Abstract
The Staphylococcus epidermidis glucose/H+ symporter (GlcPSe) is a membrane transporter highly specific for glucose and a homolog of the human glucose transporters (GLUT, SLC2 family). Most GLUTs and their bacterial counterparts differ in the transport mechanism, adopting uniport and sugar/H+ symport, respectively. Unlike other bacterial GLUT homologs (for example, XylE), GlcPSe has a loose H+/sugar coupling. Asp22 is part of the proton-binding site of GlcPSe and crucial for the glucose/H+ co-transport mechanism. To determine how pH variations affect the proton site and the transporter, we performed surface-enhanced IR absorption spectroscopy on the immobilized GlcPSe. We found that Asp22 has a pKa of 8.5 ± 0.1, a value consistent with that determined previously for glucose transport, confirming the central role of this residue for the transport mechanism of GlcPSe. A neutral replacement of the negatively charged Asp22 led to positive charge displacements over the entire pH range, suggesting that the polarity change of the WT reflects the protonation state of Asp22. We expected that the substitution of the residue Ile105 for a serine, located within hydrogen-bonding distance to Asp22, would change the microenvironment, but the pKa of Asp22 corresponded to that of the WT. A167E mutation, selected in analogy to the XylE, introduced an additional protonatable site and perturbed the protonation state of Asp22, with the latter now exhibiting a pKa of 6.4. These studies confirm that Asp22 is the proton-binding residue in GlcPSe and show that charged residues in its vicinity affect the pKa of glucose/H+ symport.
Keywords: transport proteins, membrane proteins, proton transport, surface-enhanced infrared spectroscopy, major facilitator superfamily, transport, membrane transport, spectroscopy, glucose transport, major facilitator family, pK value
The cellular uptake of monosaccharides, polyols, and other small carbon compounds across the membranes of eukaryotic cells is an essential physiological process. In humans, their transport is facilitated by specialized transporters that are members of the GLUT family (SLC2 gene family), which belongs to the major facilitator superfamily (MFS), one of the largest protein families with over 10,000 members (1).
There are 14 human GLUT proteins; they share sequence homology with 19–65% identity but differ in tissue distribution, substrate selectivity, and substrate affinity to meet local physiological needs. GLUTs have been implicated in several diseases, including cancer (2, 3) and diabetes (4). In plants, monosaccharide and sucrose transporters play a fundamental role in stress responses and developmental processes, including seed germination and balanced growth (5, 6). Yeast glucose transporters are essential for glucose uptake and metabolism during sugar fermentation and alcohol production (7). Most GLUT proteins catalyze the facilitative (energy-independent) bidirectional transfer of their substrates across membranes, and they may exhibit either symmetric or asymmetric transport kinetics. On the other hand, most bacterial GLUT homologs, such as GlcPSe (8), AraE (9), GalP (10), and XylE (11), are H+ symporters and depend on the electrochemical proton gradient (ΔμH+).
GLUTs and their homologs have 12 transmembrane α-helices organized as two domains (N- and C-halves), which form a central, solvent-accessible, cavity: the substrate-binding site. MFS proteins presumably share an alternating access mechanism of transport in which the substrate-binding site alternatively opens to either side of the membrane (12). The available X-ray crystal structures of GLUTs and their homologs are consistent with this model; they include structures for the inward-facing (open to the cytoplasmic side) (13–15), outward-facing (open to the periplasmic side) (11, 16), and occluded conformations (17–19).
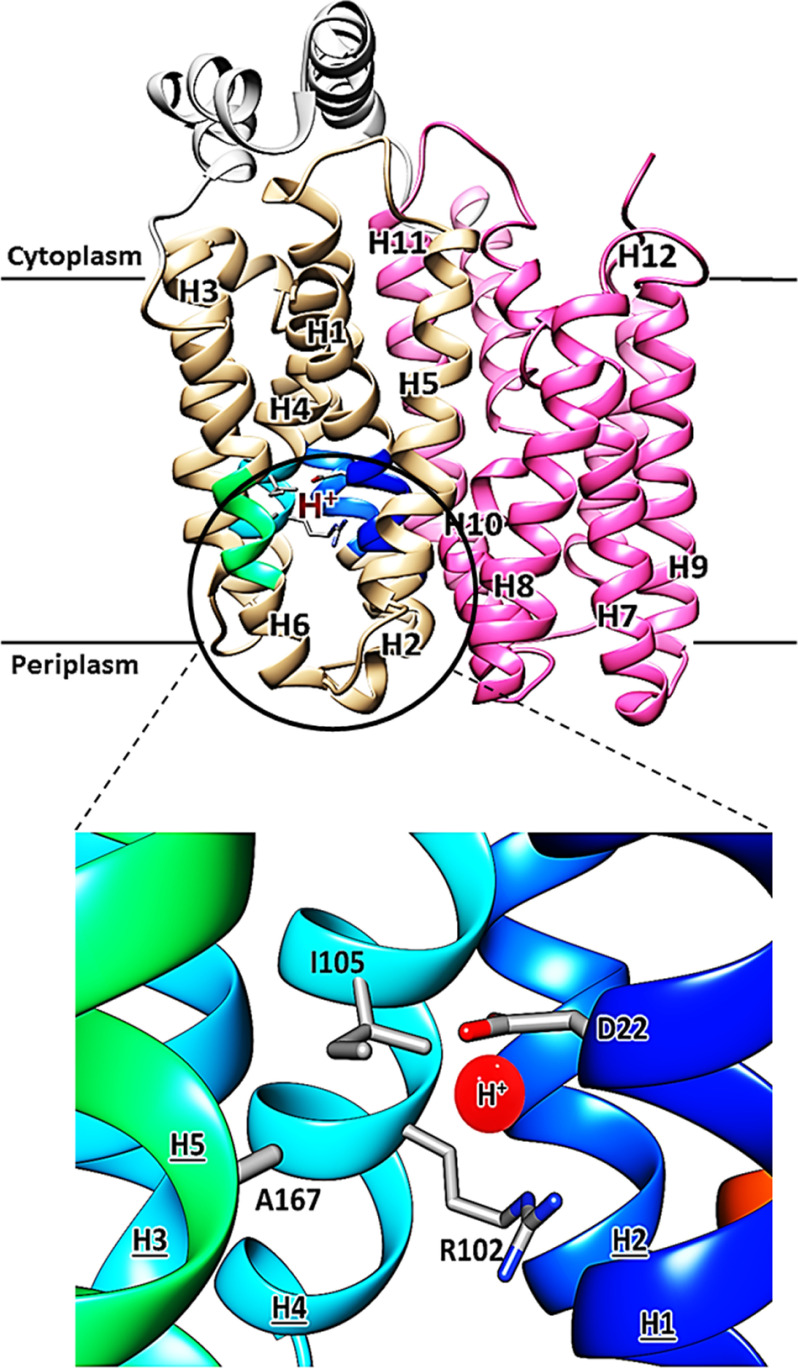
The crystal structure of the Staphylococcus epidermidis glucose/H+ symporter, a bacterial GLUT homolog, was solved in the inward-facing conformation (8). GlcPSe shares high sequence identity (27–34%) and homology (49–58%) with the human GLUTs, is highly specific for glucose, and is inhibited by the well-characterized inhibitors of human GLUTs phloretin, cytochalasin B, and forskolin (8). Residues with potential H+-binding capabilities were identified; however, unlike some bacterial MFS transporters, for example, XylE, GlcPSe has only two charged residues in the H+-binding site: Asp22 in helix 1, involved in the proton-binding site, and Arg102 from helix 4, which may form a salt bridge with Asp22 when the proton is absent (Fig. 1). Indeed, crystal structures of XylE obtained at acidic (pH 5.8) and alkaline (pH 9.6) pH were in the inward- and outward-facing conformations, respectively, with Asp27 (the equivalent of Asp22 in GlcPSe) interacting with Arg133 (the equivalent of Arg103 in GlcPSe) in the outward-facing conformation but not in the inward-facing conformation (11, 20).
