Abstract
The incidence of hepatocellular cancer (HCC) is gradually rising. HCC occurs as a sequela to various chronic liver diseases and ensuing cirrhosis. There have been many therapies approved for unresectable HCC in the last 5 years, including immune checkpoint inhibitors, and the overall response rates have improved. However, there are many cases that do not respond, and personalized medicine is lacking, making HCC an unmet clinical need. Generation of appropriate animal models have been key to our understanding of HCC. Based on the overall concept of hepatocarcinogenesis, two major categories of animal models are discussed herein that can be useful to address specific questions. One category is described as the “outside-in” model of HCC and is based on the premise that it takes decades of hepatocyte injury, death, wound healing, and regeneration to eventually lead to DNA damage and mutations in a hepatocyte, which initiates tumorigenesis. Several animal models have been generated, which attempt to recapitulate this complex tissue damage and cellular interplay through genetics, diets, and toxins. The second category is the “inside-out” model of HCC, where clinically relevant genes can be coexpressed in a small subset of hepatocytes to yield a tumor, which matches HCC subsets in gene expression. This model has been made possible in part by the widely available molecular characterization of HCC, and in part by modalities like sleeping beauty transposon/transposase, Crispr/Cas9, and hydrodynamic tail vein injection. These two categories of HCC have distinct pros and cons, which are discussed in this Thinking Out Loud article.
Key words: Hepatocellular cancer (HCC), Animal models, “Outside-in” model, “Inside-out” model
Chronic liver diseases due to a variety of etiologies including viral hepatitis, nonalcoholic fatty liver disease, alcoholic liver disease, and others remain a major cause of morbidity and mortality worldwide. In fact, around 2 million patients succumb every year due to some form of liver disease1. A major cause of liver-related mortality is cirrhosis, which is the end result of progressive liver fibrosis due to any underlying etiology. In 2017, cirrhosis caused more than 1.32 million deaths in females and 883,000 in males globally2. Currently, cirrhosis is the 11th leading cause of death worldwide1. Another major sequela of chronic liver diseases and cirrhosis is hepatocellular cancer (HCC). Almost 90% of HCCs develop in the background of ongoing chronic liver injury, advanced fibrosis, or cirrhosis3. The rare development of a de novo HCC in a healthy liver is mostly due to malignant transformation of benign liver tumor such as hepatocellular adenoma4. As the incidence of chronic liver diseases increases albeit due to distinct etiologies such a viral hepatitis in Asia, and nonalcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD) in the US and Europe, the incidence of HCC has also increased in tandem1. Worldwide, for both sexes together, HCC is the sixth most commonly diagnosed cancer and fourth leading cause of death related to cancers5. It occurs two to three times more in men and thus is the fifth commonest malignancy in men and the ninth commonest cancer in women. In the US, while it only constitutes 2.4% of all tumors, its incidence has been increasing gradually for the last 3 decades6. Also, the 5-year survival rate of patients with liver tumors is less than 20%. In fact, around 32,000 patients died due to liver tumors in 2019 in the US. While the advent of immunotherapy and approval of additional agents for medical management of unresectable HCC have improved the overall prognosis, the treatments are suboptimal and nonpersonalized, despite the knowledge of key drivers of hepatocarcinogenesis7. Thus, HCC remains a major unmet clinical need requiring additional understanding through generation of better animal models representing human HCC subsets.
