Abstract
Objective
Oral squamous cell carcinoma (OSCC) is one of the most common cancers, accounting for over 90% of malignant lesions in the oral cavity. Long non-coding RNAs play an important role in the development of OSCC. This study aimed to investigate the effects of lncRNA XIST on the malignant behaviors of OSCC cells and its possible molecular mechanisms.
Methods
Real-time quantitative PCR and Western blot were used to detect the RNA and protein level, respectively. CAL27 and SCC25 cells with the lowest expression level of XIST were used for further study. MTT, transwell assay, colony formation, and xenograft model were applied to examine the effect of XIST on the progression of OSCC. FISH assay was performed to investigate the co-location of XIST and miR-455-3p in OSCC cells. The bioinformatics analysis, luciferase, and RNA pull down assay were utilized to predict and verify the target genes of miR-455-3p.
Results
XIST was downregulated in OSCC tissues and cell lines. Overexpression of XIST inhibited the proliferation, migration, and invasion ability of OSCC cells. Bioinformatics analysis and luciferase reporter assay confirmed XIST could bind to miR-455-3p. Besides, miR-455-3p directly targeted BTG2 in OSCC cells. Rescue experiments further confirmed the positive interaction between miR-455-3p and XIST as well as between miR-455-3p and BTG2.
Conclusion
XIST was down-regulated in OSCC. XIST regulated the expression of BTG2 via sponging miR-455-3p. XIST/miR-455-3p/BTG2 signal axis inhibited the malignant progression of OSCC.
Keywords: XIST, miR-455-3p, BTG2, OSCC
Introduction
Oral squamous cell carcinoma (OSCC) is a common disease in oral oncology.1 It has a high degree of malignancy and is prone to lymph node metastasis, leading to poor prognosis and low survival rate.2 Recently, the prevalence rate of OSCC has tended to become younger and increased year by year.3 At present, a wide range of comprehensive treatment methods, such as expanded surgical resection and lymphadenectomy, supplemented with radiotherapy and chemotherapy, are widely applied in clinical practice, but the survival time and life quality of patients are still not ideal.4 The occurrence and development of oral squamous cell carcinoma is a complex biological process involving multiple genetic and epigenetic changes. Therefore, in-depth study of the molecular mechanism of the development of OSCC is the key to develop new effective strategy for OSCC.
LncRNAs are considered as by-products produced by RNA polymerase II during transcription. LncRNAs are a group of non-coding RNAs that are longer than 200 nt.5,6 Although they lack open reading frames encoding proteins, it is still proposed that LncRNAs perform multiple functions, such as direct or indirect transcriptional regulation, transcription factor isolation, and protein, or RNA regulation. LncRNAs are abnormally expressed in tumors such as colorectal cancer,7 renal cancer,8 breast cancer,9 and esophageal cancer.10 Sun et al evidence that lncRNAs are abnormally expressed in oral precancerous lesions and OSCC. For the first time, the expression level and mechanism of lncRNAs in oral diseases are described.11 In recent years, LncRNAs can be used as diagnostic markers or therapeutic targets for OSCC. However, the potential roles of lncRNA XIST in OSCC have not been fully elucidated.
microRNAs (miRNAs) are a type of single-chain small molecule RNAs which collectively participate tumor proliferation and metastasis.12 Numerous studies have demonstrated the critical role of miRNAs in the development of OSCC.13
In this study, we found that dysregulated lncRNA XIST induced the malignant evolution of OSCC. XIST directly regulated the expression of miR-455-3p. Silencing of XIST can promote tumor proliferation, migration, and invasion by upregulating miR-455-3p. Moreover, BTG2 was predicted and proved to be a target of miR-455-3p. In short, XIST modulated the malignant progression of OSCC by miR-455-3p/BTG2 axis. The XIST/miR-455-3p/BTG2 axis may be a potential biomarker for the treatment of OSCC.
Methods
Specimen Source
A total of 14 pairs of tumor tissues and the adjacent normal tissues were collected from The First Affiliated Hospital of Zhengzhou University and stored at −80 °C. All patients did not undergo chemotherapy or radiotherapy before the operation. This study was approved by the Ethics Committee of Affiliated Hospital of Hebei University of Engineering. The patients had provided the informed consent. The informed consent were obtained from the patients. All procedures were in accordance with the Helsinki declaration.
Cell Culture
Human OSCC cell lines CAL27, SCC15, SCC25, and TSCCA and oral mucosa epithelium cell line (NHOK) were purchased from Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and 100 U/mL penicillin/streptomycin (Beyotime, China) at 37°C in an atmosphere humidified with 5% CO2.
