Abstract
This study aimed to expand the competing endogenous RNA network in osteosarcoma (OS) involving hsa_circ_0085539 and its downstream target miR-526b-5p. The expression levels of circ_0085539, miR-526b-5p, and stress-associated endoplasmic reticulum protein 1 (SERP1) mRNA in OS tissues and cells were detected and analyzed by qRT-PCR. After that, the interrelationships between these three genetic materials were validated with a luciferase reporter assay system. The effect of the circ_0085539/miR-526b-5p/SERP1 axis on OS cell malignancy phenotypes was further assessed using in vitro assays, including cell counting kit-8 (CCK-8) assays, colony foci formation assays, wound-healing migration assays, and transwell invasion assays. To determine the function of circ_0085539 on OS tumor growth in vivo, a xenograft formation assay was performed. In OS tissues and cells, the expression of circ_0085539 and SERP1 was upregulated, while that of miR-526b-5p was downregulated. After experimental analyses, it was found that silencing circ_0085539 inhibited the aggression of OS in vivo and in vitro. Mechanistic investigations also revealed that circ_0085539 could sponge miR-526b-5p and that miR526b-5p could directly target SERP1. The cytological experiments in vitro demonstrated that miR-526b-5p could restore the effect of circ_0085539 in terms of promoting OS malignancy phenotypes by suppressing SERP1. Overall, the present study validated that hsa_circ_0085539 could promote the progression of OS by regulating miR-526b-5p/SERP1.
Keywords: hsa_circ_0085539, miR-526b-5p, SERP1, osteosarcoma
Graphical Abstract

This study is the first to investigate the effect of the interactions between circ_0085539, miR-526b-5p, and SERP1 in OS. The authors found that circ_0085539 accelerated OS progression by binding to and regulating miR-526b-5p, which suppressed SERP1. Their findings also revealed a novel molecular interactome of circ_0085539/miR-526b-5p/SERP1 in OS.
Introduction
Osteosarcoma (OS) is a primary bone malignancy that is prevalent among children and adolescents.1,2 Although the combination of surgical treatment and chemotherapy has been significantly developed over the years,3 therapies that can improve the survival rate of patients are still limited. The existing poor prognosis of OS can be traced to tumor progression and recurrence, and this problem has resulted in a low 5-year survival rate for people diagnosed with bone cancer.4 To improve the survival outcome of patients with OS, it is crucial to explore the molecular mechanism of OS progression.
First found in 1976 with a circular structure, circular RNAs (circRNAs) are found active in mammalian cells, and the degradation of these RNAs by RNA exonuclease can be avoided.5,6 An enormous amount of research has reported that circRNAs have multiple sites complementary to microRNAs (miRNAs) to relieve the inhibitory effect of miRNAs on their target genes.7 Accumulating studies have also found that circPVT1 plays a tumor-promoting role in the progression of cancers. For instance, hsa_circ_PVT1 was reported to be a proliferative factor in the acceleration of esophageal carcinoma.8 In another study, circ_PVT1 facilitated cell proliferation and invasion in non-small-cell lung cancer by sponging miR-125b.9 One study also reported that circ_PVT1 contributed to cell proliferation in gastric cancer by sponging the miR-125 family.10 While hsa_circ_0085539 (circ_0085539) located on chr8:12886700–129082518 was generated from PVT1, its effect has not been studied in any cancer, including OS.
In the last 6 years, miR-526b has been proven to participate in various cancers. For instance, miR-526b was found to downregulate and inhibit cell proliferation in gastric cancer by regulating the downstream KDM4A/YAP1 pathway.11 An increase in miR-526b-3p, which is a regulatory and prognostic factor, limited the development of glioma by targeting WEE1 in vivo and in vitro.12 Additionally, in colon cancer, miR-526b-3p acted as a tumor suppressor by restricting the expression of HIF-1α.13 The TargetScan algorithm predicted that miR-526-5p could specifically bind to circ_0085539. For this reason, we speculated that miR-526b-5p might be a tumor suppressor in OS by being regulated by circ_0085539.
Stress-associated endoplasmic reticulum (ER) protein 1 (SERP1) encodes a 66-aa protein. SERP1 expression is upregulated in response to hypoxia and/or reperfusion and ER stress.14 In one experiment, SERP1 knockout mice showed enhanced ER stress and impaired growth.15 It was also reported that similar to many other upstream and downstream molecules of YAP, SERP1 was associated with unfavorable survival of pancreatic cancer patients.16 In addition, SERP1 re-expression reversed the miR-124-caused cell death phenotype in glioblastoma cells grown under hypoxia.17 Given that very little had been known about SERP1 in OS, we conducted this study to explore the effects of SERP1 in OS progression.
Results
circ_0085539 Was Upregulated in OS Tissue Samples and Cell Lines
To identify the expression of circ_0085539 in OS tissues and cells, qRT-PCR was utilized. The experimental result showed that the expression of circ_0085539 was upregulated by 2.07-fold in 16 OS tissue specimens compared with matched healthy tissues (p < 0.001, Figure 1A). Similar to the trend of circ_0085539 in the OS tissues, the expression of circ_0085539 was upregulated by a 1.5-fold to 4.5-fold increase in five OS cell lines in comparison with human osteoblast cell lines (∗p < 0.05, ∗∗p < 0.001 versus hFOB1.19 cells, Figure 1B). HOS and Saos2 cell lines demonstrated the most significantly upregulated circ_0085539; hence, they were selected as the cell line models for the following experiments. It was found that circ-0085539 was resistant to RNase R digestion in HOS and Saos2 cells. This result indicated that circ_0085539 was more stable than PVT1 because of its particular cyclic structure (∗∗p < 0.001, Figure 1C). Agarose gel electrophoresis was also conducted to identify the circRNA character of circ_0085539 in HOS and Saos2 cell lines. The results revealed that circ_0085539 was blotted in cDNA rather than in genomic DNA (gDNA) in HOS and Saos2 cell lines (Figure 1D), a finding that indicated the circRNA feature of circ_0085539 in HOS and Saos2 cell lines. Cytoplasmic and nuclear RNA analyses showed that the distribution of circ_0085539 was mainly in the cell cytoplasm (∗p < 0.05, ∗∗p < 0.001, Figure 1E). After sh-circ_0085539 plasmids were used to transfect the HOS and Saos2 cells, it was observed that sh-circ_0085539 led to a 26% and 32% decrease of the circ_0085539 expression level in HOS cells and Saos2 cells, respectively (∗∗p < 0.001 versus the short hairpin RNA [shRNA] [sh-]negative control [NC] group, Figure 1F).