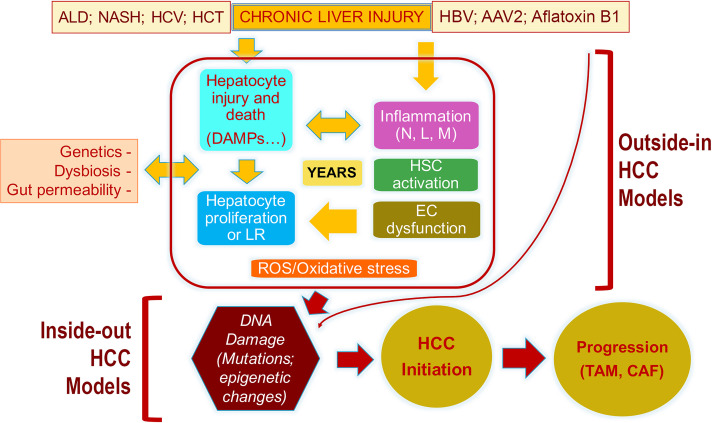
As mentioned, HCC occurs as a consequence of years of chronic liver injury. In fact, the evolution of HCC is a result of sometimes decades of chronic insult and ensuing repair. In most chronic liver diseases, the primary cell afflicted by an injurious agent, be it a virus [hepatitis B (HBV) and hepatitis C (HCV)], toxin like alcohol, or metabolic stressors as in NAFLD, is the hepatocyte. As the hepatocytes are injured, they die only to be replaced by proliferation of neighboring hepatocytes or dedifferentiate and adapt to escape injury, but lose function, which may also trigger proliferation of other hepatocytes, as overall hepatic function is compromised and realized. In addition, a well-differentiated hepatocyte is anti-inflammatory and proactively suppresses immune cell infiltration as has been shown by effect on spontaneous inflammation after conditional deletion of HNF4α or mir-122 from the liver8,9. Hepatocyte injury through generation of damaged-associated molecular patterns (DAMPs) or dedifferentiation triggers immune response, partially driven by the specific etiology (alcohol vs. metabolic vs. viral injury) of hepatocyte injury (Fig. 1).
Figure 1.
The pathogenesis of HCC and the descriptions of events captured by the “outside-in” and the “inside-out” models of HCC development. Because of any of the noted chronic injuries to the liver (ALD, alcoholic liver disease; NASH, nonalcoholic steatohepatitis; HCV, hepatitis C virus; HCT, hemochromatosis), there is hepatocyte injury, release of DAMPs, inflammation (N, neutrophils; L, lymphocytes; M, macrophages), hepatic stellate cell (HSC) activation, and endothelial cell (EC) dysfunction, all of which result in the presence of reactive oxygen species (ROS) and oxidative stress. Loss of hepatocyte viability or differentiation also induces hepatocyte replication, which in the presence of ROS/oxidative stress is prone to DNA damage and, eventually, mutations or misexpression of genes. Any such alteration that provides significant growth and survival advantage to a hepatocyte is really the initiation of malignant transformation and HCC development. Once HCC develops, it creates its own microenvironment in the form of cancer-associated fibroblasts (CAFs) and tumor-associated macrophages (TAMs), which help in further perpetuation of the tumor. There are models that are more relevant in studying events leading up to the transformation and are referred to as “outside-in” models as these address the microenvironmental changes that are upstream of DNA damage and mutations in the hepatocyte. There are models that utilize expression of sets of genes (normal or mutant) in a subset of normal hepatocytes, which lead to tumor initiation and growth and are referred to as “inside-out” models as the expression of these sets of genes/shRNA is sufficient to induce clinically relevant HCC quite promptly without the need for any previous injury or fibrosis. This suggests that once such DNA aberrations occur in a hepatocyte and the cell is transformed, it does not require any additional cues from the microenvironment to grow and propagate, and the tumor then creates its own microenvironment, which is optimal for its development.
Immune cell infiltration including macrophages can be sustained by continued insult and often cross-talks with stellate cells, leading to their activation and fibrogenesis10, which can also be triggered directly by hepatocyte injury. Stellate cell activation to myofibroblast occurs as a wound healing process, but chronic and sustained collagen deposition itself can contribute to hepatic dysfunction and eventually to cirrhosis and can also contribute to the environment that is permissive to HCC development11. Immune cells and stellate cells can also impact sinusoidal endothelial cell function and can undergo capillarization to further perpetuate immune cell infiltration, stellate cell activation, and hepatic dysfunction12. Several cell-extrinsic and cell-intrinsic factors, such as gut dysbiosis13, gut permeability14, genetic variants such as PNPLA3 polymorphisms15, and alterations in bile acid metabolism16, can also impact the overall adverse hepatic milieu. Another common feature of chronic liver injury is the ductular reaction composed of hyperproliferating cholangiocytes17. There is evidence that this ductular reaction may in part be a transdifferentiation mechanism giving rise to hepatocytes and, thus, contributing to hepatocyte repair18,19. However, these reactive ductules can also be proinflammatory and profibrogenic and thus can also contribute to an adverse microenvironment in the liver17.