Cell Transfection
miR-455-3p mimic, miR-455-3p inhibitor, and their negative controls were provided by Genepharma (Shanghai, China). The pSilencer and pcDNA3.1 plasmids for silencing or overexpressing XIST were synthesized by General biocompany (Hefei, Anhui). Cells were transfected with the miR-455-3p mimics, miR-455-3p inhibitor, negative controls, and specific plasmids using Lipofectamine2000 according to manufacturer’s instructions. After 6 to 8 h of culture, change the medium to complete medium, and continue to culture for 48 h for subsequent experiments.
MTT Assay
After 48 h transfection, cells were digested with 2.5 g/L trypsin.1 × 104 cells were seeded per well in a 96-well plate and incubated at room temperature. At the time point of 12, 24, 48, and 72 h, the culture medium was removed and replaced with the new medium containing 1% MTT solution. After cultured for 4 hours, each well was supplemented with 100 μL of dimethyl sulfoxide (DMSO). The absorbance of each well was measured with a microplate at a wavelength of 490 nm and the cell viability curve is drawn.
Transwell Cell Invasion Assay
First, the upper chamber was pre-coated with or without matrigel (BD, USA). After the matrigel was solidified, transfected CAL27 and SCC25 cells were seeded on the upper chamber (Millipore, MA, USA) and cultured in 200 μL serum-free DMEM. DMEM containing 12% FBS was added to the lower chamber. Cells were incubated at 37°C. 24 h later, cells migrated or invaded to the lower surface were fixed with methanol, followed by stained with 0.1% crystal violet at room temperature for 20 min. Finally, the migrated or invaded cells were imaged under an optical microscope (Nikon, Japan).
Colony Formation Experiment
After 48 h transfection, CAL27 and SCC25 cells were seeded to 12-well plates (2 x 102 cells/well) and cultured with DMEM medium containing 10% FBS. Cells were cultured at 37°C in an atmosphere humidified with 5% CO2 for 2 weeks. Finally, cell colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet at room temperature for 15 min. The colonies were imaged and counted with a microscope using Image J software.
Quantitative Real-Time PCR (qRT-PCR)
After extracting total RNA with Trizol reagent (Invitrogen, USA), 5 μg of RNA was reverse transcribed into cDNA with a reverse transcription kit (Takara, Dalian, China) according to the instructions. qRT-PCR reaction was carried out a Power SYBR-Green PCR Master mix (Thermo Fisher Scientific, USA) on a FAST7500 realtime-PCR system (ABI, USA). The PCR was performed under the following reaction conditions: pre-denaturation at 96 °C for 5min, 30 cycles of denaturation at 96 °C for 30s, annealing at 54 °C for 30s, extension at 72 °C for 30s; finally, extension at 72 °C was 10min and stored at 4 °C. The relative expression levels were calculated using the 2−ΔΔCt method. GAPDH and U6 were used as the internal control for the mRNA and miRNAs, respectively.
Dual Luciferase Reporter Gene Experiment
Bioinformatics software was used to predict the target genes of miR-455-3p. The wild-type and mutant sequence containing the binding region of miR-455-3p were synthesized and cloned into the pGL3.1 reporter plasmid. The wild-type and mutant plasmid along with miR-455-3p mimics and its negative control were transfected into OSCC cells. After 48 h, the luciferase activity was determined by a dual luciferase detection kit (Promega, USA). Firefly luciferase activity was normalized to Renilla.
RNA Pull Down
Biotin labeled miR-455-3p probe or the negative control were obtained from Sangon Biotech (CA, USA). Cells were transfected with specific probes for 48 h. The transfected cells were harvested and lysed. Then, the lysates were incubated with pre-coated beads overnight. RNA enrichment was evaluated by qPCR assay.
Xenografts Model
Six-week-old male nude mice (20 to 25g) used for the in vivo study were provided by Shanghai SLRC Experimental Animal Center (Shanghai, China). The mice were injected subcutaneously with 5 × 106 OSCC cells per mouse. Tumor size was measured every 3 days, volume = length (mm) × width (mm) × height (mm) × 0.5. At the end of the experiment, the mice were sacrificed and the tumor were removed, imaged, and weighed. The present study was approved by the First Affiliated Hospital of Zhengzhou University.
Statistical Analysis
SPSS21.0 software was used for the data statistical analysis. Data were expressed as mean ± standard deviation (x ± SD). The Students’ t-test was used for comparison between two groups, and ANOVA for comparison among multiple groups. P<0.05 was considered statistically significant.
