Highlights
-
•
The clonal origin and parallel evolution of the metastatic lesions and primary tumour.
-
•
The evolutionary branches of primary and metastatic clones diverge early in the development of the tumour.
-
•
Increased genome instability and specific enriched somatic copy number alteration (SCNAs) in metastatic lesions compared to primary tumour.
-
•
LOH at 14q, loss of 14q32.31 and gain of 6p22.2 are highly selected events during metastatic evolution.
Keywords: Clear cell renal cell carcinoma, Metastasis, Evolutionary trajectory, Somatic copy number alteration, Loss of heterozygosity
Abstract
The evolutionary trajectories of treatment-naïve metastatic tumour are largely unknown. Such knowledge is crucial for cancer prevention and therapeutic interventions. Herein, we performed whole genome or exome sequencing of 19 tumour specimens and 8 matched normal kidney tissues from 8 clear cell renal cell carcinoma (ccRCC) patients.
The clonal origin and parallel evolution of the metastatic lesions and primary tumour is identified in all 8 patients. But the evolutionary branches of primary and metastatic clones diverge early in the development of the tumour. More importantly, larger scale genomic aberrations including somatic copy number alteration (SCNA) or loss of heterozygosity (LOH) differentiate the metastasis lesions from primary tumour. Based on it, we identify that LOH at 14q, loss of 14q32.31 and gain of 6p22.2 are highly selected events during metastatic evolution. Further functional validations of multiple genes within the SCNA regions indicated that these selected events interact to drive metastatic risk with potential therapeutic relevance. Collectively, we described increased genome instability in metastatic ccRCC and validated it via molecular biology, providing an evolution pattern which may facilitate the translation of basic finding.
Introduction
Bone metastases are the second most common metastases (after lung) in patients with metastatic renal cell carcinoma (m-RCC) [1]. Bone metastasis has been identified as an independent prognostic factor associated with poor survival in patients with mRCC. Patients with osteolytic metastases experience severe pain, life-threatening hypercalcemia, pathologic fracture, and spinal cord compression. The clinical treatment for bone metastasis RCC patients is limited to bisphosphonates, which does not specifically target metastatic tumour cells [2]. The management of metastasis in RCC patients remains challenging.
VHL (von Hippel-Landau, on chromosome 3p25) gene inactivation, either by somatic mutation, chromosomal loss or epigenetic silencing, presents in the majority of ccRCC (the major subtype of RCC) patients. It is widely known that the inactivation of VHL protein (pVHL) can lead to the abnormal expression of genes that play crucial roles in tumorigenesis, such as hypoxia inducible factors (HIFs) and vascular endothelial growth factor (VEGF) [3,4]. Based on the discoveries, the drugs targeting the VEGF have been development, such as tyrosine kinase inhibitors (TKIs), which have improved the overall outcomes of advanced RCC patients [5]. Previous study comprising a single-centre retrospective evaluation of m-RCC patients with bone metastasis at the University of Texas MD Anderson Cancer Centre has shown that m-RCC patients demonstrate a median overall survival (OS) of 24 months with TKI treatment compared with 18 months without the TKI treatment [6]. Moreover, our previous study has shown that sorafenib monotherapy can achieve promising OS results (24 months) in Chinese patients with m-RCC, with manageable adverse events [7]. These results suggest that TKI treatment does improve overall survival in m-RCC patients. However, the rate of new bone metastasis has been found similar between the treatment and non-treatment patient groups [6]. These results suggest that TKI targeted therapies may target common pathways between primary and bone metastatic tumours, but may not effectively target metastatic tumour cells.
Therefore, understanding the genomic evolution of metastatic disease requires simultaneous analysis of the genomic heterogeneity in metastatic lesions and matched primary tumor [8]. Additional biopsy of metastases is, therefore, becoming advocated, but sampling of secondary tumours is often impractical due to surgical inaccessibility. Moreover, the extensive therapies to the metastatic patients further complicated the evolutionary trajectory of tumour cells. Relatively few genome-wide analyses of solid primary tumours including ccRCC and their metastases have been conducted to date especially in treatment-naïve metastatic patients. Herein, we systematically evaluated the evolutionary trajectories of individual treatment-naïve patients by comprehensively mapping the longitudinal genomic variations in 8 ccRCC patients with bone metastasis. We further explored exogenous mutagen exposure by evaluating the mutation spectra during tumour metastasis. Collectively, we aimed to better understand the molecular mechanisms of metastasis in ccRCC to allow for the development of better prevention and therapeutic interventions against this highly morbid disease manifestation.
Materials and methods
Patients and tumour samples
Tissue samples are collected from ccRCC patients at Peking University First Hospital and Peking University School of Oncology. Approval for the study was granted by the Clinical Research Ethics Committee of Peking University First Hospital and informed consent forms are obtained (ethical approval code: 2015 [977]). The histopathological results are evaluated by three independent pathologists before microdissection of the formalin-fixed, paraffin embedded (FFPE) samples or fresh samples (–80 °C).
DNA extraction from fresh and FFPE samples
The fresh surgical samples are stored at –80 °C post operationally, and genomic DNA was extracted according to the manufacturer's instructions (Qiagen, Mississauga, Canada). For FFPE samples, a kit (Zymo Research, Orange, CA, USA) is used with modifications. Briefly, FFPE samples are deparaffinised with xylene, digested with 20 µl Proteinase K, heated at 80 °C for 1 h, lysed, de-crosslinked (homemade de-crosslinking buffer, the patent # 201510818823.X), washed and subsequently eluted.
Whole-genome library preparation and sequencing
Briefly, genomic DNA is sheared into approximately 300-bp fragments, end-repaired, A-tailed, and ligates to Illumina paired-end adapters, according to the manufacturer's instructions, by using the NEB end repair, dA-tailing and Next Quick Ligation modules. After purification with Agencourt AMPure XP beads, the DNA fragments with adapters are enriched by PCR. The products are quality-checked and sequenced with an Illumina X-ten instrument, thus generating 2 × 150-bp pair-end (PE) reads.
Exome library preparation and sequencing
The exonic regions of the samples are captured using the SureSelect All Exon V5 platform (Agilent Technologies, Santa Clara, CA) according to the manufacturer's protocol. Briefly, genomic DNA is sheared into approximately 200-bp fragments, end-repaired, A-tailed, and ligated to Illumina paired-end adapters, according to the manufacturer's instructions, by using the NEB end repair, dA-tailing and Quick Ligation modules. After purification with Agencourt AMPure XP beads, the DNA fragments with adaptors are enriched by PCR. Then, the DNA samples are used for hybridization and post-hybridization amplification according to the manufacturer's protocol. The products are quality-checked and sequenced with an Illumina HiSeq 2000 instrument, thus generating 2 × 100-bp PE reads.
