 New fluorescent molecular tools for the muscarinic acetylcholine M2 receptor, bearing various fluorescent dyes, showed high M2 receptor affinity in flow cytometric saturation binding studies at CHO–hM2R cells (pKd > 8.3).
New fluorescent molecular tools for the muscarinic acetylcholine M2 receptor, bearing various fluorescent dyes, showed high M2 receptor affinity in flow cytometric saturation binding studies at CHO–hM2R cells (pKd > 8.3).
Abstract
A series of fluorescent dibenzodiazepinone-type muscarinic acetylcholine M2 receptor (M2R) ligands was synthesized using various fluorescent dyes (5-TAMRA, λex/λem ≈ 547/576 nm; BODIPY 630/650, λex/λem ≈ 625/640 nm; pyridinium dye Py-1, λex/λem ≈ 611/665 nm and pyridinium dye Py-5, λex/λem ≈ 465/732 nm). All fluorescent probes exhibited high M2R affinity (pKi (radioligand competition binding): 8.75-9.62, pKd (flow cytometry): 8.36–9.19), a very low preference for the M2R over the M1 and M4 receptors and moderate to pronounced M2R selectivity compared to the M3 and M5 receptors. The presented fluorescent ligands are considered useful molecular tools for future studies using methods such as fluorescence anisotropy and BRET based MR binding assays.
Introduction
In the past decades, fluorescence-based techniques have been increasingly used to study membrane receptors such as G-protein coupled receptors (GPCRs), including the analysis of ligand-receptor interactions as well as the investigation of receptor expression, structure and function.1–4 Besides fluorescently tagged receptors, fluorescent ligands represent interesting molecular tools, which can also be used to study endogenously expressed receptors.1 Therefore, there is a growing demand for suitable fluorescent GPCR ligands. Compared with radioligands, fluorescent ligands exhibit several advantages, such as fewer problems concerning safety risks, legal issues and waste disposal. Furthermore, they are applicable to techniques, which have become routine in many laboratories, for instance fluorescence microscopy and flow cytometry. Typically, fluorescent ligands are composed of a pharmacophore, mediating receptor affinity, a spacer/linker moiety and the fluorophore.1,5 The design of fluorescent ligands is not trivial, e.g. with respect to high receptor affinity and favorable physicochemical properties. Crucial factors are the type of the fluorescent dye (size, lipophilicity, spectroscopic and bleaching properties, etc.) and the point of attachment, structure and length of the linker.1,6–11
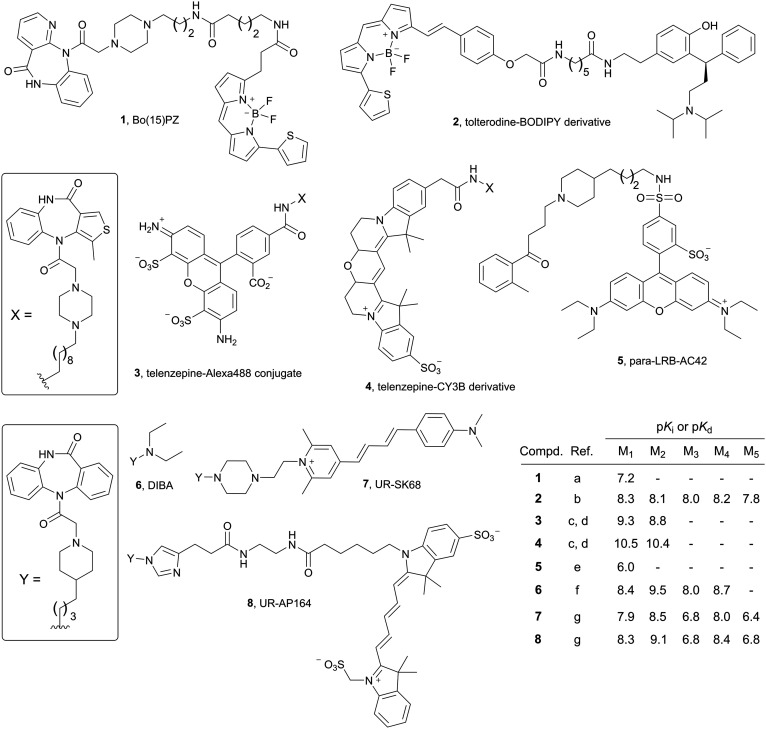
Numerous fluorescent probes have been reported for GPCRs, for instance for histamine,12–16 dopamine,17,18 opioid,19–22 neuropeptide Y,23–28 adenosine,29 and neurotensin30,31 receptors. Concerning muscarinic acetylcholine receptors (MRs), various fluorescent MR ligands were reported, e.g. the BODIPY558/568-labelled derivative 1,32 derived from the M1 subtype preferring MR antagonist pirenzepine, the BODIPY630/650-labelled tolterodine derivative 2,33 the Alexa488-labelled telenzepine derived MR antagonist 3,34,35 the Cy3B-labelled telenzepine analog 4,34–36 and the lissamine rhodamine B-labelled AC-42 derivative 5 (ref. 37) (Fig. 1; note: in the case of 1 and 3–5 further congeners, containing the same pharmacophore but different fluorophores such as Cy3, Cy5 cascade blue or 6-carboxyfluorescein and, in part, also different linkers, were reported32,36–41). The recent finding that replacement of the diethylamine moiety in the M2R preferring dibenzodiazepinone-type MR antagonist DIBA42 (6, Fig. 1) by bulky moieties was well tolerated with respect to M2R binding,43–46 gave rise to prepare a series of DIBA-derived red-emitting fluorescent MR ligands, comprising compounds 7 and 8 (Fig. 1). Whereas the indolinium-type cyanine dye containing ligands (e.g.8) were investigated in fluorescence-based techniques, the pyridinium-type cyanine dye containing probes (e.g.7) were only characterized with respect to M1R–M5R affinity in radioligand competition binding studies.47 In the present study, a new series of fluorescently labelled dibenzodiazepinone-type MR ligands was prepared using various fluorescent dyes (TAMRA, BODIPY630/650, pyridinium-type cyanine dyes Py-1 and Py-5). The probes were characterized with respect to M1R–M5R affinity (radioligand competition binding) and by flow cytometric saturation binding studies, the latter including the reported ligand 7.
Fig. 1. Structures and MR binding data of the reported fluorescent probes 1–5, being non-selective MR ligands (1–4) or showing a slight preference for the M1R (2),33 and structures and MR affinities of the M2R preferring dibenzodiazepinone-type MR ligands DIBA and the DIBA-derived fluorescent probes 7 and 8.47 (Note: binding data for all MR subtypes were not reported in all cases). a Tahtaoui et al. (the reported Ki value was converted to a pKi value);32b Jones et al. (reported Ki values were converted to pKi values);33c Hern et al. (reported Ki values were converted to pKi values);34d Nenasheva et al.;35e Daval et al.;37f Gitler et al. (reported Ki values were converted to pKi values);42g She et al.47.
