Summary
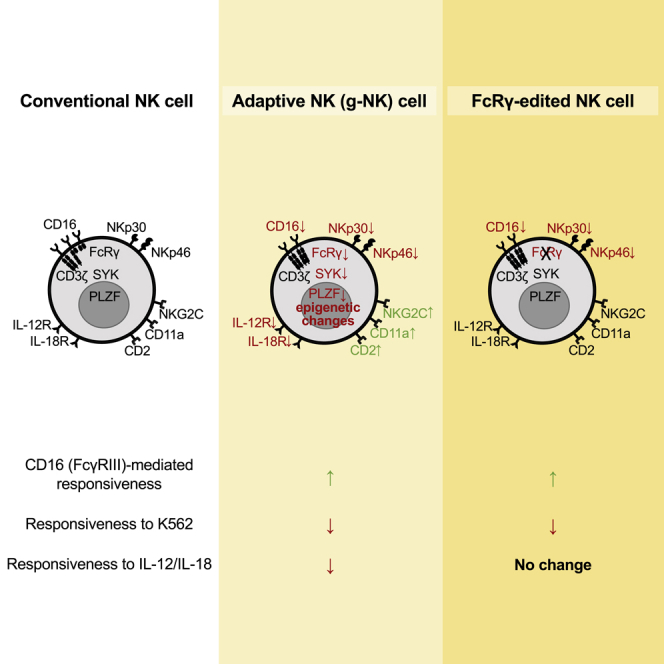
Adaptive human natural killer (NK) cells display significantly enhanced responsiveness to a broad-range of antibody-bound targets through the engagement of CD16 compared to conventional NK cells, yet direct reactivity against tumor targets is generally reduced. Adaptive NK cells also display a distinct phenotype and differential expression of numerous genes, including reduced expression of signaling adapter FcRγ and transcription factor PLZF. However, it is unclear whether differential expression of specific genes is responsible for the characteristics of adaptive NK cells. Using CRISPR-Cas9, we show deletion of FcRγ in conventional NK cells led to enhanced CD16 responsiveness, abolished cell surface expression of natural cytotoxicity receptors, NKp46 and NKp30, and dramatically reduced responsiveness to K562 and Raji tumor cells. However, deletion of PLZF had no notable effects. These results suggest multiple roles for FcRγ and identify its deficiency as an important factor responsible for the functional and phenotypic characteristics exhibited by adaptive NK cells.
Subject Areas: Molecular Biology, Immunology, Cancer
Graphical Abstract
Highlights
-
•
FcRγ deletion leads to increased cytokine production in response to CD16 stimulation
-
•
FcRγ deletion abolishes cell surface expression of NKp46 and NKp30
-
•
FcRγ deletion results in reduced responsiveness to K562 and Raji cells
-
•
PLZF deletion does not change responsiveness to CD16 and cytokine stimulation
Molecular Biology; Immunology; Cancer
Introduction
Natural killer (NK) cells constitute the third largest population of lymphocytes and play important roles in host defense against malignancy and viral infection (Vivier et al., 2011). Upon recognition of tumors or virus-infected target cells via activation receptors, NK cells can rapidly release cytotoxic granules and produce inflammatory cytokines. Unlike B and T lymphocytes that express gene-rearranged antigen-specific receptors, NK cells only express germline-encoded receptors and are considered part of the innate immune system. However, many recent studies have revealed adaptive immune features of NK cells in humans as well as in animal models (Cooper et al., 2009; Guma et al., 2004; Hammer and Romagnani, 2017; O'Leary et al., 2006; Paust et al., 2010; Reeves et al., 2015; Rolle and Brodin, 2016; Sun et al., 2009; Zhang et al., 2013).
In humans, two distinct but largely overlapping NK cell subsets have been identified that exhibit adaptive immune features, such as clonal-like expansion and long-term persistence (Beziat et al., 2013; Cichocki et al., 2018; Foley et al., 2012; Hammer et al., 2018; Lee et al., 2015; Muccio et al., 2018; Schlums et al., 2015; Zhang et al., 2013). These subsets were initially identified by either a deficiency in expression of the signaling adapter protein FcRγ or a high-level expression of the activation receptor NKG2C (Guma et al., 2004; Hwang et al., 2012). Epidemiological analyses indicate that the presence of these FcRγ-deficient NK cells (termed g-NK cells), as well as NKG2C+ NK cells that often co-express maturation marker CD57, is associated with previous infection by human cytomegalovirus (HCMV), a common herpesvirus that infects the majority of the human population (Guma et al., 2004; Zhang et al., 2013). Importantly, these adaptive NK cells display heightened potential for broad antiviral and antibacterial responses through enhanced function of CD16 (Costa-Garcia et al., 2015; Goodier et al., 2018; Hart et al., 2019; Lee et al., 2015; Schlums et al., 2015; Wu et al., 2013; Zhang et al., 2013; Zhou et al., 2015), a Fc receptor that can bind to the Fc portion of multiple subclasses of IgG (Bruhns et al., 2009). For instance, it has been shown that, compared to conventional NK cells, adaptive NK cells produce greater amounts of pro-inflammatory cytokines in response to cells infected with viruses (e.g., HCMV and HSV-1), as well as to the virus itself, in the presence of virus-specific antibodies. In contrast, adaptive NK cells generally show poor responsiveness to tumor targets, unless target-specific antibodies are provided (Beziat et al., 2012; Hwang et al., 2012; Liu et al., 2016). Moreover, adaptive NK cells show defective expression of natural cytotoxicity receptors (NCRs), NKp46 and NKp30, both of which are commonly expressed by conventional NK cells (Guma et al., 2004; Hwang et al., 2012; Moretta et al., 2001). However, the molecular basis responsible for the altered functional and phenotypic characteristics of adaptive NK cells is unclear.