Figure 1.
Overall structure of the inward-facing conformation of GlcPSe showing the mutated positions in the proton-binding site (Protein Data Bank code 4LDS). Asp22 is from helix H1, Ile105 and Arg102 are from helix H4, and Ala167 is from helix H6. The red ball indicates the putative proton-binding site.
For both GlcPSe and XylE, a transport mechanism is discussed where the transition between the inward-facing and outward-facing conformations involves a 24° relative rotation of the N- and C-domains around an axis that passes through the substrate-binding site, suggesting that translocation of glucose is coupled with conformational changes associated with the transition between the outward- and inward-facing conformations. The involvement of Asp22 in the H+-binding site was shown by the loss of a pH effect on entrance counterflow in proteoliposomes and of active glucose transport in right-side-out vesicles (8).
It was proposed that, in the absence of a proton, Asp22 and Arg102 form a salt bridge to juxtapose helices 1 and 4, thereby opening the substrate cavity wide. When the proton binds to Asp22, the salt bridge breaks, and helices 1 and 4 rearrange to decrease the size of the substrate cavity; this then lowers the energetic barrier of the transporter's conformations, and the glucose is transported (8). This proposal has been supported by structural studies and molecular dynamics simulations of XylE (11, 20). Bazzone et al. (21) suggested that H+/sugar coupling is incomplete in GlcPSe, leading to the unexpected finding that, depending on the experimental conditions, GlcPSe can function as a symporter or show partially uncoupled transport modes. It has been proposed that the existence of an H+-binding site is necessary but not a sufficient requirement for H+/sugar symport (21), leading to the hypothesis that an additional structural element for strictly coupled transport would be necessary, such as an acidic residue.
Here we study the pH- and substrate-dependent conformational changes in purified GlcPSe by IR spectroscopy for WT and several H+-binding site mutants, including A167E. This mutation is at a position proposed to play the role of the additional structure element crucial for strict symport. The suggestion is based on the analogy with the residue at position 206 in XylEEc (22). The pKa of Asp22 was determined by monitoring pH-induced changes of the COOH vibrational modes of the purified GlcPSe immobilized on a gold layer in an attenuated total reflectance (ATR) cell in analogy to the surface-enhanced absorption IR spectroscopy (SEIRAS) approach used to study the pKa value of Glu325 in LacY, the most-studied MFS transporter (23). The effect of the mutations on the pKa value of Asp22 is discussed.
Results
SEIRAS spectra and assignment of the signal for residue Asp22
IR difference spectra have been measured to monitor the protonation/deprotonation reactions and coupled conformational changes. Fig. 2 shows the difference spectra that were obtained by subtracting spectra acquired from samples equilibrated at pH 5.5 from spectra collected of samples equilibrated at a pH that completely or almost completely deprotonates Asp22 (pH 9.5 or 8.5, respectively), in the absence of glucose. The grey line shows the inverse reaction and thus a high degree of reaction reversibility.
Figure 2.

Perfusion-induced FTIR difference spectra of GlcPSe WT (A) and GlcPSe D22A (B) obtained from the sample equilibrated at pH 5.5 without sugar subtracted from the sample equilibrated at pH 8.5 (black line) and the reverse subtraction (grey line).
The difference spectra reflect structural reorganization within GlcPSe caused by the shift in pH, including changes in the protein backbone conformations and the protonation state of individual side chains; for example, the COOH vibrational mode of protonated acidic residues is typically ∼1750 cm−1 (24, 25). Distinguishable positive and negative signals reflect these conformational changes. The position of the COOH vibrational mode that is specific for the hydrophobicity of the environment of a protonated acidic residue is only 4 cm−1 higher than the one recently observed for Glu325 in LacY (23, 26), indicating a similar microenvironment (Fig. 2A). The difference spectra for the same pH step of D22A GlcPSe (Fig. 2B) is almost identical to the WT one, except for the important signal at 1750 cm−1, confirming that this signal arises from Asp22, a residue previously suggested to play a similar role to Glu325 in LacY (23, 26).
The spectra of both WT and D22A GlcPSe have signals in the so-called amide I region, between 1690 and 1620 cm−1, representing contributions of the protein backbone and individual side chains. The position of the amide I backbone signal is specific for the type of secondary structure. Signals at ∼1650 cm−1 correspond to α-helices, and those at ∼1642 cm−1 indicate disordered structures (27); β-sheets are commonly observed at ∼1636 and 1670–1690 cm−1. Depending on the environment of each secondary structure element, a shift may take place as described by so-called “doorway” modes identified by calculations on IR modes for KcsA K+ channels (28). In that study, α-helices, which reorganize during ion transport, show shifts to positions usually assigned to β-sheet structures. The signals seen in the amide I range for GlcPSe reflect similar conformational changes.