Whole-genome and exome sequencing data analysis for SNV and INDEL calling
PE reads are aligned to the human genome hg19 (UCSC) by using the Burrows-Wheeler aligner [9]. The aligned BAM files are sorted using Samtools 0.1.19 [10]. For further single nucleotide variants (SNVs) and INDEL calling, the aligned reads are realigned to the genome and base quality recalibration is performed using the Genome Analysis Toolkit (GATK 3.7) [11] based on dbSNP 138. Picard-tools 1.118 (http://broadinstitute.github.io/picard/) is applied to remove the PCR duplicates. The SNVs are called using MuTect [12] and annotated using Annovar [13] in the form of normal-tumour pairs. The following criteria are applied to the called SNVs: (i) at least 10 × coverage is required for the normal and tumour samples; (ii) variants listed in dbSNP 138 and dbsnp147 are removed, and (iii) variants listed in The 1000 Genomes Project are removed. INDELs are called using Varscan2 and annotated using the Annovar software. The following criteria are applied to the called indels: (i) At least 10 × total coverage is required for the tumour samples, (ii) variants listed in dbSNP 138 are removed, and (iii) variants listed in The 1000 Genomes Project are removed. The functional effects of the SNVs and INDELs are further annotated by using a combination of Seattleseq Annotation, MutationAssessor [14] and Annovar [13].
Phylogenetic analysis
We construct phylogenetic tree by using the Canopy software for each patient to show the clonal relationship between the primary and secondary tumours [15]. Missense somatic single-nucleotide alterations (SNAs) and copy number alterations (CNAs) are extracted to construct the phylogenetic tree. The alternative allele read depth matrix and total read depth matrix of the mutations are used as input for the Canopy software. The Sequenza software is used to generate CNA input for Canopy.
Copy number analysis based on whole-genome and exome sequencing
We use Sequenza R v.3.1.0 [16] to calculate the copy numbers across the genome on the basis of the whole-genome and exome sequencing data. Briefly, BAM files (generated from whole-genome or exome sequence data) for each tumour sample and its paired normal control are used as the inputs to calculate the depth ratio. The ratio is normalized by considering both the GC content bias and data ratio. GISTIC2.0 was used to identify the specific enriched SCNAs in metastatic lesions compared to primary tumour [17].
Ploidy and cellularity estimation
We estimate the ploidy and cellularity of the tumour samples by using ascatNgs software [18]. Briefly, we use the BAM files from each tumour sample and its paired normal control to estimate the amount of tumour DNA in the tumour sample; this is known as the aberrant cell fraction (ACF).
Mutational signature analysis
We investigate the mutational spectrum by considering both the mutated bases and the context. There are 96 possible substitution patterns when considering trinucleotide sequences, because there are six substitution patterns for each base mutation (C>A, C>T, C>G, T>A, T>C, and T>G). The mutational signature of the germline and somatic mutations in each patient was analysed using the R package deconstructSigs [19]. The mutation signature from the current dbSNP147 build is used as a control.
Cell culture
All cell lines used in our experiment were acquired from the American Type Culture Collection (ATCC; Manassas, USA). HK-2 cells are maintained in KM medium (Invitrogen, USA). 786-O, A498, Caki-1, and ACNH cells are maintained in DMEM (high glucose) medium with 10% fetal bovine serum (FBS; Life Technologies) and penicillin G (100 U/ml), and streptomycin (100 μg/ml) (Sigma-Aldrich, Germany). 769P cells are maintained in RPMI-1640 medium with 10% fetal bovine serum and penicillin G (100 U/ml), and streptomycin (100 μg/ml). All cultures were maintained at 37 °C in humidified air containing 5% CO2.
Stable cell lines
To generate stable knock down cell lines, the shHSP90AA1, shSOS2, shWDR20, or scrambled control were inserted into pLKO.1 puro vector and transfected with lentiviral vectors into ccRCC cells. shRNAs are listed in Table S1. Under the effect of the puromycin, stable polyclonal colonies are selected and expanded for further assays.
q-PCR assays
Total RNA was extracted from cells using TRIzol reagent (Invitrogen), the quantity and quality of RNA were determined by using NanoDrop 1000. Single-strand cDNA was synthesized using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific). Real-time PCR was performed using the SYBR FAST PCR master mix (KAPA) on a Multiplex Quantitative PCR System (Bio-Rad CFX96 TOUCH/GelDoc XR+). mRNA levels were normalized to respective GAPDH and correlated to the wild type. Every treatment have three replicates (n = 3). The primer sequences and siRNAs are listed in Table S1.
Transwell assay
For the transwell assays, 2 × 104 cells/well in serum-free medium are added above the transwell chamber and the lower chamber is filled with 500 μl of serum-containing medium. Cells that have migrated the wells after 24 h are fixed and stained with a 0.5% crystal violet fixative solution for 15 min and quantified. We choose five different views under microscope and calculate the average migrate cells from every single transwell. Every treatment has three replicates. Significance in all figures is indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001 and ****p < 0.0001.
Statistical analyses
Differences between two groups were assessed with two-tailed or paired Student's t-test according to the data types. Log-rank test was used for Kaplan–Meier survival analysis. Differences in the ratios between γH2AX+ and γH2AX- cells were assessed by Fisher's exact test. P values equal to 0.05 or less were considered significant. qPCR expression data were presented as mean ± SD.