Results
XIST is Highly Expressed in OSCC
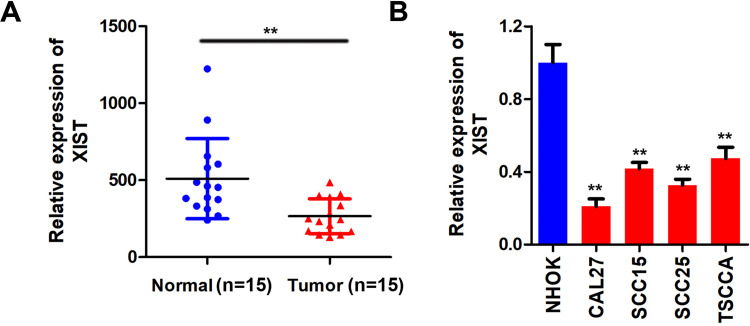
To investigate the biological role of XIST in OSCC, we firstly performed qPCR assay to assess the level of XIST in OSCC specimens. XIST was significantly downregulated in OSCC tissues (Figure 1A). Moreover, we determined the XIST expression in oral mucosa epithelium cell line and OSCC cell lines. The results showed that XIST was downregulated in OSCC cells compared to control group, which was more potent in SCC25 and CAL27 cells (Figure 1B). Therefore, SCC25 and CAL27 cells were used in subsequent experiments.
Figure 1.
XIST is lowly expressed in OSCC. (A and B) qPCR was used to detect the expression of XIST in the OSCC tissues and cell lines compare to the normal tissues and oral mucosa epithelium cells. **P<0.01.
XIST Inhibits the Malignant Behavior of OSCC Cells
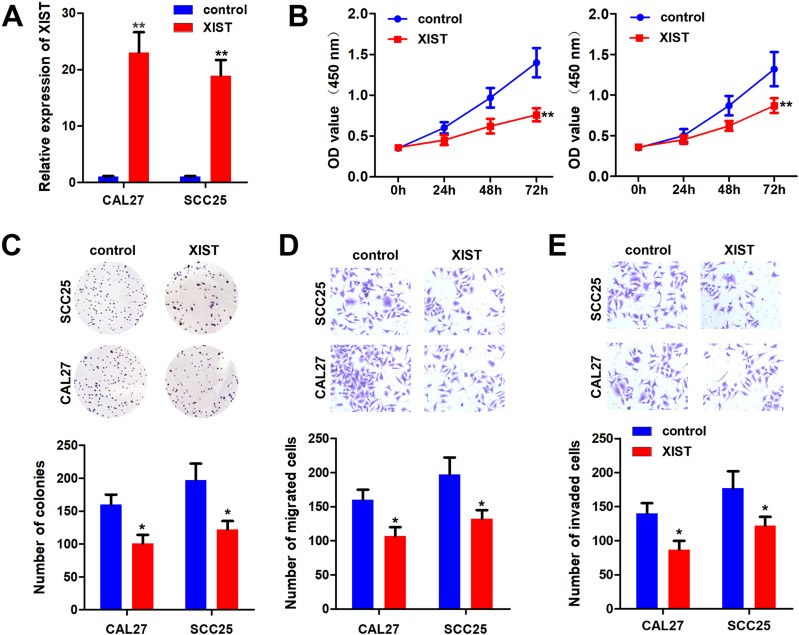
Function studies were performed to evaluate the effect of XIST on OSCC cells. First, the qPCR results showed that the overexpressing of XIST notably elevated the level of XIST, suggesting cells were successfully transfected (Figure 2A). MTT and colony formation results showed that XIST overexpression inhibited the proliferation of OSCC cells (Figure 2B and C). Transwell assay showed that overexpression of XIST inhibited the invasion ability of OSCC cells (Figure 2D and E).
Figure 2.
XIST inhibited the malignant behavior of OSCC. pcDNA3.1/XIST was transfected to OSCC cells to overexpressing XIST. (A) qPCR was used to assess the XIST expression. (B and C) MTT and colony formation assay were performed to detect the cell proliferation of OSCC cells after overexpression of XIST. (D and E) Transwell assay was used to evaluate the migration and invasion of OSCC cells. *P<0.05, **P<0.01.