H/D exchange was performed to probe the amide region (Fig. S1). In the amide II region near 1570 cm−1, where the contribution from the protein backbone includes the in-plane N–H bending (40–60%) coupled to the ν(C–N) (20–40%) vibrational mode, characteristic signals are seen at ∼1509 cm−1 (29). Upon H/D exchange (Fig. S1B), the amide II band intensity decreases, the in-plane N–H (N–D) bending mode uncouples and appears in the 940–1040 cm−1 region, and the ν(C–N) moves to 1488 and 1461 cm−1, mixing with other modes to form a new band called amide II*. The changes in the spectra upon H/D exchange confirm that most of the signals in the difference spectra at positions lower than 1700 cm−1 originate from the protein backbone.
SEIRAS spectra of mutations at the proton-binding site
Mutations of residues in critical positions for proton transfer and glucose transport activity have been studied at different pH values by SEIRAS. Fig. 3A shows the data obtained for pH 7.5, 8.5, and 9.5, with (spectra d–f) and without (spectra a–c) glucose, in WT GlcPSe. Whereas glucose does not seem to influence the signal at 1750 cm−1, major changes occur in the amide I and II regions, indicating differences in the pH-dependent conformational reorganization of the protein in the presence of glucose. In all spectra, the steps to pH 8.5 and 9.5 are very similar, whereas the steps between pH 8.5 and 7.5 show a significant difference.
Figure 3.
SEIRA difference spectra of the pH-dependent change of the GlcPSe WT (A), D22A (B), I105S (C), A167E (D), and A167E/I105G (E). Signal from sample equilibrated at pH 5.5 was subtracted from that equilibrated at pH 7.5 (spectra a), 8.5 (spectra b), and 9.5 (spectra c) in the absence of glucose or the sample equilibrated at pH 5.5 subtracted from that equilibrated at pH 7.5 (spectra d), 8.5 (spectra e), and 9.5 (spectra f) in the presence of glucose.
In an analogous study performed on LacY with the same approach (23), the presence of 4-nitrophenyl-R-d-galactopyranoside did not influence the conformational changes. However, it is noted that 4-nitrophenyl-R-d-galactopyranoside shows very different transport rates, and direct comparison is difficult.
I105S was previously studied in GlcPSe to investigate why human GLUT2, which seemingly can have a proton-binding site (Asp22 of GlcPSe is Asp27 of GLUT2), is a uniporter and not a proton symporter (8). Close to the proton-binding site, GlcPSe has Ile105, whereas in the same position, GLUT2 has Ser161, which can come within hydrogen bond distance from Asp27. Therefore, it was proposed that I105S GlcPSe would exhibit impaired active glucose transport. Indeed, this mutant had no active glucose transport in right-side-out vesicles and had an attenuated pH effect in the entrance counterflow glucose transport in proteoliposomes, although much less drastically than the D22N mutant (8). Nonetheless, we find that I105S substitution does not perturb the signal at 1750 cm−1, although some changes in the amide I and II region suggest smaller structural changes (Fig. 3C).
Interestingly, for A167E GlcPSe (Fig. 3D), the signal at 1750 cm−1 and the signal at 1724 cm−1 are lost in the absence of glucose and restored at 2 cm−1 lower position in the presence of the substrate. The introduced Glu167 changes the environment of Asp22, pointing toward an interaction between Asp22 and Glu167, eventually introduced by a conformational change. In XylE, Asp27 (the equivalent of Asp22), interacts with Glu206 (the equivalent of Ala167 in GlcPSe), at both acidic (pH 5.8) and alkaline (pH 9.6) pH, although E206A has only a slight reduction in transport activity (11, 20). Residue 167, identified in analogy to XylEEc, was suggested to play a critical role in the pK switch of the H+-binding site to acidic values, to facilitate H+ release (21, 22). Nevertheless, the environments of the proton-binding sites in XylE and GlcPSe vary significantly, with that of XylE being mostly polar (amino acid residues Ser98, Ser102, Arg133, and Glu206), including structural water molecules, whereas that in GlcPSe is mostly hydrophobic, the only charged residue being the critical Arg102. Introducing A167E mutation in GlcPSe seems very disruptive, especially given the proximity of the hydrophobic bulky residue Ile105. Therefore, in analogy to XylE proton-binding site, we also generated the double mutant A167E/I105G; structural modeling suggested a better accommodation of A167E substitution in the double mutant than in the single mutant A167E (Fig. S2).
Investigation of the double mutant A167E/I105G (Fig. 3E) pointed to major structural changes. The negative signal at 1750 cm−1 was lost in all cases and replaced by a positive feature at 1732–1720 cm−1. Because positive signals in the difference spectra corroborate with the data collected at more alkaline pH values, there is no easy explanation for this new signal that is in the spectral region typical for protonated acidic residues. It indicates the presence of a protonated residue at alkaline pH, in an environment whose pH-induced changes lead to deprotonation. This would point to a strong reorganization of the residue's microenvironment, enabling a stabilization of the protonated residue despite the buffer alkalization. The introduced Glu167 residue could be contributing here, and also Asp22 could be showing a different signature caused by the modified environment in the mutant.
pKa titration
The pKa of Asp22 in WT and different mutants, in the presence and absence of glucose, was determined by plotting the difference in signal intensity at 1750 cm−1 versus pH (Fig. 4). From the ΔIR fit observed (red and black, with and without sugar, respectively), Asp22 has a pKa of 8.5 ± 0.1, a value that agrees remarkably well with the pKa obtained previously by Bazzone et al. (21) for the glucose transport, through a different approach.
Figure 4.
Titration curves for the ν(C=O) vibrational mode of Asp22 at 1750 cm−1. A, GlcPSe WT with (red circle) and without (black circle) glucose and GlcPSe D22A (blue circle). B, GlcPSe A167E with (gray circle) and without (green circle) glucose and GlcPSe I105S with (cyan circle) and without (magenta circle) glucose. The data were triplicated and are shown as means 8.5 ± 0.1. The error bars represent the standard deviation.
The mutant I105S GlcPSe, which has a serine in the proximity of the H+-binding site, would have been a possible candidate to perturb the proton binding. However, the pKa of this mutant (8.5 ± 0.1) is similar to that of WT GlcPSe, showing that Asp22 is not interacting with Ser105. The comparison of the data with and without glucose reveals that the pKa of Asp22 in WT and I105S is independent of glucose.
For the A167E mutant, the scenario is more complex. Whereas Asp22 has a pKa of 8.5 in the presence of glucose, there is no signal from Asp22 in the absence of sugar, pointing to a major structural reorganization in the environment of the residue, which might be locked in a certain conformation because of the steric disruptions in the proton-binding site (Fig. S2).