Recent gene expression profiling and epigenetic analyses have revealed differences in the expression of approximately 400 genes and widespread epigenetic changes between conventional and adaptive NK cells (Lee et al., 2015; Schlums et al., 2015). Flow cytometric analyses have confirmed altered expression of many genes at the protein level, including FcRγ and PLZF. FcRγ is an immunoreceptor tyrosine-based activation motif (ITAM)-containing signaling adapter protein and is known to have physical association with CD16, along with CD3ζ, as homodimers or heterodimers. While CD3ζ contains 3 ITAMs, FcRγ contains only 1 ITAM. FcRγ and numerous other intracellular proteins are also deficient in adaptive g-NK cells, including transcription factors (e.g., PLZF and HELIOS) and signal transduction proteins (e.g., SYK and EAT-2). Through correlation studies, it has been suggested that deficiencies in SYK may contribute to some of the altered characteristics of adaptive NK cells (Lee et al., 2015). It was reported that epigenetic remodeling of the IFNG locus (Luetke-Eversloh et al., 2014), as well as elevated expression of the transcription regulator, ARID5B, correlates with enhanced IFN-γ production by NKG2C+ NK cells (Cichocki et al., 2018). Moreover, because of PLZF's potential involvement in chromatin structure modification and regulation of several genes differentially expressed in adaptive NK and conventional NK cells (Gleimer et al., 2012; Mathew et al., 2012), PLZF deficiency has been suggested to be an important factor contributing to the observed adaptive properties (Schlums et al., 2015). However, given the numerous epigenetic and gene expression differences in adaptive NK cells, it has been challenging to define the molecular mechanism(s) responsible for the enhanced CD16 responsiveness and reduced tumor responsiveness of adaptive NK cells.
In this study, using CRISPR/Cas9-mediated gene editing, we sought to determine the roles of FcRγ and PLZF deficiencies in the functional response and phenotypic characteristics of primary human NK cells. Our data show that deletion of FcRγ resulted in enhanced CD16 responsiveness but reduced NCR expression and tumor responsiveness. However, deletion of PLZF had no notable effects. These data provide evidence that deficiency of FcRγ is an important factor responsible for the altered functional and phenotypic characteristics exhibited by adaptive NK cells.
Results and Discussion
CRISPR/Cas9-mediated Deletion of FcRγ in Human NK Cells
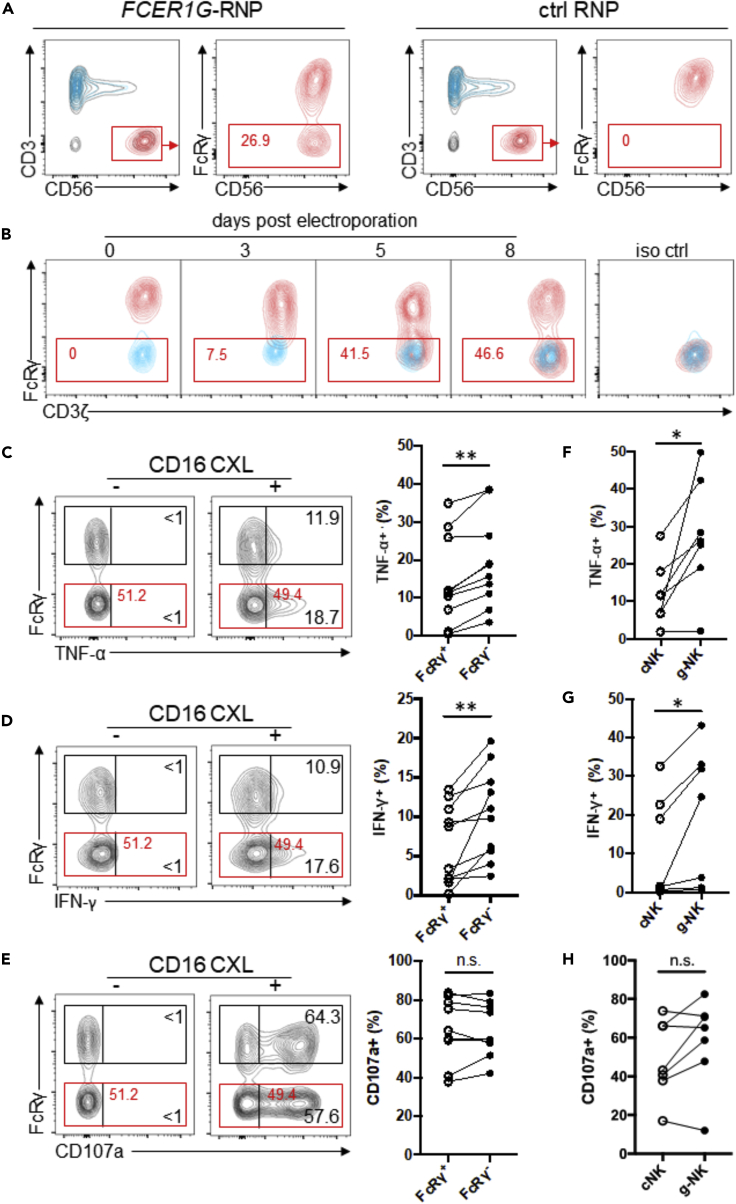
Although NK cells express a limited repertoire of germline-encoded activation receptors, they are able to recognize and respond to a broad-range of targets in the presence of target-specific antibodies through the expression of CD16 (Bruhns et al., 2009; Costa-Garcia et al., 2015; Hwang et al., 2012; Lanier, 2008; Zhang et al., 2013). To determine the role of FcRγ in the function of CD16, we initially sought to silence FcRγ expression in human NK cells using a siRNA-mediated knockdown approach. However, this strategy resulted in only transient and partial (approximately 50%) reduction of FcRγ protein level with no notable change in functional responsiveness of NK cells (data not shown). As an alternative approach to achieve more stable and more complete depletion of FcRγ, we explored CRISPR/Cas9-mediated gene editing. To obtain sufficient quantities of NK cells for gene editing and subsequent functional analysis, NK cells were enriched from peripheral blood mononuclear cell (PBMC) samples specifically chosen for their lack of detectable FcRγ-deficient (hereafter termed g-NK) cells, and these enriched samples were expanded in vitro for 10–14 days. Expanded cells were subjected to nucleofection of FCER1G (encoding FcRγ)-targeting Cas9/gRNA ribonucleoprotein (FCER1G-RNP). After subsequent culturing, flow cytometric analysis revealed the appearance of a distinct subset of CD3−CD56+ NK cells deficient for FcRγ expression (Figure 1A). This effect was specific to FCER1G-RNP, as there was no change in FcRγ expression in cells treated with non-targeting control RNP (ctrl RNP). Moreover, the expression of CD3ζ, another CD16-associated signaling adapter protein that is homologous to FcRγ in function and sequence and encoded by a gene located close to FCER1G on chromosome 1 (Eiseman and Bolen, 1992), was unaffected by