Abstract
Background
To fully elucidate the roles and mechanisms of plant hormones in leaf senescence, we adopted an integrated analysis of both non-senescing and senescing leaves from red maple with transcriptome and metabolome data.
Results
Transcription and metabolite profiles were generated through a combination of deep sequencing, third-generation sequencing data analysis, and ultrahigh-performance liquid chromatograph Q extractive mass spectrometry (UHPLC-QE-MS), respectively. We investigated the accumulation of compounds and the expression of biosynthesis and signaling genes for eight hormones. The results revealed that ethylene and abscisic acid concentrations increased during the leaf senescence process, while the contents of cytokinin, auxin, jasmonic acid, and salicylic acid continued to decrease. Correlation tests between the hormone content and transcriptional changes were analyzed, and in six pathways, genes closely linked with leaf senescence were identified.
Conclusions
These results will enrich our understanding of the mechanisms of plant hormones that regulate leaf senescence in red maple, while establishing a foundation for the genetic modification of Acer in the future.
Keywords: Acer rubrum, Transcriptome, Metabolomics, Hormone pathways, Leaf senescence
Background
The senescence of plant leaves is a proactive and genetically precise process that is regulated by a series of internal and external factors [1]. Many environmental stresses, such as extreme temperatures, drought, nutrient deficiencies, inadequate light/shadowing, or complete darkness, and biotic stress (e.g., bacterial infections) can induce senescence [2, 3]. Phytohormone levels are determined by plant age and stress during the induction and propagation of leaf senescence [4]. These internal and external factors may act alone or in combination. Among these, plant hormones are one class of organics that are generated in certain parts of plants, which can regulate almost all aspects of plant growth and development, while inducing physiological reactions at low concentrations [5]. Currently, research on the regulation and mechanisms of plant hormones in plant senescence has made clear progress [6].
According to the role of plant senescence, previous research has shown that, in general, phytohormones can be divided into two types: senescence promoters and retardants [7]. The former encompass ethylene (ET), abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA), and brassinosteroid (BR), whereas the latter include cytokinin (CK), auxin (IAA), and gibberellin (GA). A series of studies have confirmed that ethylene promotes senescence when the leaves reach a certain age or are mature enough [8]. Research has also shown that abscisic acid inhibits stomata closure during leaf senescence, which leads to significant water loss in leaves and their subsequent death. Simultaneously, it ensures that there is sufficient oxygen available in leaves for increased breathing during aging [9]. There has been considerable progress in the study of the role of jasmonic acid and salicylic acid in leaf senescence [10, 11]. The role of brassinosteroid in leaf senescence has been primarily addressed in studies of physiology and genetics. Following the treatment of leaves with brassinosteroid (24-Epibrassiolide; EBR), the malondialdehyde content of plants increases and reactive oxygen species is inhibited, which leads to leaf senescence [12]. However, its unique role in aging needs to be investigated in depth. In contrast, it has been verified that cytokinins have an inhibitory effect on leaf senescence [13]. The inhibitory effect of gibberellin on leaf senescence is not as obvious as for cytokinins [14]. Traditionally, auxin has been thought to inhibit plant senescence [15]. It is worthy of mention that these phytohormones do not work in isolation, and they are often employed in variable combinations to achieve the regulation of senescence [16, 17].
Leaf senescence is the final stage of the life history of leaves from maturity to demise, as they gradually yellow. This is accompanied by the transfer of nutrients from old leaves to new leaves and seeds [18]. Leaf senescence derives from the long-term evolution of plants, which assists them in adapting to environmental changes and maintaining efficient energy use [19]. However, the premature senescence of leaves seriously impacts photosynthesis time and nutrient transport in plants, which in turn reduces the yields and quality of crops [20]. The genus Acer L., also referred to as red maple, is among the most important tree and shrub genus in the northern hemisphere [21–25]. Maple trees are of high ornamental value and are attractive species for gardening. In addition to their outstanding landscape applications, many species of Acer also make contributions as major raw materials for maple sugar, maple syrup, furniture, and lumber [26]. Furthermore, a high number of maple phytochemicals have been found to possess potential antioxidant, antitumor, and anti-inflammatory activities [27–31]. Therefore, the study of leaf senescence not only has high scientific significance for exploring the life cycle and functional phenomena of this species, but also has important implications for red maple production.