Results
Reconstruction the routes of m-ccRCC evolution with single-nucleotide variants
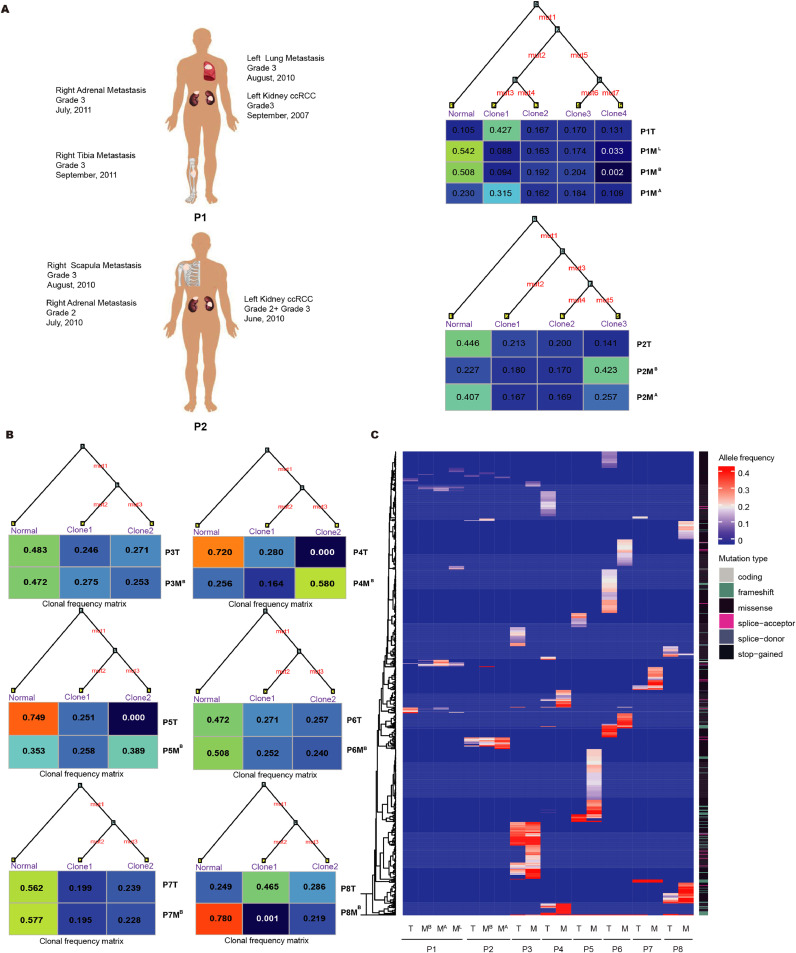
First, we performed exome sequencing of two m-ccRCC patients with multiple metastatic lesions, and metachronous metastases in patient 1 and synchronous metastasis in patient 2 (Table S2). Mean target coverages for tumours and normal exomes were from 60 × to 114 × (Table S3). The number of somatic alterations per tumour was shown in Fig. S1A and S1B. Of a total of 217 mutations found in the tumours for patient 1 (5 shared, 168 metastasis-specific, 44 primary-tumour-specific), 212 (97.7%) are different between primary tumour and metastasis, indicating a high degree of divergence and therefore parallel evolution. Similarly, of a total of 196 mutations found in the tumours for patient 2 (52 shared, 109 metastasis-specific, 35 primary-tumour-specific), 144 (73.5%) are different between primary tumour and metastasis (Table S4). Collectively, both patients generate a high degree of divergence between primary tumour and its metastases, as well as between metastases in different anatomic locations.
We further explored whether the above findings can be generalized to m-ccRCC. We focused on the selected ccRCC patients with bone metastases which metastasectomy as an alternative to systemic therapy. We performed whole-genome sequencing of 18 specimens from 6 ccRCC patients including normal kidney tissue, primary tumour and bone metastatic lesion. The depth coverages for tumours and normal tissues are from 27 × to 34 × (Table S3). The divergent between primary tumour and bone metastatic lesions is from 78.3% to 98.7% (Table S4). Taken together, by genome-wide comparisons of primary tumours and their metastases in treatment-naïve ccRCC patients, our data showed that the evolutionary branches of primary and metastatic clones diverge early in the development of the tumour.
The frequency of somatic alterations on ccRCC driver genes
Next, we focused on the subset of mutations leading to amino acid substitutions and premature stop codons. We constructed the tumour phylogeny for each patient using the missense SNAs and CNAs (Fig. 1A, 1B and 1C). The SNAs and CNAs related to each phylogenetic tree are listed in Table S5. The tumour phylogeny of the missense SNAs and CNAs further suggested the clonal origin and parallel evolution. Then, we evaluated the candidate driver mutations which carry the bulk of functional and clinical relevance. Driver genes were selected from genes that were frequently mutated in TCGA or highlighted in relevant studies [20], [21], [22] (Table S6). Only alterations in driver genes represented in all two panels were considered as ccRCC driver genes. The mutations on the ccRCC driver genes in the trunk and/or branch are highlighted in Table S5. Based on the mutation allele frequency information for each mutation, we determined whether these mutations were common or exclusive to the primary tumour and metastasis. As shown in Fig. 1B and 1C, no significant difference was observed in median number of missense SNAs between primary tumours and metastases of patient 3 to 8 (41.5 versus 56.5, Student's t-test, P = 0.58). Meanwhile, a limited number of known driver genes is identified as common or exclusive to the primary tumour and metastasis. As shown in Fig. 1C, the frequency of somatic alterations can't differ primary tumours from matched metastatic tumours.
Fig. 1.
The main routes of m-RCC evolution. (A) The phylogenetic trees with missense SNAs and CNAs along the branches are generated by Canopy software from WES in two ccRCC patients with multiple metastatic lesions. The ancestral normal cell population is at the root, and each primary tumour or metastatic lesion sample is composed of different proportions of subpopulations. The lengths of the branches are arbitrary. The sites of core biopsies harvested from nephrectomy and metastasectomy specimens were shown in the left panel. T = the primary ccRCC tumour; MB = bone metastasis; ML = lung metastasis; and MA = adrenal metastasis. (B) The phylogenetic trees with missense SNAs along the branches are generated by Canopy software from the whole-genome sequencing data for other 6 metastatic ccRCC patients with triplet samples, including matched normal, primary tumour and bone metastatic lesion. The matched normal cell population is at the root, and each primary tumour or metastatic lesion is composed of different proportions of subpopulations. The lengths of the branches are arbitrary. (C) A limited number of recurrent missense mutations is identified in 8 patients. The allele frequency changes of the missense mutations grouped using unsupervised hierarchical clustering.