FCER1G-RNP treatment (Figure 1B). Kinetic analysis showed a partial reduction of FcRγ expression detectable 3 days post-nucleofection, but approximately 1 week was needed to reveal a distinct population having complete depletion (Figure 1B). Since it is estimated that CRISPR/Cas9-mediated gene editing occurs within 12 hr post-nucleofection (Brinkman et al., 2018; Kim et al., 2014), our data suggest that pre-existing FCER1G transcripts and FcRγ proteins have relatively long lifespans. Therefore, subsequent analyses of FCER1G gene-edited cells were performed at least 10 days post-nucleofection. Under this condition, we were able to obtain on average approximately 60% efficacy in the production of FcRγ− NK cells.
Figure 1.
Deletion of FcRγ in NK Cells Leads to Enhanced Cytokine Production in Response to CD16 Cross-linking
(A) Contour plots show a mixture of NK (CD3−CD56+), T (CD3+CD56- and CD3+CD56+), and non-NK/non-T (CD3−CD56-) cells after expansion of PBMC samples enriched for NK cells. NK cells (gated, red) were further analyzed for FcRγ expression following nucleofection with FCER1G-targeting Cas9/gRNA ribonucleoprotein (FCER1G-RNP) or non-targeting Cas9/gRNA ribonucleoprotein (ctrl RNP) after 10 days of in vitro culture. Data are representative of 5 independent experiments performed on NK cells from 10 donors.
(B) Time course analysis of FcRγ depletion in FCERIG-RNP-treated NK cells (red), with CD3+CD56- T cells (blue) overlaid for comparison. The staining pattern obtained using a matched isotype control antibody (iso ctrl) is shown.
(C–E) Contour plots show flow cytometric analysis of (C) TNF-α, (D) IFN-γ production, and (E) cell surface CD107a expression by FcRγ+ and FcRγ− NK subsets from a representative donor without (−) or with (+) CD16 cross-linking (CXL). Numbers represent the relative percentages of FcRγ+ or FcRγ− subset that produced indicated cytokine or displayed CD107a. Line graphs show the relative percentages of FcRγ+ or FcRγ− subset that produced indicated cytokine or displayed CD107a from several donors in response to CD16 CXL (n = 10). Circles connected by a line designate data obtained from the same donor sample.
(F–H) Line graphs show the relative percentages of conventional NK (cNK) cells and g-NK cells that produced (F) TNF-α, (G) IFN-γ or (H) displayed CD107a following CD16 CXL (n = 7). Wilcoxon signed-rank test; ∗p < 0.05, ∗∗p < 0.01, n.s., not significant.
Deletion of FcRγ Increases Cytokine Production in Response to CD16 Stimulation
To evaluate whether FcRγ deletion affects the functional responsiveness to CD16 stimulation, we examined cytokine production by the gene-edited FcRγ-null NK cells and NK cells expressing normal levels of FcRγ (hereafter termed FcRγ− subset and FcRγ+ subset, respectively) following incubation with immobilized anti-CD16 mAb. Flow cytometric analysis showed that the FcRγ− subset produced significantly greater amounts of TNF-α compared to the corresponding FcRγ+ subset in the same culture (n = 10, p < 0.01; Figure 1C). In addition to TNF-α, CD16 stimulation resulted in significantly more IFN-γ production by FcRγ− subset (n = 10, p < 0.01; Figure 1D). Of these donors, 7 were HCMV seronegative donor samples which also exhibited statistically significant increases in TNF-α and IFN-γ production (p < 0.05). However, there was no consistent difference in the degranulation response between FcRγ− and FcRγ+ subsets, as measured by cell surface expression of CD107a (Figure 1E). These data demonstrate that FcRγ deletion in NK cells results in increased cytokine production but does not alter degranulation activity in response to CD16 stimulation, suggesting that FcRγ may function to modulate CD16-mediated cytokine production.
Given that CD16 is known to associate with either FcRγ-FcRγ or CD3ζ-CD3ζ homodimer or FcRγ-CD3ζ heterodimer in human NK cells (Lanier, 2008), exclusive association with CD3ζ-CD3ζ homodimer may yield biochemical signaling that is more effective for cytokine production. Thus, when both adapters are co-expressed, FcRγ may negatively regulate the CD16-CD3ζ signaling pathway by competing for association with CD16 and/or interaction with downstream signaling molecules. The lack of any detectable difference in CD107a expression between FcRγ− and FcRγ+ subsets could be explained by a lower activation threshold for degranulation (Bryceson et al., 2009).