Results and discussion
Chemistry
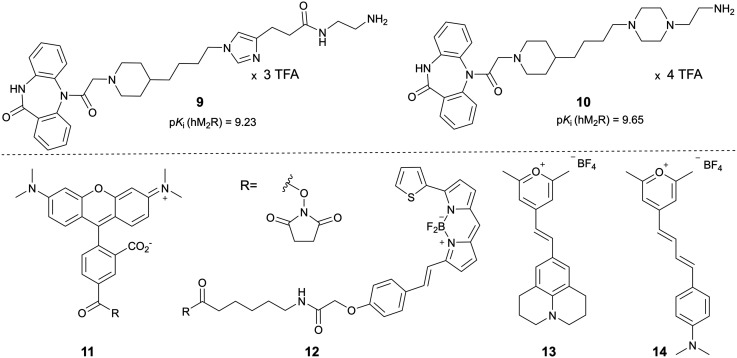
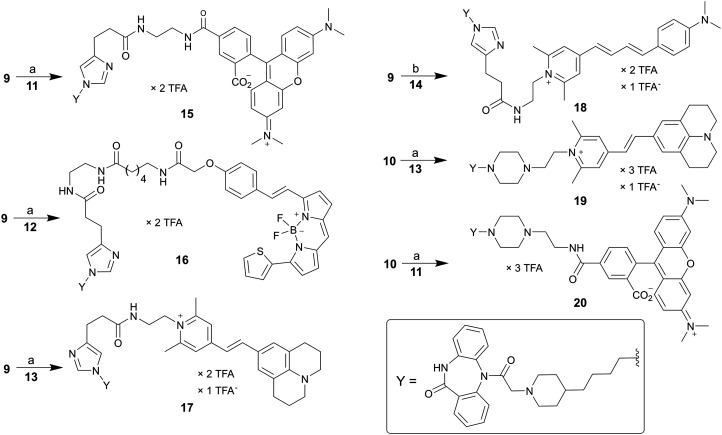
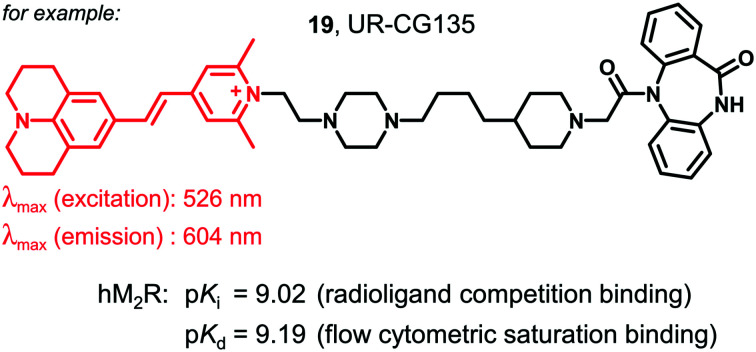
A set of six fluorescent dibenzodiazepinone-type MR ligands (15–20) was prepared from the previously reported amine-functionalized dibenzodiazepinone derivatives 9 (ref. 43) and 10 (ref. 46) using four different fluorescent dyes: 5-TAMRA succinimidyl ester (11), BODIPY 630/650 succinimidyl ester (12), and the pyrylium dyes Py-1 (13)48 and Py-5 (14)48 (cf.Fig. 2). Treatment of 9 with the fluorescent dyes 11–14 in the presence of DIPEA or triethylamine yielded the fluorescent ligands 15–18 (Scheme 1). Likewise, treatment of 10 with 13 or 11 gave the fluorescent probes 19 and 20, respectively (Scheme 1). Purification by preparative RP-HPLC afforded 15–20 with high purities (≥96%, HPLC analysis at 220 nm). All fluorescent ligands were investigated with respect to their chemical stability in PBS pH 7.4. Whereas compounds 15, 16, 19 and 20 showed no decomposition within the incubation period of 48 h (Fig. S2, ESI†), compounds 17 and 18 showed minor decomposition after 24 h (after 24 h, peak areas (220 nm) of 17 and 18 amounted to 94% and 90%, respectively, of the total peak area).
Fig. 2. Structures and M2R affinities of the previously described DIBA derived amine-functionalized precursors 9 and 10,43,46 and structures of the fluorescent dyes 11–14, which were used for the preparation of the fluorescent MR ligands 15–20 (note: M2R binding data of 9, reported as pIC50 value,43 were re-analyzed to obtain the pKi value).
Scheme 1. Synthesis of the fluorescent dibenzodiazepinone-type MR ligands 15–20, labelled with TAMRA (15, 20), BODIPY630/650 (16), pyrylium/pyridinium dye Py-1 (17, 19) or Py-5 (18). Reagents and conditions: (a): DIPEA, DMF, rt, 2 h, 31% (15), 23% (16), 16% (17), 23% (19), 66% (20); (b) triethylamine, DMF, rt, 2 h, 28% (18).
Radioligand competition binding studies with [3H]NMS
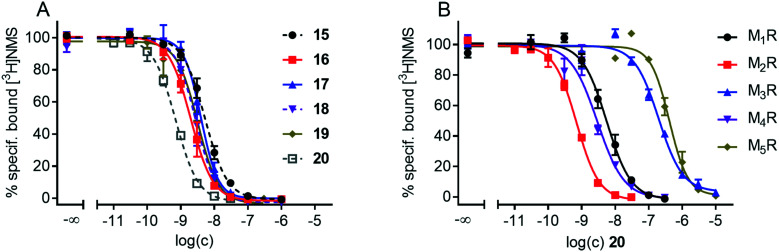
M1R–M5R affinities of 15–20 were determined at intact CHO–hMxR cells (x = 1–5) using the orthosteric non-selective MR antagonist [3H]NMS as radioligand. Compounds 15–20 were capable of completely displacing [3H]NMS from all MR subtypes (Fig. 3 and Fig. S1, ESI†). The corresponding pKi values are listed in Table 1. All fluorescent ligands 15–20 exhibited high M2R affinity (pKi values >8.7). The TAMRA-labelled compound 20 showed the highest M2R affinity (pKi: 9.62). For this fluorescent ligand, sigmoidal [3H]NMS displacement curves (M1R–M5R) are shown in Fig. 3B. As in the case of the recently reported series of dibenzodiazepinone-type fluorescent ligands including 7 and 8,4715–20 showed no or very low preference for the M2R compared to the M1 and M4 receptor, but moderate to pronounced M2R selectivity towards the M3 and M5 subtypes (Table 1). Compound 20 exhibited the highest, but still moderate preference for the M2R over the M1R and M4R.
Fig. 3. (A) Concentration-dependent effects of 15–20 on [3H]NMS (c = 0.2 nM) equilibrium M2R binding determined at intact CHO–hM2R cells. (B) Radioligand displacement curves obtained from competition binding experiments with [3H]NMS (0.2 nM (M1R, M2R, M3R), 0.1 nM (M4R) or 0.3 nM (M5R)) and compound 20 performed at intact CHO–hMxR cells (x = 1–5). M2R data are the same as in A. Data in A and B represent mean values ± SEM from two (20, M5R) or at least three independent experiments (each performed in triplicate).
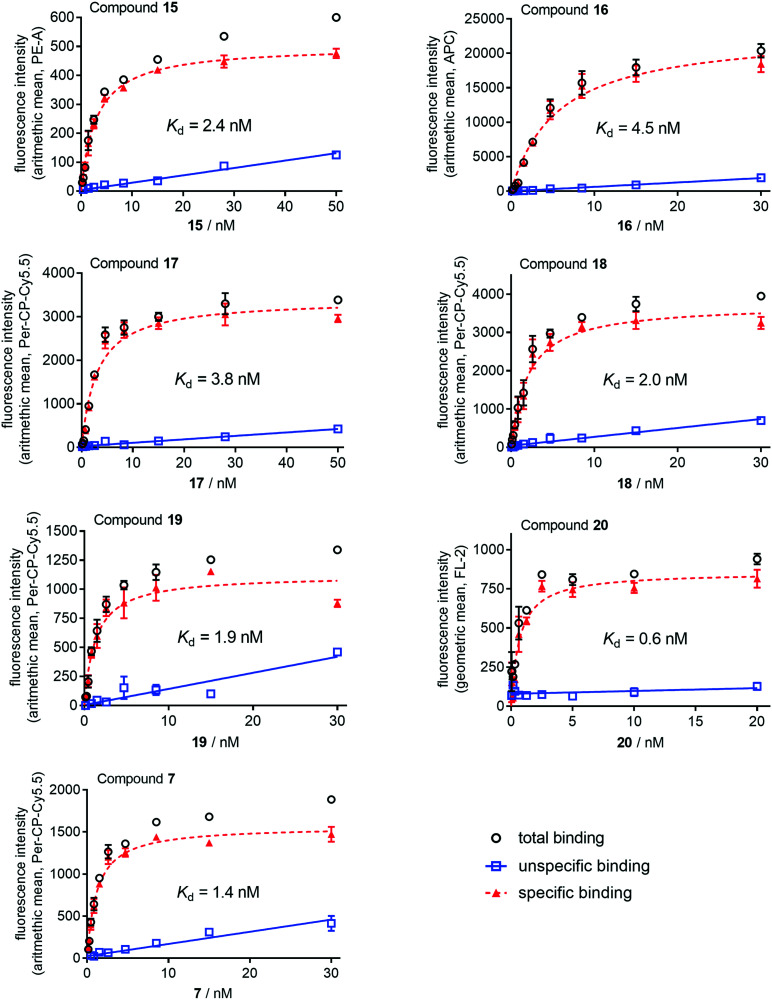
Table 1. M1–M5 receptor binding data (pKi values) of compounds 7 and 15–20 obtained from radioligand competition binding studies, and M2R binding data (pKd values) from flow cytometric saturation binding experiments.