Mutational spectra remain stable during metastasis
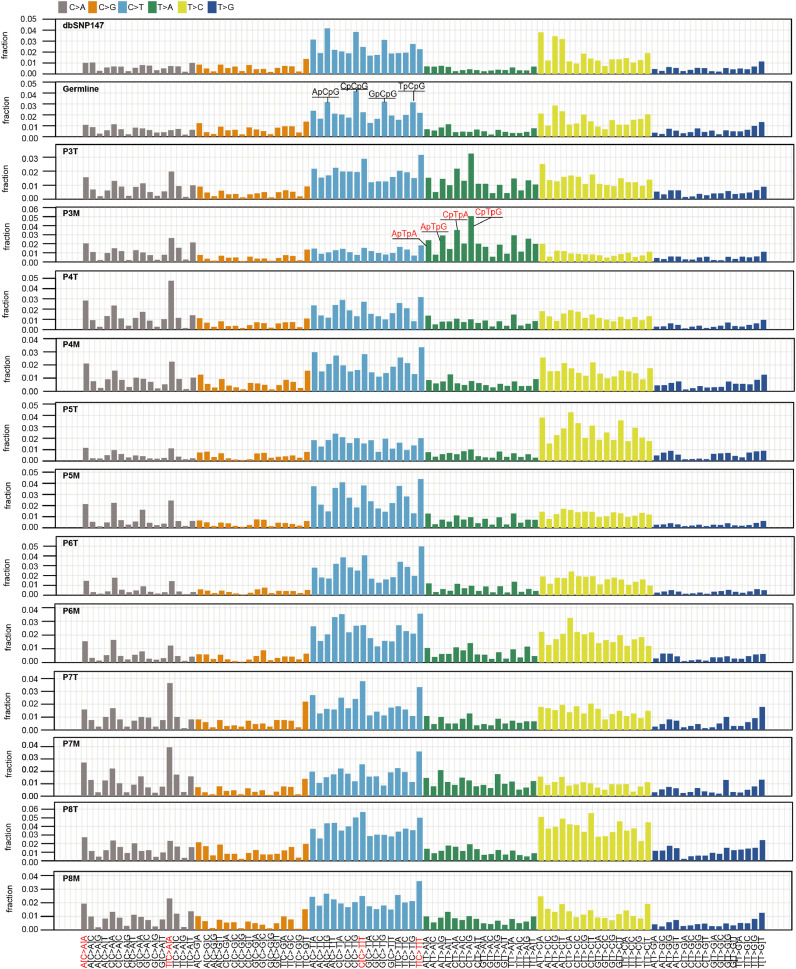
Since the primary tumours and metastatic lesions diverge early, next we explored whether the DNA damage and the DNA repair or replicate mechanisms will change especially for the metastatic cells proliferating in ectopic environment. Each mutational process leaves a characteristic imprint a mutational signature on the cancer genome. So, there are 6 classed of case substitutions (C>A, C>G, C>T, T>A, T>C, T>G) and 16 possible sequence contexts for each mutated base, 96-trinucleotide are possible [23,24]. Therefore, the presence of mutation signatures will inform the etiology of an individual's primary tumour cells and the metastatic cells. Strikingly, the mutational spectra remains relatively stable between the metastatic lesion and matched primary tumour in each patient (Fig. 2). The C>T transition, especially in the NpCpA and NpCpT context, is the most dominant mutated spectrum, followed by T>C in the NpTpA and NpTpT context. C>A in the NpCpA context is also frequently identified in the samples with unknown causes (Fig. 2 and Fig. S1C-1D). An unexpectedly high frequency of T>A transversion is identified in patient 3; this result is consistent with exposure to aristolochic acid (AA), which leads to a very high mutational burden in urothelial carcinoma [25,26]. Similar mutation spectra have also been found in ccRCC patients from Romania [20]. These findings indicate that similar mutational processes foster the generation of genetic diversity in both primary and metastatic tumours in each patient.
Fig. 2.
Mutational spectra remained stable during metastasis in ccRCC patients. The height of each bar (the y-axis) represents the proportion of all the 96 observed mutational spectrum that fall in a particular trinucleotide mutational class. The mutation signature of the current dbSNP147 build is used as a control. A known mutation signature associated with aristolochic acid (AA) exposure is identified in patient 3. Four signatures that are prominent are also highlighted in red.
The shared and metastasis-selected loss of heterozygosity (LOH) and somatic copy number alterations (SCNAs) at arm level and clinical implications in m-ccRCC
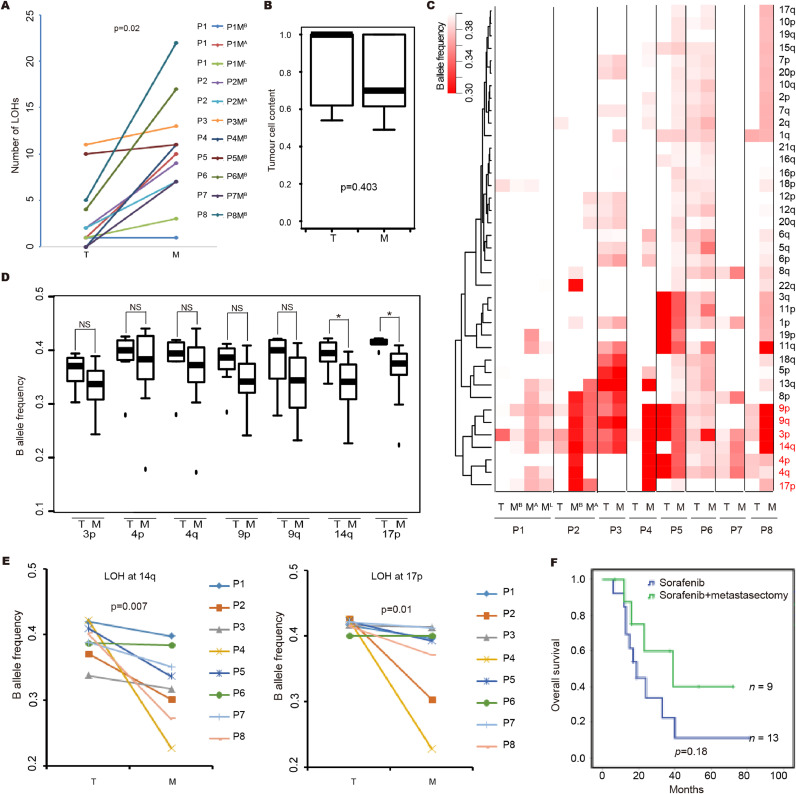
More larger scale loss of heterozygosity (LOH) events is a hallmark of metastasis compared to primary tumour. As shown in Fig. 3A, metastatic lesions have larger number of LOHs at arm-level compared to matched primary tumour in all 8 patients. Although the estimated tumour cell contents are comparable between metastatic lesions and primary tumours (Fig. 3B and Table S7). Notably, the frequent of arm-level events involved LOH at 3p, 4p, 4q, 9p, 9q, 14q and 17p are usually shared events among both the primary and metastatic tumours (Fig. 3C). Similarly, the frequent arm-level events involved loss of 3p, gain of 5q, loss of 9p and loss of 14q, which are usually shared events among both the primary and metastatic tumours (Fig. S2A). We discovered LOH at 14q and 17p are significantly enrichment in metastatic lesions compared to the primary tumours (Fig. 3D and 3E). The wider involvement of LOH mutations in cancer is assumed to be related to unmasking a somatically mutated tumour suppressor gene through loss of the wild type allele. These results indicated that LOH at 14q and 17p were selected events in metastasis. Consistently, 14q loss and 17p loss was significantly associated with metastatic stage (Fig. S2B and S2C) and 14q loss was associated with shorter overall survival in ccRCC patients (Fig. S2D). Moreover, consistent with previous studies, LOH at 3p as potential truncal driver events which spans all the known driver alterations in ccRCC including VHL, PBRM1, BAP1 and SETD2 for all 8 patients. This observation supported targeted therapy for m-ccRCC patients. VHL loss increases the expression of HIFs and their targets, including vascular endothelial growth factor (VEGF), and platelet-derived growth factor (PDGF). Therefore, TKIs treatment which target the angiogenesis pathways may also target the metastatic lesions. Consistently, the two patients, patients 1 and 7, who received sorafenib after metastasectomy, a TKI treatment, achieve 117 months and 40 months overall survival, respectively. The other treatment-free patients had a median overall survival of 14 months after nephrectomy and metastasectomy (Table S2). Additionally, if applicable, metastasectomy combined with TKIs treatment should be an effective management for m-ccRCC. To validate the hypothesis, 72 m-RCC patients at the Department of Urology, Peking University First Hospital were selected. among these patients, 22 patients had bone metastases (13 for targeted therapy and 9 for both targeted and metastasectomy therapies); the median survival for targeted therapy was 19 months, whereas that for metastasectomy combined with targeted therapy was 39 months (Fig. 3F). However, with the limited number of patients, statistical significance was not achieved (P = 0.18). And we can not exclude other clinical variables, such as the general health conditions of the related patients may contribute to the different outcome.