Figure 1.
circ_0085539 Was Upregulated in Osteosarcoma Tissue and Cell Lines
(A) In 16 cases of osteosarcoma patients, circ_0085539 was significantly upregulated in osteosarcoma tissue compared to adjacent non-tumor tissue. (B) circ_0085539 expression was detected by qRT-PCR in osteosarcoma cell lines (HOS, Saos2, U2OS, and SJSA-1) and a human osteoblastic cell line (hFOB1.19). ∗p < 0.05, ∗∗p < 0.001 versus hFOB1.19 cells. (C) The expression of circ-0085539 and PVT1 were detected by qRT-PCR after RNase R intervened in HOS and Saos2 cells. PVT1 was the symbol gene of circ_0085539. ∗p < 0.05, ∗∗p < 0.001. (D) Agarose gel electrophoresis was used to identify the circRNA feature of circ_0085539, and GAPDH was taken as a reference substance. (E) qRT-PCR analysis of circ_0085539 and PVT1 in the cytoplasm or the nucleus in HOS and Saos2 cells. PVT1 was the symbol gene of circ_0085539. ∗p < 0.05, ∗∗p < 0.001. (F) The transfection efficiency of sh-circ_0085539 was identified by qRT-PCR. Con, the HOS and Saos2 cells were cultured without any treatments; NC, the HOS and Saos2 cells were transfected with negative control vectors; sh-circ, the HOS and Saos2 cells were transfected with sh-circ_0085539. ∗p < 0.05, ∗∗p < 0.001 versus the sh-NC group. Three independent repeated experiments were implemented in all of the assays.
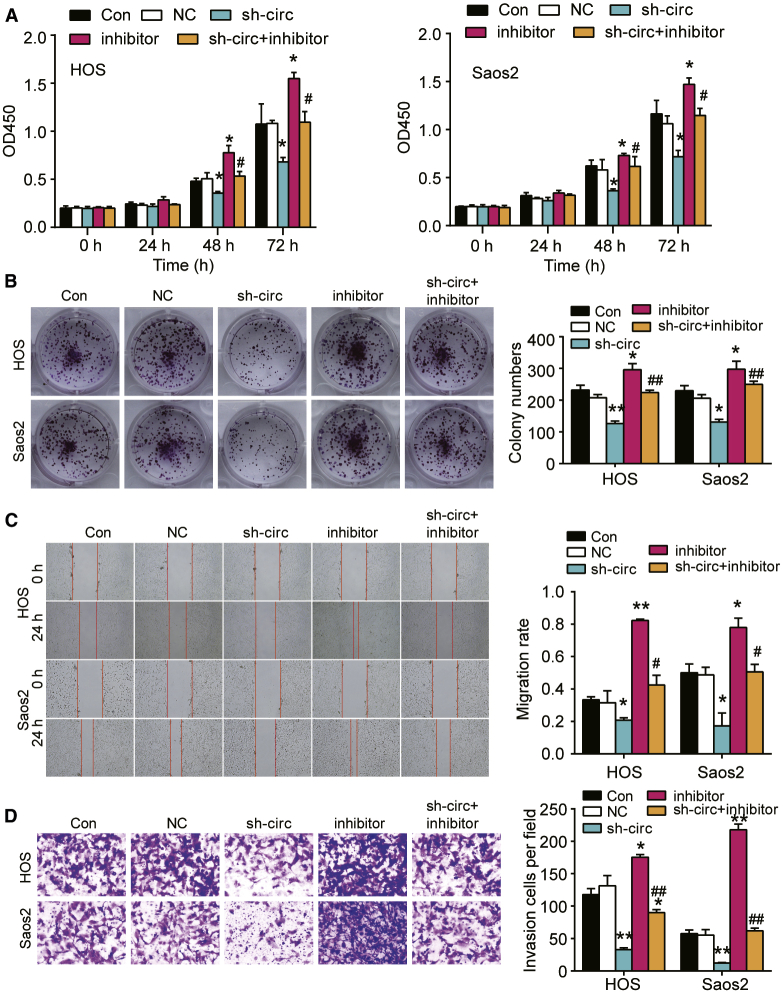
circ_0085539 Acted as the Tumor Suppressor in OS Cells
A cell counting kit-8 (CCK-8) assay, colony foci formation assay, wound-healing assay, and transwell assay were performed to identify the function of circ_0085539 in OS cells. The CCK-8 assay result showed that the circ_0085539 knockdown impaired cell proliferation in both HOS and Saos2 cells at 48 and 72 h (∗p < 0.05, Figure 2A). The colony numbers were reduced by 28% and 15% in HOS and Saos2 cells transfected with sh-circ_0085539, respectively (∗p < 0.05, Figure 2B). The wound-healing assay result showed a 20% decline in the migration rate when the HOS cells were transfected with sh-circ_0085549 and a 32% decrease in the migration rate when the Saos2 cells were treated the same way as HOS cells (∗∗p < 0.001, Figure 2C). Additionally, sh-circ_0085539 transfection impaired cell invasion in both HOS cells and Saos2 cells (∗∗p < 0.001, Figure 2D). These data demonstrated that knocking down circ_0085539 suppressed cell proliferation, migration, and invasion of OS.
Figure 2.
circ_0085539 Contributed to Cell Proliferation, Migration, and Invasion In Vitro
(A) The CCK-8 assay was used to assess the cell proliferation ability when HOS and Saos2 cells were transfected with sh-circ_0085539. (B) The proliferation was further evaluated by a colony formation assay after the HOS and Saos2 cells were transfected with sh-circ_0085539. (C) The migration rate of the transfected HOS and Saos2 cells was measured using the wound-healing assay. Migration rate was represented by the ratio of the migration distance at 24 h to the wound width at 0 h. (D) The transwell invasion assay was performed to analyze the cell invasion ability in the transfected HOS and Saos2 cells. Con, the HOS and Saos2 cells were cultured without any treatments; NC, the HOS and Saos2 cells were transfected with negative control vectors; sh-circ, the HOS and Saos2 cells were transfected with sh-circ_0085539. ∗p < 0.05, ∗∗p < 0.001 versus the sh-NC group. Three independent repeated experiments were implemented in all of the assays.