We have previously demonstrated that g-NK cells in PBMC produced greater quantities of cytokines compared to the corresponding conventional NK cells upon CD16 cross-linking, whereas degranulation responses between these groups of cells were similar (Hwang et al., 2012). Given that the current study tested in vitro expanded NK cells to examine the effects of FcRγ deletion, we also tested CD16 responsiveness of g-NK and conventional NK cells after expansion in the same culture conditions as the gene-edited cells. Consistent with observations of fresh NK cell populations, in vitro expanded g-NK cells produced significantly greater amounts of both TNF-α and IFN-γ compared to the corresponding conventional NK cells that had been expanded in the same culture conditions (n = 7, p < 0.05; Figures 1F and 1G), whereas no significant difference in degranulation was observed (Figure 1H). When these data were compared with gene-edited FcRγ - and FcRγ + subsets, a similar trend in cytokine production between g-NK and conventional NK cells was observed, although g-NK cells in certain donors appeared to produce dramatically higher levels of cytokine (Figures 1C, 1D, 1F, and 1G). In light of the genetic heterogeneity among donors, as well as differential expression of numerous genes between g-NK and conventional NK cells, determining the specific contribution of FcRγ loss to the enhanced CD16-mediated cytokine production of g-NK cells is challenging with currently available technology. However, given the notable positive impact of CRISPR/Cas9-mediated FcRγ deletion in the absence of other gene expression differences (Figures 1C and 1D), our data suggest that FcRγ-deficiency is an important factor responsible for the enhanced CD16-mediated cytokine production by g-NK cells.
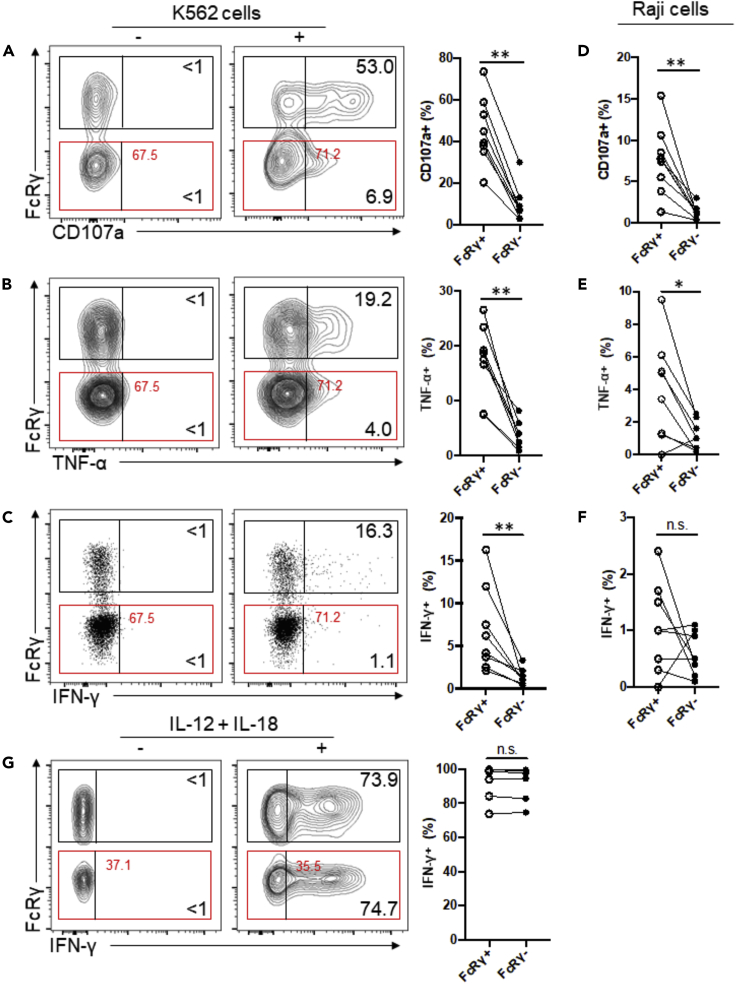
Deletion of FcRγ Decreases Functional Responsiveness to Tumor Target Cells
In contrast to enhanced CD16 responsiveness, g-NK cells respond relatively poorly to the prototypical NK cell-sensitive leukemic cell line K562, as evidenced by a lower degranulation response and lower cytokine production compared to conventional NK cells (Hwang et al., 2012). Similarly, NKG2C+ NK cells were also shown to respond poorly to this tumor target relative to conventional NKG2C− NK cells (Beziat et al., 2012; Liu et al., 2016). We evaluated the potential impact of FcRγ deletion on tumor responsiveness by examining FcRγ− and FcRγ+ subsets for degranulation and cytokine production following incubation with K562 tumor cells. Compared to the FcRγ+ subset, the FcRγ− subset showed dramatic reduction in both degranulation activity and cytokine production (Figures 2A–2C). Similar results were obtained following incubation with Raji tumor cells; FcRγ− subset showed a reduced degranulation response compared to the FcRγ+ subset (Figure 2D), as well as a reduced cytokine response (Figures 2E and 2F). Taken together, these results indicate that FcRγ plays an important role in functional response of conventional NK cells to both K562 and Raji tumor cells. Thus, FcRγ deficiency is likely a major factor responsible for the reduced responsiveness of g-NK cells to tumor cells.
Figure 2.
Deletion of FcRγ Yields Reduced NK Cell Responsiveness to K562 and Raji Tumor Cells
(A) Contour plots show flow cytometric analysis of CD107a expression by FcRγ+ and FcRγ− NK subsets from a representative donor following incubation in the absence (−) or presence (+) of K562 tumor cells. Numbers represent the relative percentages of FcRγ+ or FcRγ− subset that displayed CD107a. Line graph shows the relative percentages of FcRγ+ or FcRγ− subsets that displayed CD107a from several donors in response to K562 stimulation (n = 8). Circles connected by a line designate data obtained from the same donor sample.
