Abstract
North American porcine reproductive and respiratory syndrome virus (NA-PRRSV), especially NADC30-like PRRSV, has evolved and is prevalent in China. We collected 503 samples from pig breeding farms across 4 provinces in northern China from 2016 to 2018. The samples were screened by PCR testing with specific primers that could differentiate groups of NA-PRRSV; phylogenetic trees were constructed and analyzed. Overall, 175 of 503 (34.8%) samples were positive for NA-PRRSV. Dual (NADC30-like and highly pathogenic [HP]-PRRSV; NADC30-like and typical PRRSV; HP and typical PRRSV) and triple (NADC30-like, HP, and typical PRRSV) infections (92 of 175, 52.6%) were common in coinfections by NADC30-like and HP-PRRSV. Notably, 18 of 125 (14.4%) semen samples were positive for PRRSV, and 17 of the 18 positive semen samples contained NADC30-like PRRSV. Phylogenetic analysis based on GP5 amino acids revealed that the novel NADC30-like PRRSV with a unique single amino acid deletion at position 34 has become widespread and has evolved into a new subgroup.
Keywords: NADC30-like porcine reproductive and respiratory syndrome viruses, novel deletion
Porcine reproductive and respiratory syndrome (PRRS), which is caused by porcine reproductive and respiratory syndrome virus (PRRSV; Betaarterivirus suid), impacts severely the global swine industry. PRRSV strains can be divided into 2 types based on genetic distances, type 1 (European-like, EU) and type 2 (North American-like, NA-PRRSV).6 NA-PRRSV was first confirmed in China in 1996 with the characterization of the CH-1a isolate, which was designated as typical PRRSV in China. In 2006, highly pathogenic PRRSV (HP-PRRSV) with a unique discontinuous deletion of 30 amino acids in nonstructural protein 2 (NSP2), of which JXA1 is the representative strain, emerged in China and caused high mortality with immense economic losses for the swine industry.5,10 HP-PRRSV has spread throughout China and has become the dominant strain circulating in Chinese swine herds. Since 2013, the NADC30 strain, a NA-PRRSV that was isolated in the United States in 2008, emerged in southeastern China and spread rapidly throughout China.2,3,11 Based on sequence information, PRRSV isolates with a unique discontinuous deletion of 131 amino acids in NSP2 were designated as NADC30-like PRRSV in China.
NADC30-like PRRSV, which led to the reemergence of new circulating variants with different levels of virulence, has been detected by many researchers in China.3 The virus has not only led to clinical PRRS outbreaks but also was involved in recombination events, resulting in the generation of new strains of virus.1,12,13 Additionally, varieties of commercial type 2 PRRSV modified-live virus (MLV) vaccines, including typical PRRSV (Ingelvac PRRS MLV-VR2332, Ch-1R, and R98) and HP-PRRSV (JXA1-R, TJM-F92, HuN4-F112, and GDr180), were widely used throughout China, and have evolved into MLV-like VR2332 virus and MLV-like HP-PRRSV (e.g., JXA1-R–like), and have been identified frequently under field conditions in China.8,14 As well, the co-circulation of different groups of NA-PRRSV strains in swine herds further causes complicated clinical signs. Semen also plays an important role in the PRRSV epidemic situation in China.4,7,12
To increase our knowledge about the epidemic prevalence and molecular evolution of NA-PRRSV in pig breeding farms, we collected samples across 4 provinces in China from 2016 to 2018 and screened them by PCR testing with specific primers that could differentiate different groups of NA-PRRSVs. We sequenced complete open reading frame 5 (ORF5) nucleotide sequences and used representative samples for virus isolation on MARC-145 (cloned African green monkey kidney) cells or porcine alveolar macrophages (PAMs). The complete genomes were also sequenced for phylogenetic and recombinant analysis, which could provide valuable information to further evaluate the relationship between coinfection and recombination status in China.