In the A167E/I105G mutant, the signal of the acidic residue at 1750 cm−1 is lost irrespective of the glucose presence. Most likely Asp22 is deprotonated even at pH 5.5, the lowest pH applied in our experiments.
Isothermal titration calorimetry (ITC) of WT and proton-binding site mutants
We examined the glucose binding affinity to purified GlcPSe WT and proton-binding site mutants, at pH 7.5 and 9.5, using isothermal calorimetry (ITC). Previous studies showed a lower Kd of glucose in GlcPSe at alkaline pH compared with that at acidic pH (21). We found a similar trend by ITC, with Kd of glucose at pH 7.5 being ∼7-fold higher than that at pH 9.5 for the WT transporter (Fig. S3 and Table S1). Nevertheless, glucose dissociation constants measured by ITC (Kd 11.7 ± 1.7 μm at pH 7.5 and 1.75 ± 0.93 μm at pH 9.5; Table S1) were ∼1000-fold lower than those obtained in solid support membrane (SSM) experiments (Kd of 12.3 mm at pH 5.5 and 2.6 mm at pH 10) (22) and comparable with the Km of glucose previously reported (∼30 μm) (8). Interestingly, mutations in the proton-binding site significantly reduced the difference in glucose affinity between pH 7.5 and 9.5, reversing the trend found with the WT. Thus, D22A, D22N, and A167E GlcPSe had 1.4–2-fold higher Kd at pH 9.5 than at pH 7.5 (Table S1). Intriguingly, compared with WT, all of these proton-binding site mutants had better glucose affinity at pH 7.5, whereas the reverse was true at pH 9.5 (Table S1).
SSM-based electrophysiology
The pH dependence of the transient currents generated by WT GlcPSe after a sugar concentration jump has previously shown an atypical behavior: a transient current of positive polarity at acidic pH and negative polarity at alkaline pH (21). The GlcPSe mutant A167E/I105G shows a very similar behavior, although the pKa is shifted to the acidic region at pH 6.8 (Fig. 5B). Unlike the WT GlcPSe, in the mutant A167E/I105G, the biphasic characteristics of the transient currents at alkaline pH are much stronger. Transient currents with both positive and negative amplitudes are not affected by the lipid-to-protein ratio (LPR) (Fig. S4), indicating a characteristic of pre–steady-state reactions. Unlike in experiments with the WT GlcPSe, in the mutant A167E/I105G, the positive phase overcompensates the amplitude with the negative polarity also at alkaline pH. Consequently, the normalized total charge does not change the polarity (21).
Figure 5.
Transient currents recorded at the SSM after a substrate concentration jump, at symmetrical pH as indicated, in GlcPSe A167E/I105G mutant. A, pH dependence at symmetrical pH as indicated. Transient currents were induced by 30 mm d-glucose at different pH values, as indicated. B, means and S.D. of the respective charge translocations (peak integrals) from at least three data sets, the apparent pKa is indicated. C, substrate dependence at symmetrical pH as indicated. Sugar-induced currents were measured at pH 8.5. The apparent Kd = 11.4 ± 3.3 mm. The error bars represent the standard deviation.
One feature that distinguishes the GlcPSe from the more classical Escherichia coli sugar transporters, e.g. LacY (30, 31), XylE, and FucP, is that the affinity toward sugar decreases with higher H+ concentration (lower pH). For the WT GlcPSe, a Kd of 2.6 mm at an alkaline pH of 10 is reported and 12.3 mm at an acidic pH of 5.5 (21). The mutant A167E/I105G shows a comparable Kd of 11.4 ± 3.3 mm at an alkaline pH of 8.5 (Fig. 5C).
Glucose transport activity of GlcPSe
WT, D22A, A167E, and I105S/A167E GlcPSe were expressed in JM-1100 E. coli cells, which lack original glucose transporters (32), and the transport activity was measured as glucose uptake in the whole cells. We found that D22A was a dead mutant, whereas A167E and I105S/A167E mutants retained 13.6 and 34.5% of WT activity, respectively (Fig. S5).
Discussion
The molecular basis of the coupling between the proton and substrate transport in MFS transporters has been the object of intense research (20, 21, 33, 34), with LacY featuring prominently as a model. In LacY, the two residues critical for proton symport are Glu325 and Arg302, located in TM helices 10 and 9, respectively, (13, 23, 35), and the proton and substrate sites are adjacent, providing a structural basis for the proton–substrate coupling. Arg302 of LacY is important for deprotonation of Glu325 (24, 36). In GlcPSe and related bacterial sugar porters, like XylE, AraE, and GalP, the proton-binding residue (Asp22 in GlcPSe) is located in TM helix 1, and the substrate and proton sites are separated (Fig. S6), making the proton–substrate coupling more obscure (8, 11).
SEIRAS investigations of the proton site in GlcPSe revealed that the pKa of Asp22 is 8.5, confirming previous studies (21). Unlike XylE, whose proton-binding site (Asp27) is surrounded by polar residues (Ser98, Ser102, Arg133, and Glu206) and even water molecules (11, 20), the only polar residue near Asp22 is Arg102 (corresponding to Arg133 in XylE), which presumably plays the role of Arg302 in LacY (8, 20). Buried acidic or basic residues can have markedly different pKa compared with when they are solvent-accessible (36). For instance, the pKa of E325 in LacY was 10.5 (27), or that of Glutamates at various internal positions in a staphylococcal nuclease ranged from 5.2 to 9.4 (37). Buried Lys residues can have a pKa as low as 5.3 (38), raising the possibility that Arg102 may have a lower pKa, as required by its proposed role in deprotonating Asp22.
When discussing the effect of pH on a proton transporter, there are two considerations. First, the pH may stabilize particular transporter conformations by affecting polar interactions and salt bridges. Second, the proton binding or release can favor or induce conformational changes in the transporter. SEIRAS studies here capture the pH-induced conformational changes independent of proton binding. Thus, D22A GlcPSe lacks the proton-binding site, but variations in pH still induce conformational changes in the transporter, which get significantly abated in the presence of glucose (Fig. 3B, spectra a–c). ITC shows an attenuation of the pH effect on glucose affinity in D22A versus the WT transporter, although it has the opposite impact on glucose affinity (Table S1). D22A abrogates ΔμH+-dependent transport, and its glucose transport activity is abolished (Fig. S4a). Nevertheless, it binds glucose better than the WT transporter at pH 7.5 (Table S1), implying that glucose binding alone is insufficient to trigger the conformational change necessary for transport. The increase in glucose affinity with pH lead to the proposal that the proton and substrate are loosely coupled in GlcPSe and that an additional element, like Glu206 in XylE, may be required for tight symport (21).