| Compd. | pKi
a
|
pKd b | ||||
| M1R | M2R | M3R | M4R | M5R | M2R | |
| 7 (ref. 47) | 7.86 | 8.52 | 6.83 | 8.02 | 6.41 | 8.63 ± 0.08 |
| 15 (UR-CG072) | 8.21 ± 0.02 | 8.75 ± 0.07 | 6.89 ± 0.06 | 8.43 ± 0.07 | 6.56 ± 0.10 | 8.36 ± 0.09 |
| 16 (UR-CG073) | 9.00 ± 0.13 | 9.16 ± 0.10 | 7.43 ± 0.13 | 9.38 ± 0.05 | 8.00 ± 0.08 | 8.41 ± 0.10 |
| 17 (UR-CG074) | 8.15 ± 0.05 | 8.87 ± 0.08 | 7.16 ± 0.01 | 8.58 ± 0.08 | 7.39 ± 0.07 | 8.70 ± 0.04 |
| 18 (UR-AP175) | 8.31 ± 0.04 | 9.04 ± 0.15 | 7.07 ± 0.04 | 8.37 ± 0.04 | 7.15 ± 0.06 | 8.74 ± 0.13 |
| 19 (UR-CG135) | 8.32 ± 0.03 | 9.02 ± 0.10 | 7.20 ± 0.15 | 8.57 ± 0.01 | 6.90 ± 0.03 | 9.19 ± 0.03 |
| 20 (UR-MK342) | 8.59 ± 0.10 | 9.62 ± 0.04 | 7.09 ± 0.06 | 9.01 ± 0.03 | 6.75 ± 0.05 | 8.86 ± 0.06 |
aDetermined by competition binding with [3H]NMS (Kd values/applied concentrations: M1, 0.17/0.2 nM; M2, 0.10/0.2 nM; M3, 0.12/ 0.2 nM; M4, 0.052/0.1 nM; M5, 0.20/0.3 nM) at whole CHO–hMxR cells (x = 1-5) at 23 °C. Means ± SEM from two (20, M5R) or at least three independent experiments (each performed in triplicate).
bDetermined by flow cytometric saturation binding experiments at intact CHO–hM2R cells at 22 °C. Means ± SEM from two (20) or at least three independent experiments (performed in duplicate).
Fluorescence properties
Excitation and corrected emission spectra of 15–20, recorded in PBS containing 1% bovine serum albumin (BSA), are shown in ESI† Fig. S3 and the respective excitation and emission maxima are summarized in Table 2. It should be noted that compounds 7 and 18, both labelled with the pyrylium/pyridinium-type fluorescent dye Py-5 are perfectly suited for an excitation with an argon laser (488 nm), belonging to the standard equipment of many instruments. Compound 16 is well suited for an excitation with the commonly used red diode laser (ca. 635 nm), and 15, 17, 19 and 20 require green light for an optimal excitation (cf.Table 2).
Table 2. Excitation and emission maxima of the fluorescent ligands 7, 15–20 determined in PBS containing 1% BSA.
| Compd. | Dye | λ ex/λem |
| 7 (ref. 47) | Pyridinium cyanine (Py-5) | 484/643 |
| 15 | 5-TAMRA | 557/583 |
| 16 | BODIPY 630/650 | 639/647 |
| 17 | Pyridinium cyanine (Py-1) | 526/602 |
| 18 | Pyridinium cyanine (Py-5) | 483/634 |
| 19 | Pyridinium cyanine (Py-1) | 526/604 |
| 20 | 5-TAMRA | 550/583 |
Flow cytometric M2R saturation binding studies
M2R affinities of the fluorescent ligands 7 and 15–20 were also determined by flow cytometric saturation binding studies at intact CHO–hM2R cells at 22 °C using a FACSContoII (7, 15–19) or a FACSCalibur (20) flow cytometer, both equipped with two light sources (argon and red diode laser) (cf.Fig. 4). For these experiments, an incubation period of 2 h was applied because association binding experiments with recently reported radio- and fluorescence labelled dibenzodiazepinone-type MR ligands at intact CHO–hM2R cells or cell homogenates revealed that equilibrium was reached within 2 h.45–47 The obtained pKd values of compounds 7 and 15–20 were in good agreement with the corresponding pKi values obtained from competition binding studies with [3H]NMS (Table 1).
Fig. 4. Representative saturation isotherms (specific binding, dashed line) obtained from flow cytometric saturation binding experiments performed with 7 and 15–20 at intact CHO–hM2R cells. Unspecific binding was determined in the presence of atropine (21, for structure see Fig. S4, ESI†) used in 500- or 1000-fold excess. Cells were incubated with the fluorescent ligands at 22 °C in the dark for 2 h. Experiments were performed in duplicate. Measurements were performed on a FACSCantoII (7, 15–19) or a FACSCalibur (20) flow cytometer (Becton Dickinson). Used laser lines/emission filters: 15, 488 nm/585 ± 21 nm (PE channel); 16, 488 nm/660 ± 10 nm (APC channel); 7 and 17–19, 488 nm/670 ± 65 nm (PerCP–Cy5 channel); 20, 488 nm/585 ± 21 nm (channel FL-2). Data represent mean values ± SEM (total and unspecific binding) or calculated values ± propagated error (specific binding). Note: total and unspecific binding data represent autofluorescence-corrected data.
Conclusion
The synthesized series of fluorescence-labelled dibenzodiazepinone-type M2R ligands represents an extension of a recently reported set of fluorescent MR ligands47 with respect to the type of attached fluorescent dyes. As the 5-TAMRA fluorophore exhibits a fluorescence life-time (ca. 2.4 ns), which is well compatible with fluorescence anisotropy measurements,49 the TAMRA-labelled MR ligands (15, 20) will be characterized in fluorescence anisotropy-based assays in future studies including real-time kinetic measurements. Furthermore, the Py-5 labelled probes (7, 18), exhibiting an excitation maximum around 480 nm and a large Stokes shift (emission maximum ≈ 630 nm) should be ideal probes for a recently reported BRET-based GPCR binding assay,4 requiring fusion of the NanoLuciferase (λmax (bioluminescence) ≈ 460 nm) to the M2R.