Fig. 3.
The shared and metastasis-selected LOHs and clinical implications in m-ccRCC. (A) The metastatic lesions have larger number of LOH at arm-level compared to matched primary tumour in all 8 patients. T = the primary tumour; MB = bone metastasis; ML = lung metastasis; and MA = adrenal metastasis. Statistical significance is evaluated by paired Student's t-test. (B) Box plot showing tumour cell content evaluated by ascatNgs between primary tumours and metastatic tumours. Statistical significance was evaluated by Student's t-test. (C) The B allele frequency changes at arm-level grouped using unsupervised hierarchical clustering. The scale bar ranges from white to red and represents the allele frequency from high to low. (D) Shared and metastasis selected LOH regions. (E) Metastasis-selected LOH regions at 14q and 17p. Statistical significance was evaluated by Student's t-test. (F) Kaplan-Meier survival analysis of different treatments for ccRCC patients with bone metastasis (log-rank test).
Increased chromosomal instability characterizes metastatic renal cell carcinoma evolution with therapeutic relevance
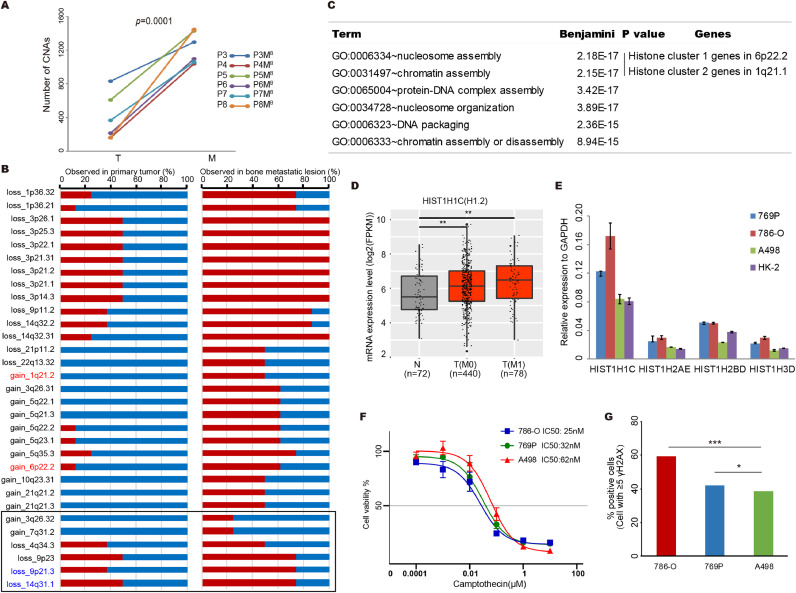
Additionally, with whole-genome sequencing data, the focal SCNAs can be more accurately evaluated in the patients 3 to 8. We found that the number of focal SCNAs in metastatic lesion is also larger than that of matched primary tumours in all 6 patients (Fig. 4A). These results support that chromosome instability increase during metastasis. Next, we compared the rates of each driver SCNA event in metastatic and primary tumours. Consistent with previous reports [27], we noted that multiple focal SCNAs, such as loss of 14q31.1 and loss of 9p21.3, were selected in metastatic tumors (Fig. 4B). Strikingly, we also identified multiple novel selected SCNA events during metastasis, such as gain of 1q21.2 and gain of 6p22.2, which may reflect the greater sensitivity of WGS profiling compared to targeted sequencing (Fig. 4B).
Fig. 4.
Increased chromosomal instability characterizes metastatic renal cell carcinoma evolution with therapeutic relevance. (A) The metastatic lesions have larger number of focal SCNAs compared to matched primary tumour in patient 3 to 8. Statistical significance is evaluated by Student's t-test. (B) Selected SCNAs during metastatic progression. For each SCNA event, the percentage of observed numbers in primary and metastatic tumors in the 8 m-ccRCC patients are plotted. The red box represents the observed pencentage of each SCNA event in primary or metastatic tumors of all patients. Previous reported enriched SCNA events in metastatic tumors are highlight. (C) The GO pathway analysis with all the genes within the selected gain copy number regions presented in (B). The enriched GO names, adjusted p value and enriched gene clusters of each pathway are listed. Histone cluster 1 and 2 genes are involved in nucleosome assembly and chromatin assembly pathways. (D) Expression levels of HIST1H1C (H1.2) gene in normal kidney tissues, primary tumor tissues of M0 or M1 stage ccRCC patients are plotted. Expression of HIST1H1C (H1.2) in primary tumor tissue (M0 and/or M1) of ccRCC patients are significantly higher than normal kidney tissues. The gene expression datasets are downloaded from TCGA. N presents the normal kidney tissues, T (M0) presents primary tumors from non-metastatic ccRCC patients and T (M1) presents primary tumors from metastatic ccRCC patients. The sample size of each group was indicated. Statistical significance was evaluated by Student's t-test. * p<0.05, **p<0.01.(E) mRNA levels of histone cluster genes in ccRCC cell lines and human renal tubular cells HK-2. Expression levels are shown as fold differences (mean ± SD) compared to GAPDH. (F) Growth curves for ccRCC cells with varying expression of histone cluster genes after 72hr exposure to camptothecin. (G) DNA double stand break induced by camptothecin treatment for ccRCC cells with varying expression of histone cluster genes.