Like D22A, D22N and A167E also display reversed glucose affinities at pH 7.5 and 9.5, compared with the WT. Furthermore, all these mutants have 3- or 5-fold higher glucose affinity than the WT at pH 7.5, whereas glucose uptake in whole cells is abolished (D22A) or 86% decreased (A167E) (Fig. S5). Thus, the increase in glucose affinity does not translate in higher transport activity, even when the proton site is present, as in A167E. Nevertheless, structural modeling predicts that substitution of Ala167 with the charged long side chain of Glu disrupts the proton-binding site significantly, given the neighboring Ile105 (Fig. S2). This perturbation is reduced in the double mutant A167E/I105G, where the bulky hydrophobic side chain is removed.
The IR spectra of A167E mutant in the absence of glucose do not change significantly with pH (Fig. 3D, spectra a–c), possibly indicating that the transporter is blocked in a particular conformation. Glucose restores pH-dependent conformational variability, as evidenced by IR data (Fig. 3D, spectra d–f) and the ITC results (Fig. S3 and Table S1). On the other hand, A167E/I105G mutant responds to pH changes, whether glucose is present or not, suggesting that the double mutant is better behaved than the A167E mutant. Indeed, the transport assay showed that A167E/I105G mutant had better glucose transport activity than A167E (Fig. S5A). Expectedly, the pKa of transport for A167E/I105G of 6.8 (Fig. 5) is almost 2 pH units lower than that of the WT (Fig. 4), suggesting higher solvent exposure of the proton-binding site in this mutant.
LacY, FucP, and XylE, all show the decrease of substrate affinity with pH displayed by A167E mutant (Table S1) (22), presumably a hallmark of tightly coupled symporter. However, the same relationship between pH and glucose affinity is also present in D22A, lacking the proton-binding residue. Thus, even before proton binding, the proton gradient may play a role in selecting transporter conformations with particular affinities for the substrate.
Interestingly, similarly to GlcPSe, AraE, a pentose/proton symporter (9), and GalP, a hexose-proton symporter (10), both bacterial GLUT homologs, lack Glu206 of XylE (Ala167 in GlcPSe) having instead Leu184 or Ile177, respectively. The proton-binding sites of AraE and GalP resemble that of GlcPSe, although they do have the equivalent of Ser102 of XylE, Ser106 (AraE), or Ser99 (GalP) (Fig. S6). Given the overall proton environment, AraE and GalP may also exhibit high pKa for their carbohydrate transport. Whether they behave like GlcPSe (increased sugar affinity with increased pH) or like XylE and LacY (decreased sugar affinity with higher pH) remains to be established.
In humans, GLUT2 has D47 as the corresponding residue for Asp22 of GlcPSe, and the only significant change in the proton-binding site is Ser161 instead of Ile105. Nevertheless, I105S GlcPSe data (Fig. 3C) and a previous report (21) indicated no changes in the pKa of transport compared with that of WT GlcPSe. This raises the possibility that GLUT2 may be capable of proton symport in certain conditions, as shown for GLUT12, which has glutamate in the position of Asp22 of GlcPSe.
Conclusion
The presence of a residue with a high pKa value can be crucial to transport proteins (39, 40). The high pKa value is based on the hydrophobicity of the microenvironment and will be manipulated because of conformational changes during transport. The presence of such residues with a high pKa was observed before in other membrane protein, for example Rhodopsins (21) and cytochrome c oxidase (41). Here Asp22 seems to play this role with a pK value of 8.5. Interestingly this pKa can be corroborated with that of glucose transport. Residues within the hydrogen bonding distance to the proton site actively interact with Asp22 and change the pKa of the transport as well as the affinity of glucose and its pH dependence. These residues including an arginine, often found as a partner of acidic residues when the pKa is upshifted (41). SEIRAS of MFS proton symporters can monitor directly the protonation/deprotonation of the transporters and the backbone conformational changes associated with substrate and proton binding, providing invaluable insight into the MFS transport mechanism.
Experimental procedures
Protein purification
Site-directed mutagenesis GlcPSe was performed on the pBAD plasmid constructs of WT with C-terminal hexahistidine-tagged proteins and verified by DNA sequencing (42). WT, D22A, D22N, I105S, and A167E GlcPSe were expressed in C41 E. coli cells and purified as previously described (8).
Glucose transport assay
Glucose transport assay of WT and mutant GlcPSe in whole cells was performed as previously described (8). GlcPSe WT and D22A, A167E, and I105S/A167E mutants in pBAD vector and empty pBAD vector were transformed into E. coli JM-1100 cells. The cells were grown in 10 ml of LB medium with 50 µg/ml ampicillin at 37 °C with shaking (200 rpm). At cell A600 nm of 0.6–0.8, GlcPSe expression was induced with 0.3 mm l-arabinose, and cells were grown for 2–3 h. The cells were collected by centrifugation at 4000 × g, 5 min, and the cell pellet was resuspended in 5–10 ml of PBS buffer (10 mm Na2HPO4, 1.8 mm KH2PO4, 2.7 mm KCl, 137 mm NaCl, pH 7.4) to wash the cells. This was followed by another centrifugation and resuspension of the cell pellet to A600 nm ∼3.0 in 0.1 m KPi, pH 7.5, 10 mm MgSO4. Each assay had 50 µl of this cell solution. The transport was initiated by the addition of 50 μm [C14]glucose and stopped by the addition of 3-ml ice-chilled Quench buffer (0.1 m KPi, 0.1 m LiCl, pH 5.5), followed by filtration on cellulose nitrate membrane filters (catalog no. 7184-002; Whatman plc), and two more washes with 3 ml of quench buffer. The filters were then transferred into scintillation vials, combined with 10-ml scintillation mixture (BioSafeII; Research Products International, Mount Prospect, IL, USA), and after brief vortexing, the radioactivity was measured with a scintillation counter (Tri-carb 2900TR; PerkinElmer). The cells expressing the pBAD vector without GlcPSe provided the control for the background glucose transport activity. To quantify protein expression in JM-1100 cells, cell pellets from 3 ml of cell solution A600 nm 3.0 (same as that used for transport assay) were resuspended in 300 μl of PBS buffer, combined with 300 μl of 2× SDS-PAGE sample loading buffer, and disrupted by sonication. For Western blotting, 10 μl of the broken cell solution were loaded in each SDS-PAGE gel well. We used horseradish peroxidase–conjugated pentahistidine antibody (Qiagen) and visualized the Western blotting with chemiluminescent substrate (Thermo Fisher Scientific), according to the manufacturer's instructions.