Thus, chronic liver diseases are characterized by cycles of hepatocyte injury, immune cell infiltration, stellate cell activation, and endothelial cell dysfunction, which provide a signal to relatively healthy hepatocytes due to innate heterogeneity within these cells owing to zonation and ploidy, to proliferate and allow liver regeneration. Chronic hepatocyte proliferation in an adverse milieu of inflammation and fibrosis, which are associated with oxidative stress (partially dependent on primary etiology) from generation of reactive oxygen species, can lead to DNA damage, errors in DNA repair, and mutations, especially in vulnerable replicating cells20. Most of the DNA damage is repaired through a host of mechanisms, and only a rare cell that is unable to repair may undergo senescence. However, DNA repair mechanisms can sometimes be deficient, leading to an error that can eventually give rise to mutations or epigenetic alterations. If such a change, especially a mutation, yields a survival or a proliferative advantage to a hepatocyte within this adverse microenvironment of existing inflammation, oxidative stress, fibrosis, and endothelial cell dysfunction, this cell will outgrow and outcompete other cells and thus mark the beginning of neoplasia. Over time, these cells may gain additional mutational events and will continue to grow and expand and lead to development of HCC. Thus, HCC can very well be defined as an adaptation and survival response of a subset of hepatocytes during chronic wound healing and may very well represent the dark side of the liver’s ability and will regenerate to maintain its functional mass. It is relevant to mention that some injuries to the liver can bypass these chronic bouts of injury, regeneration, and fibrosis by directly impacting DNA and inducing mutations that provide cells with replicative and survival advantage as in HBV, adeno-associated virus 2 (AAV2), and aflatoxin exposure.
The mutations in hepatocytes are key to the origin of HCC. Gain-of-function (GOF) mutations in TERT promoter are the earliest mutations to be mapped through analysis of very early preneoplastic nodules in patients with cirrhosis21. Other earlier mutations in HCC include loss-of function (LOF) mutations in P53 and GOF mutations in CTNNB1, the gene encoding for β-catenin21,22. Further, these mutations in TERT, P53, and CTNNB1 have also been ascribed as trunk mutations based on their presence in early lesions, their presence in sampling of more than one part of a large tumor, their presence in all nodules in multinodular tumors, and their presence in both primary and metastatic site22. As tumors evolve, they gain additional mutations, and the spectra of common mutations in HCC is now well described as mostly limited to around 30–35 genes with varying frequency. The top few leading mutations are those affecting TERT promoter, P53, CTNNB1, ARID1A, ALB, AXIN1, APOB, CDKN2A, EEF1A1, ARID2, RPS6KA3, SMARCA4, and NFE2L2, among others23,24. These various mutations have been uniformly observed through whole-exome sequencing (WES) of several independent HCCs in various databases, suggesting their potential relevance in disease initiation and progression. Intriguingly, most studies have been limited to analysis of relatively early stage lesions by virtue of sampling bias since the available materials for WES are often tumors available in explanted or hepatectomized livers. A recent study analyzed HCCs ranging from early to advanced stages for various well-known mutations25. This analysis showed persistence of the commonly mutated genes (TERT promoter, TP53, CTNNB1, ARID1A, etc.) irrespective of the stage of the disease. Intriguingly, this study identified enrichment of P53 mutations in a large series of HCCs in their cohort, while mutations in TERT promoter or CTNNB1 were more or less comparably distributed across HCCs at various BCLC stages25. Lastly, as now shown in multiple studies, there are some mutations in HCC that are mutually exclusive, while others show significant association and often coexist. A classic example is that of the significant association of CTNNB1 mutations with TERT promoter, NFE2L2, ARID2, and RPS6KA3, while CTNNB1 mutations never occur with mutations in AXIN1 23–25. All of these clinical observations are highly relevant and should be considered when generating animal models of human HCC.