The metabolism of plants is a networked relationship. The regulation of plant responses to environmental factors not only impacts individual metabolic pathways, but also influences the balance of the overall metabolic network [32]. Compared with traditional physiological studies and genetic phenotypic analyses, high-throughput omics research techniques can more systematically observe the physiological changes in plants and may have a stronger potential for discovering new genes [33, 34]. Recently, the association analyses of different omics platforms (transcriptomics and metabolomics) have gradually emerged as a new trend in the area of plant metabolism research [35–41]. For this study, we merged transcriptomics and metabolomics and studied eight common hormones in Acer rubrum (ethylene, abscisic acid, cytokinin, auxin, jasmonic acid, salicylic acid, brassinosteroid, and gibberellin). Changes in the levels of metabolites and expression levels of pathway genes in leaf senescence demonstrated the mechanisms through which different hormones regulate the process of senescence in plant leaves. This comprises the first ever complete transcriptomics and metabolomics analysis of the regulation of phytohormones involved in leaf senescence in Acer rubrum. The results of this study can enrich our understanding of the active mechanisms of phytohormones in Acer.
Methods
Plant materials and sample preparation
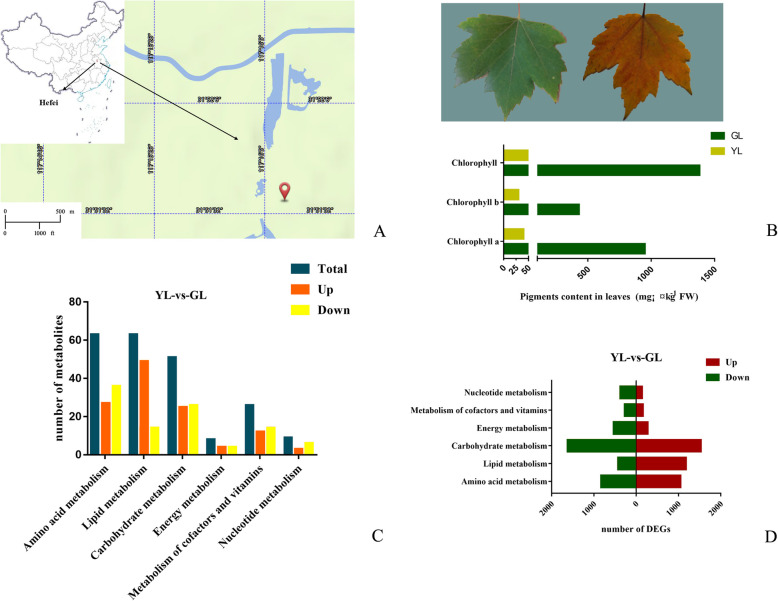
Plants were planted in our test plots at the Anhui Academy of Agricultural Sciences, Hefei, China (Fig. 1a) (31.86 N latitude, 117.27 E longitude). Plant seeds from Canada Forest Service, presented by Dr. Shiyou Li. Non-senescing leaves (fully expanded green leaves) and senescing leaves (100% yellowing leaf surface, ~ 95% chlorophyll loss) were collected in middle-to-late November. Samples (20 leaves of Acer rubrum constituting one biologicalreplicate) for non-senescing leaves and senescing leaves were subjected to freeze drying, and then stored at − 80 °C in a deep freezer. The voucher specimens were deposited in Institute of Agricultural Engineering, Anhui Academy of Agricultural Sciences. Dr. Qian Zhu undertook the formal identification of the samples. This research examines a native plant species that does not require any special permit.
Fig. 1.