We collected 503 samples, including 125 semen samples, from breeding farms in 4 provinces (Beijing, Tianjin, Hebei, and Shandong) in China from 2016 to 2018. Clinical signs in these pigs were typical of PRRS, including labored breathing, pyrexia, lethargy, and anorexia. Total RNA was extracted from samples (RNeasy mini kit; Qiagen) according to the manufacturer’s instructions, and complementary DNA (cDNA) was produced (Reverse transcription system; Promega). The cDNA was screened for the presence of NA-PRRSV RNA using a new PCR assay that could rapidly differentiate NADC30-like, HP, and typical PRRSV. The primers (F: 5′-TTGATTGGGATGTTGTGCTTC-3′, R: 5′-CAATGATGGCTTGAGCTGAGT-3′) were designed based on the NSP2 region, and the sizes of the final amplicons were 628 bp (NADC30-like PRRSV), 931 bp (HP-PRRSV), and 1,021 bp (typical PRRSV). Additionally, the complete ORF5 nucleotide sequences of mono-infection samples were also amplified by PCR assay (F: 5′-ATG TTGGGGAAATGCTTGACC-3′; R: 5′-CTAGAGACGACCCCATTGTTC-3′). The cycling conditions were: 94°C for 5 min, followed by 30 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 7 min. This yielded a 603-bp PCR product containing a complete ORF5 gene of PRRSV.
Samples were inoculated on PAMs and MARC-145 cells for virus isolation depending on sequence analysis results, and complete genomes were sequenced for further analysis of the genetic evolution and the state of recombination as described previously.2 Inoculated cells were maintained and monitored daily for cytopathic effect (CPE). Cultures were harvested when CPE reached 70% and were stored at −80°C. The virus was purified by plaque assay, and total RNA was extracted from cell cultures. The complete genomic sequences of various strains were amplified with overlapping fragments.1 The PCR products were purified using a PCR purification kit (Tiangen), according to the manufacturer’s instructions, and cloned into the pMD18-T vector (TaKaRa). The plasmid was transformed into Escherichia coli JM 109. Positive clones were sequenced (Sangon Biotech). Multiple alignments were performed and compared with those of reference strains of PRRSV in GenBank (Table 1). Phylogenetic trees were constructed from aligned nucleotide sequences using the neighbor-joining method in the MEGA6.09 software package, with 1,000 bootstrap replicates of the alignment. Recombination was further analyzed by Bootscan analysis in SimPlot software v.3.5.1 to confirm possible recombination, according to methods described previously.1
Table 1.
PRRSV strains used in our study of phylogenetic trees.
| No. | Strain | Country | Year | Accession |
|---|---|---|---|---|
| 1 | CH-1a | China | 1996 | AY032626 |
| 2 | CH-1R | China | 2008 | EU807840 |
| 3 | BJ-4 | China | 2000 | AF331831 |
| 4 | HB-1(sh)/2002 | China | 2002 | AY150312 |
| 5 | R98 | China | 2006 | DQ355796 |
| 6 | JXA1 | China | 2006 | EF112445 |
| 7 | JXwn06 | China | 2007 | EF641008 |
| 8 | HuN4 | China | 2007 | EF635006 |
| 9 | BJ-4 | China | 2007 | EU825723 |
| 10 | CH-1R | China | 2008 | EU807840 |
| 11 | JXA1 P80 | China | 2009 | FJ548853 |
| 12 | TJ | China | 2008 | EU860248 |
| 13 | TJbd1401 | China | 2014 | KP742986 |
| 14 | VR-2332 | USA | 1992 | AY150564 |
| 15 | MN184A | USA | 2005 | DQ176019 |
| 16 | MN184B | USA | 2006 | DQ176020 |
| 17 | MN184C | USA | 2007 | EF488739 |
| 18 | NADC30 | USA | 2008 | JN654459 |
| 19 | Chsx1401 | China | 2014 | KP861625 |
| 20 | HENAN-HEB | China | 2012 | KJ143621 |
| 21 | HENAN-XINX | China | 2013 | KF611905 |
| 22 | Lelystad virus | Netherlands | 1991 | M96262 |
Samples were positive for NA-PRRSV (175 of 503, 34.8%). Dual (NADC30-like and HP-PRRSV; NADC30-like and typical PRRSV; HP and typical PRRSV) and triple (NADC30-like, HP, and typical PRRSV) infections (92 of 175, 52.6%) were common in coinfections by NADC30-like and HP-PRRSV. Among the dual and triple coinfected cases, 60 of 92 (65.2%) samples had infection with NADC30-like and HP-PRRSV (Table 2). Notably, 18 of 125 (14.4%) semen samples were positive for PRRSV, and 17 of the 18 positive semen samples contained NADC30-like PRRSV, which may be one of the most important reasons causing or accelerating NADC30-like PRRSV prevalence in pig breeding farms. These findings showed that NADC30-like and HP-PRRSV have become the dominant pathogens in China.