Chemical carcinogenesis using agents such as diethylnitrosamine (DEN), which leads to DNA adducts, has been a popular means to study HCC in rodents. Around 70% of DEN-induced HCC in C3H/He mice, however, have been shown to be due to mutations in genes such a Ha-ras and B-raf and hence very dissimilar from clinical mutational spectra26. Since HCC in patients is almost always associated with ongoing injury, inflammation, and fibrosis, mouse models have been generated to induce such chronic insult by high doses of repeated DEN or combining DEN with toxicants like alcohol27, carbon-tetrachloride28,29, or Western diet30,31. Others have combined even three forms of injury or more to create an even optimal environment and accelerate HCC development in rodents30,32. Several genetic knockout or transgenic mice can also demonstrate spontaneous injury, fibrosis, and HCC, such as hepatocyte-specific PDGF-CC transgenic mice33 and hepatocyte-specific miRNA-122 knockout mice9, or can be combined with insults such as feeding high-fat diet to the major urinary protein-urokinase-type plasminogen activator transgenic or the MUP-uPA mice, to yield chronic injury and HCC development34. However, while these “outside-in” models lend themselves well to studying various specific events leading up to tumorigenesis such as mechanisms of hepatocyte injury, induction of immune response, and mechanisms of stellate cell activation and fibrosis, the mutational spectra of the observed tumors are heterogeneous and distinct from what is observed in patients (Table 1). Further complicating these studies, other than chronicity of events, are how mice differ from patients in their immune cell composition and signaling35, relative resistance, and differences in fibrosis36, and other modifiers of chronic liver injury such as gut microbiota37 and bile acid metabolism16. Thus, while the “outside-in” models of HCC have distinct advantages in terms of studying the underlying mechanisms of injury involving various cell types that lead to initiation of dysplasia, the molecular underpinnings in the process of hepatocarcinogenesis itself, from initiation to progression, are distinct and disparate from humans. Thus, these models should be cautiously used to study tumor biology, tumor microenvironment, and therapeutics.
Table 1.
Examples of “Outside-In” and “Inside-Out” Models of Liver Tumors
| Type | Phenotype | Reference(s) | |
|---|---|---|---|
| Outside-in models | |||
| DEN + carbon tetrachloride | Chemical carcinogen + injury | Liver inflammation, fibrosis, and hepatocellular carcinoma | 27,28 |
| DEN + alcohol | Chemical carcinogen + injury | Liver inflammation, fibrosis, and hepatocellular carcinoma | 27 |
| DEN + high-fat diet | Chemical carcinogen + injury | Obesity, liver inflammation, and hepatocellular carcinoma | 30,31 |
| DEN + alcohol + carbon tetrachloride | Chemical carcinogen + injury + injury | Liver inflammation, fibrosis, cirrhosis, and hepatocellular carcinoma | 32 |
| PDGF-CC transgenic mice | Spontaneous genetic mouse model | Fibrosis, adenomas, and hepatocellular carcinoma | 33 |
| Liver-specific miR-122 knockout mice | Spontaneous genetic mouse model | Fatty liver, liver inflammation, and hepatocellular cancer | 9 |
| MUP-uPA transgenic mice + high fat diet | Genetic mouse model + injury | Fatty liver, liver inflammation, and hepatocellular cancer | 34 |
| Outside-in models | |||
| S127AYap-S45Y/S33Y-β-catenin or S127AYap-Δ90-β-catenin | SBTT-HDTVI | Hepatoblastoma | 49,50 |
| Met-S45Y/S33Y-β-catenin or Met-Δ90-β-catenin | SBTT-HDTVI | Hepatocellular cancer | 45,47,52 |
| Met-sgAxin1 | SBTT-HDTVI | Hepatocellular cancer | 47 |
| MYC-lucOS-N90-CTNNB1 | SBTT-HDTVI (immunogenic) | Liver inflammation and hepatocellular cancer | 41 |
| MYC-lucOS-sgp53 | SBTT (immunogenic), and CRISPR based HDTVI | Liver inflammation and hepatocellular cancer | 41 |
| myr-AKT-YapS127A | SBTT-HDTVI | Intrahepatic cholangiocarcinoma | 53 |
| myr-AKT-NICD | SBTT-HDTVI | Intrahepatic cholangiocarcinoma | 51 |
DEN, diethylnitrosamine; MUP-uPA, urokinase plasminogen activator overexpression under hepatocyte-specific major urinary protein promoter; PDGF-CC, platelet derived growth factor-CC; SBTT-HDTVI, sleeping beauty transposon-transposase and hydrodynamic tail vein injection; NICD, notch intracellular domain.