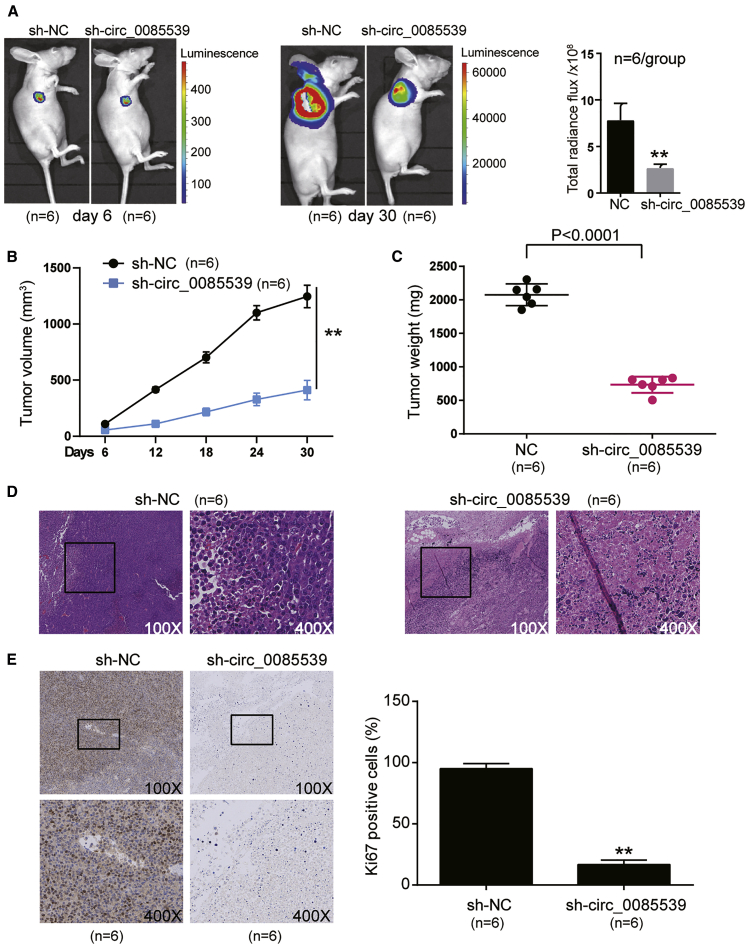
circ_0085539 Silence Impaired the Growth of OS In Vivo
Our in vivo xenograft assay showed that after 30-day injections, the bioluminescence imaging showed that unlike the NC transfection group, the transfection of sh-circ_0085539 inhibited tumor growth. It could be seen from the images that sh-circ_0085539 decreased the tumor growth in mice after 30 days compared with the NC group (∗∗p < 0.001, Figure 3A). This indicated that circ_0085539 inhibition could reduce OS progression. As for the tumor volume and weight, the tumor volume in the sh-circ_0085539 group was 33.03% of that in the NC group at day 30 (∗∗p < 0.001, Figure 3B), and the tumor weight in the sh-circ_0085539 group was 35.37% of that in the NC group at day 30 (p < 0.001, Figure 3C), also indicating that circ_0085539 knockdown could reduce OS progression. The H&E staining results demonstrated that circ_0085539 silence damaged the tissue structure of the xenografted tumor (Figure 3D). Moreover, the Ki67 staining result showed that circ_0085539 silence significantly inhibited tumor cell growth compared with the sh-NC group (∗∗p < 0.001 versus the sh-NC group, Figure 3E). Taken together, these data implied that circ_0085539 knockdown resulted in the inhibition of tumor growth in vivo.
Figure 3.
circ_0085539 Promotes Osteosarcoma Tumor Genesis In Vivo
(A) The representative images of bioluminescence imaging were taken from the nude mice after subcutaneous injection with sh-circ_0085539-transfected Saos2 cells. sh-NC, the six nude mice were subcutaneously injected with the HOS cells transfected with negative control vectors; sh-circ_0085539, the six nude mice were subcutaneously injected with the HOS cells transfected with sh-circ_0085539 vectors. (B) The tumor volume was measured 6 days apart. sh-NC, the six nude mice were subcutaneously injected with the HOS cells transfected with negative control vectors; sh-circ_0085539, the six nude mice were subcutaneously injected with the HOS cells transfected with sh-circ_0085539 vectors. ∗p < 0.05, ∗∗p < 0.001. (C) Tumor weight was calculated when the mice were euthanized after 30-day injections. sh-NC, the six nude mice were subcutaneously injected with the HOS cells transfected with negative control vectors; sh-circ_0085539, the six nude mice were subcutaneously injected with the HOS cells transfected with sh-circ_0085539 vectors. ∗p < 0.05, ∗∗p < 0.001. (D) H&E staining was used to evaluated the histology at ×100 and ×400 original magnification. sh-NC, the six nude mice were subcutaneously injected with the HOS cells transfected with negative control vectors; sh-circ_0085539, the six nude mice were subcutaneously injected with the HOS cells transfected with sh-circ_0085539 vectors. (E) The effect of sh-circ-0085539 on cell proliferation was evaluated by Ki67 staining. Representative images are shown at ×100 and ×400 original magnification. sh-NC, the six nude mice were subcutaneously injected with the HOS cells transfected with negative control vectors; sh-circ_0085539, the six nude mice were subcutaneously injected with the HOS cells transfected with sh-circ_0085539 vectors. ∗p < 0.05, ∗∗p < 0.001 versus sh-NC group. Three independent repeated experiments were implemented in all of the assays.
circ_0085539 Negatively Regulated miR-526b-5p
The TargetScan algorithm predicted a binding site between circ_0085539 and miR-526b-5p (Figure 4A). Then, the dual-luciferase reporter assay results indicated that the co-transfection of circ_0085539-wild-type (WT) and miR-526b-5p mimic resulted in an approximately 50% reduction of the relative luciferase activity in both OS cell lines (∗∗p < 0.001 versus co-transfection of circ_0085539-WT and the miR-NC group, Figure 4B). The RNA immunoprecipitation assay (RIPA) results revealed that circ_0085539 and miR-526b-5p directly interacted with each other in an Ago2-dependent manner in HOS and Saos2 cells (∗∗p < 0.001, Figure 4C). The expression of miR-526b-5p with a 60.7% decrease in tumor tissues was observed (p < 0.001, Figure 4D). The correlation analysis further revealed that the expression of circ_0085539 was negatively correlated with the expression of miR-526b-5p (p < 0.001, Figure 4E). While the expression of miR-526b-5p was upregulated by more than 1.5-fold in the sh-circ_0085539 group, the expression of miR-526b-5p decreased by approximately 75% in the miR-526b-5p inhibitor group versus the control group. It was also observed that the co-transfection of miR-526b-5p inhibitor with sh-circ_0085539 reversed the effects of circ_0085539 silence on the expression of miR-526b-5p (∗p < 0.05, ∗∗p < 0.001 versus the control group, Figure 4F). Overall, these findings confirmed that circ_0085539 inhibited the expression of miR-526b-5p directly.
Figure 4.
circ-0085539 Was a Sponge of miR-526b-5p in OS Cells
(A) The TargetScan was used to predict the binding site of circ_0085539 for miR-526b-5p. (B) The dual-luciferase reporter assay was used to prove the binding site between circ_0085539 and miR-526b-5p. circ_0085539-WT, the plasmids contained the binding site of circ_0085539 for miR-526b-5p; circ_0085539-MUT, the plasmids did not have any biding site of circ_0085539 for miR-526b-5p. ∗p < 0.05, ∗∗p < 0.001 versus co-transfection of circ_0085539-WT and NC plasmids. (C) RNA immunoprecipitation (RIP) assay displayed abundances of circ_0085539 and miR-526b-5p in Ago2 or IgG. ∗p < 0.05, ∗∗p < 0.001. (D) In 16 cases of osteosarcoma patients, miR-526b-5p was significantly downregulated in osteosarcoma tissue compared to adjacent non-tumor tissue. (E) The negative correlation between the expression of circ_0085539 and miR-526b-5p in 16 OS tissues is shown. (F) The expression of circ_0085539 and miR-526b-5p in HOS and Saos2 cells after stable transfection of Con, NC, sh-circ_0085539, miR-526b-5p inhibitor, or sh-circ_0085539+miR-526b-5p inhibitor. Con, the HOS and Saos2 cells were cultured without any treatments; NC, the HOS and Saos2 cells were transfected with negative control vectors; sh-circ, the HOS and Saos2 cells were transfected with sh-circ_0085539; inhibitor, the HOS and Saos2 cells were transfected with miR-526b-5p inhibitor; sh-circ+inhibitor, the HOS and Saos2 cells were co-transfected with sh-circ_0085539 and miR-526b-5p inhibitor. ∗p < 0.05, ∗∗p < 0.001 versus control group. Three independent repeated experiments were implemented in all of the assays.