Table 2.
NA-PRRSV detection in pig breeding farms in 4 regions in northern China from 2016 to 2018.
| Region/Sample | No. of samples | Infection |
NADC30-like PRRSV |
HP-PRRSV |
Typical PRRSV |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MI | DI | TI | MI | DI | TI | MI | DI | TI | MI | DI | TI | ||
| Tianjin | |||||||||||||
| Serum | 139 | 28 | 21 | 5 | 7 | 16 | 5 | 14 | 19 | 5 | 7 | 7 | 5 |
| Semen | 60 | 2 | 5 | 2 | 2 | 5 | 2 | 0 | 5 | 2 | 0 | 0 | 2 |
| Beijing | |||||||||||||
| Serum | 74 | 13 | 17 | 0 | 3 | 12 | 0 | 7 | 17 | 0 | 3 | 5 | 0 |
| Semen | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Shandong | |||||||||||||
| Serum | 75 | 16 | 11 | 4 | 6 | 9 | 4 | 7 | 11 | 4 | 3 | 2 | 4 |
| Semen | 25 | 1 | 3 | 0 | 1 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| Hebei | |||||||||||||
| Serum | 90 | 21 | 19 | 2 | 6 | 17 | 2 | 11 | 17 | 2 | 4 | 4 | 2 |
| Semen | 30 | 2 | 3 | 0 | 2 | 2 | 0 | 0 | 3 | 0 | 0 | 1 | 0 |
| Total | 503 | 83 | 79 | 13 | 27 | 64 | 13 | 39 | 75 | 13 | 17 | 19 | 13 |
| 175/503 (34.8%) | 104/175 (59.4%) | 127/175 (72.6%) | 49/175 (28.0%) | ||||||||||
DI = dual infection; HP-PRRSV = highly pathogenic porcine reproductive and respiratory syndrome virus; MI = mono-infection; NA-PRRSV = North American porcine reproductive and respiratory syndrome virus; TI = triple infection.
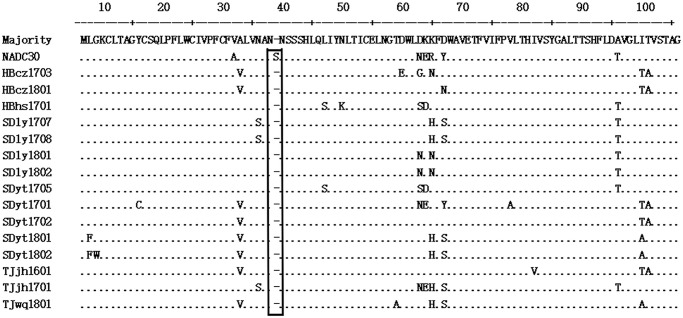
Amino acid alignment of GP5 revealed that 15 of the 81 ORF5 gene sequences from mono-infection NADC30-positive samples by PCR testing contained a unique amino acid deletion at position 34 (Fig. 1). Furthermore, 36 representative isolates, including 8 deletion isolates, were selected for further phylogenetic analysis. All of the isolates belonged to type 2 and could be further subdivided into 3 groups (Fig. 2): group 1 is comprised of our 13 representative isolates that are closely related to the MLV-like HP-PRRSV existing in China; group 2 consists of isolates closely related to VR-2332 and BJ-4 isolates; group 3 consists of 12 PRRSV isolates and can be further divided into 2 subgroups; our 8 deletion isolates are included in subgroup 2. Hence, different groups of PRRSV isolates coexist in pigs in China, and it is noteworthy that all deletion isolates have evolved into a new subgroup.