ITC of GlcPSe
The experiment was performed on Affinity ITC (TA Instruments, New Castle, DE, USA). The sample cell contained 300 µl of purified protein at 7.5 mg/ml (0.15 mm) in 0.1 m KPi buffer at pH 7.5 or 9.5, with 0.1 m NaCl, and 0.05% (w/v) dodecylmaltoside (DDM). Glucose, at 1.5 mm concentration, dissolved in the corresponding buffer, was titrated as 3 µl/injection in 30 injection steps. The data were processed with NanoAnalyze (TA Instruments).
Surface modification of the silicon crystal and protein immobilization
Before the formation of the gold film, the crystal was polished with finer grade 0.3-μm alumina, follow by rinsing with copious amounts of Millipore water, acetone, and water again. On the surface of a silicon ATR crystal, a thin gold layer was formed by chemical deposition as described previously (43). The crystal was dried under an argon stream, and 40% NH4F (w/v) was added for 1 min to remove the silicon oxide layer and to terminate with hydrogen; finally the surface was rinsed and dried again. The crystal was heated at 65 °C for 10 min together with the plating solution. The composition of the solution was a 1:1:1 mix (v/v/v) of 15 mm NaAuCl4, 150 mm Na2SO3, 50 mm Na2S2O3, 50 mm NH4Cl, and 2% HF (w/v: 1 ml). After reaching the temperature, the prism was covered with the solution for 40 s and stopped by washing the plating solution with water, followed by drying with a stream of argon.
After formation of gold layer on the silicon crystal, a nickel–nitrilotriacetic acid self-assembled monolayer was adapted from Refs. 43 and 44. First, 1 mg/ml of 3,3-dithiodipropionic acid di(N-hydroxysuccinimide ester) in DMSO was allowed to self-assemble for 1 h. After monolayer formation, excess 3,3-dithiodipropionic acid di(N-hydroxysuccinimide ester) was washed away with DMSO, and the crystal was dried under an argon stream. Afterward, the self-assembled monolayer was immersed with 100 mm Nα,Nα-bis(carboxymethyl)-l-lysine in 0.5 m K2CO3 at pH 9.8 for 3 h and then rinsed with water. Finally, the surface was incubated in 50 mm Ni(ClO4)2 for 1 h. After washing with water, 2–35 mg/ml of the protein GlcPSe dissolved in 50 mm phosphate buffer containing 0.05% (w/v) n-dodecyl-β-maltoside was deposited on the modified gold surface for 1 h.
IR spectroscopy
A simultaneous acquisition of FTIR spectra in the ATR mode with perfusion of solutions was used. A silicon crystal with 3-mm surface diameter was used as a single reflection ATR unit. The experiments were carried out with a Bruker Vertex 70 FTIR spectrometer (Globar source, KBr Beamsplitter, mercury cadmium telluride detector) at 8-mm aperture and 40-kHz scanner velocity. The measurements were carried out at ∼7 °C, and the solutions were kept on ice before use. Before each perfusion step, the input tube was carefully washed with water and buffer, and the pump speed was kept constant at a flow rate of 0.2 ml/min.
Secondary structure determination and protein orientation on the surface
SEIRAS spectra have been obtained for the protein immobilized via its His tag on a silicon crystal, covered with a gold nanostructure. The immobilization ensures that the protein concentration remains stable during the experiments. The deconvolution of the amide I band of the absorbed protein (Fig. S7) confirmed the structural integrity of the protein after immobilization. For each experiment the immobilization was followed spectroscopically to ensure a high degree of reproducibility.
The orientation of the amide I mode was studied with polarized IR light, showing that at pH 7.5 the protein is oriented perpendicular to the ATR crystal, and at pH values higher and lower the protein shows some inclination, most likely because of the charges on the surface of the protein (Table S2). Nevertheless, the secondary structures of each pH value and of the mutants were found to be unchanged within the experimental error of the technique (3–5%) (Fig. S6).
Difference spectra of GlcPSe in the presence or absence of glucose
To monitor pH-induced difference spectra, we used one perfusion buffer with constant pH value 5.5 (50 mm KPi, 200 mm NaCl, 0.05% (w/v) DDM) and a second perfusion solution with the same composition but at different pH values ranging from 7.5 to 10.5. First, the system was equilibrated with the KPi (pH 5.5) for 20 min, then a spectrum was recorded as background, and the perfusion solution was changed to the second solution (pH 7.5–10.5). After 20 min (pH 7.5–10.5) minus pH 5.5 difference spectra were recorded. The new state of the protein was recorded as background, and the solution was changed to pH 5.5. Again, after 20 min, the pH 5.5 minus (pH 7.5–10.5) difference spectra were obtained. The same experiments that have been performed in the absence of glucose were performed in the presence of glucose. Glucose was dissolved in 50 mm KPi, 200 mm NaCl, 0.05% (w/v) DDM (pH 5.5) at a final concentration of 100 μm. The data were normalized based on the absorbance spectra obtained at the beginning of each experiment, when the data from different samples had to be compared.
Reconstitution of GlcPSe into proteoliposomes
Reconstitution of purified proteins (2 mg/ml) was carried out with E. coli phospholipids (E. coli polar lipid extract; Avanti Polar Lipids, Alabaster, AL, USA). Preformed liposomes (0.2–2 ml, 10 mg/ml) dissolved in 1% (w/v) octyl-glucoside and the protein suspension were mixed on ice to their respective concentration, to result in different LPR. The LPR 5 sample was used for all SSM measurements of the pH and glucose titrations.
All proteins were reconstituted using overnight incubation in 400 mg/ml BioBeads (SM-2 adsorbent media; Bio-Rad) at 4 °C. After reconstitution, the samples were diluted to 2.5 mg/ml lipid concentration, frozen in liquid nitrogen, and stored at −80 °C.
SSM-based electrophysiology
SSM measurements were performed as described previously (21, 30, 45, 46). The nonactivating buffer and activating solutions were prepared in 100 mm KPi, 1 mm DTT at pH 7.5. The nonactivating solution contained 30 mm xylose, the activating solution contained 30 mm glucose for the pH jumps, and the pH of 8.5 was applied for the glucose titration. Glucose concentration and pH jumps experiments were carried out respectively on the same sensor.
Data availability
All data shown are available in the article and the supporting information.