XIST Can Be targetedbymiR-455-3p in OSCC Cells
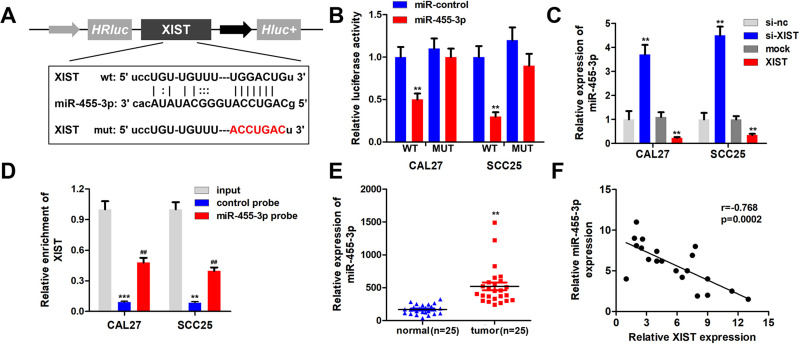
Bioinformatics analysis revealed the binding sites between miR-455-3p and XIST (Figure 3A). Luciferase assay revealed that miR-455-3p significantly inhibited the luciferase activity of the reporter vector carrying the wild type region of XIST but not the mutant one (Figure 3B). XIST inhibited the expression of miR-455-3p while XIST silencing upregulated miR-455-3p (Figure 3C). RNA pull down assay further showed that XIST could bind with miR-455-3p directly (Figure 3D). Moreover, miR-455-3p was notably upregulated in OSCC tissues (Figure 3E). Additionally, Pearson analysis showed that the expression of XIST and miR-455-3p was negatively correlated (Figure 3F).
Figure 3.
XIST sponges miR-455-3p in OSCC cells. (A) Bioinformatics analysis showed the binding site between XIST and miR-455-3p. (B) Luciferase assay was carried out to verify the combination of XIST and miR-455-3p. (C) qPCR was used to assess the expression of miR-455-3p. (D) RNA pulled down experiments was performed to confirm the interaction between XIST and miR-455-3p. (E) qPCR was used to detect the expression of miR-455-3p in OSCC tissues. (F) Pearson analysis was performed to evaluate the correlation between XIST and miR-455-3p. **P<0.01, ***P<0.001, ##P<0.01.
miR-455-3p Reverses the Effect of XIST on OSCC Cells
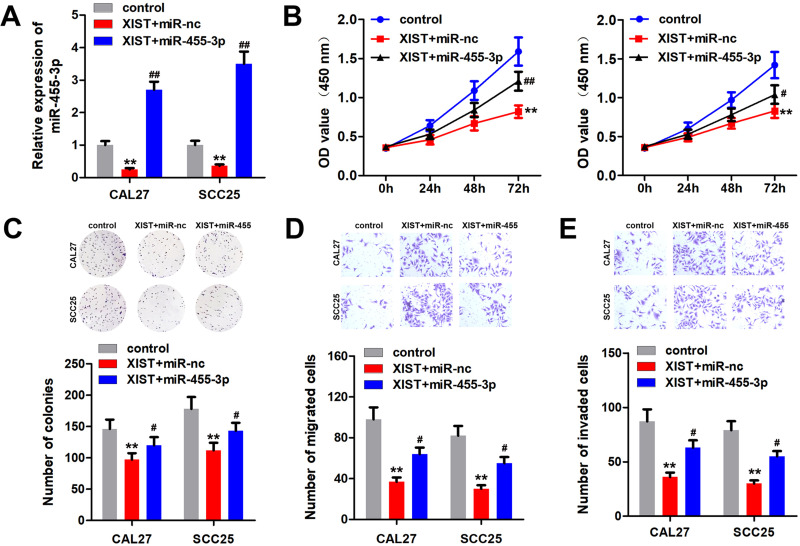
We performed rescue experiment to confirm the relationship between XIST and miR-455-3p. The qPCR results showed that overexpression of XIST reduced the level of miR-455-3p, which was abated by upregulation of miR-455-3p (Figure 4A). The results of MTT and colony formation experiments indicated that overexpression of XIST inhibited the proliferation of OSCC cells, which was antagonized by miR-455-3p (Figure 4B and C). Moreover, overexpressed miR-455-4p alleviated the inhibition of OSCC cell migration and invasion induced by XIST (Figure 4D and E).
Figure 4.
miR-455-3p reversed the effect of XIST in OSCC cells. OSCC cells were transfected with pcDNA3.1/XIST, miR-455-3p mimic or their combination. (A) miR-455-3p expression was detected by qPCR. (B and C) MTT and colony formation assay were performed to detect the cell proliferation of OSCC cells. (D and E) Transwell assay was used to evaluate the migration and invasion of OSCC cells. **P<0.01, #P<0.05, ##P<0.01.