We further explored the functional mechanisms underlying the selected SCNA events. Since the gain of copy number in oncogenes may promote the gene expression change involving tumorigenesis, we performed KEGG pathways analysis using genes within the selected copy number gain regions to explore the underlying oncogenic pathways. Strikingly, two of the most enriched biological processes are nucleosome assembly and chromatin assembly which included two clusters of histone genes, histone cluster 1 at 6p21.1 and histone cluster 2 at 1p21.2 (Fig. 4C). Histones are basic proteins which are responsible for the assembly and maintenance of the nucleosomal structure within the chromosomal fiber in eukaryotes. To further validate gain of copy number would promote the gene expression of histone genes, we downloaded the gene expression dataset of ccRCC from TCGA. The expression of four genes from histone cluster 1, HIST1H1C, HIST1H2AE, HIST1H2BD and HIST1H3D increased in tumor tissues compared to normal kidney tissues (Fig. 4D and Fig. S3A-S3C). Similarly, the expression of these four genes are also higher in ccRCC cell lines compared to human renal tubular HK-2 cells (Fig. 4E).
DNA repair pathways are well known to be critical to maintain genome stability. The synthetic lethality data have identified several opportunities for using existing therapies in novel genetic backgrounds. For example, in yeast, the homologues of topoisomerase 1 (TOP1) and the chromatin remodelling factor, PBRM1, show synthetic lethality. PBRM1 is mutated and/or copy number loss in more than 80% of ccRCC [28]. Thus, we hypothesis that the TOP1 inhibitor, camptothecin, could be effective in ccRCC. Consistent with this scenario, camptothecin displayed nanomolar potency in cytotoxicity against ccRCC cell lines, 786-O, 769P and A498, with IC50 values from 25 nM to 62 nM (Fig. 4F). Camptothecin can induce double strand break in ccRCC cell lines (Fig. S3D) as expected. Consistent with previous study, deletion of HIST1H1C (H1.2) has been shown to render cancer cells resistant to DNA damaging agents [29], the lower level of H1.2 expression in ccRCC cell lines, the lower number of double strand break foci inducing by camptothecin (Fig. 4G).
Functional validation the candidate driver genes in selected SCNA events during metastatic evolution
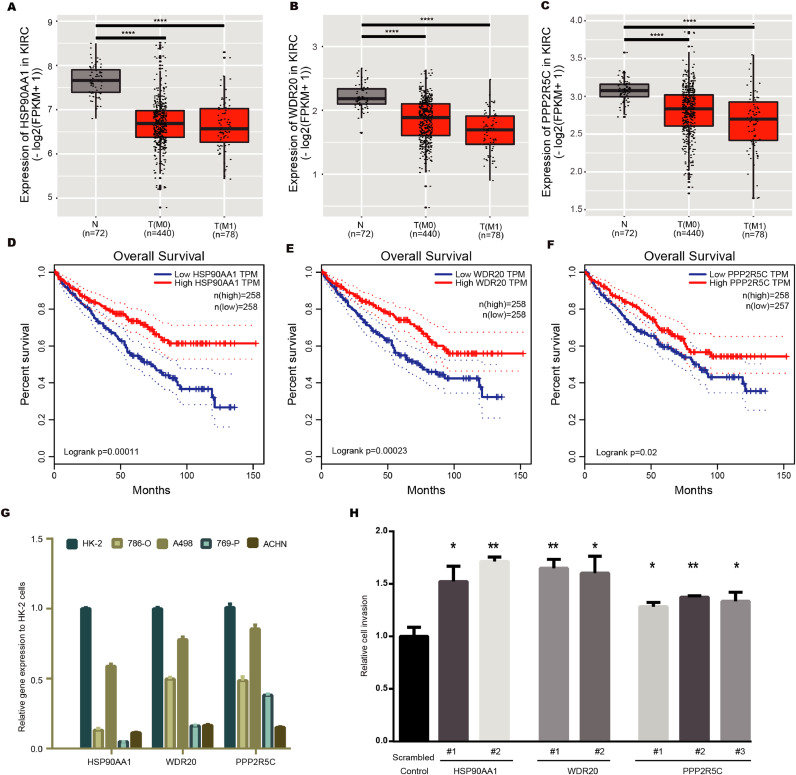
Since we discovered multiple novel copy number loss regions, such as 14q32.31, 14q32.2, 3p14.3 and 3p21.1, which were selected during metastasis (Fig. S4A). These SCNAs were more strongly selected by metastasis compared to previous metastatic-associated SCNAs, such as loss of 14q31.1 and loss of 9p21.3 [27] which may reflect the greater sensitivity of WGS data. Next, we selected one novel strongly selected event, loss of 14q32.31 to demonstrate whether knockdown the involved genes will promote tumor cell invasion. Using Level 3 RNA sequencing data from The Cancer Genome Atlas (TCGA). We performed statistical analysis with the following criterias: (i) the genes express more weakly in the tumour than in normal kidney tissue; and (ii) the expression of the gene was associated with clinical outcome. Three genes, HSP90AA1, WDR20 and PPP2R5C, fit these two criteria (two-tailed Student's T test, P ≤ 0.05) (Fig. 5A-5F). Consistently, all three genes showed downregulated in ccRCC cell lines compared to human renal tubular HK-2 cells (Fig. 5G). We therefore examined the relationship between knockdown these three genes and in vitro invasion capacity in 786-O cell line (Fig. S4B-S4D). As expected, knockdown either of the three genes promote tumor cell invasion capacity (Fig. 5H). Collectively, these results indicated that multiple genes especially HSP90AA1 in chromosome 14q32.31 may involve in metastatic phenotype.
Fig. 5.