Silencing miR-526b-5p Reversed the Inhibitory Influence of sh-circ_0085539 in OS Cells
To thoroughly investigate the mechanism of circ_0085539 in OS cells, sh-circ_0085539, miR-526b-5p inhibitor, or both were transfected into HOS and Saos2 cells. The cell proliferation in the circ_0085539 silence group was weaker than that in the control group in HOS and Saos2 cells. However, the co-transfection with miR-526b-5p inhibitor reversed the changes in cell proliferation induced by sh-circ_0085539 (∗p < 0.05, ∗∗p < 0.001 versus control; #p < 0.05, ##p < 0.001 versus sh-circ_0085539, Figures 5A and 5B). The wound-healing assay results showed that the migration rate of HOS cells declined by 39% in the sh-circ_0085539 group, but it increased by 2.48-fold in the miR-526b-5p inhibitor group. Furthermore, the HOS cells co-transfected with sh-circ_0085539 and miR-526b-5p inhibitor showed no significant difference from the control group. Put simply, the results in Saos2 cells were similar to those in HOS cells (∗p < 0.05, ∗∗p < 0.001 versus control; #p < 0.05 versus sh-circ_0085539, Figure 5C). In the same transfection pattern, the transwell invasion assay results indicated that the numbers of invaded cells were reduced in OS cells with the transfection of sh-circ_0085539. Nonetheless, this anti-invasion effect of sh-circ_0085539 was reversed by miR-526b-5p inhibition (∗p < 0.05, ∗∗p < 0.001 versus control; ##p < 0.001 versus sh-circ_0085539, Figure 5D).
Figure 5.
Inhibition of miR-526b-5p Reversed the Effect of si-circ_0085539 on Cell Proliferation, Migration, and Invasion in HOS and Saos2 Cells
(A) Proliferation of HOS and Saos2 cells was measured using a CCK-8 assay. (B) Proliferation was evaluated by a colony formation assay. (C) The cell migration ability was evaluated by a wound-healing assay. The migration rate represented the ratio of the migration distance at 24 h to the wound width at 0 h. (D) The cell invasion ability was evaluated by a transwell invasion assay. NC, the HOS and Saos2 cells were transfected with negative control vectors; sh-circ, the HOS and Saos2 cells were transfected with sh-circ_0085539; inhibitor, the HOS and Saos2 cells were transfected with miR-526b-5p inhibitor; sh-circ+inhibitor, the HOS and Saos2 cells were co-transfected with sh-circ_0085539 and miR-526b-5p inhibitor. ∗p < 0.05, ∗∗p < 0.001 versus control; #p < 0.05, ##p < 0.001 versus sh-circ_0085539. Three independent repeated experiments were implemented in all of the assays.
SERP1 Was the Target Gene of miR-526b-5p
The differentially expressed mRNAs (selection criteria: log2 fold change [FC] ≥ 1.5, p ≤ 0.05) of the GEO: GSE37552 data series and the predicted targets of miR-526b-5p by the TargetScan Human 7.2 database were obtained. The 11 common mRNAs from the two datasets included WDR72, FAM134B, ACADL, HHIP, SERP1, CALML4, PDE3A, TEX9, FRMD4B, FAXC, and UPP1 (Figure 6A). Among the five top significant mRNAs, that is, WDR72, FAM134B, ACADL, HHIP, and SERP1, SERP1 arouse our interest. As a stress-associated ER protein, SERP1 is widely expressed in the cytoskeleton, ER, and cytosol. During ER stress, it protects unfolded proteins from degradation. After ER stress, it may facilitate the glycosylation of target proteins. ER stress has been confirmed to be significant during tumorigenesis.18,19 For these reasons, we speculated that SERP1 might be a tumorigenesis-related gene. The TargetScan Human 7.2 database predicted a binding site between the SERP1 mRNA 3′ UTR and miR-526b-5p (Figure 6B). The dual-luciferase reporter assay was performed to study whether miR-526b-5p could bind to the SERP1 mRNA 3′ UTR. Findings showed that the miR-526b-5p mimic transfection resulted in almost a 50% decrease in the relative luciferase activity of the WT SERP1 mRNA 3′ UTR vectors in HOS and Saos2 cells (∗p < 0.05 versus co-transfection of SERP1-WT and miR-NC plasmids, Figure 6C). Furthermore, SERP1 was found to be upregulated in OS tissues, unlike the adjacent healthy tissues (3.364 ± 1.577 versus 1.043 ± 0.508, p < 0.001, Figure 6D). Additionally, the expression of SERP1 was negatively correlated with that of miR-526b-5p (p < 0.001, Figure 6E). The qRT-PCR assay results further proved that miR-526b-5p suppressed the expression of SERP1. Furthermore, sh-SERP1 transfection led to a significant downregulation of SERP1 (∗p < 0.05, ∗∗p < 0.001 versus control; ##p < 0.001 versus sh-SERP1, Figure 6F). Furthermore, western blot assay results also demonstrated that the protein level of SERP1 was inhibited by 0.4–0.5 by sh-SERP1 transfection but elevated by 20%–30% by silencing miR-526b-5p (∗p < 0.05, ∗∗p < 0.001 versus control; #p < 0.05, ##p < 0.001 versus sh-SERP1, Figure 6G). This outcome indicated that miR-526b-5p could modulate the protein level of SERP1. Hence, we identified that miR-526b-5p could inhibit the mRNA and protein expression of SERP1 by specifically binding to the SERP1 mRNA 3′ UTR in OS cells.
Figure 6.
SERP1 Was the Target Gene of miR-526b-5p in OS Cells
(A) The intersection of the differentially expressed mRNAs (log2FC ≥ 1.5 with p ≤ 0.05) of the GEO: GSE37552 dataset and the predicted targets of miR-526b-5p by the TargetScan Human 7.2 database. (B) TargetScan Human 7.2 was used to predict the binding site of the SERP1 3′ UTR for miR-526b-5p. (C) The dual-luciferase reporter assay was used to prove the binding site between the SERP1 3′ UTR and miR-526b-5p. SERP1-WT, the plasmids contained the binding site of SERP1 3′ UTR for miR-526b-5p; SERP1-MUT, the plasmids did not have any biding site of the SERP1 3′ UTR for miR-526b-5p. ∗p < 0.05, ∗∗p < 0.001 versus co-transfection of SERP1-WT and NC plasmids. (D) The expression of SERP1 mRNA was detected in normal and tumor tissues. (E) The negative correlation between the expression of SERP1 and miR-526b-5p in 16 OS tissues is shown. (F) Expression of SERP1 in HOS and Saos2 cells after stable transfection of Con, NC, sh-SERP1, miR-526b-5p inhibitor, or sh-SERP1+miR-526b-5p inhibitor. Con, the HOS and Saos2 cells were cultured without any treatments; NC, the HOS and Saos2 cells were transfected with negative control vectors; sh-SERP1, the HOS and Saos2 cells were transfected with sh-SERP1; inhibitor, the HOS and Saos2 cells were transfected with miR-526b-5p inhibitor; sh-SERP1+inhibitor, the HOS and Saos2 cells were co-transfected with sh-SERP1 and miR-526b-5p inhibitor. ∗p < 0.05, ∗∗p < 0.001. (G) The protein level of SERP1 was assessed via western blot assay in HOS and Saos2 cells after stable transfection of Con, NC, sh-SERP1, miR-526b-5p inhibitor, or sh-SERP1+miR-526b-5p inhibitor. Three independent repeated experiments were implemented in all of the assays.