(B and C) Contour and dot plots show the percentages of FcRγ+ and FcRγ− subsets that produced (B) TNF-α or (C) IFN-γ in response to K562 stimulation.
(D–F) Line graphs show the relative percentages of FcRγ+ and FcRγ− subsets that displayed (D) CD107a or (E) produced TNF-α or (F) IFN-γ in response to Raji stimulation. (G) Contour plots and line graphs show the percentages of FcRγ+ and FcRγ− subsets that produced IFN-γ in response to stimulation with IL-12 plus IL-18. Wilcoxon signed-rank test; ∗p < 0.05, ∗∗p < 0.01, n.s., not significant.
In addition to altered responsiveness to CD16 and tumor stimulation, g-NK cells display altered responsiveness to stimulation with IL-12 and IL-18, i.e., although conventional NK cells can produce copious amounts of IFN-γ following incubation with these cytokines, IFN-γ production by g-NK cells is dramatically reduced (Schlums et al., 2015). However, both FcRγ+ and FcRγ− subsets showed comparable IFN-γ production with respect to percentage and median fluorescence intensity in response to stimulation with IL-12 and IL-18 by generating ample amounts of IFN-γ (n = 6; Figure 2G), indicating that FcRγ deletion does not affect IL-12 and IL-18 driven cytokine production. Thus, the impaired IL-12/18-mediated responsiveness of g-NK cells is likely controlled by a mechanism that is independent of FcRγ deficiency.
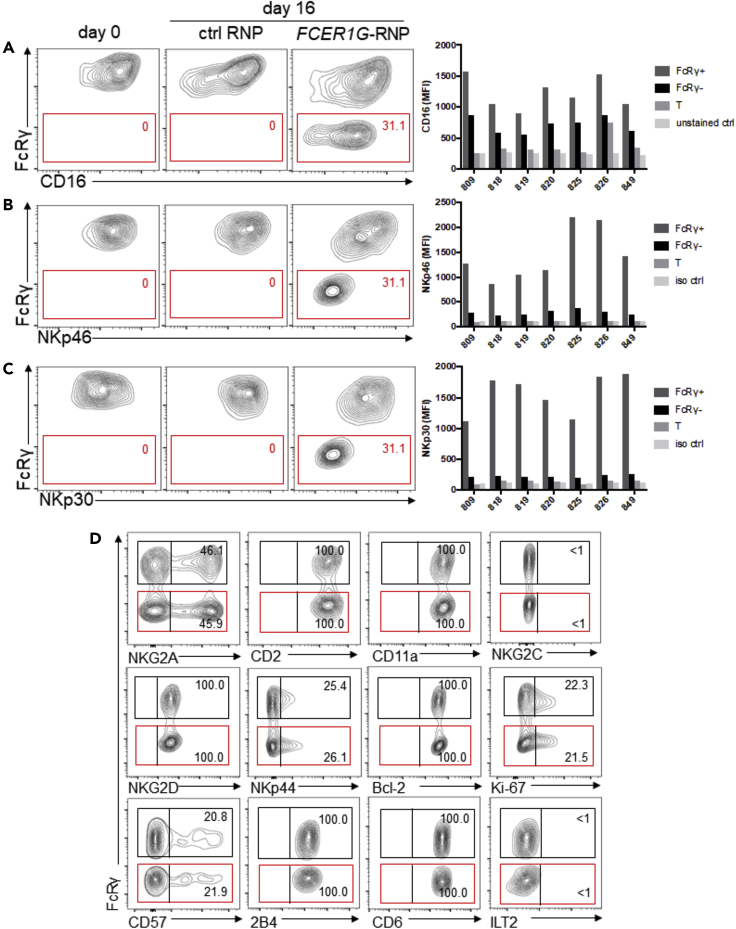
Role of FcRγ in the Expression of CD16, NKp46, and NKp30
It has been shown that co-expression of CD16 with either FcRγ or CD3ζ in heterologous expression systems can support cell surface expression of CD16 (Blazquez-Moreno et al., 2017; Lanier et al., 1991). To evaluate the specific contribution of FcRγ to cell surface expression of CD16 on primary NK cells, we compared CD16 expression on FcRγ− and FcRγ+ subsets. Flow cytometric analysis showed that CD16 expression is lower on the FcRγ− subset compared to the FcRγ+ subset for all donors tested (Figure 3A). Based on the median fluorescence intensities, CD16 expression was reduced on average by 53% in the FcRγ− subset (Figure S1A), indicating that normal CD3ζ expression in the FcRγ− subset (Figure 1B) cannot compensate for the absence of FcRγ and does not support normal levels of CD16 expression. This, coupled with the previous observation of lower CD16 expression on g-NK cells relative to conventional NK cells (Hwang et al., 2012), supports the conclusion that FcRγ deficiency is also responsible for reduced expression of CD16 on g-NK cells. Of note, NK cells in FcRγ knockout mice do not express any detectable cell surface CD16 (Takai et al., 1994), suggesting a differential role of FcRγ in humans and mice. Importantly, the enhanced CD16-mediated cytokine production of the FcRγ− subset (Figures 1C and 1D) was not due to higher CD16 expression. In the absence of FcRγ bearing 1 ITAM, the potential exclusive association between CD16 and homodimer of CD3ζ bearing 3 ITAMs may induce higher cytokine production. This possibility warrants further investigation.
Figure 3.
Loss of FcRγ Leads to Reduced Cell Surface Expression of CD16, NKp46, and NKp30
(A–C) Contour plots show (A) CD16, (B) NKp46, and (C) NKp30 expression with respect to FcRγ status in NK cells from one representative donor. Bar graphs summarize (A) CD16, (B) NKp46, and (C) NKp30 median fluorescence intensity (MFI) of FcRγ+, FcRγ− NK subsets, T cells, and staining controls from 7 donors.