Figure 1.
The amino acid alignment of ORF5 for NADC30-like porcine reproductive and respiratory syndrome virus. The amino acid sequences were aligned by the ClustalW method in MEGA6.0. The boxed region was the missing amino acid at position 34 of the isolates with deletion in ORF5.
Figure 2.
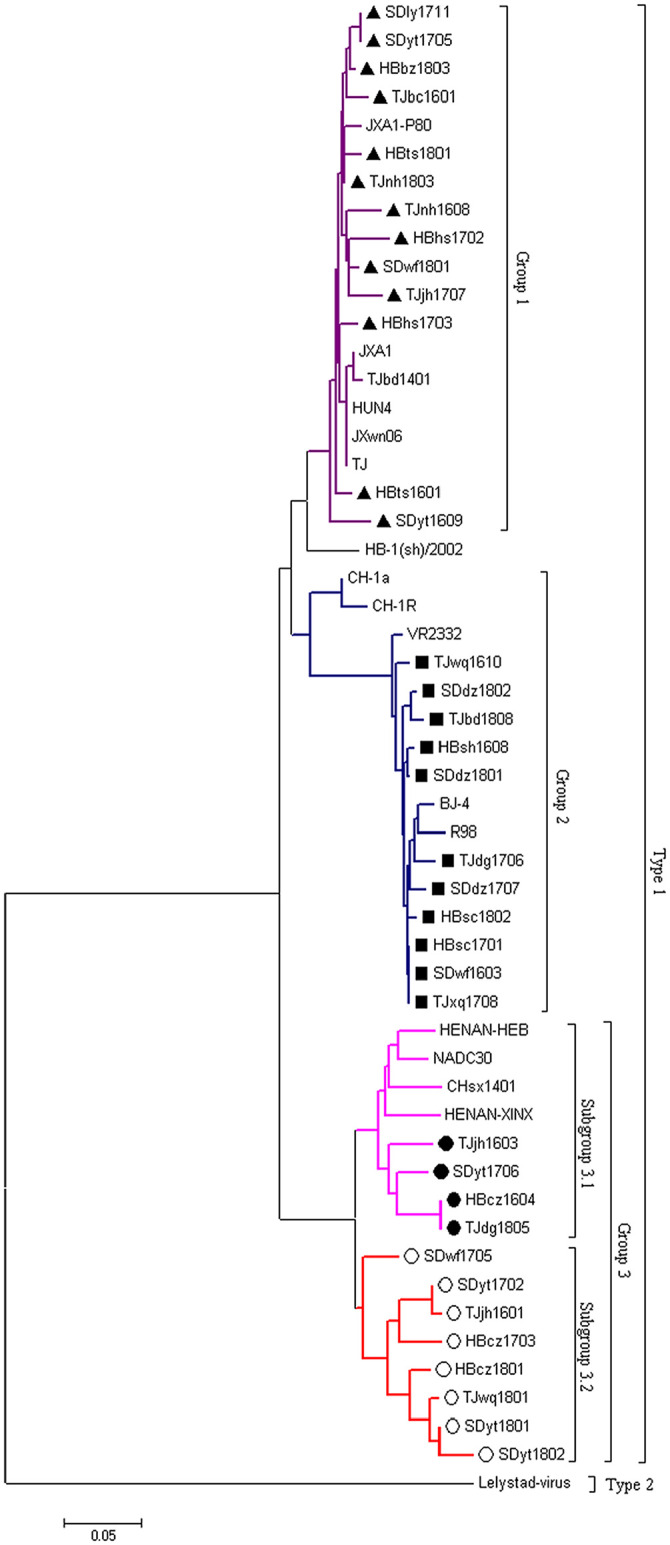
Phylogenetic tree constructed based on the full-length ORF5 gene of 36 representative porcine reproductive and respiratory syndrome virus isolates. The tree was constructed using the neighbor-joining method with bootstrap values calculated from 1,000 replicates. Group 1 isolates are indicated by a filled triangle; group 2 isolates are indicated by a filled square; group 3 isolates are indicated by a circle; isolates with a deletion at position 34 in GP5 are indicated by a hollow circle.