Analysis of TCGA data assessing the effects of 14q32.31 loss on mRNA gene expression of potential metastasis-associated genes. (A-C) Lower expression of HSP90AA1 (A), WDR20 (B) and PPP2R5C (C) in primary tumor tissue (M0 and/or M1) of ccRCC patients compared to normal kidney tissues. The gene expression dataset of ccRCC was downloaded from TCGA. Statistical significance was evaluated by Student's t-test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.(D-F) Lower expression of HSP90AA1 (D), WDR20 (E) and PPP2R5C (F) in primary tumor tissue was associated with shorter overall survival. (G) mRNA levels of HSP90AA1, WDR20 and PPP2R5C in ccRCC cell lines and human renal tubular cells HK-2. (H) 786-O cells were transiently transfected with scrambled control or shRNAs (#1 to #3) targeting HSP90AA1, WDR20 and PPP2R5C, after 24 h, cell invasion capacity was evaluated through transwell assays. Data are represented as mean ± SD. Statistical significance was evaluated by Student's t-test. * p<0.05, **p<0.01.
Discussion
Whole-genome sequencing has been widely used for tracing cell lineage. However, the study of genome wide comprehensive analysis of paired primary tumours and metastases is very limited because of the absence of samples. Herein, we found that primary tumours and metastatic lesions display clonal origins and parallel evolution in all 8 treatment-naïve metastatic ccRCC patients. Although, similar patterns have also been identified in previous studies with multiregion exome sequencing (M-seq) in two patients with metastatic lesions after everolimus treatments [30,31]. Our data provide direct evidence of the evolutionary trajectory for m-ccRCC without the potential selection pressure from the targeted therapy.
Additionally, whole-genome sequencing is also the only way to assess comprehensively and comparatively how driver and passenger mutations evolve during cancer development and metastasis. We identify that larger scale genomic variations, but not somatic mutations differentiate metastasis from matched primary tumour. These results are consistent with a recent study which shows chromosome instability driving metastasis in lung cancers [32]. We also found that copy number gain of histone genes may facilitate to maintain the genome stability in metastatic tumor. Previous study showed that destabilization of H1.2 is essential for in activating DNA damage repair pathway [33]. Moreover, deletion of H1.2 render cancer cells resistant to DNA damaging agents [29]. Classically, defects in the DNA damage repair have been exploited therapeutically in the treatment of cancer with radiation therapies or genotoxic chemotherapies. Synthetic lethality analysis suggested that TOP1 inhibitor might be effective in ccRCC with defective PBRM1 [34]. We also showed ccRCC cell lines are very sensitive to TOP1 inhibitor, camptothecin, treatment. 786-O cells with highest H1.2 level showed the strongest cytotoxicity of camptothecin, indicating even stronger sensitivity to TOP inhibitor in metastatic ccRCC tumors. Consistently, in a phase Ib/2 trial, the combination treatment of CRLX101, a novel nanoparticle–drug conjugate containing camptothecin, and bevacizumab was well tolerated and showed promising efficacious in mRCC [35]. However, further randomized phase II trial showed that the CRLX101+bevacizumab combination treatment did not prove improvement in PFS compared to standard of care (SOC) in heavily pretreated m-ccRCC patients (refractory mRCC to 2–3 prior lines of therapy) [36]. But, in the study, within the SOC arm, patients received single-agent bevacizumab (n = 19), axitinib (n = 18), everolimus (n = 7), pazopanib (n = 4), sorafenib (n = 4), Sunitinib (n = 2), or temsirolimus (n = 2). When comparing 50 ccRCC patients treated with CRLX101+bevacizumab with 19 ccRCC patients having received bevacizumab monotherapy, median PFS was higher in combined therapy (3.7 versus 3.4 months) [36]. Although the statistical significance was not achieved which may be due to limited number of patients. Collectively, further clinical study should not focus on CRLX101 in the third-/four- line setting. Further studies exploring the role and extent of chromosome instability during ccRCC metastatic evolution would aid the development of better prevention and therapeutic interventions against m-ccRCC.
Additionally, we also identified multiple selected SCNAs with copy number loss during ccRCC metastasis, such as 14q32.31, 14q32.2, 9p11.1, 3p14.3, 3p21.1 and 3p21.2. We selected one representative loci to evaluate the functional consequences of these SCNAs in tumor invasive capacity. As expected, knockdown either of the three genes within this region promotes tumor cell invasion. Further investigation of function mechanisms for the genes in these selected SCNAs may identify other candidate driver genes for ccRCC metastasis. Taken together, these results supported that tumour phylogenetic data from the same patient could identify the roots of metastasis and provide a resource to further explore the molecular mechanisms of metastasis.
Limitations of this study include the relatively small sample size, owing to constraints of the availability of primary, metastatic tissues and germline tissue from the same treatment-naïve ccRCC patient. Therefore, we firstly focused on the ccRCC patients with bone metastatic lesions. Whether our findings can expand to ccRCC patients with lung or other organs metastasis remain exclusive. Consistent with our conclusions, a recent evolutionary study of matched primary metastasis biopsies from 100 ccRCC cases with targeted driver panel sequencing also find that loss of 14q is one of the hallmark genomic driver of ccRCC metastasis [27]. But the novel SCNAs we discovered was not identified by the targeted sequencing [27]. In summary, our observations serve as important hypothesis-generating findings of how tumour genomics evolved from primary to metastatic disease. These evidences will initiate further studies of m-ccRCC prevention and therapeutic interventions, such as a new clinical trial for treatment of the bone metastatic ccRCC patients by targeting DNA damage repair pathway.
Author contributions statement
Qin Ma: Methodology, Data analysis, Experiments, Writing-Reviewing and Editing. Jilu Wang: Data analysis, Experiments, Writing-Reviewing and Editing. Jie Qi: Experiments, Writing- Reviewing and Editing. Ding Peng: Experiments, Sample collections. Bao Guan: Sample collections. Jianye Zhang: Sample collections. Zhongwu Li: Sample collections. Hongxian Zhang: Sample collections. Ting Li: Sample collections and pathological reviewing. Yue Shi: Writing-Reviewing. Xuesong Li: Study supervision. Liqun Zhou: Study supervision. Ke Chen: Study supervision, Funding acquisition. Weimin Ci: Conceptualization, Study supervision, Funding acquisition, Methodology, Writing-Original draft preparation.
Accession number
The sequence data reported in this paper have been deposited in the genome sequence archive of the Beijing Institute of Genomics, Chinese Academy of Sciences, gsa.big.ac.cn (accession no: PRJCA000352).