Experimental section
General experimental conditions
All chemicals and solvents were purchased from commercial suppliers and were used without further purifications. Acetonitrile for HPLC (gradient grade) was obtained from Merck (Darmstadt, Germany) or Sigma-Aldrich (Taufkirchen, Germany). Atropine (21) was purchased from Sigma-Aldrich. The 5-carboxytetramethylrhodamine succinimidyl ester (11) was purchased from ABCR (Karlsruhe, Germany) and BODIPY 630/650 succinimidyl ester (12) was purchased from Lumiprobe (Hannover, Germany). The pyrylium dyes 9 and 10 were prepared according to described procedures.48 [3H]NMS (specific activity = 80 Ci/mmol) was purchased from American Radiolabeled Chemicals Inc. (St. Louis, MO) via Hartman Analytics (Braunschweig, Germany). Millipore water was used throughout for the preparation of stock solutions and HPLC eluents. Polypropylene reaction vessels (1.5 or 2 mL) with screw cap (Süd-Laborbedarf, Gauting, Germany) were used for the synthesis of all fluorescent ligands and for the preparation and storage of stock solutions. NMR spectra were recorded on a Bruker Avance III HD 600 equipped with a cryogenic probe (14.1 T; 1H: 600 MHz) (Bruker, Karlsruhe, Germany). Abbreviations for the multiplicities of the signals are s (singlet), d (doublet), t (triplet), m (multiplet) and brs (broad singlet). High-resolution mass spectrometry (HRMS) analysis was performed on an Agilent 6540 UHD Accurate-Mass Q-TOF LC/MS system (Agilent Technologies, Santa Clara, CA) using an ESI source. Preparative HPLC of compounds 15–20 was performed with a Prep 150 LC system from Waters (Eschborn, Germany) consisting of a binary gradient module, a 2489 UV/visible detector, and a Waters fraction collector III. Compounds 18 and 20 were purified with a preparative HPLC system from Knauer (Berlin, Germany) consisting of two K-1800 pumps and a K-2001 detector. For both systems a Kinetex-XB C18 (5 μm, 250 mm × 21 mm; Phenomenex, Aschaffenburg, Germany) served as stationary phases at a flow rate of 20 mL min–1. Mixtures of 0.1% aq TFA and acetonitrile were used as mobile phase. The detection wavelength was set to 220 nm throughout. The solvent of collected fractions was removed by lyophilization using a Scanvac freeze drying apparatus (Labogene, Allerød, Denmark) equipped with a RZ 6 rotary vane vacuum pump (Vacuubrand, Wertheim, Germany). Analytical HPLC analysis was performed with a system from Agilent Technologies composed of a 1290 Infinity binary pump equipped with a degasser, a 1290 Infinity autosampler, a 1290 Infinity thermostated column compartment, a 1260 Infinity diode array detector, and a 1260 Infinity fluorescence detector. A Kinetex-XB C18 (2.6 μm, 100 × 3 mm; Phenomenex) was used as stationary phase at a flow rate of 0.6 mL min–1. Mixtures of 0.04% aq TFA (A) and acetonitrile (B) were used as mobile phase. The following linear gradients were applied: compounds 15–17, 19 and 20 (purity): 0–12 min: A/B 90 : 10 to 55 : 45, 12–16 min: 55 : 45 to 5 : 95, 16–20 min: 5 : 95; compound 18 (purity): 0–15 min: A/B 90 : 10 to 50 : 50, 15–19 min: 50 : 50 to 5 : 95; 20–22 min: 5 : 95; compounds 15–20 (chemical stabilities): 0–20 min: A/B 90 : 10 to 35 : 65, 20–22 min: 35 : 65 to 5 : 95, 22–26 min: 5 : 95. For all analytical HPLC runs the oven temperature was set to 25 °C, the injection volume was 20 μL and detection was performed at 220 nm. The stock solutions (final concentration: 1 mM) of fluorescent ligands were prepared in DMSO and were stored at –78 °C.
Annotation concerning the 1H-NMR spectra of the dibenzodiazepinone derivatives 15–20: due to a slow rotation about the exocyclic amide group of the diazepinone ring on the NMR time scale, two isomers (ratios provided in the experimental protocols) were evident in the 1H-NMR spectra.
Compounds characterization
All target compounds (15–20) were characterized by 1H-NMR spectroscopy, HRMS, and RP-HPLC analysis. The HPLC purity of all fluorescent ligands amounted to ≥96% (220 nm) (chromatograms shown in the electronic ESI†).
Experimental synthetic protocols and analytical data of compounds 15–20
2-(6-(Dimethylamino)-3-(dimethyliminio)-3H-xanthen-9-yl)-5-((2-(3-(1-(4-(1-(2-oxo-2-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-5-yl)ethyl)piperidin-4-yl)butyl)-1H-imidazol-4-yl)propanamido)ethyl)carbamoyl)benzoate bis(hydrotrifluoroacetate) (15)
The reaction was carried out in a 1.5 mL propylene reaction vessel equipped with a micro stir bar. Amine precursor 9 (tris(hydrotrifluoroacetate), 14.5 mg, 0.016 mmol) and DIPEA (10.3 mg, 14 μL, 0.089 mmol) were dissolved in anhydrous DMF (90 μL) followed by the addition of 11 (4.2 mg, 0.008 mmol) dissolved in anhydrous DMF (50 μL). After stirring at room temperature in the dark for 2 h, 10% aq TFA (corresponding to 0.09 mmol of TFA) were added.
Purification of the product by preparative HPLC (gradient: 0–40 min: 0.1% aq TFA/acetonitrile 81 : 19-28 : 72, tR = 13 min) yielded compound 15 as a red solid (3.0 mg, 31%). Ratio of configurational isomers evident in the 1H-NMR spectrum: ca. 1.5 : 1. 1H-NMR (600 MHz, MeOH-d4): δ (ppm) 1.33–1.40 (m, 4H), 1.44–1.57 (m, 3H), 1.85–1.93 (m, 3H), 1.93–1.99 (m, 1H), 2.62 (t, 2H, J 7.2 Hz), 2.89–2.98 (m, 1H), 3.00 (t, 2H, J 7.2 Hz), 3.02-3.10 (m, 1H), 3.45(t, 3H, J 6.0 Hz, interfering with the 13C satellite of the solvent residual peak), 3.56 (t, 2H, J 6.1 Hz), 3.72 (d, 1.4H, J 7.2 Hz), 3.80 (d, 0.60H, J 17 Hz), 4.18 (t, 2H, J 7.2 Hz), 4.37–4.46 (m, 1H), 7.0–7.02 (m, 2H), 7.03–7.08 (m, 2H), 7.11–7.14 (m, 2H), 7.24–7.36 (m, 2H), 7.36–7.43 (m, 1.4H), 7.46–7.55 (m, 3.2H), 7.59–7.65 (m, 1.4H), 7.65–7.70 (m, 0.6H), 7.72–7.77 (m, 0.4H), 7.87–7.92 (m, 0.6H), 7.95–7.99 (m, 0.4H), 8.22–8.26 (m, 1H), 8.73–8.77 (m, 1H), 8.80 (s, 1H). Note: exchangeable protons (NH, OH) were not apparent. The proton signals of the four methyl groups (12 protons) were not apparent due to an interference with the solvent residual peak. HRMS (ESI): m/z [M + H]+ calcd. for [C57H62N9O7]+: 984.4767, found: 984.4757. RP-HPLC (220 nm): 97% (tR = 7.40 min, k = 8.7). C57H61N9O7·C4H2F6O4 (984.17 + 228.04).
(E)-6-(2-(4-(2-(5,5-Difluoro-7-(thiophen-2-yl)-5H-4λ4,5λ4-dipyrrolo[1,2-c:2′,1′-f][1,3,2]diazaborinin-3-yl)vinyl)phenoxy)acetamido)-N-(2-(3-(1-(4-(1-(2-oxo-2-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-5-yl)ethyl)piperidin-4-yl)butyl)-1H-imidazol-4-yl)propanamido)ethyl)hexanamide bis(hydrotrifluoroacetate) (16)
Compound 16 was prepared from 9 (tris(hydrotrifluoroacetate), 8.6 mg, 0.009 mmol) and 12 (3.1 mg, 0.0047 mmol) according to the procedure used for the synthesis of 15. DIPEA: 6.1 mg, 8.2 μL, 0.047 mmol. Purification of the product by preparative HPLC (gradient: 0-40 min: 0.1% aq TFA/acetonitrile 81:19-19:81, tR = 22 min) yielded 16 as a blue solid (1.35 mg, 23%). Ratio of configurational isomers evident in the 1H-NMR spectrum: ca. 1.5 : 1. 1H-NMR (600 MHz, MeOH-d4): δ (ppm) 1.25–1.33 (m, 7H), 1.42–1.51 (m, 2H), 1.52–1.64 (m, 4H), 1.77–1.93 (m, 4H), 2.16 (t, 2H, J 7.2 Hz), 2.52 (t, 2H, J 7.2 Hz), 2.81–2.88 (m, 1H), 2.92 (t, 2H, J 7.2 Hz), 2.95–3.02 (m, 1H), 3.21–3.25 (m, 4H, interfering with the solvent peak), 3.26–3.28 (m, 3H, interfering with the solvent peak), 3.36–3.41 (m, 1H, interfering with the 13C satellite of the solvent residual peak), 3.66–3.72 (m, 1H); 4.09 (t, 2H, J 7.2 Hz), 4.33–4.42 (m, 1H), 4.58 (s, 2H), 6.87 (d, 1H, J 4.2 Hz), 7.03-7.07 (m, 2H), 7.15 (d, 2H, J 4.2 Hz), 7.19–7.23 (m, 2H), 7.25–7.35 (m, 3H), 7.39 (s, 1H), 7.39–7.42 (m, 0.4H), 7.46–7.51 (m, 1.6H), 7.52–7.66 (m, 7H), 7.66–7.71 (m, 0.6H), 7.71–7.76 (m, 0.4H), 7.89–7.94 (m, 0.6H), 7.95–8.00 (m, 0.4H), 8.11 (d, 1H, J 3.9 Hz), 8.72 (s, 1H). Note: exchangeable protons (NH) were not apparent. HRMS (ESI): m/z [M + H]+ calcd. for [C61H68BF2N10O6S]+: 1117.5100, found: 1117.5110. RP-HPLC (220 nm): 99% (tR = 7.62 min, k = 9.0). C61H67BF2N170O6S·C4H2F6O4 (1117.14 + 228.04).