miR-526b-5p Impeded the Progression of OS by Inhibiting SERP1 In Vitro
To further explore the mechanism of the miR-526b-5p/SERP1 axis in OS, the sh-SERP1 and miR-526b-5p inhibitor were used to transfect or co-transfect the HOS and Saos2 cells. After transfection, the cell proliferation in the sh-SERP1 group significantly dropped at 48 and 72 h compared with that of the control group. The cell proliferation in the co-transfection group (sh-SERP1+inhibitor group) was compromised at 48 and 72 h compared with that of the miR-526b-5p inhibitor group in HOS and Saos2 cells (∗p < 0.05 versus the control group and #p < 0.05 versus the inhibitor group, Figure 7A). The results of the colony formation assay displayed a similar trend with those of the CCK-8 assay (∗p < 0.05 versus the control group and #p < 0.05 versus the inhibitor group, Figure 7B). The migration rate of HOS cells decreased by 18.5% in the co-transfection group compared with that of the miR-526b-5p inhibitor group. Similarly, the migration rate of Saos2 cells decreased by 13.1% compared with that of the miR-526b-5p inhibitor group (∗p < 0.05 versus the control group and #p < 0.05 versus the inhibitor group, Figure 7C). The number of invaded cells declined in the co-transfection group (121 ± 9.54) versus the miR-526b-5p inhibitor group (278 ± 9.78) in HOS cells. It was also found that the number of invaded cells declined in the co-transfection group (127 ± 8.08) versus the miR-526b-5p inhibitor group (320 ± 19.93) in Saos2 cells (∗∗p < 0.001 versus the control group and ##p < 0.001 versus the inhibitor group, Figure 7D). Thus, circ_0085539 inhibited the expression of miR-526b-5p in OS, thereby causing the impaired suppression of miR-526b-5p on SERP1 mRNA and resulting in enhanced cell proliferation, migration, and invasion. The data mentioned above demonstrated that miR-526b-5p acted as the tumor suppressor factor in OS via targeting SERP1.
Figure 7.
Inhibition of SERP1 Reversed miR-526b-5p Inhibitor-Induced Cell Proliferation, Migration, and Invasion in OS Cells
(A) The proliferation of the transfected HOS and Saos2 cells was evaluated by a CCK-8 assay. Con, the HOS and Saos2 cells were cultured without any treatments. NC, the HOS and Saos2 cells were transfected with negative control vectors. Sh-SERP1, the HOS and Saos2 cells were transfected with sh-SERP1. Inhibitor, the HOS and Saos2 cells were transfected with miR-526b-5p inhibitor. Sh-SERP1+inhibitor, the HOS and Saos2 cells were co-transfected with sh-SERP1 and miR-526b-5p inhibitor. ∗p < 0.05, ∗∗p < 0.001 versus control group. #p < 0.05, ##p < 0.001 versus inhibitor group. (B) The proliferation of HOS and Saos2 cells was assessed by colony formation assay. Con, the HOS and Saos2 cells were cultured without any treatments. NC, the HOS and Saos2 cells were transfected with negative control vectors. Sh-SERP1, the HOS and Saos2 cells were transfected with sh-SERP1. Inhibitor, the HOS and Saos2 cells were transfected with miR-526b-5p inhibitor. Sh-SERP1+inhibitor, the HOS and Saos2 cells were co-transfected with sh-SERP1 and miR-526b-5p inhibitor. ∗p < 0.05, ∗∗p < 0.001 versus control group; #p < 0.05, ##p < 0.001 versus inhibitor group. (C) Wound-healing assay was performed to assess the migration ability of the transfected HOS and Saos2 cells. Con, the HOS and Saos2 cells were cultured without any treatments; NC, the HOS and Saos2 cells were transfected with negative control vectors; sh-SERP1, the HOS and Saos2 cells were transfected with sh-SERP1; inhibitor, the HOS and Saos2 cells were transfected with miR-526b-5p inhibitor; sh-SERP1+inhibitor, the HOS and Saos2 cells were co-transfected with sh-SERP1 and miR-526b-5p inhibitor. ∗p < 0.05, ∗∗p < 0.001 versus control group; #p < 0.05, ##p < 0.001 versus inhibitor group. (D) The invasion ability was evaluated by a transwell invasion assay in HOS and Saos2 cells. Con, the HOS and Saos2 cells were cultured without any treatments; NC, the HOS and Saos2 cells were transfected with negative control vectors; sh-SERP1, the HOS and Saos2 cells were transfected with sh-SERP1; inhibitor, the HOS and Saos2 cells were transfected with miR-526b-5p inhibitor; sh-SERP1+inhibitor, the HOS and Saos2 cells were co-transfected with sh-SERP1 and miR-526b-5p inhibitor. ∗p < 0.05, ∗∗p < 0.001 versus control group; #p < 0.05, ##p < 0.001 versus inhibitor group.
Discussion
The experimental results demonstrated that the knockdown of circ_0085539 hindered the aggression of OS by releasing its sponging miRNA, miR-526b-5p. The released miR-526b-5p, as displayed in the Results, suppressed its downstream target SERP1 to inhibit OS cell malignancy, such as proliferation and mobility. In recent years, the functions of circRNAs have attracted the attention of researchers, and a growing number of studies have reported that circRNA can play a key role in cancers, including OS. For instance, Liu et al.20 used a microarray expression profiling and functional analysis method to prove that the expression of many circRNAs in tumor tissues was different from that in non-tumor tissues. Their research suggested the potential role of these circRNAs in the progression of OS. In another experiment, hsa_circ_0001564 with high expression was observed in OS tissues and cells. It was also found that silencing circ_0001564 inhibited cell proliferation by inducing a cell cycle arrest in the G1 phase.21 Another significant circRNA, hsa_circ_0009910, was found to act as a sponge of miR-449a, meaning it could contribute to the carcinogenesis of OS.22 The previous studies also suggested that circRNA could be instrumental in the development of OS. A circRNA generated from its host gene PVT1, circ_0085539, has not been investigated in OS so far, however. In this study, we found that circ_0085539 was significantly upregulated in OS tissues and cells and later proved that circ_0085539 knockdown not only suppressed the tumor growth in vivo but also impaired OS cell proliferation and mobility in vitro.