Seven strains (HBts1601, HBhs1701, SDyt1702, SDyt1706, TJwq1610, TJnh1501, and TJjh1707) were isolated and purified successfully. The 5 isolated strains (HBts1601, HBhs1701, TJwq1610, TJnh1501, and TJjh1707) grew on PAMs or MARC-145 cells, but 2 isolated strains (SDyt1702 and SDyt1706) would only grow on PAMs. The genomic sequences show that SDyt1702 and SDyt1706 had 3 discontinuous 131 amino acid deletions in NSP2 and shared 99.7% nucleotide similarity with NADC30. Compared with SDyt1706 and NADC30, only a unique amino acid deletion at position 34 in GP5 in the SDyt1702 isolate was confirmed. HBhs1701 and HBts1601 have discontinuous 30 and 120 amino acid deletions in NSP2 as reported previously for MLV-like HP-PRRSV (TJM-F92) and shared 99.5% nucleotide similarity with TJbd14-1 (TJM-F92–like). Phylogenetic analysis based on the full-genome sequences (Fig. 3) show that TJjh1707, HBhs1701, and HBts1601 clustered in group 1 and all are MLV-like HP-PRRSV; TJwq1610 clustered in group 2, and is a MLV-like VR2332 virus; SDyt1702 and SDyt1706 are more closely related to NADC30 and can be clustered in group 3. Further recombination analysis revealed that TJnh1501 was a recombinant type 2 PRRSV,1 which indicated that we could not ignore the potential risk of a recombination event, leading to a new variation of a virulent strain.
Figure 3.
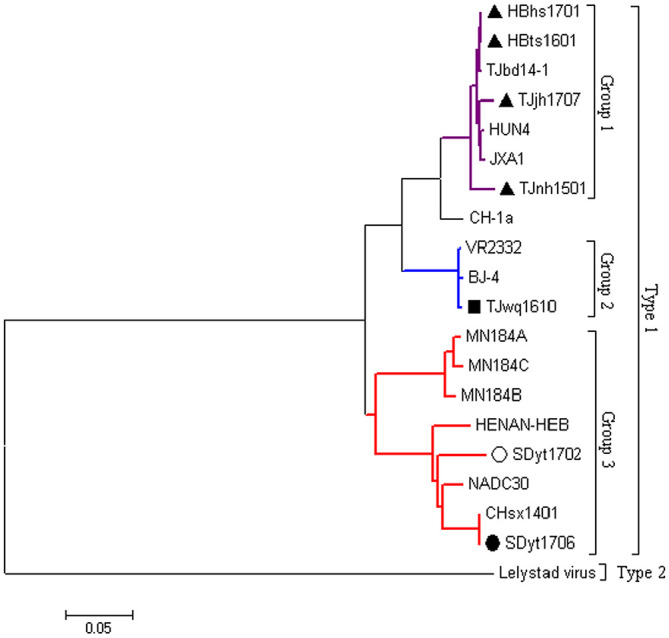
Phylogenetic tree based on the complete genomes. The phylogenetic tree was constructed using a distance-based neighbor-joining method with 1,000 bootstrap replicates in MEGA6.0. HBhs1701, Hbts1601, TJjh1707, and TJnh1501 isolates are marked with a filled triangle; TJwq1610 isolate is marked with a filled square; SDyt1702 isolate is marked with a filled circle; SDyt1702 isolate with deletion at position 34 in GP5 is indicated by a hollow circle.
In our study, NADC30-like PRRSV and MLV-like HP-PRRSV were identified as the predominant PRRSVs, and coinfections were common in China during 2016–2018. Importantly, novel NADC30-like PRRSV strains with a unique deletion at position 34 in GP5 were isolated from pigs, implying that NADC30-like PRRSV continuously evolved toward greater genetic diversification and a potential hazard to farms. This evolution highlights the importance of continuous monitoring for PRRSV, especially after the emergence of NADC30-like NA-PRRSV with a novel deletion in China.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This work was supported by grants from the Natural Science Foundation of Tianjin (19JCYBJC29 800), the key technologies R&D program of Tianjin (17YFZCNC00240), Tianjin sci-tech projects (19ZXBTSN00240), Project of Tianjin “131” innovative talent team (JRC2018044), and Technical System of Pig Industry of Tianjin (TTPRS2017004).