(D) Contour plots show expression of indicated markers with respect to FcRγ status in NK cells. Data are representative of at least three donors.
As FcRγ is also known to be associated with NKp46 and NKp30 (Sivori et al., 2019), we examined cell surface expression of these activation receptors on FcRγ+ and FcRγ− subsets. The FcRγ− subset showed a dramatic reduction in NKp46 expression (87% reduction on average), compared to FcRγ+ subset (Figures 3B and S1B). Moreover, the FcRγ− subset also showed a dramatic reduction in NKp30 expression (92% reduction) (Figures 3C and S1C). These results indicate that FcRγ plays a crucial role in supporting cell surface expression of both NKp46 and NKp30. Given that both NKp46 and NKp30 were shown to associate with CD3ζ as well (Pende et al., 1999; Vitale et al., 1998), it is likely that CD3ζ alone in the FcRγ− subset cannot compensate for the absence of FcRγ and can only support a minimal level of expression of these receptors. Nonetheless, our results provide strong evidence that FcRγ deficiency is responsible for reduced expression of these receptors by g-NK cells, as well as by NKG2C+ NK cells (Guma et al., 2004; Hwang et al., 2012; Schlums et al., 2015). Since NKp46 and NKp30 are considered major tumor recognition receptors, the abolished expression of these receptors likely resulted in the reduced responsiveness of FcRγ− subset to K562 and Raji tumor cells. Consistent with this possibility, both K562 and Raji cells were shown to express B7-H6, a ligand for NKp30 (Brandt et al., 2009). Taken together, our results suggest that FcRγ deficiency in g-NK cells is a major factor contributing to their reduced responsiveness toward tumor target cells through limiting the cell surface expression of NKp30 and NKp46.
In contrast to CD16, NKp46, and NKp30, we observed that the expression of other markers, including NKG2A, CD2, CD11a, NKG2C, CD57, ILT2, and Bcl-2, all of which are differentially expressed between g-NK and conventional NK cells (Hwang et al., 2012; Lee et al., 2015; Zhang et al., 2013), were unaffected by FcRγ deletion (Figure 3D), suggesting that altered expression of these molecules in g-NK cells is controlled by other mechanisms.
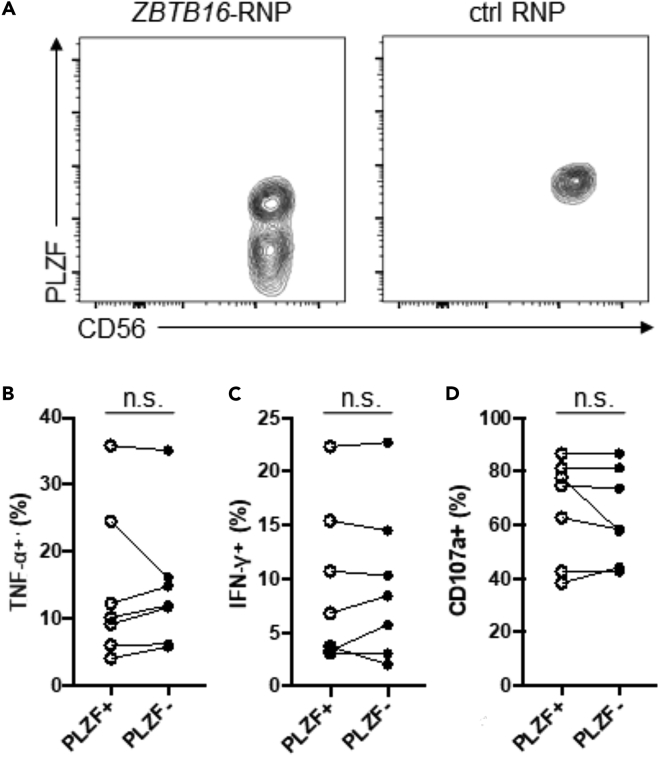
Deletion of PLZF Does Not Alter Responsiveness to CD16 or IL-12/18 Stimulation
In addition to FcRγ deficiency, altered expression of other genes may also contribute to the altered functional responsiveness exhibited by adaptive NK cells. Among such alterations, PLZF deficiency has been suggested to be an important factor contributing to the observed adaptive properties (Schlums et al., 2015). To explore the role of PLZF in NK cells, we sought to delete ZBTB16 (encoding PLZF) from conventional NK cells by gene editing. NK cells enriched from PBMC samples that did not have detectable PLZF-deficient NK cells were expanded and then subjected to nucleofection of ZBTB16-targeting Cas9/gRNA ribonucleoprotein (ZBTB16-RNP). Flow cytometry analysis showed that a distinct NK cell subset deficient in expression of PLZF appeared approximately 1 week post-nucleofection (Figure 4A). The cytokine production assay demonstrated that following incubation with immobilized anti-CD16 mAb, PLZF− NK cells produced similar levels of IFN-γ and TNF-α as PLZF+ NK cells (n = 7; Figures 4B and 4C). Degranulation responses were also comparable (Figure 4D). In addition, stimulation by IL-12 and IL-18 resulted in equivalently high levels of IFN-γ by both PLZF− and PLZF+ NK cells (data not shown). We also observed no difference in the expression of components for IL-12 or IL-18 receptors (Figure S2). Finally, we observed that FcRγ, CD16, NKp46, and NKp30 were all normally expressed by PLZF− NK cells (data not shown). Taken together, unlike the deletion of FcRγ, the available data do not demonstrate any notable functional or phenotypic changes resulting from the deletion of PLZF.
Figure 4.
Deletion of PLZF in NK Cells Does Not Affect Responsiveness to CD16
(A) Contour plots show the expression of PLZF in NK cells following nucleofection with ZBTB16-targeting Cas9/gRNA ribonucleoprotein (ZBTB16-RNP). Data are representative of 4 independent experiments performed on NK cells from 7 donors.