(E)-2,6-Dimethyl-1-(2-(3-(1-(4-(1-(2-oxo-2-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-5-yl)ethyl)piperidin-4-yl)butyl)-1H-imidazol-4-yl)propanamido)ethyl)-4-(2-(2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-9-yl)vinyl)pyridin-1-ium bis(hydrotrifluoroacetate) trifluoroacetate (17)
Compound 17 was prepared from 9 (tris(hydrotrifluoroacetate), 28 mg, 0.031 mmol) and 13 (17 mg, 0.043 mmol) according to the procedure used for the synthesis of 15. DIPEA: 40 mg, 53 μL, 0.31 mmol. Purification of the product by preparative HPLC (gradient: 0–40 min: 0.1% aq TFA/acetonitrile 85 : 15–38 : 62, tR = 23 min) yielded 17 as a red solid (5.4 mg, 16%). Ratio of configurational isomers evident in the 1H-NMR spectrum: ca. 1.5 : 1. 1H-NMR (600 MHz, MeOH-d4): δ (ppm) 1.32-1.39 (m, 4H), 1.41–1.56 (m, 3H), 1.81–1.92 (m, 3H), 1.93–2.00 (m, 5H), 2.60 (t, 2H, J 7.3 Hz), 2.75 (t, 4H, J 6.6 Hz), 2.82 (s, 6H), 2.89–2.96 (m, 3H), 2.99–3.06 (m, 1H), 3.45 (t, 1H, J 13 Hz, interfering with the 13C satellite of the solvent residual peak), 3.62 (t, 2H, J 7.2 Hz), 3.69–3.81 (m, 2H), 4.14 (t, 2H, J 7.3 Hz), 4.38 (s, 0.4 H), 4.40–4.42 (m, 0.6H), 4.43–4.48 (m, 2H), 6.83 (d, 1H, J 16 Hz), 7.13 (s, 2H), 7.25–7.41 (m, 3.6H), 7.47–7.55 (m, 2.4H), 7.61–7.67 (m, 4H), 7.67–7.71 (m, 0.6H), 7.73–7.77 (m, 0.4H), 7.89–7.92 (m, 0.6H), 7.97 (d, 0.4H, J 7.6 Hz), 8.79 (s, 1H). Note: exchangeable protons (NH) were not apparent. The proton signals of two methylene groups (4 protons) of the fluorophore were not apparent due to an interference with the solvent residual peak. HRMS (ESI): m/z [M]+ calcd. for [C53H63N8O3]+: 859.5018, found: 859.5026. RP-HPLC (220 nm): 96% (tR = 9.5 min, k = 12.0). C53H63N8O3+·C2F3O2–·C4H2F6O4 (860.14 + 341.07).
4-((1E,3E)-4-(4-(Dimethylamino)phenyl)buta-1,3-dien-1-yl)-2,6-dimethyl-1-(2-(3-(1-(4-(1-(2-oxo-2-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-5-yl)ethyl)piperidin-4-yl)butyl)-1H-imidazol-4-yl)propanamido)ethyl)pyridin-1-ium bis(hydrotrifluoroacetate)trifluoroacetate (18)
Compound 18 was prepared from 9 (tris(hydrotrifluoroacetate), 6.4 mg, 0.007 mmol) and 14 (7.7 mg, 0.021 mmol) following the procedure used for the synthesis of 15, but triethylamine (7.1 mg, 9.8 μL, 0.07 mmol) was used instead of DIPEA. Purification of the product by preparative HPLC (gradient: 0–40 min: 0.1% aq TFA/acetonitrile 81 : 19–40 : 60, tR = 16 min) yielded 18 as a red solid (2.3 mg, 28%). Ratio of configurational isomers evident in the 1H-NMR spectrum: ca. 1.5 : 1. 1H-NMR (600 MHz, MeOH-d4): δ (ppm) 1.32–1.38 (m, 4H), 1.39–1.55 (m, 3H), 1.82–1.89 (m, 3H), 1.89–1.97 (m, 1H), 2.60 (t, 2H, J 7.3 Hz), 2.85 (s, 6H), 2.89–2.95 (m, 3H), 3.02 (s, 7H), 3.44 (t, 1H, J 13 Hz, interfering with the 13C satellite of the solvent residual peak), 3.64 (t, 2H, J 7.2 Hz), 3.68–3.80 (m, 2H), 4.15 (t, 2H, J 7.4 Hz), 4.36–4.45 (m, 1H), 4.50 (t, 2H, J 7.1 Hz), 6.57 (d, 1H, J 15 Hz), 6.76 (d, 2H, J 8.8 Hz), 6.92–6.99 (m, 1H), 7.02 (d, 1H, J 15 Hz), 7.25–7.41 (m, 3.6H), 7.44 (d, 2H, J 8.9 Hz), 7.47–7.55 (m, 2.4H), 7.61–7.66 (m, 1.6H), 7.66–7.72 (m, 3H), 7.73–7.78 (m, 0.4H), 7.89–7.92 (m, 0.6H), 7.98 (d, 0.4H, J 8.4 Hz), 8.79 (s, 1H). Note: exchangeable protons (NH) were not apparent. HRMS (ESI): m/z [M]+ calcd. for [C51H61N8O3]+: 833.4861, found 833.4879. RP-HPLC (220 nm): 98% (tR = 8.4 min, k = 10.0). C51H61N8O3+·C2F3O2–·C4H2F6O4 (834.10 + 341.07).
(E)-2,6-Dimethyl-1-(2-(4-(4-(1-(2-oxo-2-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-5-yl)ethyl)piperidin-4-yl)butyl)piperazin-1-yl)ethyl)-4-(2-(2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-9-yl)vinyl)pyridin-1-ium tris(hydrotrifluoroacetate)trifluoroacetate (19)
Compound 19 was prepared from 10 (17 mg, 0.018 mmol), and 13 (10 mg, 0.026 mmol) according to the procedure used for the synthesis of 15. DIPEA: 23 mg, 31 μL, 0.18 mmol. Purification of the product by preparative HPLC (gradient: 0–40 min: 0.1% aq TFA/acetonitrile 81 : 19–29 : 71, tR = 18 min) yielded 19 as a red solid (5.1 mg, 23%). Ratio of configurational isomers evident in the 1H-NMR spectrum: ca. 1.5 : 1. 1H-NMR (600 MHz, MeOH-d4): δ (ppm) 1.33–1.57 (m, 7H), 1.68–1.77 (m, 2H), 1.90–2.00 (m, 6H), 2.56–2.63 (m, 2H), 2.75 (t, 4H, J 6.3 Hz), 2.80 (s, 6H), 2.92 (t, 3H, J 6.7 Hz), 2.98–3.18 (m, 8H), 3.45 (t, 1H, J 12 Hz, interfering with the 13C satellite of the solvent residual peak), 3.52–3.59 (m, 2H), 3.70–3.82 (m, 2H), 4.37–4.46 (m, 1H), 4.51 (t, 2H, J 6.6 Hz), 6.84 (d, 1H, J 16 Hz), 7.13 (s, 2H), 7.24–7.32 (m, 1H), 7.32–7.36 (m, 1H), 7.40 (t, 0.4H, J 7.7 Hz), 7.47–7.56 (m, 2.2H), 7.60–7.72 (m, 5H), 7.76 (t, 0.4H, J 7.8 Hz), 7.91 (d, 0.6H, J 8.2 Hz), 7.98 (d, 0.4H, J 8.2 Hz). Note: exchangeable protons (NH) were not apparent. The proton signals of two methylene groups (4 protons) of the fluorophore were not apparent due to an interference with the solvent residual peak. HRMS (ESI): m/z [M]+ calcd. for [C51H64N7O2]+: 806.5116, found 806.5121. RP-HPLC (220 nm): 97% (tR = 9.6 min, k = 12.0). C51H64N7O2+·C2F3O2–·C6H3F9O6 (807.12 + 455.09).