Leaf color and biochemical analysis of changes in red maple during leaf senescence. a Sampling location. The source of map depicted in (a) is from BIGMAP Company. b Stages of leaf senescence. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, 100% yellowing of the leaf surface; ~ 95% chlorophyll loss. c Changes in the relative populations of biological macromolecules during leaf senescence. “Total” refers to the total number of metabolites changed from senescent to non-senescent leaves. d Statistics of differential genes in macromolecular metabolic pathways during leaf senescence. “GL-vs-YL” means “non-senescing leaves compared senescing leaves”
Metabolic analysis
Metabolite extraction
We used a mixture comprising of 40% methanol (LC–MS grade, CNW Technologies): 40% acetonitrile (LC–MS grade, CNW Technologies): 20% water (v: v: v) as an extract solvent. The ratio was 0.05 g of leaves (each sample set consisted of six biological replicates) to 1 mL of solvent, and the solutions were vortexed for 30 s. The samples were homogenized (45 Hz, 4 min, JXFSTPRP-24; Jingxin Technology), and sonicated (5 min in ice-water bath). This step was repeated three times, after which the samples were left to stand at − 20 °C for 1 h, and underwent centrifugation (Heraeus Fresco17; Thermo Fisher Scientific) for 15 min at 12000 rpm at 4 °C. The supernatants were introduced to LC-MS vials for UHPLC-QE Orbitrap/MS detection. Equivalent amounts of supernatants from all samples were mixed as quality-control (QC) samples for testing.
LC-MS/MS analysis
LC-MS/MS analysis was conducted using a UHPLC system (1290; Agilent, Technologies) and Q Exactive Orbitrap (Thermo Fisher Scientific). Chromatographic separation was performed using a UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm) [42].
The mobile phase was comprised of the following components:
Positive mode: mobile phase A: formic acid (0.1%) in water solution; mobile phase B: acetonitrile.
Negative mode: mobile phase A: ammonium acetate (5 mmol/L) in water solution; mobile phase B: acetonitrile.
The flow rate was maintained at 500 μL/min. The gradient parameters of t elution were set as follows: 0 min, 99% A, 1% B; 1 min, 99% A, 1% B; 8 min, 1% A, 99% B; 10 min, 1% A, 99% B; 10.1 min, 99% A, 1% B; 12 min, 99% A, 1% B. For the LC/MS experiments, QE mass spectrometer were able to perform MS/MS acquisition using information-dependent acquisition (IDA). In this mode, the acquisition software (Xcalibur 4.0.27, Thermo) continuously evaluates and triggers the acquisition of MS/MS spectra based on pre-selected criteria when acquiring full scan measurement MS data. Mass data acquisition was achieved in two ways: using ESI+ (electrospray ionization-positive ion mode) and ESI− (electrospray ionization-negative ion mode). The following settings were used (sheath gas flow rate: 45 Arb, auxiliary gas flow rate: 15 Arb, capillary temperature:320 °C, full ms resolution:70,000, MS/MS resolution:17500, collision energy:20/40/60 eV, spray voltage:3.8kv (positive) or − 3.1kv (negative), respectively.
Data preprocessing and annotation
ProteoWizard software was employed to convert the raw MS data (.raw) to mzML format, after which the R package XCMS (version 3.2) was used to perform retention time correction, peak identification, peak extraction, peak integration, and peak alignment [43]. OSI-SMMS (version 1.0, Dalian Dashuo Information Technology Co., Ltd.) software was used in conjunction with an in-house MS/MS database for substance identification. Multivariate statistical analyses (PCA analysis, PLS-DA analysis, opls-da analysis) were carried out using the normalized data matrix. Differential metabolites were screened through the combination of multivariate statistical analysis opls-da and single variable statistical analysis (student’s t test), whereas metabolic pathway KEGG enrichment analysis was conducted using non-commercial databases (Supplementary Table S1).
Combination sequencing analysis
RNA sequencing experiment
Transcriptome sequencing analyses were assessed as previously described [44]. A combination of next-generation sequencing (NGS) and single-molecule real-time (SMRT) sequencing were adopted for this study. The experiment involved total RNA isolation (using the RNAprep kit DP441; Tiangen,) followed by conversion to the Iso-Seq library of short cDNA fragments according to the Isoform Sequencing protocol (Iso-Seq) using the Clontech SMARTer PCR cDNA Synthesis Kit. Sequencing used the BluePippin Size Selection System protocol as described by Pacific Biosciences (PN 100–092–800-03).