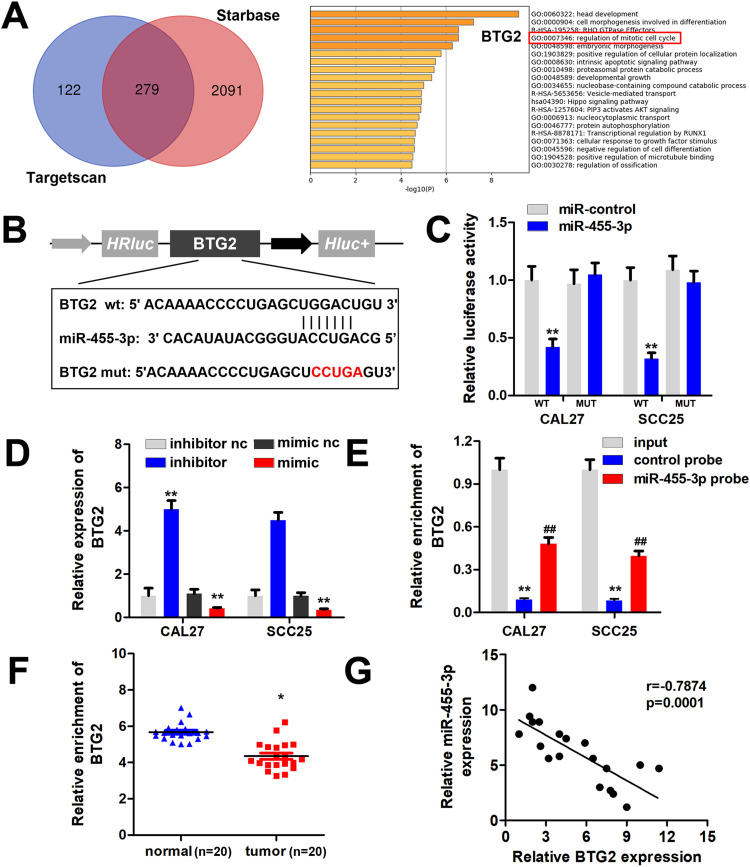
miR-455-3p Directly Targets BTG2 in OSCC Cells
To elucidate the precise mechanism underlying the effect of XIST/miR-455-3p signaling, we predicted the potential targets of miR-455-3p using TargetScan and Starbase. As showed in Figure 5A, there were 279 target genes that were predicted by these two software. Then, enrichment analysis was performed based on the Metaspace software (Figure 5B). We found that BTG2 is involved in mitotic cell cycle modulation. Thus, BTG2 was chosen for further study. Figure 5B shows the targeting region between BTG2 and miR-455-3p. Further luciferase experiment results showed that miR-455-3p inhibited the luciferase activity of the reporter carrying wild-type BTG2 3ʹ-UTR (Figure 5C). Moreover, miR-455-3p inhibited the expression of XIST, while knockdown of miR-455-3p promoted the expression of XIST (Figure 5D). RNA pull down assay showed that there was a negative interaction between miR-455-3p and BTG2 (Figure 5E). We then evaluated the level of BTG2 in the OSCC tissues. As showed by Figure 5F, BTG2 was significantly downregulated in OSCC tissues. Moreover, the expression of miR-455-3p was negatively correlated with BTG2 (Figure 5G).
Figure 5.
miR-455-3p targets BTG2 in OSCC cells. (A) Bioinformatics analysis based on the TargetScan and Starbase database was performed to screen the targets of miR-455-3p. Enrichment analysis of these 279 potential target genes was performed using Metaspace software. (B) The binding site between miR-455-3p and BTG2 was showed. (C) Luciferase assay was carried out to verify the combination of miR-455-3p and BTG2. (D) qPCR was used to assess the BTG2 expression in each group. (E) RNA pulled down experiments was performed to confirm the interaction between miR-455-3p and BTG2. (F) qPCR was used to assess the expression level BTG2 in OSCC tissues. (G) Pearson analysis was performed to evaluate the correlation between miR-455-3p and BTG2 expression level. *P<0.05, **P<0.01, ##P<0.01.