(B–D) Line graphs show the relative percentages of PLZF+ and PLZF− subsets that produced (B) TNF-α, (C) IFN-γ or (D) displayed CD107a from several donors in response to CD16 CXL (n = 7). Circles connected by a line designate data obtained from the same donor. n.s., not significant.
Concluding Remarks
Collectively, our results demonstrate that CRISPR/Cas9-mediated deletion of FcRγ in human NK cells led to enhanced CD16 responsiveness but reduced tumor responsiveness in association with reduced expression of NCRs. However, deletion of PLZF did not elicit notable changes. Importantly, our results also provide strong evidence that FcRγ deficiency is an important factor responsible for both enhanced CD16-mediated cytokine production and reduced tumor responsiveness exhibited by adaptive human NK cells. However, it should be noted that our data do not exclude the possibility that factors other than FcRγ deficiency may further contribute to the altered functional responsiveness of adaptive NK cells. Future studies utilizing the CRISPR-Cas9 gene editing approach will be helpful in identifying such factors, thereby deepening our understanding of the mechanism(s) contributing to adaptive NK cell functionality and illuminating potential development for clinical application.
Limitations of the Study
In the gene knock-out experiments, NK cells were cultured in vitro before gene editing to expand the number of available cells and then again after editing to completely deplete any pre-existing target proteins before analysis. It is likely that during in vitro culture, expression of many genes, including those involved in functional responses of NK cells, may be altered. Therefore, it is possible that in addition to the disrupted expression of the target genes being edited in this study, other genes with altered expression may have contributed to the phenotypic and functional characteristics exhibited by the gene-edited NK cells.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sungjin Kim (sjikim@ucdavis.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate datasets or code.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Jaewon Lee, Rachel Baek, and Tracey Rourke for technical assistance. W.L was partially supported by the China Scholarship Council. This work was supported by the NIH grant (AI110894) to S.K.
Author Contributions
W.L. conceived the study, performed experiments, analyzed data, and wrote the manuscript; J.M.S. analyzed data and wrote the manuscript; E.L., H.C., and P.H.P performed experiments; S.K. conceived the study, analyzed data, and wrote the manuscript.
Declaration of Interests
S.K. is a consultant, stock owner, and advisory board member of Indapta Therapeutics, Inc. The other authors declare no competing interests.
Published: November 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101709.
Supplemental Information
References
- Beziat V., Dalgard O., Asselah T., Halfon P., Bedossa P., Boudifa A., Hervier B., Theodorou I., Martinot M., Debre P. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur. J. Immunol. 2012;42:447–457. doi: 10.1002/eji.201141826. [DOI] [PubMed] [Google Scholar]
- Beziat V., Liu L.L., Malmberg J.A., Ivarsson M.A., Sohlberg E., Bjorklund A.T., Retiere C., Sverremark-Ekstrom E., Traherne J., Ljungman P. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez-Moreno A., Park S., Im W., Call M.J., Call M.E., Reyburn H.T. Transmembrane features governing Fc receptor CD16A assembly with CD16A signaling adaptor molecules. Proc. Natl. Acad. Sci. U S A. 2017;114:e5645–e5654. doi: 10.1073/pnas.1706483114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt C.S., Baratin M., Yi E.C., Kennedy J., Gao Z., Fox B., Haldeman B., Ostrander C.D., Kaifu T., Chabannon C. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 2009;206:1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E.K., Chen T., de Haas M., Holland H.A., Akhtar W., van Steensel B. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol. Cell. 2018;70:801–813.e6. doi: 10.1016/j.molcel.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhns P., Iannascoli B., England P., Mancardi D.A., Fernandez N., Jorieux S., Daeron M. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- Bryceson Y.T., Ljunggren H.G., Long E.O. Minimal requirement for induction of natural cytotoxicity and intersection of activation signals by inhibitory receptors. Blood. 2009;114:2657–2666. doi: 10.1182/blood-2009-01-201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichocki F., Wu C.Y., Zhang B., Felices M., Tesi B., Tuininga K., Dougherty P., Taras E., Hinderlie P., Blazar B.R. ARID5B regulates metabolic programming in human adaptive NK cells. J. Exp. Med. 2018;215:2379–2395. doi: 10.1084/jem.20172168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.A., Elliott J.M., Keyel P.A., Yang L., Carrero J.A., Yokoyama W.M. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. U S A. 2009;106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Garcia M., Vera A., Moraru M., Vilches C., Lopez-Botet M., Muntasell A. Antibody-mediated response of NKG2Cbright NK cells against human cytomegalovirus. J. Immunol. 2015;194:2715–2724. doi: 10.4049/jimmunol.1402281. [DOI] [PubMed] [Google Scholar]
- Eiseman E., Bolen J.B. Signal transduction by the cytoplasmic domains of Fc epsilon RI-gamma and TCR-zeta in rat basophilic leukemia cells. J. Biol. Chem. 1992;267:21027–21032. [PubMed] [Google Scholar]
- Foley B., Cooley S., Verneris M.R., Curtsinger J., Luo X., Waller E.K., Anasetti C., Weisdorf D., Miller J.S. Human cytomegalovirus (CMV)-induced memory-like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J. Immunol. 2012;189:5082–5088. doi: 10.4049/jimmunol.1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleimer M., von Boehmer H., Kreslavsky T. PLZF controls the expression of a limited number of genes essential for NKT cell function. Front. Immunol. 2012;3:374. doi: 10.3389/fimmu.2012.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier M.R., Jonjic S., Riley E.M., Juranic Lisnic V. CMV and natural killer cells: shaping the response to vaccination. Eur. J. Immunol. 2018;48:50–65. doi: 10.1002/eji.201646762. [DOI] [PubMed] [Google Scholar]