2-(6-(Dimethylamino)-3-(dimethyliminio)-3H-xanthen-9-yl)-5-((2-(4-(4-(1-(2-oxo-2-(11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepin-5-yl)ethyl)piperidin-4-yl)butyl)piperazin-1-yl)ethyl)carbamoyl)benzoate tris(hydrotrifluoroacetate) (20)
Compound 20 was prepared from 10 (8.9 mg, 0.010 mmol) and 11 (4.1 mg, 0.0077 mmol) according to the procedure used for the synthesis of 15. DIPEA: 11 mg, 14 μL, 0.083 mmol. Purification of the product by preparative HPLC (gradient: 0–40 min: 0.1% aq TFA/acetonitrile 81 : 19–30 : 40, tR = 13 min) yielded 20 as a red solid (6.5 mg, 66%). Ratio of configurational isomers evident in the 1H-NMR spectrum: ca. 1.5 : 1. 1H-NMR (600 MHz, MeOH-d4): δ (ppm) 1.35–1.43 (m, 4H), 1.44–1.61 (m, 3H), 1.68–1.77 (m, 2H), 1.89–2.02 (m, 2H), 2.61 (brs, 2H), 2.83 (s, 2H), 2.86–3.00 (m, 1H), 3.00–3.12 (m, 4H), 3.45 (t, 2H, J 12 Hz, interfering with the 13C satellite of the solvent residual peak), 3.66 (t, 2H, J 6.2 Hz), 3.70–3.83 (m, 2H), 4.40 (d, 0.6H, J 17 Hz), 4.44 (d, 0.4H, J 17 Hz), 7.00 (d, 2H, J 2.2 Hz), 7.05 (d, 0.8H, J 2.2 Hz), 7.07 (d, 1.2H, J 2.3 Hz), 7.13 (d, 2H, J 9.4 Hz), 7.25–7.36 (m, 2H), 7.39 (t, 0.4H, J 7.8 Hz), 7.47–7.56 (m, 3.2H), 7.60–7.67 (m, 1.4H), 7.70 (t, 0.6H, J 7.4 Hz), 7.76 (t, 0.4H, J 7.6 Hz), 7.91 (d, 0.6H, J 7.8 Hz), 7.98 (d, 0.4H, J 7.7 Hz), 8.23–8.28 (m, 1H), 8.77 (s, 1H). Note: exchangeable protons (NH, OH) were not apparent. The proton signals of the four methyl groups (12 protons) and four protons of the piperazine ring (CH2 groups) were not apparent due to an interference with the solvent residual peak. HRMS (ESI): m/z [M + H]+ calcd. for [C55H63N8O6]+: 931.4865, found 931.4868. RP-HPLC (220 nm): 99% (tR = 6.9 min, k = 8.1). C55H62N8O6·C6H3F9O6 (931.15 + 342.06).
Investigation of the chemical stability
The chemical stabilities of 15–20 were investigated in PBS pH 7.4 at 22 ± 1 °C using propylene vessels. The incubation was started by the addition of 10 μL of 1 mM solution of the fluorescent ligand in DMSO to PBS (90 μL) to yield a final concentration of 100 μM. After 0, 24 and 48 h, aliquots (20 μL) were taken and added to 1% aq TFA/acetonitrile (8 : 2 v/v) (20 μL). The resulting solutions were analyzed by RP-HPLC (analytical HPLC system and conditions see general experimental conditions; tR: 7.8 min (15), 13.5 min (16), 9.95 min (17), 8.1 min (18), 10.0 min (19), 7.3 min (20)).
Cell culture
CHO–K9 cells, stably transfected with the DNA of human muscarinic receptors M1–M5 (obtained from Missouri S&T cDNA Resource Center; Rolla, MO) were cultured in HAM's F12 medium supplemented with fetal calf serum (Biochrom, Berlin, Germany) (10%) and G418 (Biochrom) (750 μg mL–1).
Determination of excitation and emission spectra
Excitation and emission spectra of compounds 15–20 were recorded in PBS, pH 7.4, containing 1% BSA (Serva, Heidelberg, Germany), at 22 °C with a Cary Eclipse spectrofluorimeter (Varian Inc., Mulgrave, Victoria, Australia) using acryl cuvettes (10 × 10 mm, Ref. 67.755, Sarstedt, Nümbrecht, Germany). The slit adjustments (excitation/emission) were 5/10 nm for excitation spectra and 10/5 nm in case of emission spectra. Net spectra were calculated by subtracting the respective vehicle reference spectrum, and corrected emission spectra were calculated by multiplying the net emission spectra with the respective lamp corrections spectrum (same slit adjustments, etc.).
Radioligand competition binding assay
Equilibrium competition binding studies with [3H]NMS were performed at intact CHO–hMxR cells (x = 1–5) at 23 ± 1 °C in white 96-wells plates with clear bottom (Corning Life Science, Tewksbury, MA; Corning cat. No. 3610) using Leibovitz's L-15 medium (Gibco, Life Technologies, Darmstadt, Germany) supplemented with 1% BSA (Serva) as binding buffer (in the following referred to as L15 medium). Experiments were performed using a previously described protocol for MR binding studies with [3H]NMS,43 but the total volume per well was 200 μL, i.e. wells were pre-filled with 180 μL of L15 medium followed by the addition of L15 medium (20 μL) containing [3H]NMS (10-fold concentrated), to determine total binding, or pre-filled with 160 μL of L15 medium followed by the addition of L15 medium (20 μL) containing atropine or the compound of interest (10-fold concentrated) and L15 medium (20 μL) containing [3H]NMS (10-fold-concentrated), to determine unspecific binding and the displacing effect of a compound of interest, respectively. The concentrations of [3H]NMS were 0.2 nM (M1R, M2R, M3R), 0.1 nM (M4R) or 0.3 nM (M5R). Samples were incubated in the dark under gentle shaking for 3 h. Prior to the competition binding experiments, the Kd values of [3H]NMS were determined by saturation binding applying the same conditions (buffer, temperature, incubation time, unspecific binding, etc.). The obtained Kd values amounted to 0.17 ± 0.01 nM (M1R), 0.10 ± 0.01 nM (M2R), 0.12 ± 0.01 nM (M3R), 0.052 ± 0.01 nM (M4R) and 0.20 ± 0.02 nM (M5R) (mean value ± SEM from at least four independent determinations performed in triplicate), being in excellent agreement with previously determined Kd values of [3H]NMS.43
Flow cytometric saturation binding experiments
Flow cytometric M2R binding studies were performed with a FACSCantoII flow cytometer (Becton Dickinson, Heidelberg, Germany) (compounds 7 and 15–19) or with a FACSCalibur flow cytometer (Becton Dickinson) (20), both equipped with an argon laser (488 nm) and a red diode laser (640 and 635 nm, respectively). Fluorescence signals were recorded using the following instrument settings: compound 15, excitation: 488 nm, emission: 585 ± 21 nm (PE channel), gain: 385–440 V; compound 16, excitation: 633 nm, emission: 660 ± 10 nm (APC channel), gain: 480–510 V; compounds 7 and 17–19, excitation: 488 nm, emission: 670 ± 65 nm (PerCP–Cy5.5 channel), gain: 430 V (7) or 465–485 V (17-19); compound 20, excitation: 488 nm, emission: 585 ± 21 nm (FL-2), gain: 750 V. Measurements were stopped after counting of 10 000 gated events at medium (7, 17–19) or high (20) flow rate.