Supplementary Material
Acknowledgments
P. H. is grateful to the late H. Ronald Kaback who introduced her to the field of transport proteins and shared his enthusiasm.
This article contains supporting information.
Author contributions—A. F. S. S., B. P., and M. G. M. investigation; A. F. S. S. visualization; A. F. S. S. and J.-Y. C. writing-review and editing; data curation; C. V. I. and J.-Y. C. formal analysis; M. G. M. and P. H. validation; J.-Y. C. and P. H. supervision; J.-Y. C. and P. H. project administration; P. H. funding acquisition; P. H. writing-original draft.
Funding and additional information—This work was supported by National Institutes of Health Grant R01-GM123103 (to J.-Y. C.), University of Strasbourg Institute for Advanced Studies (USIAS) Grant USIAS-2018-060 (to P. H.), and Grant ANR-10-LABX-0026_CSC (to P. H.). A. F. S. S. was supported by the programme doctoral international - initiative d'excellence (PDI-IDEX) program from the University of Strasbourg. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- GlcPSe
- glucose permease from S. epidermidis
- SEIRAS
- surface-enhanced absorption infrared spectroscopy
- ATR
- attenuated total reflectance
- MFS
- major facilitator superfamily
- SSM
- solid support membrane
- H/D
- hydrogen/deuterium
- ITC
- isothermal calorimetry
- DDM
- dodecylmaltoside
- LPR
- lipid-to-protein ratio.
References
- 1. Mueckler M., and Thorens B. (2013) The SLC2 (GLUT) family of membrane transporters. Mol. Aspects Med. 34, 121–138 10.1016/j.mam.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Medina R. A., and Owen G. I. (2002) Glucose transporters: expression, regulation and cancer. Biol. Res. 35, 9–26 10.4067/s0716-97602002000100004 [DOI] [PubMed] [Google Scholar]
- 3. Airley R. E., and Mobasheri A. (2007) Hypoxic regulation of glucose transport, anaerobic metabolism and angiogenesis in cancer: novel pathways and targets for anticancer therapeutics. Chemotherapy 53, 233–256 10.1159/000104457 [DOI] [PubMed] [Google Scholar]
- 4. Watson R. T., and Pessin J. E. (2001) Intracellular organization of insulin signaling and GLUT4 translocation. Recent Prog. Horm. Res. 56, 175–193 10.1210/rp.56.1.175 [DOI] [PubMed] [Google Scholar]
- 5. Williams L. E., Lemoine R., and Sauer N. (2000) Sugar transporters in higher plants: a diversity of roles and complex regulation. Trends Plant. Sci. 5, 283–290 10.1016/S1360-1385(00)01681-2 [DOI] [PubMed] [Google Scholar]
- 6. Slewinski T. L. (2011) Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. Mol. Plant 4, 641–662 10.1093/mp/ssr051 [DOI] [PubMed] [Google Scholar]
- 7. Kruckeberg A. L., and Dickinson J. R. (2004) Carbon metabolism. Metabolism and Molecular Physiology of Saccharomyces cerevisiae (Dickinson JR, Schweizer M, eds) 2nd Ed., pp. 42–103, CRC Press, London [Google Scholar]
- 8. Iancu C. V., Zamoon J., Woo S. B., Aleshin A., and Choe J. Y. (2013) Crystal structure of a glucose/H+ symporter and its mechanism of action. Proc. Natl. Acad. Sci. U.S.A. 110, 17862–17867 10.1073/pnas.1311485110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maiden M. C., Jones-Mortimer M. C., and Henderson P. J. (1988) The cloning, DNA sequence, and overexpression of the gene araE coding for arabinose-proton symport in Escherichia coli K12. J. Biol. Chem. 263, 8003–8010 [PubMed] [Google Scholar]
- 10. Sanderson N. M., Qi D., Steel A., and Henderson P. J. (1998) Effect of the D32N and N300F mutations on the activity of the bacterial sugar transport protein, GalP. Biochem. Soc. Trans. 26, S306 10.1042/bst026s306 [DOI] [PubMed] [Google Scholar]
- 11. Sun L., Zeng X., Yan C., Sun X., Gong X., Rao Y., and Yan N. (2012) Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 490, 361–366 10.1038/nature11524 [DOI] [PubMed] [Google Scholar]
- 12. Smirnova I., Kasho V., and Kaback H. R. (2011) Lactose permease and the alternating access mechanism. Biochemistry 50, 9684–9693 10.1021/bi2014294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., and Iwata S. (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615 10.1126/science.1088196 [DOI] [PubMed] [Google Scholar]
- 14. Huang Y., Lemieux M. J., Song J., Auer M., and Wang D.-N. (2003) Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301, 616–620 10.1126/science.1087619 [DOI] [PubMed] [Google Scholar]
- 15. Newstead S., Drew D., Cameron A. D., Postis V. L., Xia X., Fowler P. W., Ingram J. C., Carpenter E. P., Sansom M. S., McPherson M. J., Baldwin S. A., and Iwata S. (2011) Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 30, 417–426 10.1038/emboj.2010.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dang S., Sun L., Huang Y., Lu F., Liu Y., Gong H., Wang J., and Yan N. (2010) Structure of a fucose transporter in an outward-open conformation. Nature 467, 734–738 10.1038/nature09406 [DOI] [PubMed] [Google Scholar]
- 17. Yin Y., He X., Szewczyk P., Nguyen T., and Chang G. (2006) Structure of the multidrug transporter EmrD from Escherichia coli. Science 312, 741–744 10.1126/science.1125629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan H., Huang W., Yan C., Gong X., Jiang S., Zhao Y., Wang J., and Shi Y. (2013) Structure and mechanism of a nitrate transporter. Cell Rep. 3, 716–723 10.1016/j.celrep.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 19. Hirai T., Heymann J. A. W., Shi D., Sarker R., Maloney P. C., and Subramaniam S. (2002) Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol. 9, 597–600 10.1038/nsb821 [DOI] [PubMed] [Google Scholar]