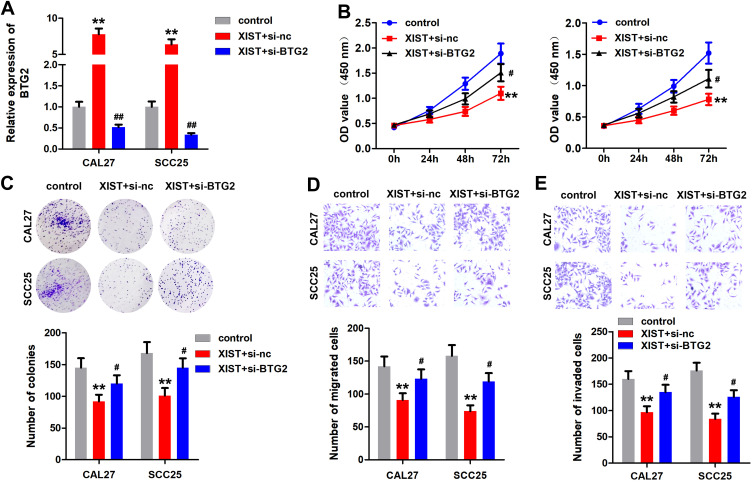
BTG2 Knockdown Reverses the Effect of XIST
First, qPCR results showed that overexpression of XIST promoted the expression of BTG2, which was alleviated by BTG2 knockdown (Figure 6A). Additionally, the suppression of cell proliferation induced by XIST was antagonized by BTG2 knockdown (Figure 6B and C). This was paralleled with the results from transwell assay. As showed in Figure 6D and EE, knockdown of BTG2 significantly abated XIST-induced decrease of OSCC cell migration and invasion ability.
Figure 6.
Knockdown of BTG2 reverses the anti-cancer effect of XIST. OSCC cells were transfected with pcDNA3.1/XIST, si-BTG2 or their combination. (A) The expression of BTG2 was detected by qPCR. (B and C) MTT and colony formation assay were performed to detect the cell proliferation of OSCC cells. (D and E) Transwell assay was used to evaluate the migration and invasion of OSCC cells. **P<0.01, #P<0.05, ##P<0.01.
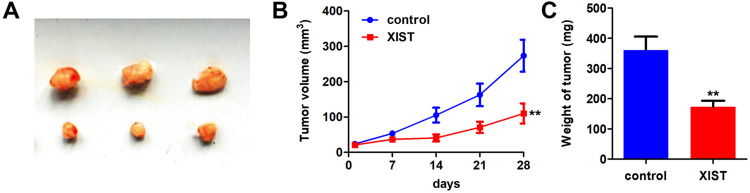
Knockdown of XIST Inhibited the Growth and Metastasis of OSCC
In order to study the biological effects of XIST, we further studied the effects of XIST on the progression of OSCC in vivo. The results showed that XIST overexpression reduced tumor size, volume, and weight (Figure 7A–C).
Figure 7.
XIST inhibited tumor proliferation of OSCC. (A) CAL27 cell line stably expressing pcDNA3.1/XIST was used to establish the Xenografts mice model. (A) The image of the tumors in each group. (B) The growth curve of the tumors was made. (C) The wight of tumor in each group was detected. **P<0.01.
Discussion
The purpose of this study was to explore the roles of XIST/miR-455-3p axis in the progression of OSCC and the underlying cellular/molecular mechanisms. Previously, XIST functions as an anti-cancer gene. For instance, XIST inhibits the progression of breast,14 liver,15 and prostate cancer,16 pancreatic,17,18 esophageal cancer,19 non-small cell lung cancer,20 colon cancer,21 ovarian cancer,22 laryngeal squamous cell carcinoma.23 However, the effect of XIST in OSCC development has not been elucidated before. To our knowledge, this is the first study to investigate roles of XIST in OSCC. We found that XIST was notably downregulated in OSCC tissues and cell lines. Additionally, XIST overexpression significantly inhibited the proliferation, migration, and invasion of OSCC cells. However, the mechanism underlying its effect remains to be further explored.
lncRNAs modulate gene expression through transcription level modification, post-transcriptional modification, and translational regulation. XIST bind with TET1 to downregulate p53 expression in bladder cancer.24 Meanwhile, it promotes the transcription of PHLPP1 by increasing the HDAC3 recruitment to the promoter and modulates the phosphorylation of AKT.25 Interestingly, XIST interacts directly with SHARP to silence transcription through HDAC3.26 Besides, lncRNA are capable of sponging miRNAs to block their functions. lncRNA–miRNA–mRNA interactions play a crucial role in tumorigenesis which is called the ceRNA mechanism.27,28 miRNA plays an important regulatory role in the occurrence and development of various tumors.29 Recent studies have identified that some miRNAs promote the invasion or metastasis of OSCC by adjusting related genes, which provides potential therapeutic targets for anti-metastatic strategies.30 However, the roles of miR-455-3p have not been elucidated in OSCC. In this study, miR-455-3p is significantly upregulated in OSCC tissues and cell lines. Moreover, miR-455-3p could reverse the effect of XIST on OSCC progression. Thus, XIST may regulate the progression of OSCC via degrading miR-455-3p. However, the underlying mechanisms are not clear.
