV, Fradinho N, Carmo S, de Sousa JM, Rasteiro D, Duarte R, Leal MJ. 2018. Associated malformations in children with orofacial clefts in Portugal: a 31-year study. Plast Reconstr Surg Glob Open. 6(2):e1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K, Kumari P, Sepulveda Rincon L, Gu R, Ji Y, Kumar S, Zhou CJ. 2019. Wnt signaling in orofacial clefts: crosstalk, pathogenesis and models. Dis Model Mech. 12(2):dmm037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh HJ, Ahmed Hassan MH, Innes NPT, Baik AA, Mossey PA. 2014. Parental consanguinity and nonsyndromic orofacial clefts in children: a systematic review and meta-analyses. Cleft Palate Craniofac J. 51(5):501–513. [DOI] [PubMed] [Google Scholar]
- Sabbagh HJ, Hassan MHA, Innes NPT, Elkodary HM, Little J, Mossey PA. 2015. Passive smoking in the etiology of non-syndromic orofacial clefts: a systematic review and meta-analysis. PLoS One. 10(3):e0116963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarper SE, Inubushi T, Kurosaka H, Ono Minagi H, Murata Y, Kuremoto KI, Sakai T, Taniuchi I, Yamashiro T. 2019. Anterior cleft palate due to Cbfb deficiency and its rescue by folic acid. Dis Model Mech. 12(6):dmm038851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H, Faschingbauer F, Schuepbach-Mallepell S, Korber I, Wohlfart S, Dick A, Wahlbuhl M, Kowalczyk-Quintas C, Vigolo M, Kirby N, et al. 2018. Prenatal correction of X-linked hypohidrotic ectodermal dysplasia. N Engl J Med. 378(17):1604–1610. [DOI] [PubMed] [Google Scholar]
- Schuffenhauer S, Leifheit HJ, Lichtner P, Peters H, Murken J, Emmerich P. 1999. De novo deletion (14)(q11.2q13) including PAX9: clinical and molecular findings. J Med Genet. 36(3):233–236. [PMC free article] [PubMed] [Google Scholar]
- Shimomura T, Kawakami M, Tatsumi K, Tanaka T, Morita-Takemura S, Kirita T, Wanaka A. 2019. The role of the Wnt signaling pathway in upper jaw development of chick embryo. Acta Histochem Cytochem. 52(1):19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JO, Song J, Choi HS, Lee J, Lee K, Ko HW, Bok J. 2019. Activation of sonic hedgehog signaling by a smoothened agonist restores congenital defects in mouse models of endocrine-cerebro-osteodysplasia syndrome. EBioMedicine. 49:305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivertsen A, Wilcox AJ, Skjaerven R, Vindenes HA, Abyholm F, Harville E, Lie RT. 2008. Familial risk of oral clefts by morphological type and severity: population based cohort study of first degree relatives. BMJ. 336(7641):432–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Wu D, Wang Y, Li H, Yin N, Zhao Z. 2013. SNPs and interaction analyses of IRF6, MSX1 and PAX9 genes in patients with nonsyndromic cleft lip with or without palate. Mol Med Rep. 8(4):1228–1234. [DOI] [PubMed] [Google Scholar]
- Spivak RM, Endo M, Zajac A, Zoltick P, Ang B, Horn R, Flake A, Kirschner R, Nah H-D. 2007. In utero gene delivery of adenovirus encoded tgf-beta3 restores physiologic palatal fusion and rescues cleft palate in a tgf-beta3 knockout mouse. J Am Coll Surg. 205(3 Suppl):S92. [Google Scholar]
- Stelnicki EJ, Lee S, Hoffman W, Lopoo J, Foster R, Harrison MR, Longaker MT. 1999. A long-term, controlled-outcome analysis of in utero versus neonatal cleft lip repair using an ovine model. Plast Reconstr Surg. 104(3):607–615. [DOI] [PubMed] [Google Scholar]
- Thevenon J, Duffourd Y, Masurel-Paulet A, Lefebvre M, Feillet F, El Chehadeh-Djebbar S, St-Onge J, Steinmetz A, Huet F, Chouchane M, et al. 2016. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet. 89(6):700–707. [DOI] [PubMed] [Google Scholar]
- Thomae TL, Stevens EA, Bradfield CA. 2005. Transforming growth factor-beta3 restores fusion in palatal shelves exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 280(13):12742–12746. [DOI] [PubMed] [Google Scholar]
- Tian M, Xiao L, Jian N, Wei X, Liu S, Zhao H, Li G, Zhang S, Liang W, Lin N, et al. 2019. Accurate diagnosis of fetal cleft lip/palate by typical signs of magnetic resonance imaging. Prenat Diagn. 39(10):883–889. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Voet T, Devriendt K. 2016. Prenatal and pre-implantation genetic diagnosis. Nat Rev Genet. 17(10):643–656. [DOI] [PubMed] [Google Scholar]
- Veroniki AA, Cogo E, Rios P, Straus SE, Finkelstein Y, Kealey R, Reynen E, Soobiah C, Thavorn K, Hutton B, et al. 2017. Comparative safety of anti-epileptic drugs during pregnancy: a systematic review and network meta-analysis of congenital malformations and prenatal outcomes. BMC Med. 15(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapner RJ, Babiarz JE, Levy B, Stosic M, Zimmermann B, Sigurjonsson S, Wayham N, Ryan A, Banjevic M, Lacroute P, et al. 2015. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am J Obstet Gynecol. 12(3):332.e1–9. [DOI] [PubMed] [Google Scholar]
- Wehby GL, Murray JC. 2010. Folic acid and orofacial clefts: a review of the evidence. Oral Dis. 16(1):11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SM, Cornell R, Leslie EJ. 2018. Craniofacial genetics: where have we been and where are we going? PLoS Genet. 14(6):e1007438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcicki P, Drozdowski PH. 2011. In utero surgery—current state of the art—part II. Med Sci Monit. 17(12):RA262–RA270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Zhou Y, Fan W, Han P, Chang J, Yuen J, Zhang M, Xiao Y. 2012. Hypoxia-mimicking mesoporous bioactive glass scaffolds with controllable cobalt ion release for bone tissue engineering. Biomaterials. 33(7):2076–2085. [DOI] [PubMed] [Google Scholar]
- Yu K, Deng M, Naluai-Cecchini T, Glass IA, Cox TC. 2017. Differences in oral structure and tissue interactions during mouse vs. Human palatogenesis: implications for the translation of findings from mice. Front Physiol. 8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N, Boutros M. 2017. Wnt signaling in cancer. Oncogene. 36(11):1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hao L, Li S, Bailey LB, Tian Y, Li Z. 2010. MTHFR, TGFB3, and TGFA polymorphisms and their association with the risk of non-syndromic cleft lip and cleft palate in China. Am J Med Genet A. 152A(2):291–298. [DOI] [PubMed] [Google Scholar]