All samples were prepared and incubated in 1.5 mL reaction vessels (Sarstedt). Cells were seeded in a 175 cm2 culture flask 5–6 days prior to the experiment. On the day of the experiment, cells were treated with trypsin, detached and suspended in culture medium followed by centrifugation. The cell pellet was re-suspended in Leibovitz's L15 culture medium (Gibco, Life Technologies) supplemented with 1% BSA (Serva) (in the following referred to as L15 medium). The cell density was adjusted to 1 × 106 cells per mL. For the determination of total binding, 2 μL of a solution of fluorescent ligand (100-fold concentrated compared to the final concentration) in DMSO/H2O (1 : 1 v/v) and 2 μL of DMSO/H2O (1 : 4 v/v) were added to 200 μL of the cell suspension. For the determination of unspecific binding (in the presence of atropine at 500-fold access to the fluorescent ligand), 2 μL of a solution of atropine (100-fold concentrated) in DMSO/H2O (1 : 4 v/v) were added instead of neat DMSO/H2O (1 : 4 v/v) (note: in case of 20, the sample volume was 500 μL, i.e. 5 μL instead of 2 μL of ligand solution was added, and the excess of atropine was 1000-fold). Compound 15 was used at final concentrations of 0.23–50 nM, 20 was used at final concentrations of 0.04–20 nM, compounds 7 and 16–19 were used at final concentrations of 0.15–30 nM. Samples were incubated at 22 °C in the dark under gentle shaking for 2 h. All experiments were performed in duplicate.
Data processing
Retention (capacity) factors were calculated from retention times (tR) according to k = (tR – t0)/t0 (t0 = dead time). Raw data from flow cytometric experiments were processed with FACSDiva Software (Becton Dickinson) (7, 15–19) or with FlowJo Software (FlowJo LLC, Ashland, OR) (20) to obtain arithmetic mean values of the areas of the signals detected in the respective channel (FACSCantoII) and geometric mean values of the height of the signals detected in channel FL-2 (FACSCalibur), respectively. Specific binding data from flow cytometric saturation binding experiments, obtained by subtracting unspecific binding data from total binding data, were plotted against the fluorescent ligand concentration and analyzed by a two-parameter equation describing hyperbolic binding (one site-specific binding; GraphPad Prism 5, GraphPad Software, San Diego, CA) in order to obtain Kd and Bmax values. Kd values of individual experiments were transformed to pKd values. Unspecific binding data were fitted by linear regression. Data from the radioligand ([3H]NMS) saturation43 and competition44 binding assays were processed as reported previously. pIC50 values were converted to pKi values according to the Cheng–Prusoff equation50 (logarithmic form). Propagated errors were calculated as described previously.47
Author contributions
C. G. G., A. P. and X. S. synthesized the fluorescent ligands. C. G. G. performed radiochemical and flow cytometric binding experiments and analysed the data. M.K. initiated the project and supervised the research. C. G. G. and M. K. wrote the manuscript. All authors have given approval to the final version of the manuscript.
Abbrevations used
- CHO-cells
Chinese hamster ovary cells
- DIPEA
Diisopropylethylamine
- GPCR
G-Protein coupled receptor
- k
Retention (or capacity) factor (HPLC)
- Kd
Dissociation constant obtained from saturation binding experiment
- MR
Muscarinic receptor
- NMS
N-Methylscopolamine
- PBS
Phosphate buffered saline
- pKd
Negative logarithm of the Kd in M
- pKi
Negative logarithm of the dissociation constant Ki (in M) obtained from a competition binding experiment
- TFA
Trifluoracetic acid
- tR
Retention time
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
The authors thank Maria Beer-Krön, Brigitte Wenzl and Susanne Bollwein for excellent technical assistance. This work was funded by the Graduate Training Program GRK 1910 of the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
†Electronic supplementary information (ESI) available: Fig. S1–S4; RP-HPLC chromatograms of compounds 15–20; 1H-NMR spectra of compounds 15–20. See DOI: 10.1039/d0md00137f
References
- Briddon S. J., Kellam B., Hill S. J. Methods Mol. Cell. Biol. 2011;746:211–236. doi: 10.1007/978-1-61779-126-0_11. [DOI] [PubMed] [Google Scholar]
- Stoddart L. A., Kilpatrick L. E., Briddon S. J., Hill S. J. Neuropharmacology. 2015;98:48–57. doi: 10.1016/j.neuropharm.2015.04.033. [DOI] [PubMed] [Google Scholar]
- Stoddart L. A., Kilpatrick L. E., Hill S. J. Trends Pharmacol. Sci. 2018;39:136–147. doi: 10.1016/j.tips.2017.10.006. [DOI] [PubMed] [Google Scholar]
- Stoddart L. A., White C. W., Nguyen K., Hill S. J., Pfleger K. D. G. Br. J. Pharmacol. 2016;173:3028–3037. doi: 10.1111/bph.13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briddon S. J., Hill S. J. Trends Pharmacol. Sci. 2007;28:637–645. doi: 10.1016/j.tips.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J. G., Middleton R., Adams L., May L. T., Briddon S. J., Kellam B., Hill S. J. Br. J. Pharmacol. 2010;159:772–786. doi: 10.1111/j.1476-5381.2009.00488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuder K., Kiec-Kononowicz K. Curr. Med. Chem. 2008;15:2132–2143. doi: 10.2174/092986708785747599. [DOI] [PubMed] [Google Scholar]
- Kuder K. J., Kiec-Kononowicz K. Curr. Med. Chem. 2014;21:3962–3975. doi: 10.2174/0929867321666140826120058. [DOI] [PubMed] [Google Scholar]
- Middleton R. J., Briddon S. J., Cordeaux Y., Yates A. S., Dale C. L., George M. W., Baker J. G., Hill S. J., Kellam B. J. Med. Chem. 2007;50:782–793. doi: 10.1021/jm061279i. [DOI] [PubMed] [Google Scholar]
- Middleton R. J., Kellam B. Curr. Opin. Chem. Biol. 2005;9:517–525. doi: 10.1016/j.cbpa.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Vernall A. J., Hill S. J., Kellam B. Br. J. Pharmacol. 2014;171:1073–1084. doi: 10.1111/bph.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon M., Ligneau X., Schwartz J. C., Stark H. Bioorg. Med. Chem. Lett. 2006;16:1938–1940. doi: 10.1016/j.bmcl.2005.12.084. [DOI] [PubMed] [Google Scholar]
- Li L., Kracht J., Peng S., Bernhardt G., Buschauer A. Bioorg. Med. Chem. Lett. 2003;13:1245–1248. doi: 10.1016/s0960-894x(03)00113-6. [DOI] [PubMed] [Google Scholar]
- Li L., Kracht J., Peng S., Bernhardt G., Elz S., Buschauer A. Bioorg. Med. Chem. Lett. 2003;13:1717–1720. doi: 10.1016/s0960-894x(03)00235-x. [DOI] [PubMed] [Google Scholar]