To further explore the precise mechanism, we investigated the downstream genes regulated by miR-455-3p in OSCC. B-cell translocation gene 2 (BTG2) was predicted and proved to be a target of miR-455-3p. BTG2 is the first identified gene of the BTG/TOB family.31 BTG2 plays an anti-cancer role in various human cancers.32,33 It suppresses the activity of cyclin D and cyclin E and attenuates the cell proliferation through G1 to S cell cycle arrest.34,35 In OSCC, BTG2 inhibits the proliferation of OSCC cells via modulating p38 and MAPK pathways.36 In this study, BTG2 was decreased in OSCC tissues and cells. BTG2 was overexpressed by XIST, while downregulated by miR-455-3p. Moreover, downregulation of BTG2 reversed the effects of XIST on the proliferation, migration, and invasion of OSCC cells. Therefore, XIST may upregulate BTG2 to inhibit the progression of OSCC via sponging miR-455-3p. The XIST/miR-455-3p/BTG2 axis may be a promising biomarker for OSCC.
Conclusion
In this study, XIST targeted miR-455-3p and inhibited the effects of oncomiRNA miR-455-3p in OSCC. It acted as a ceRNA to regulate the expression of BTG2 via sponging miR-455-3p. Therefore, our study revealed new aspects of the cellular function and pathophysiological role of XIST and miR-455-3p, both of which might be considered as potential molecular targets for the treatment of OSCC.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143. doi: 10.1016/j.cellsig.2017.10.009 [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Chen D, Liu H, Yang K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10(2):41. doi: 10.1038/s41419-018-1280-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazawa M, Lin DC, Handral H, et al. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene. 2017;36(16):2243–2254. doi: 10.1038/onc.2016.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Zhang X, Wang Z, et al. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating glut1-mediated glycolysis. Cancer Lett. 2018;434:172–183. doi: 10.1016/j.canlet.2018.07.016 [DOI] [PubMed] [Google Scholar]
- 5.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi: 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bian Z, Zhang J, Li M, et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24(19):4808–4819. doi: 10.1158/1078-0432.CCR-17-2967 [DOI] [PubMed] [Google Scholar]
- 8.Wang A, Bao Y, Wu Z, et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019;10(3):154. doi: 10.1038/s41419-019-1331-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong H, Wang W, Mo S, et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37(1):202. doi: 10.1186/s13046-018-0875-3 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Kang M, Ren M, Li Y, Fu Y, Deng M, Li C. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J Exp Clin Cancer Res. 2018;37(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Sun CC, Zhang L, Li G, et al. The lncRNA PDIA3P interacts with miR-185-5p to modulate oral squamous cell carcinoma progression by targeting cyclin D2. Mol Ther Nucleic Acids. 2017;9:100–110. doi: 10.1016/j.omtn.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh P-L, Liao Y-W, Pichler M, Yu C-C. MicroRNAs as theranostics targets in oral carcinoma stem cells. Cancers (Basel. 2020;12(2):2. doi: 10.3390/cancers12020340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorenchtein M, Poh CF, Saini R, Garnis C. MicroRNAs in an oral cancer context - from basic biology to clinical utility. J Dent Res. 2012;91(5):440–446. doi: 10.1177/0022034511431261 [DOI] [PubMed] [Google Scholar]
- 14.Zheng R, Lin S, Guan L, et al. Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem Biophys Res Commun. 2018;498(4):1002–1008. doi: 10.1016/j.bbrc.2018.03.104 [DOI] [PubMed] [Google Scholar]
- 15.Lin XQ, Huang ZM, Chen X, Wu F, Wu W. XIST induced by JPX suppresses hepatocellular carcinoma by sponging miR-155-5p. Yonsei Med J. 2018;59(7):816–826. doi: 10.3349/ymj.2018.59.7.816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Y, Weng XD, Wang L, et al. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget. 2017;8(55):94358–94370. doi: 10.18632/oncotarget.21719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem. 2017;118(10):3349–3358. doi: 10.1002/jcb.25988 [DOI] [PubMed] [Google Scholar]