As for the regulatory mechanisms of circRNAs, several studies have proved that circRNAs have the ability to regulate the progression of OS. For instance, hsa_circ_0000285 overexpression promoted the proliferative and migratory capabilities of OS cells by sponging miR-599.23 circMYO10, generated from host gene MYO10, could promote OS development by inhibiting miR-370-3p.24 Also reported was that circ_0001658 could promote OS malignancy by regulating miR-382-5p.25 In addition, circ_0003998 enhanced OS progression by targeting miR-197-3p.26 We validated in this research that miR-526b-5p could directly bind with circ_0085539. According to a previous study on miR-526b, miR-526b could inhibit epithelial-mesenchymal transition (EMT) in cervical cancer cells.27 Therefore, we suspected that miR-526b-5p might be a tumor suppressor factor in OS and that circ_00085539 could possibly promote OS progression by targeting miR-526b-5p. Our cytological experiment results confirmed our conjecture: circ_00085539 could contribute to OS progression by sponging miR-526b-5p.
SERP1 was first investigated due to its effect on ER stress. Its role in cancers has begun to draw researchers’ attention in recent years. Ma et al.28 suggested that the high expression of SERP1 was associated with the poor prognosis of pancreatic ductal adenocarcinoma using univariate and multivariate Cox regression analysis. In their research, Rozengurt et al.16 showed that SERP1, which belongs to the YAP/TEAD-regulated genes, was related to the unfavorable survival of patients with pancreatic ductal adenocarcinoma. Taken together with the previous studies on SERP1, we hypothesized that SERP1 might be an oncogene in OS even though neither clinical nor basic experiments had investigated the effect of SERP1 on OS. We herein found that miR-526b-5p could specially bind to the SERP1 mRNA 3′ UTR. Our findings also showed that the upregulation of miR-526b-5p was accompanied by the downregulation of SERP1. Silencing SERP1 inhibited the malignancy phenotypes of OS cells, and miR-526-5p inhibition reversed the effect of SERP1 knockdown.
The results of this research are compelling. However, a major drawback of the present study lies in the ER stress-related experiments, which we did not conduct due to experimental condition limitations. We have mentioned earlier in the Results that ER stress has been confirmed to be associated with the cancer progression of many human cancers, including OS.29, 30, 31 Nonetheless, further research is needed to investigate whether and how SERP1 regulates OS development via the ER stress pathway.
Our research is the first to investigate the effect of the interactions between circ_0085539, miR-526b-5p, and SERP1 in OS. We found that circ_0085539 accelerated OS progression by binding to and regulating miR-526b-5p, which suppressed SERP1. Our findings also revealed a novel molecular interactome of circ_0085539/miR-526b-5p/SERP1 in OS. We think that this knowledge might provide a novel target for OS clinical treatments.
Materials and Methods
Clinical Samples
The OS tissues and matched adjacent healthy tissue specimens were collected from 16 OS patients who underwent complete resection surgery at The First Hospital of Jilin University. None of the patients who participated in this study received any chemical therapies or radiotherapies before the resection surgery. All relevant procedures regarding specimens in this study were approved by the Ethics Committee of The First Hospital of Jilin University. The pathology of the clinical samples was characterized by three independent pathologists according to the guidelines recommended by the World Health Organization. The fresh tissue specimens obtained from the participants were promptly placed in liquid nitrogen immediately after the complete resection surgery, and they were stored at −80°C for the following experiments. The baseline characteristics of OS patients used in this study are listed in Table S1.
Cell Source and Cell Culture
Human OS cell lines (HOS, U2OS, SJSA-1, and Saos2) and a human osteoblast cell line (hFOB1.19) were purchased from ATCC (USA). All the OS cells were kept in DMEM supplemented with 10% fetal bovine serum (FBS; Gibco, USA), 100 U/mL streptomycin, and penicillin (Fubo Biotechnology, Beijing, China). The hFOB1.19 cell line was sustained in the DMEM/nutrient mixture F-12 (DMEM/F-12) augmented with 10% FBS and 0.3 mg/mL G418 (Kanglang Biotechnology, China). All of the cell lines were kept in an incubator with 5% CO2 at 37°C, and they were carefully scrutinized to ensure that they were not contaminated by bacteria, chlamydia, mycoplasma, or other malicious cells.
Transfection Constructs and Cell Transfection
All the vectors used in this study were designed and obtained from GeneChem (Shanghai, China), such as shRNAs targeting circ_0085539 or SERP1 (sh-circ_0085539 or sh-SERP1), miR-526b-5p inhibitor, and NC vectors. These vectors were transfected or co-transfected into HOS and Saos2 cells using Lipofectamine 3000 (Life Technologies, USA). The HOS cells transfected with sh-circ0085539, which could stably silence the expression of circ_0085539, were selected under 14 days of treatment with medium containing 2 μg/mL puromycin to perform xenograft formation in vivo.
RNA Extraction, RNase R Digestion, Nuclear and Cytoplasm Fractionation, and qRT-PCR
All the RNAs were isolated from clinical specimens and cells using TRIzol reagents (Invitrogen, USA). The isolation was carried out according to the kit specifications. For RNA digestion using RNase R (Epicenter Technologies, USA), 4 mg of total RNA was incubated with 6 U/mg RNase R for 20 min. For cellular fractionation, NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific, USA) were used to separate the nuclear and cytoplasmic fractions. To detect circRNA and mRNA, two kits were used, that is, SYBR Premix Ex Taq II (TaKaRa, China) and RT reagent kit (TaKaRa, China). Subsequently, the reactions were measured with the Roche LightCycler 480 PCR instrument (Roche, Basel, Switzerland) according to the manufacturer’s specifications. For miRNA qRT-PCR, DNase I was utilized to treat the samples and eliminate gDNA in the samples. The Mir-X miR first-strand synthesis kit (TaKaRa, China) was later used to synthesize cDNA. Lastly, SYBR Premix Ex Taq II (TaKaRa, China) was utilized to perform fluorescence PCR. The primers are listed in Table S2.
Gel (2%) Electrophoresis
Divergent primers and convergent primers were synthesized using GenePharma (Shanghai, China). They were respectively used to amplify circ_00085539, PVT1, and GAPDH (the control) in cDNA and gDNA (extracted using the DNeasy Blood & Tissue kit from QIAGEN) from the HOS cell line and Saos2 cell line. A molecular weight ladder was carefully loaded to the first lane of every gel. The DNA samples in the loading buffer were then added to the remaining lanes of the gel. The electrophoresis was subsequently run at 5 V/cm. Afterward, the gel was placed to 1× TAE buffer for 30 min and in water for 5 min. Lastly, the DNA fragments were visualized under a Bio-Rad gel imager. The sequences of the divergent primers are shown in Table S2.
Dual-Luciferase Reporter Assay
The TargetScan algorithm was first used to predict the binding site between circ_0085539 and miR-526b-5p or between miR-526b-5p and SERP1. Following that, the dual-luciferase reporter assay was used to validate the binding relationship between them. The next step was the insertion of the WT or mutant (MUT) circ_0085539 (or SERP1) into the pGL3 vector. HOS and Saos2 cells (1 × 105) were subsequently seeded in 24-well plates and were cultured until 30% confluence. After that, they were co-transfected with the constructed vectors described above with miR-526b-5p mimic (or mimic NC). After co-transfection for 48 h, the relative luciferase activity of different treatment groups was measured using the Dual-Luciferase reporter assay system (Promega, USA).