- Malan S. F., van Marle A., Menge W. M., Zuliani V., Hoffman M., Timmerman H., Leurs R. Bioorg. Med. Chem. 2004;12:6495–6503. doi: 10.1016/j.bmc.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Xie S. X., Petrache G., Schneider E., Ye Q. Z., Bernhardt G., Seifert R., Buschauer A. Bioorg. Med. Chem. Lett. 2006;16:3886–3890. doi: 10.1016/j.bmcl.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Leopoldo M., Lacivita E., Passafiume E., Contino M., Colabufo N. A., Berardi F., Perrone R. J. Med. Chem. 2007;50:5043–5047. doi: 10.1021/jm070721+. [DOI] [PubMed] [Google Scholar]
- Tabor A., Weisenburger S., Banerjee A., Purkayastha N., Kaindl J. M., Hubner H., Wei L., Gromer T. W., Kornhuber J., Tschammer N., Birdsall N. J., Mashanov G. I., Sandoghdar V., Gmeiner P. Sci. Rep. 2016;6:33233. doi: 10.1038/srep33233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arttamangkul S., Alvarez-Maubecin V., Thomas G., Williams J. T., Grandy D. K. Mol. Pharmacol. 2000;58:1570–1580. doi: 10.1124/mol.58.6.1570. [DOI] [PubMed] [Google Scholar]
- Balboni G., Salvadori S., Dal Piaz A., Bortolotti F., Argazzi R., Negri L., Lattanzi R., Bryant S. D., Jinsmaa Y., Lazarus L. H. J. Med. Chem. 2004;47:6541–6546. doi: 10.1021/jm040128h. [DOI] [PubMed] [Google Scholar]
- Drakopoulos A., Koszegi Z., Lanoiselee Y., Hubner H., Gmeiner P., Calebiro D., Decker M. J. Med. Chem. 2020;63:3596–3609. doi: 10.1021/acs.jmedchem.9b02011. [DOI] [PubMed] [Google Scholar]
- Houghten R. A., Dooley C. T., Appel J. R. Bioorg. Med. Chem. Lett. 2004;14:1947–1951. doi: 10.1016/j.bmcl.2004.01.090. [DOI] [PubMed] [Google Scholar]
- Dukorn S., Littmann T., Keller M., Kuhn K., Cabrele C., Baumeister P., Bernhardt G., Buschauer A. Bioconjugate Chem. 2017;28:1291–1304. doi: 10.1021/acs.bioconjchem.7b00103. [DOI] [PubMed] [Google Scholar]
- Dumont Y., Gaudreau P., Mazzuferi M., Langlois D., Chabot J. G., Fournier A., Simonato M., Quirion R. Br. J. Pharmacol. 2005;146:1069–1081. doi: 10.1038/sj.bjp.0706425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M., Erdmann D., Pop N., Pluym N., Teng S., Bernhardt G., Buschauer A. Bioorg. Med. Chem. 2011;19:2859–2878. doi: 10.1016/j.bmc.2011.03.045. [DOI] [PubMed] [Google Scholar]
- Liu M., Richardson R. R., Mountford S. J., Zhang L., Tempone M. H., Herzog H., Holliday N. D., Thompson P. E. Bioconjugate Chem. 2016;27:2166–2175. doi: 10.1021/acs.bioconjchem.6b00376. [DOI] [PubMed] [Google Scholar]
- Schneider E., Keller M., Brennauer A., Hoefelschweiger B. K., Gross D., Wolfbeis O. S., Bernhardt G., Buschauer A. ChemBioChem. 2007;8:1981–1988. doi: 10.1002/cbic.200700302. [DOI] [PubMed] [Google Scholar]
- Ziemek R., Brennauer A., Schneider E., Cabrele C., Beck-Sickinger A. G., Bernhardt G., Buschauer A. Eur. J. Pharmacol. 2006;551:10–18. doi: 10.1016/j.ejphar.2006.08.075. [DOI] [PubMed] [Google Scholar]
- Kozma E., Jayasekara P. S., Squarcialupi L., Paoletta S., Moro S., Federico S., Spalluto G., Jacobson K. A. Bioorg. Med. Chem. Lett. 2013;23:26–36. doi: 10.1016/j.bmcl.2012.10.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure M. P., Gaudreau P., Shaw I., Cashman N. R., Beaudet A. J. Histochem. Cytochem. 1994;42:755–763. doi: 10.1177/42.6.8189037. [DOI] [PubMed] [Google Scholar]
- Keller M., Kuhn K. K., Einsiedel J., Hubner H., Biselli S., Mollereau C., Wifling D., Svobodova J., Bernhardt G., Cabrele C., Vanderheyden P. M., Gmeiner P., Buschauer A. J. Med. Chem. 2016;59:1925–1945. doi: 10.1021/acs.jmedchem.5b01495. [DOI] [PubMed] [Google Scholar]
- Tahtaoui C., Parrot I., Klotz P., Guillier F., Galzi J. L., Hibert M., Ilien B. J. Med. Chem. 2004;47:4300–4315. doi: 10.1021/jm040800a. [DOI] [PubMed] [Google Scholar]
- Jones L. H., Randall A., Napier C., Trevethick M., Sreckovic S., Watson J. Bioorg. Med. Chem. Lett. 2008;18:825–827. doi: 10.1016/j.bmcl.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Hern J. A., Baig A. H., Mashanov G. I., Birdsall B., Corrie J. E., Lazareno S., Molloy J. E., Birdsall N. J. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenasheva T. A., Neary M., Mashanov G. I., Birdsall N. J., Breckenridge R. A., Molloy J. E. J. Mol. Cell. Cardiol. 2013;57:129–136. doi: 10.1016/j.yjmcc.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A., Cox S., Burns D., Norey C. J. Biomol. Screening. 2003;8:410–420. doi: 10.1177/1087057103256319. [DOI] [PubMed] [Google Scholar]
- Daval S. B., Valant C., Bonnet D., Kellenberger E., Hibert M., Galzi J. L., Ilien B. J. Med. Chem. 2012;55:2125–2143. doi: 10.1021/jm201348t. [DOI] [PubMed] [Google Scholar]
- Daval S. B., Kellenberger E., Bonnet D., Utard V., Galzi J. L., Ilien B. Mol. Pharmacol. 2013;84:71–85. doi: 10.1124/mol.113.085670. [DOI] [PubMed] [Google Scholar]
- Jacobson K. A., Fischer B., van Rhee A. M. Life Sci. 1995;56:823–830. doi: 10.1016/0024-3205(95)00016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karton Y., Baumgold J., Handen J. S., Jacobson K. A. Bioconjugate Chem. 1992;3:234–240. doi: 10.1021/bc00015a006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Gu Q., Mao F., Haugland R. P., Cynader M. S. J. Neurosci. 1994;14:4147–4158. doi: 10.1523/JNEUROSCI.14-07-04147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler M. S., Reba R. C., Cohen V. I., Rzeszotarski W. J., Baumgold J. Brain Res. 1992;582:253–260. doi: 10.1016/0006-8993(92)90141-u. [DOI] [PubMed] [Google Scholar]
- Keller M., Trankle C., She X., Pegoli A., Bernhardt G., Buschauer A., Read R. W. Bioorg. Med. Chem. 2015;23:3970–3990. doi: 10.1016/j.bmc.2015.01.015. [DOI] [PubMed] [Google Scholar]
- Pegoli A., She X., Wifling D., Hubner H., Bernhardt G., Gmeiner P., Keller M. J. Med. Chem. 2017;60:3314–3334. doi: 10.1021/acs.jmedchem.6b01892. [DOI] [PubMed] [Google Scholar]
- Pegoli A., Wifling D., Gruber C. G., She X., Hubner H., Bernhardt G., Gmeiner P., Keller M. J. Med. Chem. 2019;62:5358–5369. doi: 10.1021/acs.jmedchem.8b01967. [DOI] [PubMed] [Google Scholar]
- She X., Pegoli A., Mayr J., Hubner H., Bernhardt G., Gmeiner P., Keller M. ACS Omega. 2017;2:6741–6754. doi: 10.1021/acsomega.7b01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Pegoli A., Gruber C. G., Wifling D., Carpenter J., Hübner H., Wan J., Bernhardt G., Gmeiner P., Holliday N. D., Keller M. J. Med. Chem. 2020;63:4133–4154. doi: 10.1021/acs.jmedchem.9b02172. [DOI] [PubMed] [Google Scholar]
- Hoefelschweiger B. K., PhD thesis, University of Regensburg, 2005, http://d-nb.info/975903071/34. [Google Scholar]
- Rinken A., Lavogina D., Kopanchuk S. Trends Pharmacol. Sci. 2018;39:187–199. doi: 10.1016/j.tips.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Cheng Y.-C., Prusoff W. H. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.