Another way to study human HCC in preclinical models is the sleeping beauty transposon/transposase (SBTT)-mediated stable expression of clinically relevant combination of genes found to be overexpressed or mutated in patients38. Delivered to a subset of hepatocytes in vivo in mice by a hydrodynamic tail vein injection (HTVI), this methodology is able to directly address the relevance of specific genes in the carcinogenesis. This “inside-out” model relies on the selection of gene or combinations of genes for expression in 1%–5% of hepatocytes, based on data from Whole Genome Sequencing (WGS) or WES studies performed on HCCs from patients. Several publically available databases including The Cancer Genome Atlas (TCGA)39, Catalogue of Somatic Mutations in Cancer (COSMIC)40, and others have curated information on several well-characterized HCC patient cohorts. Other groups have collected and characterized their own HCC cohorts and hence generated their own databases, which may be available through collaborations24. However, multiple analyses have now validated the presence of common alterations including mutations and changes in gene expression across multiple cohorts of HCC cases23,24. Having such validated data from patients provides a unique and impactful opportunity to generate these “inside-out” models and also allows to test the relevance of such alterations if they are truly an oncogenic and driver, or just secondary, bystander, and passenger events. The premise behind these models is that irrespective of upstream events, if the DNA errors or mutations are the end result of years of damage due to chronic liver injury and are truly the initiator events for HCC, then expression of those aberrations, singly or in combination (based on clinical data), would be sufficient to lead to tumorigenesis even in a normal liver and without the need of any other microenvironmental cues. Indeed, many studies using SB-HTVI have now conclusively demonstrated the feasibility of this “inside-out” model and the lack of requirement of an adverse microenvironment to induce or propagate a tumor, as long as the initiator event of expression of specific DNA aberrations has occurred in a healthy hepatocyte. Once these mutations or misexpression occurs and a tumor is induced, the tumor cells will create their own microenvironment, such as generating cancer-associated fibroblasts or tumor-associated macrophages, which will be conducive to the survival and growth of tumor cells via autocrine, paracrine, or endocrine signals. The disadvantage of this model is the lack of its suitability to study mechanisms of injury leading up to the cancer, but the advantage of this model is its appropriateness to study tumor biology, biomarker discovery, and therapies since these tumors are induced by clinically relevant genes and show high similarity in gene expression to the respective human HCC subsets. Since these models are typically immune naive, more recently, use of artificial antigens in plasmids has even enabled studies of tumor immunology in these SB-HTVI-induced HCC models41.