KEGG enrichment and differential expression analysis
To capture the phytohormone-related genes, we utilized the KEGG (http://www.genome.jp/kegg/) online database for the pathway mapping of target genes. These included tryptophan metabolism (auxin synthesis pathway; pathway ID 00380), zeatin biosynthesis (cytokinin synthesis pathway; pathway ID 00908), diterpenoid biosynthesis (gibberellin synthesis pathway; pathway ID 00904), carotenoid biosynthesis (abscisic acid synthesis pathway; pathway ID 00906), cysteine and methionine metabolism (ethylene synthesis pathway; pathway ID 00270), brassinosteroid biosynthesis (brassinosteroid synthesis pathway; pathway ID 00905), alpha-Linolenic acid metabolism (jasmonic acid synthesis pathway; pathway ID 00592), phenylalanine metabolism (salicylic acid synthesis pathway; pathway ID 00360), plant hormone signal transduction (plant hormone signal transduction; pathway ID 04075). By comparing the values (|log2(FoldChange)| > 1 & P value < 0.01) of fragments per kilobase of transcript per million mapped reads (FPKM), the differentially expressed genes (DEGs) were selected. Heatmaps of these DEGs were generated using Rpackage (Supplementary Table S2).
Correlation analysis
The obtained metabolome and transcriptome data were applied to the calculation of correlation coefficients (Spearman’s rank correlation test, |R| > 0.8) by using GraphPad Prism (8.0) software. The metabolome and transcriptome relationships in red maple were visualized using Cytoscape (version 3.6.1) (Supplementary Table S3).
Results
Leaf senescence
Leaf senescence is the last stage in the life history of red maple leaves, from maturation tosenescence, where the most obvious external change is yellowing. Our previous research revealed that the yellowing of the senescing leaves of red maple is primarily attributed to the degradation of chlorophyll, not the emergence of yellow plant-based pigments such as carotenoids [44]. We used ultrahigh-performance liquid chromatograph Q extractive mass spectrometry (UHPLC-QE-MS) to analyze the metabolism of non-senescing leaves (fully expanded non-senescing leaves) and senescing leaves (100% yellowing of the leaf surface; ~ 95% chlorophyll loss) (Fig. 1b). The test results showed that there are many different metabolites in the metabolic pathways of amino acids, lipids, and other biological macromolecules in the non-senescing leaves compared to senescing leaves (Fig. 1c, Supplementary Table S4).
Leaf senescence is a genetically regulated recycling process in the lifecycle of plants in functional organs with photosynthesis to the degradation stage. To evaluate the leaf senescence process in Acer rubrum, we performed combination sequencing with non-senescing leaves and senescing leaves. Based on the sequencing results, we compared the numbers of differentially expressed genes in related pathways such as amino acid metabolism, lipid metabolism, carbohydrate metabolism, energy metabolism, the metabolism of cofactors and vitamins, and nucleotide metabolism in non-senescing and senescing leaves. For the comparison (non-senescing vs senescing leaves), the greatest number of DEGs were found in carbohydrate metabolism (1623 up- and 1530 down-regulated). Conversely, the metabolism of cofactors and vitamins represented the smallest group, with 269 up-and 163 down-regulated unigenes (Fig. 1d, Supplementary Table S5).