ORCID iDs: Ying-Feng Sun  https://orcid.org/0000-0001-5913-7771
https://orcid.org/0000-0001-5913-7771
Liu-an Li  https://orcid.org/0000-0002-4042-4946
https://orcid.org/0000-0002-4042-4946
Contributor Information
Ying-Feng Sun, Tianjin Key Laboratory of Agricultural Animal Breeding and Healthy Husbandry, College of Animal Science and Veterinary Medicine, Tianjin Agricultural University, Tianjin, China.
Hai Yu, Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China; Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou, China.
Xuan Jiang, Tianjin Key Laboratory of Agricultural Animal Breeding and Healthy Husbandry, College of Animal Science and Veterinary Medicine, Tianjin Agricultural University, Tianjin, China.
Ji-Fei Ma, Tianjin Key Laboratory of Agricultural Animal Breeding and Healthy Husbandry, College of Animal Science and Veterinary Medicine, Tianjin Agricultural University, Tianjin, China.
Cheng-Qian Xu, Tianjin Key Laboratory of Agricultural Animal Breeding and Healthy Husbandry, College of Animal Science and Veterinary Medicine, Tianjin Agricultural University, Tianjin, China.
Xiao-Xue Yu, Tianjin Key Laboratory of Agricultural Animal Breeding and Healthy Husbandry, College of Animal Science and Veterinary Medicine, Tianjin Agricultural University, Tianjin, China.
References
- 1. Bian T, et al. A recombinant type 2 porcine reproductive and respiratory syndrome virus between NADC30-like and a MLV-like: genetic characterization and pathogenicity for piglets. Infect Genet Evol 2017;54:279–286. [DOI] [PubMed] [Google Scholar]
- 2. Brockmeier SL, et al. Genomic sequence and virulence comparison of four type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res 2012;169:212–221. [DOI] [PubMed] [Google Scholar]
- 3. Li C, et al. Outbreak investigation of NADC30-like PRRSV in south-east China. Transbound Emerg Dis 2016;63:474–479. [DOI] [PubMed] [Google Scholar]
- 4. Liu JK, et al. Emergence of a novel highly pathogenic porcine reproductive and respiratory syndrome virus in China. Transbound Emerg Dis 2017;14:1–16. [DOI] [PubMed] [Google Scholar]
- 5. Li Y, et al. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the mid-eastern region of China. Vet J 2007;174:577–584. [DOI] [PubMed] [Google Scholar]
- 6. Nelsen CJ, et al. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol 1999;73:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reicks DL. Effective biosecurity to protect North American studs and clients from emerging infectious disease. Theriogenology 2019;137:82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun YF, et al. Efficacy evaluation of two commercial modified-live virus vaccines against a novel recombinant type 2 porcine reproductive and respiratory syndrome virus. Vet Microbiol 2018;216:176–182. [DOI] [PubMed] [Google Scholar]
- 9. Tamura K, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tong GZ, et al. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis 2007;13:1434–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang LJ, et al. Molecular epidemiology of porcine reproductive and respiratory syndrome virus in central China since 2014: the prevalence of NADC30-like PRRSVs. Microb Pathog 2017;109:20–28. [DOI] [PubMed] [Google Scholar]
- 12. Zhao H, et al. Emergence of mosaic recombinant strains potentially associated with vaccine JXA1-R and predominant circulating strains of porcine reproductive and respiratory syndrome virus in different provinces of China. Virol J 2017;14:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao K, et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J Virol 2015;89: 10712–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou L, et al. Genetic characterization and pathogenicity of a novel recombined porcine reproductive and respiratory syndrome virus 2 among NADC30-like, Jxa1-like, and MLV-like strains. Viruses 2018;10:551. [DOI] [PMC free article] [PubMed] [Google Scholar]