Our group along with collaborators have focused on CTNNB1 mutations as these have been described to be consistently present in early and advanced HCC in around 25%–35% of all HCC cases. Knowing that expression of CTNNB1 (point mutant or deletion mutant) alone is insufficient to induce HCC in murine models42–44, and knowing that CTNNB1 mutations occur frequently with other mutations such as TERT promoter, NFE2L2, and others23–25, or overexpression of genes such as Met (or downstream Ras) and Myc41,45,46, we have used SB-HTVI to coexpress mutant CTNNB1 and the “second-hit.” Similarly, LOF mutations in AXIN1, which are mutually exclusive from CTNNB1 in their occurrence, have also been coexpressed as shRNA along with Met to yield HCC47. All of these models demonstrate a clear cooperation of these abnormal genes in HCC pathogenesis, which significantly resemble subsets of human HCCs at a molecular level. This has also been extended to other liver tumors like hepatoblastoma and intrahepatic cholangiocarcinoma. Since around 80% of all hepatoblastomas in patients showed activation of β-catenin because of mutations or deletions, and yes associated protein-1 (Yap1) due to unidentified reasons48, coexpression of these two genes in mouse liver using SB-HTVI led to the development of hepatoblastoma49,50. Also, coexpression of myristoylated-Akt and notch intracellular domain (NICD) led to the development of intrahepatic cholangiocarcinoma51. All of these findings clearly suggest that coexpression of specific clinically observed mutant genes or aberrantly expressed genes in a subset of hepatocytes in a normal adult liver is sufficient to lead to development of liver tumors.
To conclude, the preclinical models of HCC that exist have been highly useful in elucidating the cellular and molecular basis of this dreadful disease. Based on the question that needs to be addressed, an investigator should select the most appropriate model that may help in answering a specific question of interest after weighing the many pros and cons of each model. The “outside-in” models are of clear value in investigating the mechanisms leading up to the cancer and even in devising chemopreventive strategies. However, one must be cognizant of limitations imposed by innate differences between a mouse model and humans. The “inside-out” model is of value in using clinically relevant genes identified in HCCs in patients to address tumor biology specific to the genetic aberrations and to study biomarkers and therapy. This reductionist approach leads to clinically relevant tumors in mice while precluding opportunities to study mechanisms leading up to tumor initiation. Both categories of HCC models thus have unique applications in oncology research.
ACKNOWLEDGMENTS
This work was supported by NIH grants 1R01DK62277, 1R01DK100287, R01CA204586, R01CA251155, and Endowed Chair for Experimental Pathology to S.P.M., and by NIH grant 1P30DK120531-01 to Pittsburgh Liver Research Center (PLRC). Dr. Monga has grant funding from Revolution Medicines, Vicero Biologicals. He is a Scientific Advisory Board member for Vicero Biologicals and Surrozen. He is a consultant for Aligos Therapeutics.
REFERENCES
- 1. Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol. 2019;70(1):151–71. [DOI] [PubMed] [Google Scholar]
- 2. Collaborators GBDC. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5(3):245–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat Rev Cancer 2006;6(9):674–87. [DOI] [PubMed] [Google Scholar]
- 4. Nault JC, Paradis V, Cherqui D, Vilgrain V, Zucman-Rossi J. Molecular classification of hepatocellular adenoma in clinical practice. J Hepatol. 2017;67(5):1074–83. [DOI] [PubMed] [Google Scholar]
- 5.2018 New Global Cancer Data: GLOBOCAN 2018. Available from https://www.uicc.org/news/new-global-cancer-data-globocan-2018 [Google Scholar]
- 6.2018 Cancer stat facts: Liver and intrahepatic bile duct cancer. Available from https://seer.cancer.gov/statfacts/html/livibd.html [Google Scholar]