Ethylene
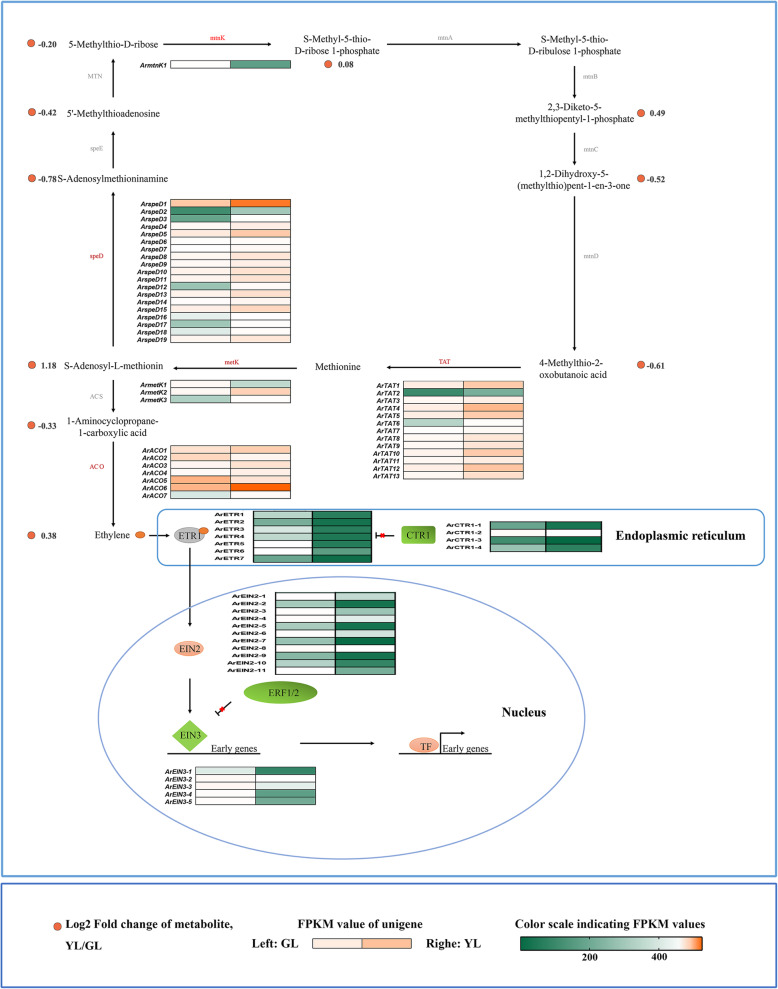
The phytohormone ethylene is extensively involved in many plant processes, where previous studies have found that ethylene levels were linked to leaf senescence. In many species, ethylene treatment can initiate alterations in aging characteristics including promoting the reduction of chlorophyll, chlorophyll–protein complexes, and other macromolecular substances, while increasing the activity of proteases and other metabolic enzymes [45, 46]. Considerable research has summarized the synthesis pathway of ethylene as the conversion of methionine to S-adenosylmethionine (SAM), SAM to 1-aminocyclopropane-1-carboxylic acid (ACC), and ACC to ethylene [47]. As there is alimited quantity of methionine in plants, the methylthio group, is recycled to replenish methionine levels and sustain ethylene biosynthesis. The detected ethylene and related compounds in the metabolic pathway are arranged in corresponding positions (Fig. 2). As displayed, S-Adenosyl-L-methionine, S-Methyl-5-thio-D-ribose 1-phosphate, 2,3-Diketo-5-methyl thiopentyl-1-phosphate, and ethylene were shown to be most abundant in senescing leaves, whereas S-Adenosylmethioninamine, 5′-Methylthioadenosine, 5-Methylthio-D-ribose, 1,2-Dihydroxy-5-(methylthio)pent-1-en-3-one, 4-methylthio-2-oxobutanoic acid, and 1-Aminocyclopropane-1-carboxylic acid were shown to be the more abundant in non-senescing leaves.
Fig. 2.
Biosynthetic and signal transduction pathways of ethylene. mtnK, 5-methylthioribose kinase; mtnA, Methylthioribose-1-phosphate isomerase; MTN, 5′-methylthioadenosine nucleosidase; mtnB, Methylthioribulose-1-phosphate dehydratase; speE, Spermidine synthase; mtnC, 2,3-diketo-5-methylthiopentyl-1-phosphate enolase-phosphatase; speD, S-adenosylmethionine decarboxylase proenzyme; mtnD, 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase; metK, S-adenosylmethionine synthetase; TAT, tyrosine aminotransferase; ACS, 1-aminocyclopropane-1-carboxylate synthase; ACO, aminocyclopropane carboxylate oxidase; ETR, ethylene receptor; CTR, constitutive; EIN2, ethylene insensitive 2; EIN3, ethylene insensitive 3, ERF1/2, ethylene-responsive transcription factor 1/2; TF, transcription factor. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, completely yellow; ~ 95% chlorophyll loss. Colored cells represent the expression profiles of differentially expressed genes (DEGs). Heatmaps were generated with the Rpackage and the color bar at the lower right. Orange dots show the fold change of the content of compounds in non-senescing and senescing leaves
Of the 43 differential expression genes (DEGs) involved in ethylene biosynthesis, 39 are highly expressed in senescing leaves (Fig. 3). ACC synthase (ACS) and ACC oxidase (ACO) are rate-limiting enzymes in the ethylene biosynthesis pathway. In our study, no detectable DEGs of ACS were observed. Seven DEGs of ArACOs were detected, where most (5/7) exhibited higher expression in senescing leaves. All of the 27 ET-signaling genes are up-regulated in non-senescing leaves.
Fig. 3.