RIPA
To perform the RIPA, we purchased the Magna RIP RNA-binding protein immunoprecipitation kit from Millipore (USA). The first step was the collection and lysis of 1.5 × 106 HOS and Saos2 cells using RIP lysis buffer. After that, 50 μL of magnetic beads was resuspended, washed with 100 μL of RIP wash buffer, and conjugated with 5 μg of antibodies against immunoglobulin G (IgG) or Ago2 in 500 μL of RIP buffer. The RIP buffer mentioned above was later added into 100 μL of the lysate. After the treatment of proteinase K buffer, the immunoprecipitated RNAs were extracted for qRT-PCR analyses.
CCK-8 Assay
The HOS and Saos2 cells (5,000 cells/well) were first seeded in the 96-well plates and cultured overnight. Then, 10 μL of CCK-8 solution (Dojindo, Japan) was added to each well after the cells were transfected for 0, 24, 48, and 72 h. The CCK-8 solution was subsequently incubated with the cells for another 4 h. Lastly, the absorbance value was detected at 450-nm wavelength using a microplate reader.
Colony Formation Assessment
Transfected cells were seeded in a 12-well plate at a density of 600 cells/well in the medium containing 10% FBS. The cell media were then replaced with fresh media every day and the cells were allowed to grow for approximately 10 days until colonies were formed. After the medium was removed, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution, respectively. Next, the cells were imaged under a microscope. Lastly, the number of colonies in every well was counted.
The Detection of Cell Migration Capability
1 × 105 HOS and Saos2 cells were first seeded in six-well plates to grow to 90% confluence and then starved in media without serum for 1 day. After that, the 200-μL pipette tip was used to scratch a wound on the cell layer. The exfoliated cells were later washed away with PBS buffer. Next, the media were refreshed to culture the cells continuously for 24 h. The images of the cell layers were later taken at 0 and 24 h. The migratory rate was eventually calculated as (gap at 0 h − gap at 24 h)/gap at 0 h, a computation that reflected the cell migration capability.
Transwell Invasion Assay
This assay was conducted in the transwell (8-μm pore size). The transwell upper chambers were precoated with Matrigel (Millipore, USA). Briefly, 400 μL of serum-free medium containing 1 × 105 transfected cells was added to the upper chambers. At the same time, 300 μL of medium containing 10% FBS was added to the lower chambers. After a 24-h incubation, the cells invading the downside of the chamber membrane were fixed using 4% paraformaldehyde for 30 min. After staining the invaded cells, images of the downside of the membranes were taken under a microscope. Importantly, note that at least three random fields of each chamber were selected.
Xenograft Formation Assay
Five to 6-week-old female BALB/c nude mice (n = 12) purchased from Charles River Laboratories (China) were indiscriminately divided into two groups as follows: the negative control group (NC, n = 6) and the sh-circ_0085539 group (n = 6). The mice in the NC group were injected subcutaneously with 1 × 106 transfected HOS cells, whereas the mice in the sh-circ_0085539 group were subcutaneously injected with 1 × 106 stable sh-circ_0085539 HOS cells mentioned above in the right armpit. After injection for 30 days, the mice were sacrificed, and the tumors were isolated and weighed. The tumors were later fixed in 4% paraformaldehyde and embedded in paraffin for the next experiments.
Bioluminescence Imaging
The growing tumors were measured using a caliper every 6 days. For bioluminescence imaging, d-luciferin (150 mg/kg) was injected into the mice 5 min before the bioluminescence imaging. The images were taken at day 6 and day 30 after cell plantation. After that, the mice were anesthetized using a 1.5%–2.5% isoflurane-oxygen mixture and then put into the camera box of the Xenogen in vivo imaging system (IVIS) Lumina series III imaging system (PerkinElmer, Waltham, MA, USA) for imaging. The Living Image software package (version 4.5.5) was eventually used to analyze the total flux of mice in the two groups at day 30 after the tumor cell grafting.
H&E Staining and Ki67 Immunohistochemistry (IHC) Staining
H&E staining was conducted for histological examinations. The tumor tissues embedded in the paraffin were cut into 4-μm sections. Then, the slices were stained with hematoxylin solution for 10 min. After that, they were rinsed with running water until the water was colorless. Then, the slices were dehydrated with 70% and 90% alcohol for 10 min to remove the excess hematoxylin staining from the cytoplasm. After the dehydration process, they were dyed with the eosin solution for 2 min. In the end, the slices were observed at ×100 and ×400 magnifications. As for Ki67 IHC staining, the slices were incubated with antibodies against Ki67 (catalog no. ab15580, Abcam, USA) and secondary antibodies (catalog no. ab205718, Abcam) as previously described.32 The Ki67-positive cells were counted in at least five randomly selected fields at ×40 magnification.
Western Blotting
RIPA buffer solution (Beyotime Biotechnology, China) was used to extract the total proteins. Then, the protein was quantified, separated by 5% SDS-PAGE, and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, USA). After blocking the membranes in 5% skim milk, the membranes were exposed to the primary antibodies against SERP1 (catalog no. ab254839, 1:1,000, Abcam, USA) and GAPDH (catalog no. ab8245, 1:1,000, Abcam, USA) overnight. After that, the membranes were incubated with the secondary goat anti-rabbit IgG antibodies for 3 h. The Odyssey infrared scanner (LI-COR Biosciences) was then used to visualize the immunoreactive bands. The blot intensity was eventually read using ImageJ software.
Statistical Analysis
SPSS 20.0 software (SPSS, USA) was used for statistical difference analyses. The Student’s t test was applied to analyze the difference between two groups. For the analysis of multiple groups, ANOVA was utilized. The data obtained in this research are shown in the format of mean ± SD (standard deviation). Three independent experiments were repeatedly implemented for all of the assays, and all p values less than 0.05 were deemed to have a statistically significant difference.
Author Contributions
W.L. and D.W. performed the experiments. M.Y. designed the research study. X.W. and P.L. analyzed the data and wrote the manuscript. All authors reviewed and approved the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We appreciate the help of our hospital —The First Hospital of Jilin University.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.09.009.