- 7. Onuma AE, Zhang H, Huang H, Williams TM, Noonan A, Tsung A. Immune checkpoint inhibitors in hepatocellular cancer: Current understanding on mechanisms of resistance and biomarkers of response to treatment. Gene Expr. 2020;20(1):53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 2011;147(6):1233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuda M, Seki E. Hepatic stellate cell–macrophage crosstalk in liver fibrosis and carcinogenesis. Semin Liver Dis. 2020;40(3):307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baglieri J, Brenner DA, Kisseleva T. The role of fibrosis and liver-associated fibroblasts in the pathogenesis of hepatocellular carcinoma. Int J Mol Sci. 2019;20(7):1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poisson J, Lemoinne S, Boulanger C, Durand F, Moreau R, Valla D, Rautou PE. Liver sinusoidal endothelial cells: Physiology and role in liver diseases. J Hepatol. 2017;66(1):212–27. [DOI] [PubMed] [Google Scholar]
- 13. Hartmann P, Chu H, Duan Y, Schnabl B. Gut microbiota in liver disease: Too much is harmful, nothing at all is not helpful either. Am J Physiol Gastrointest Liver Physiol. 2019;316(5):G563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta B, Liu Y, Chopyk DM, Rai RP, Desai C, Kumar P, Farris AB, Nusrat A, Parkos CA, Anania FA, Western diet-induced increase in colonic bile acids compromises epithelial barrier in nonalcoholic steatohepatitis. FASEB J. 2020;34(5):7089–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singal AG, Manjunath H, Yopp AC, Beg MS, Marrero JA, Gopal P, Waljee AK. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: A meta-analysis. Am J Gastroenterol. 2014;109(3):325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chiang JYL, Ferrell JM. Bile acid metabolism in liver pathobiology. Gene Expr. 2018;18(2):71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sato K, Marzioni M, Meng F, Francis H, Glaser S, Alpini G. Ductular reaction in liver diseases: Pathological mechanisms and translational significances. Hepatology 2019;69(1):420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O’Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R, Koteliansky V, Corrigendum: Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 2018;555(7696):402. [DOI] [PubMed] [Google Scholar]
- 19. Russell JO, Lu WY, Okabe H, Abrams M, Oertel M, Poddar M, Singh S, Forbes SJ, Monga SP. Hepatocyte-specific beta-catenin deletion during severe liver injury provokes cholangiocytes to differentiate into hepatocytes. Hepatology 2019;69(2):742–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, Laurent A, Cherqui D, Balabaud C, Zucman-Rossi J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Torrecilla S, Sia D, Harrington AN, Zhang Z, Cabellos L, Cornella H, Moeini A, Camprecios G, Leow WQ, Fiel MI, Trunk mutational events present minimal intra- and inter-tumoral heterogeneity in hepatocellular carcinoma. J Hepatol. 2017;67(6):1222–31. [DOI] [PubMed] [Google Scholar]
- 23. Cancer Genome Atlas Research Network. Electronic address web, Cancer Genome Atlas Research N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169(7):1327–41 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nault JC, Martin Y, Caruso S, Hirsch TZ, Bayard Q, Calderaro J, Charpy C, Copie-Bergman C, Ziol M, Bioulac-Sage P, Clinical impact of genomic diversity from early to advanced hepatocellular carcinoma. Hepatology 2020;71(1):164–82. [DOI] [PubMed] [Google Scholar]
- 26. Jaworski M, Buchmann A, Bauer P, Riess O, Schwarz M. B-raf and Ha-ras mutations in chemically induced mouse liver tumors. Oncogene 2005;24(7):1290–5. [DOI] [PubMed] [Google Scholar]
- 27. Ambade A, Satishchandran A, Gyongyosi B, Lowe P, Szabo G. Adult mouse model of early hepatocellular carcinoma promoted by alcoholic liver disease. World J Gastroenterol 2016;22(16):4091–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Preziosi M, Poddar M, Singh S, Monga SP. Hepatocyte Wnts are dispensable during diethylnitrosamine and carbon tetrachloride-induced injury and hepatocellular cancer. Gene Expr. 2018;18(3):209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012;21(4):504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakagawa H. Recent advances in mouse models of obesity- and nonalcoholic steatohepatitis-associated hepatocarcinogenesis. World J Hepatol. 