- 18.Sun Z, Zhang B, Cui T. Long non-coding RNA XIST exerts oncogenic functions in pancreatic cancer via miR-34a-5p. Oncol Rep. 2018;39(4):1591–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, Hu X, Wu Y, et al. Long non-coding RNA XIST promotes the development of esophageal cancer by sponging miR-494 to regulate CDK6 expression. Biomed Pharmacother. 2019;109:2228–2236. doi: 10.1016/j.biopha.2018.11.049 [DOI] [PubMed] [Google Scholar]
- 20.Li C, Wan L, Liu Z, et al. Long non-coding RNA XIST promotes TGF-beta-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–195. doi: 10.1016/j.canlet.2018.01.036 [DOI] [PubMed] [Google Scholar]
- 21.Sun N, Zhang G, Liu Y. Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/beta-catenin signaling pathway. Gene. 2018;665:141–148. doi: 10.1016/j.gene.2018.04.014 [DOI] [PubMed] [Google Scholar]
- 22.Zuo K, Zhao Y, Zheng Y, et al. Long non-coding RNA XIST promotes malignant behavior of epithelial ovarian cancer. Onco Targets Ther. 2019;12:7261–7267. doi: 10.2147/OTT.S204369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao D, Cui X, Wang X. Long noncoding RNA XIST increases the aggressiveness of laryngeal squamous cell carcinoma by regulating miR-124-3p/EZH2. Exp Cell Res. 2019;381(2):172–178. doi: 10.1016/j.yexcr.2019.04.034 [DOI] [PubMed] [Google Scholar]
- 24.Hu B, Shi G, Li Q, Li W, Zhou H. Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. J Cell Biochem. 2019;120(4):6330–6338. doi: 10.1002/jcb.27920 [DOI] [PubMed] [Google Scholar]
- 25.Huang YS, Chang CC, Lee SS, Jou YS, Shih HM. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget. 2016;7(28):43256–43266. doi: 10.18632/oncotarget.9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McHugh CA, Chen CK, Chow A, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng ZQ, Li ZX, Zhou GQ, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79(18):4612–4626. doi: 10.1158/0008-5472.CAN-19-0799 [DOI] [PubMed] [Google Scholar]
- 28.Zhan H, Tu S, Zhang F, Shao A, Lin J. MicroRNAs and long non-coding RNAs in c-Met-regulated cancers. Front Cell Dev Biol. 2020;8:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira CM, Sehnem D, Da FE, et al. miRNAs: important targets for oral cancer pain research. Biomed Res Int. 2017;2017:4043516. doi: 10.1155/2017/4043516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouronte-Roibas C, Leiro-Fernandez V, Ruano-Ravina A, et al. Predictive value of a series of inflammatory markers in COPD for lung cancer diagnosis: a case-control study. Respir Res. 2019;20(1):198. doi: 10.1186/s12931-019-1155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stupfler B, Birck C, Seraphin B, Mauxion F. BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat Commun. 2016;7:10811. doi: 10.1038/ncomms10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devanand P, Sundaramoorthy S, Ryu MS, Jayabalan AK, Ohn T, Lim IK. Translational downregulation of twist1 expression by antiproliferative gene, B-cell translocation gene 2, in the triple negative breast cancer cells. Cell Death Dis. 2019;10(6):410. doi: 10.1038/s41419-019-1640-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Farre B, Ramis-Zaldivar JE, Salmeron-Villalobos J, et al. Burkitt-like lymphoma with 11q aberration: a germinal center-derived lymphoma genetically unrelated to burkitt lymphoma. Haematologica. 2019;104(9):1822–1829. doi: 10.3324/haematol.2018.207928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim IK. TIS21 (/BTG2/PC3) as a link between ageing and cancer: cell cycle regulator and endogenous cell death molecule. J Cancer Res Clin Oncol. 2006;132(7):417–426. doi: 10.1007/s00432-006-0080-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu M, Wu H, Liu T, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19(7):828–837. doi: 10.1038/cr.2009.72 [DOI] [PubMed] [Google Scholar]
- 36.Lee JC, Chung LC, Chen YJ, Feng TH, Chen WT, Juang HH. Upregulation of B-cell translocation gene 2 by epigallocatechin-3-gallate via p38 and ERK signaling blocks cell proliferation in human oral squamous cell carcinoma cells. Cancer Lett. 2015;360(2):310–318. [DOI] [PubMed] [Google Scholar]