Funding
This work was supported by CAS Strategic Priority Research Program (XDA16010102 to W.C.), the National Key R&D Program of China (2018YFC2000100, 2019YFA0110900 to W.C.), CAS (QYZDB-SSW-SMC039 to W.C.), the National Natural Science Foundation of China (81422035 and 81672541 to W.C., 81772731 to K.C.), Wong Education Foundation to W.C., and the Youth Innovation Promotion Association of Chinese Academy of Sciences to K.C..
Declarations of Competing Interest
We certify that there are no conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100929.
Contributor Information
Xuesong Li, Email: pineneedle@sina.com.
Liqun Zhou, Email: zhoulqmail@sina.com.
Ke Chen, Email: chenke@big.ac.cn.
Weimin Ci, Email: ciwm@big.ac.cn.
Appendix. Supplementary materials
References
- 1.Woodward E., Jagdev S., McParland L., Clark K., Gregory W., Newsham A. Skeletal complications and survival in renal cancer patients with bone metastases. Bone. 2011;48(1):160–166. doi: 10.1016/j.bone.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Keizman D., Ish-Shalom M., Pili R., Hammers H., Eisenberger M.A., Sinibaldi V. Bisphosphonates combined with sunitinib may improve the response rate, progression free survival and overall survival of patients with bone metastases from renal cell carcinoma. Eur. J. Cancer. 2012;48(7):1031–1037. doi: 10.1016/j.ejca.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 3.Gossage L., Eisen T., Maher E.R. VHL, the story of a tumour suppressor gene. Nat. Rev. Cancer. 2015;15(1):55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 4.Greef B., Eisen T. Medical treatment of renal cancer: new horizons. Br. J. Cancer. 2016;115(5):505–516. doi: 10.1038/bjc.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arlen M., Saric O., Wang X., Dubeykovskiy A., Arlen P. Nanocytology vs. immunohistochemistry of intestinal colonocytes to assess the risk of colon cancer based on field cancerization—a preliminary report. J. Cancer. 2013;4(2):165–169. doi: 10.7150/jca.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalra S., Verma J., Atkinson B.J., Matin S.F., Wood C.G., Karam J.A. Outcomes of patients with metastatic renal cell carcinoma and bone metastases in the targeted therapy era. Clin. Genitourin Cancer. 2017 doi: 10.1016/j.clgc.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Yu X., Guo G., Li X., Zhang C., Huang L., Fang D. Retrospective analysis of the efficacy and safety of sorafenib in chinese patients with metastatic renal cell carcinoma and prognostic factors related to overall survival. Medicine (Baltimore) 2015;94(34):e1361. doi: 10.1097/MD.0000000000001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naxerova K., Jain R.K. Using tumour phylogenetics to identify the roots of metastasis in humans. Nat. Rev. Clin. Oncol. 2015;12(5):258–272. doi: 10.1038/nrclinonc.2014.238. [DOI] [PubMed] [Google Scholar]
- 9.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl. Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucl. Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y., Qiu Y., Minn A.J., Zhang N.R. Assessing intratumor heterogeneity and tracking longitudinal and spatial clonal evolutionary history by next-generation sequencing. Proc. Natl. Acad. Sci. USA. 2016;113(37):E5528–E5537. doi: 10.1073/pnas.1522203113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Favero F., Joshi T., Marquard A.M., Birkbak N.J., Krzystanek M., Li Q. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol.: Off. J. Eur. Soc. Med. Oncol. 2015;26(1):64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raine K.M., Van Loo P., Wedge D.C., Jones D., Menzies A., Butler A.P. ascatNgs: identifying somatically acquired copy-number alterations from whole-genome sequencing data. Curr. Protocol. Bioinform. 2016;56:15. doi: 10.1002/cpbi.17. 9 1- 9 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenthal R., McGranahan N., Herrero J., Taylor B.S., Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scelo G., Riazalhosseini Y., Greger L., Letourneau L., Gonzalez-Porta M., Wozniak M.B. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat. Commun. 2014;5:5135. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- 21.Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45(8):860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 22.Arai E., Sakamoto H., Ichikawa H., Totsuka H., Chiku S., Gotoh M. Multilayer-omics analysis of renal cell carcinoma, including the whole exome, methylome and transcriptome. Int. J. Cancer. 2014;135(6):1330–1342. doi: 10.1002/ijc.28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helleday T., Eshtad S., Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014;15(9):585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoang M.L., Chen C.H., Sidorenko V.S., He J., Dickman K.G., Yun B.H. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci. Transl. Med. 2013;5(197) doi: 10.1126/scitranslmed.3006200. 197ra02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C.H., Dickman K.G., Moriya M., Zavadil J., Sidorenko V.S., Edwards K.L. Aristolochic acid-associated urothelial cancer in Taiwan. Proc. Natl. Acad. Sci. U.S.A. 2012;109(21):8241–8246. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turajlic S., Xu H., Litchfield K., Rowan A., Chambers T., Lopez J.I. Tracking cancer evolution reveals constrained routes to metastases: tRACERx renal. Cell. 2018;173(3):581–594. doi: 10.1016/j.cell.2018.03.057. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konishi A., Shimizu S., Hirota J., Takao T., Fan Y., Matsuoka Y. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 2003;114(6):673–688. doi: 10.1016/s0092-8674(03)00719-0. [DOI] [PubMed] [Google Scholar]
- 30.Gerlinger M., Horswell S., Larkin J., Rowan A.J., Salm M.P., Varela I. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014;46(3):225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakhoum S.F., Ngo B., Laughney A.M., Cavallo J.A., Murphy C.J., Ly P. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553(7689):467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Li Y., Tang M., Peng B., Lu X., Yang Q. Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair. Cell Res. 2018;28(7):756–770. doi: 10.1038/s41422-018-0048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearl L.H., Schierz A.C., Ward S.E., Al-Lazikani B., Pearl F.M. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer. 2015;15(3):166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- 35.Keefe S.M., Hoffman-Censits J., Cohen R.B., Mamtani R., Heitjan D., Eliasof S. Efficacy of the nanoparticle-drug conjugate CRLX101 in combination with bevacizumab in metastatic renal cell carcinoma: results of an investigator-initiated phase I-IIa clinical trial. Ann. Oncol.: Off. J. Eur. Soc. Med. Oncol. 2016;27(8):1579–1585. doi: 10.1093/annonc/mdw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voss M.H., Hussain A., Vogelzang N., Lee J.L., Keam B., Rha S.Y. A randomized phase II trial of CRLX101 in combination with bevacizumab versus standard of care in patients with advanced renal cell carcinoma. Ann. Oncol.: Off. J. Eur. Soc. Med. Oncol. 2017;28(11):2754–2760. doi: 10.1093/annonc/mdx493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.