Supplemental Information
References
- 1.Sampson V.B., Kamara D.F., Kolb E.A. Xenograft and genetically engineered mouse model systems of osteosarcoma and Ewing’s sarcoma: tumor models for cancer drug discovery. Expert Opin. Drug Discov. 2013;8:1181–1189. doi: 10.1517/17460441.2013.817988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X., Zhang C., Wang C., Sun J., Wang D., Zhao Y., Xu X. miR-210 promotes human osteosarcoma cell migration and invasion by targeting FGFRL1. Oncol. Lett. 2018;16:2229–2236. doi: 10.3892/ol.2018.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrari S., Mercuri M., Bacci G. Comment on “Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols”. J. Clin. Oncol. 2002;20:2910–, author reply 2910–2911. doi: 10.1200/JCO.2002.20.12.2910. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida K., Okamoto M., Sasaki J., Kuroda C., Ishida H., Ueda K., Okano S., Ideta H., Kamanaka T., Sobajima A. Clinical outcome of osteosarcoma and its correlation with programmed death-ligand 1 and T cell activation markers. OncoTargets Ther. 2019;12:2513–2518. doi: 10.2147/OTT.S198421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Kulcheski F.R., Christoff A.P., Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Zhong R., Chen Z., Mo T., Li Z., Zhang P. Potential role of circPVT1 as a proliferative factor and treatment target in esophageal carcinoma. Cancer Cell Int. 2019;19:267. doi: 10.1186/s12935-019-0985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Zhang Z., Jiang H., Li Q., Wang R., Pan H., Niu Y., Liu F., Gu H., Fan X., Gao J. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell. Physiol. Biochem. 2018;51:2324–2340. doi: 10.1159/000495876. [DOI] [PubMed] [Google Scholar]
- 10.Chen J., Li Y., Zheng Q., Bao C., He J., Chen B., Lyu D., Zheng B., Xu Y., Long Z. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Chen L.H., Wang L.P., Ma X.Q. circ_SPECC1 enhances the inhibition of miR-526b on downstream KDM4A/YAP1 pathway to regulate the growth and invasion of gastric cancer cells. Biochem. Biophys. Res. Commun. 2019;517:253–259. doi: 10.1016/j.bbrc.2019.07.065. [DOI] [PubMed] [Google Scholar]
- 12.Wu M., Li X., Liu Q., Xie Y., Yuan J., Wanggou S. miR-526b-3p serves as a prognostic factor and regulates the proliferation, invasion, and migration of glioma through targeting WEE1. Cancer Manag. Res. 2019;11:3099–3110. doi: 10.2147/CMAR.S192361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang R., Zhao J., Xu J., Wang J., Jia J. miR-526b-3p functions as a tumor suppressor in colon cancer by regulating HIF-1α. Am. J. Transl. Res. 2016;8:2783–2789. [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaguchi A., Hori O., Stern D.M., Hartmann E., Ogawa S., Tohyama M. Stress-associated endoplasmic reticulum protein 1 (SERP1)/ribosome-associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J. Cell Biol. 1999;147:1195–1204. doi: 10.1083/jcb.147.6.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hori O., Miyazaki M., Tamatani T., Ozawa K., Takano K., Okabe M., Ikawa M., Hartmann E., Mai P., Stern D.M. Deletion of SERP1/RAMP4, a component of the endoplasmic reticulum (ER) translocation sites, leads to ER stress. Mol. Cell. Biol. 2006;26:4257–4267. doi: 10.1128/MCB.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozengurt E., Sinnett-Smith J., Eibl G. Yes-associated protein (YAP) in pancreatic cancer: at the epicenter of a targetable signaling network associated with patient survival. Signal Transduct. Target. Ther. 2018;3:11. doi: 10.1038/s41392-017-0005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mucaj V., Lee S.S., Skuli N., Giannoukos D.N., Qiu B., Eisinger-Mathason T.S., Nakazawa M.S., Shay J.E., Gopal P.P., Venneti S. microRNA-124 expression counteracts pro-survival stress responses in glioblastoma. Oncogene. 2015;34:2204–2214. doi: 10.1038/onc.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cubillos-Ruiz J.R., Bettigole S.E., Glimcher L.H. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urra H., Dufey E., Avril T., Chevet E., Hetz C. Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer. 2016;2:252–262. doi: 10.1016/j.trecan.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Liu W., Zhang J., Zou C., Xie X., Wang Y., Wang B., Zhao Z., Tu J., Wang X., Li H. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell. Physiol. Biochem. 2017;43:969–985. doi: 10.1159/000481650. [DOI] [PubMed] [Google Scholar]
- 21.Song Y.Z., Li J.F. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem. Biophys. Res. Commun. 2018;495:2369–2375. doi: 10.1016/j.bbrc.2017.12.050. [DOI] [PubMed] [Google Scholar]
- 22.Deng N., Li L., Gao J., Zhou J., Wang Y., Wang C., Liu Y. hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem. Biophys. Res. Commun. 2018;495:189–196. doi: 10.1016/j.bbrc.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z., Pu F., Wang B., Wu Q., Liu J., Shao Z. hsa_circ_0000285 functions as a competitive endogenous RNA to promote osteosarcoma progression by sponging hsa-miRNA-599. Gene Ther. 2020;27:186–195. doi: 10.1038/s41434-019-0112-5. [DOI] [PubMed] [Google Scholar]
- 24.Chen J., Liu G., Wu Y., Ma J., Wu H., Xie Z., Chen S., Yang Y., Wang S., Shen P. circMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling. Mol. Cancer. 2019;18:150. doi: 10.1186/s12943-019-1076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Wang P., Su X., Zhao B. circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell Biochem. Funct. 2020;38:77–86. doi: 10.1002/cbf.3452. [DOI] [PubMed] [Google Scholar]
- 26.Wang L., Du Z.G., Huang H., Li F.S., Li G.S., Xu S.N. circ-0003998 promotes cell proliferative ability and invasiveness by binding to miR-197-3p in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2019;23:10638–10646. doi: 10.26355/eurrev_201912_19761. [DOI] [PubMed] [Google Scholar]
- 27.Li H., Wang J., Xu F., Wang L., Sun G., Wang J., Yang Y. By downregulating PBX3, miR-526b suppresses the epithelial-mesenchymal transition process in cervical cancer cells. Future Oncol. 2019;15:1577–1591. doi: 10.2217/fon-2018-0575. [DOI] [PubMed] [Google Scholar]
- 28.Ma Q., Wu X., Wu J., Liang Z., Liu T. SERP1 is a novel marker of poor prognosis in pancreatic ductal adenocarcinoma patients via anti-apoptosis and regulating SRPRB/NF-κB axis. Int. J. Oncol. 2017;51:1104–1114. doi: 10.3892/ijo.2017.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Yin F., Xu J., Zhang T., Wang G., Mao M., Wang Z., Sun W., Han J., Yang M. CYT997(lexibulin) induces apoptosis and autophagy through the activation of mutually reinforced ER stress and ROS in osteosarcoma. J. Exp. Clin. Cancer Res. 2019;38:44. doi: 10.1186/s13046-019-1047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Q., Mao Y., Liu M., Luo R., Jiang R., Guo F. The active nuclear form of SREBP1 amplifies ER stress and autophagy via regulation of PERK. FEBS J. 2020;287:2348–2366. doi: 10.1111/febs.15144. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L., Wang Y., Zhang L., Xia X., Chao Y., He R., Han C., Zhao W. ZBTB7A, a miR-663a target gene, protects osteosarcoma from endoplasmic reticulum stress-induced apoptosis by suppressing lncRNA GAS5 expression. Cancer Lett. 2019;448:105–116. doi: 10.1016/j.canlet.2019.01.046. [DOI] [PubMed] [Google Scholar]
- 32.Niu Y.N., Wang K., Jin S., Fan D.D., Wang M.S., Xing N.Z., Xia S.J. The intriguing role of fibroblasts and c-Jun in the chemopreventive and therapeutic effect of finasteride on xenograft models of prostate cancer. Asian J. Androl. 2016;18:913–919. doi: 10.4103/1008-682X.167714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.