- 20. Wisedchaisri G., Park M. S., Iadanza M. G., Zheng H., and Gonen T. (2014) Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat. Commun. 5, 4521 10.1038/ncomms5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bazzone A., Zabadne A. J., Salisowski A., Madej M. G., and Fendler K. (2017) A loose relationship: incomplete H+/sugar coupling in the MFS sugar transporter GlcP. Biophys. J. 113, 2736–2749 10.1016/j.bpj.2017.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bazzone A., Madej M. G., Kaback H. R., and Fendler K. (2016) pH regulation of electrogenic sugar/H+ symport in MFS sugar permeases. PLoS One 11, e0156392 10.1371/journal.pone.0156392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grytsyk N., Sugihara J., Kaback H. R., and Hellwig P. (2017) pKa of Glu325 in LacY. Proc. Natl. Acad. Sci. U.S.A. 114, 1530–1535 10.1073/pnas.1621431114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zscherp C., Schlesinger R., Tittor J., Oesterhelt D., and Heberle J. (1999) In situ determination of transient pKa changes of internal amino acids of bacteriorhodopsin by using time-resolved attenuated total reflection Fourier-transform infrared spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 96, 5498–5503 10.1073/pnas.96.10.5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wolpert M., and Hellwig P. (2006) Infrared spectra and molar absorption coefficients of the 20 α amino acids in aqueous solutions in the spectral range from 1800 to 500 cm−1. Spectrochim. Acta A Mol. Biomol. Spectrosc. 64, 987–1001 10.1016/j.saa.2005.08.025 [DOI] [PubMed] [Google Scholar]
- 26. Grytsyk N., Seiça A. F. S., Sugihara J., Kaback H. R., and Hellwig P. (2019) Arg302 governs the pKa of Glu325 in LacY. Proc. Natl. Acad. Sci. U.S.A. 116, 4934–4939 10.1073/pnas.1820744116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barth A. (2007) Infrared spectroscopy of proteins. Biochim. Biophys. Acta 1767, 1073–1101 10.1016/j.bbabio.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 28. Stevenson P., Götz C., Baiz C. R., Akerboom J., Tokmakoff A., and Vaziri A. (2015) Visualizing KcsA conformational changes upon ion binding by infrared spectroscopy and atomistic modeling. J. Phys. Chem. B 119, 5824–5831 10.1021/acs.jpcb.5b02223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vigano C., Smeyers M., Raussens V., Scheirlinckx F., Ruysschaert J. M., and Goormaghtigh E. (2004) Hydrogen–deuterium exchange in membrane proteins monitored by IR spectroscopy: a new tool to resolve protein structure and dynamics. Biopolymers 74, 19–26 10.1002/bip.20035 [DOI] [PubMed] [Google Scholar]
- 30. Garcia-Celma J. J., Smirnova I. N., Kaback H. R., and Fendler K. (2009) Electrophysiological characterization of LacY. Proc. Natl. Acad. Sci. U.S.A. 106, 7373–7378 10.1073/pnas.0902471106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Viitanen P. V., Garcia M. L., Foster D. L., Kaczorowski G. J., and Kaback H. R. (1983) Mechanism of lactose translocation in proteoliposomes reconstituted with lac carrier protein purified from Escherichia coli: 2. Deuterium solvent isotope effects. Biochemistry 22, 2531–2536 10.1021/bi00279a034 [DOI] [PubMed] [Google Scholar]
- 32. Henderson P. J., Giddens R. A., and Jones-Mortimer M. C. (1977) Transport of galactose, glucose and their molecular analogues by Escherichia coli K12. Biochem. J. 162, 309–320 10.1042/bj1620309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zilberstein D., Schuldiner S., and Padan E. (1979) Proton electrochemical gradient in Escherichia coli cells and its relation to active transport of lactose. Biochemistry 18, 669–673 10.1021/bi00571a018 [DOI] [PubMed] [Google Scholar]
- 34. Kaback H. R. (2015) A chemiosmotic mechanism of symport. Proc. Natl. Acad. Sci. U.S.A. 112, 1259–1264 10.1073/pnas.1419325112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sahin-Toth M., and Kaback H. R. (2001) Arg-302 facilitates deprotonation of Glu-325 in the transport mechanism of the lactose permease from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 98, 6068–6073 10.1073/pnas.111139698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Isom D. G., Cannon B. R., Castañeda C. A., Robinson A., and García-Moreno B. (2008) High tolerance for ionizable residues in the hydrophobic interior of proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 17784–17788 10.1073/pnas.0805113105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Isom D. G., Castañeda C. A., Cannon B. R., Velu P. D., and García-Moreno E. B. (2010) Charges in the hydrophobic interior of proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 16096–16100 10.1073/pnas.1004213107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Isom D. G., Castañeda C. A., Cannon B. R., and García-Moreno B. (2011) Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl. Acad. Sci. U.S.A. 108, 5260–5265 10.1073/pnas.1010750108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gutteridge A., and Thornton J. M. (2005) Understanding nature's catalytic toolkit. Trends Biochem. Sci. 30, 622–629 10.1016/j.tibs.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 40. Harris T. K., and Turner G. J. (2002) Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life 53, 85–98 10.1080/15216540211468 [DOI] [PubMed] [Google Scholar]
- 41. Brändén G., Pawate A. S., Gennis R. B., and Brzezinski P. (2006) Controlled uncoupling and recoupling of proton pumping in cytochrome c oxidase. Proc. Natl. Acad. Sci. U.S.A. 103, 317–322 10.1073/pnas.0507734103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Braman J., Papworth C., and Greener A. (1996) Site-directed mutagenesis using double-stranded plasmid DNA templates. Methods Mol. Biol. 57, 31–44 [DOI] [PubMed] [Google Scholar]
- 43. Ataka K., Giess F., Knoll W., Naumann R., Haber-Pohlmeier S., Richter B., and Heberle J. (2004) Oriented attachment and membrane reconstitution of His-tagged cytochrome c oxidase to a gold electrode: in situ monitoring by surface-enhanced infrared absorption spectroscopy. J. Am. Chem. Soc. 126, 16199–16206 10.1021/ja045951h [DOI] [PubMed] [Google Scholar]
- 44. Kriegel S., Uchida T., Osawa M., Friedrich T., and Hellwig P. (2014) Biomimetic environment to study E. coli complex I through surface-enhanced IR absorption spectroscopy. Biochemistry 53, 6340–6347 10.1021/bi500955a [DOI] [PubMed] [Google Scholar]
- 45. Schulz P., Garcia-Celma J. J., and Fendler K. (2008) SSM-based electrophysiology. Methods Enzymol. 46, 97–103 10.1016/j.ymeth.2008.07.002 [DOI] [PubMed] [Google Scholar]
- 46. Garcia-Celma J. J., Dueck B., Stein M., Schlueter M., Meyer-Lipp K., Leblanc G., and Fendler K. (2008) Rapid activation of the melibiose permease MelB immobilized on a solid-supported membrane. Langmuir 24, 8119–8126 10.1021/la800428h [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data shown are available in the article and the supporting information.