- Guma M., Angulo A., Vilches C., Gomez-Lozano N., Malats N., Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- Hammer Q., Romagnani C. About training and memory: NK-cell adaptation to viral infections. Adv. Immunol. 2017;133:171–207. doi: 10.1016/bs.ai.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Hammer Q., Ruckert T., Romagnani C. Natural killer cell specificity for viral infections. Nat. Immunol. 2018;19:800–808. doi: 10.1038/s41590-018-0163-6. [DOI] [PubMed] [Google Scholar]
- Hart G.T., Tran T.M., Theorell J., Schlums H., Arora G., Rajagopalan S., Sangala A.D.J., Welsh K.J., Traore B., Pierce S.K. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J. Exp. Med. 2019;216:1280–1290. doi: 10.1084/jem.20181681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Zhang T., Scott J.M., Kim A.R., Lee T., Kakarla T., Kim A., Sunwoo J.B., Kim S. Identification of human NK cells that are deficient for signaling adaptor FcRgamma and specialized for antibody-dependent immune functions. Int. Immunol. 2012;24:793–802. doi: 10.1093/intimm/dxs080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L.L. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L.L., Yu G., Phillips J.H. Analysis of Fc gamma RIII (CD16) membrane expression and association with CD3 zeta and Fc epsilon RI-gamma by site-directed mutation. J. Immunol. 1991;146:1571–1576. [PubMed] [Google Scholar]
- Lee J., Zhang T., Hwang I., Kim A., Nitschke L., Kim M., Scott J.M., Kamimura Y., Lanier L.L., Kim S. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. 2015;42:431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.L., Landskron J., Ask E.H., Enqvist M., Sohlberg E., Traherne J.A., Hammer Q., Goodridge J.P., Larsson S., Jayaraman J. Critical role of CD2 co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep. 2016;15:1088–1099. doi: 10.1016/j.celrep.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetke-Eversloh M., Hammer Q., Durek P., Nordstrom K., Gasparoni G., Pink M., Hamann A., Walter J., Chang H.D., Dong J. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. 2014;10:e1004441. doi: 10.1371/journal.ppat.1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Seiler M.P., Scanlon S.T., Mao A.P., Constantinides M.G., Bertozzi-Villa C., Singer J.D., Bendelac A. BTB-ZF factors recruit the E3 ligase cullin 3 to regulate lymphoid effector programs. Nature. 2012;491:618–621. doi: 10.1038/nature11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A., Bottino C., Vitale M., Pende D., Cantoni C., Mingari M.C., Biassoni R., Moretta L. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- Muccio L., Falco M., Bertaina A., Locatelli F., Frassoni F., Sivori S., Moretta L., Moretta A., Della Chiesa M. Late development of FcepsilonRgamma(neg) adaptive natural killer cells upon human cytomegalovirus reactivation in umbilical cord blood transplantation recipients. Front. Immunol. 2018;9:1050. doi: 10.3389/fimmu.2018.01050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary J.G., Goodarzi M., Drayton D.L., von Andrian U.H. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- Paust S., Gill H.S., Wang B.Z., Flynn M.P., Moseman E.A., Senman B., Szczepanik M., Telenti A., Askenase P.W., Compans R.W. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat. Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende D., Parolini S., Pessino A., Sivori S., Augugliaro R., Morelli L., Marcenaro E., Accame L., Malaspina A., Biassoni R. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R.K., Li H., Jost S., Blass E., Li H., Schafer J.L., Varner V., Manickam C., Eslamizar L., Altfeld M. Antigen-specific NK cell memory in rhesus macaques. Nat. Immunol. 2015;16:927–932. doi: 10.1038/ni.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolle A., Brodin P. Immune adaptation to environmental influence: the case of NK cells and HCMV. Trends Immunol. 2016;37:233–243. doi: 10.1016/j.it.2016.01.005. [DOI] [PubMed] [Google Scholar]
- Schlums H., Cichocki F., Tesi B., Theorell J., Beziat V., Holmes T.D., Han H., Chiang S.C., Foley B., Mattsson K. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 2015;42:443–456. doi: 10.1016/j.immuni.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivori S., Vacca P., Del Zotto G., Munari E., Mingari M.C., Moretta L. Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell Mol. Immunol. 2019;16:430–441. doi: 10.1038/s41423-019-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Beilke J.N., Lanier L.L. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T., Li M., Sylvestre D., Clynes R., Ravetch J.V. FcR gamma chain deletion results in pleiotrophic effector cell defects. Cell. 1994;76:519–529. doi: 10.1016/0092-8674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Vitale M., Bottino C., Sivori S., Sanseverino L., Castriconi R., Marcenaro E., Augugliaro R., Moretta L., Moretta A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex-restricted tumor cell lysis. J. Exp. Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E., Raulet D.H., Moretta A., Caligiuri M.A., Zitvogel L., Lanier L.L., Yokoyama W.M., Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Sinzger C., Frascaroli G., Reichel J., Bayer C., Wang L., Schirmbeck R., Mertens T. Human cytomegalovirus-induced NKG2C(hi) CD57(hi) natural killer cells are effectors dependent on humoral antiviral immunity. J. Virol. 2013;87:7717–7725. doi: 10.1128/JVI.01096-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Scott J.M., Hwang I., Kim S. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J. Immunol. 2013;190:1402–1406. doi: 10.4049/jimmunol.1203034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Amran F.S., Kramski M., Angelovich T.A., Elliott J., Hearps A.C., Price P., Jaworowski A. An NK cell population lacking FcRgamma is expanded in chronically infected HIV patients. J. Immunol. 2015;194:4688–4697. doi: 10.4049/jimmunol.1402448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets or code.