2015;7(17):2110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010;140(2):197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xin B, Cui Y, Wang Y, Wang L, Yin J, Zhang L, Pang H, Zhang H, Wang RA. Combined use of alcohol in conventional chemical-induced mouse liver cancer model improves the simulation of clinical characteristics of human hepatocellular carcinoma. Oncol Lett. 2017;14(4):4722–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, Odell MM, Bauer RL, Ren HP, Haugen HS, Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci USA 2005;102(9):3389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakagawa H, Umemura A, Taniguchi K, Font-Burgada J, Dhar D, Ogata H, Zhong Z, Valasek MA, Seki E, Hidalgo J and others. ER stress cooperates with hypernutrition to trigger TNF-dependent spontaneous HCC development. Cancer Cell 2014;26(3):331–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mestas J, Hughes CC. Of mice and not men: Differences between mouse and human immunology. J Immunol. 2004;172(5):2731–8. [DOI] [PubMed] [Google Scholar]
- 36. Delire B, Starkel P, Leclercq I. Animal models for fibrotic liver diseases: What we have, what we need, and what is under development. J Clin Transl Hepatol. 2015;3(1):53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol. 2014;184(4):912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. The Cancer Genome Atlas. Available from http://cancergenome.nih.gov/
- 40. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ruiz de Galarreta M, Bresnahan E, Molina-Sanchez P, Lindblad KE, Maier B, Sia D, Puigvehi M, Miguela V, Casanova-Acebes M, Dhainaut M, beta-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019;9(8):1124–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cadoret A, Ovejero C, Terris B, Souil E, Levy L, Lamers WH, Kitajewski J, Kahn A, Perret C. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene 2002;21(54):8293–301. [DOI] [PubMed] [Google Scholar]
- 43. Harada N, Miyoshi H, Murai N, Oshima H, Tamai Y, Oshima M, Taketo MM. Lack of tumorigenesis in the mouse liver after adenovirus-mediated expression of a dominant stable mutant of beta-catenin. Cancer Res. 2002;62(7):1971–7. [PubMed] [Google Scholar]
- 44. Nejak-Bowen KN, Thompson MD, Singh S, Bowen WC Jr., Dar MJ, Khillan J, Dai C, Monga SP. Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine-45 mutant beta-catenin. Hepatology 2010;51(5):1603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tao J, Xu E, Zhao Y, Singh S, Li X, Couchy G, Chen X, Zucman-Rossi J, Chikina M, Monga SP. Modeling a human HCC subset in mice through co-expression of Met and point-mutant beta-catenin. Hepatology 2016;64(5):1587–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tao J, Zhang R, Singh S, Poddar M, Xu E, Oertel M, Chen X, Ganesh S, Abrams M, Monga SP. Targeting beta-catenin in hepatocellular cancers induced by coexpression of mutant beta-catenin and K-Ras in mice. Hepatology 2017;65(5):1581–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qiao Y, Wang J, Karagoz E, Liang B, Song X, Shang R, Evert K, Xu M, Che L, Evert M and others. Axis inhibition protein 1 (Axin1) deletion-induced hepatocarcinogenesis requires intact beta-catenin but not notch cascade in mice. Hepatology 2019;70(6):2003–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bell D, Ranganathan S, Tao J, Monga SP. Novel advances in understanding of molecular pathogenesis of hepatoblastoma: A Wnt/beta-catenin perspective. Gene Expr. 2017;17(2):141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Min Q, Molina L, Li J, Adebayo Michael AO, Russell JO, Preziosi ME, Singh S, Poddar M, Matz-Soja M, Ranganathan S, Beta-catenin and yes-associated protein 1 cooperate in hepatoblastoma pathogenesis. Am J Pathol. 2019;189(5):1091–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, Jiang L, Fan B, Terracciano L, Armeanu-Ebinger S, Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology 2014;147(3):690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fan B, Malato Y, Calvisi DF, Naqvi S, Razumilava N, Ribback S, Gores GJ, Dombrowski F, Evert M, Chen X, Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest. 2012;122(8):2911–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shang N, Arteaga M, Zaidi A, Stauffer J, Cotler SJ, Zeleznik-Le NJ, Zhang J, Qiu W. FAK is required for c-Met/beta-catenin-driven hepatocarcinogenesis. Hepatology 2015;61(1):214–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang J, Dong M, Xu Z, Song X, Zhang S, Qiao Y, Che L, Gordan J, Hu K, Liu Y, Notch2 controls hepatocyte-derived cholangiocarcinoma formation in mice. Oncogene 2018;37(24):3229–42. [DOI] [PMC free article] [PubMed] [Google Scholar]