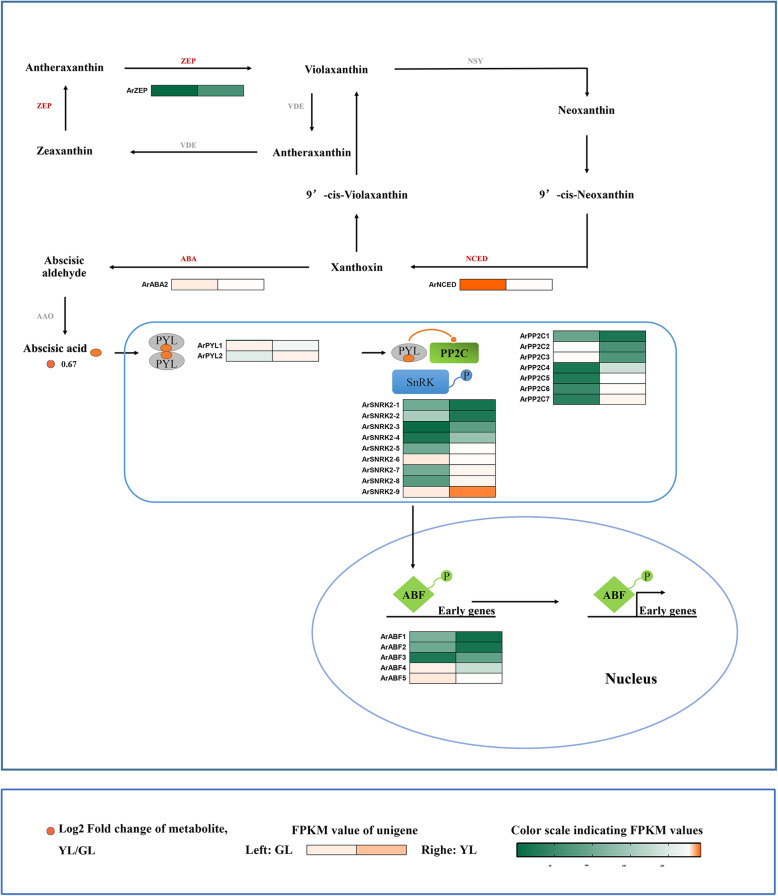
Biosynthetic and signal transduction pathways of abscisic acid. ZEP, zeaxanthin epoxidase; NSY, neoxanthin synthase; VDE, violaxanthin de-epoxidase; ABA, xanthoxin dehydrogenase; NCED, 9-cis-epoxycarotenoid dioxygenase; AAO, abscisic-aldehyde oxidase; PYL, pyrabactin-resistance 1-like; PP2C, protein phosphatase type 2C; SnRK, serine/threonine-protein kinase; ABF, abscisic acid responsive element binding factor. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, completely yellow; ~ 95% chlorophyll loss. The colored cells represent the expression profiles of differentially expressed genes (DEGs). The heatmaps were generated using the Rpackage and the color bar at the lower right. Orange dots show the fold change of the content of compounds in non-senescing and senescing leaves
Abscisic acid
Abscisic acid (ABA) was once thought to be the most effective hormone in the promotion of leaf senescence in addition to ethylene. We investigated the enrichment of metabolites and genes involved in ABA synthesis, and the ABA-signaling pathway (Fig. 3). Our results showed that the content of abscisic acid was high in senescing leaves and exceeding that in non-senescing leaves. At the ABA synthesis stage, three genes demonstrated differential expression. Among them, ArZEP had a higher expression level in senescing leaves, while ArNCED and ArABA2 were highly expressed in non-senescing leaves. Of the 23 ABA-signaling genes, 12 were up-regulated in senescing leaves (ArPYL2, ArPP2C4/5/6/7, ArSNRK2–3/4/5/7/8/9, ArABF3) and 11 were down-regulated (ArPYL1, ArPP2C1/2/3, ArSNRK2–1/2/6, ArABF1/2/4/5) (Fig. 3).
Cytokinins
Cytokinins (CKs) are phytohormones, which play an important role in plant growth and development. The administration of exogenous cytokinins on ex vivo and living leaves can prevent leaf senescence [48]. By analyzing the levels of cytokinins in leaves prior to and following senescence, it was revealed that there was a negative correlation between cytokinins levels and aging processes in various tissues and plants [7, 49, 50]. Furthermore, genetic manipulation of cytokinin production in transgenic plants has provided very solid evidence for the inhibition role of cytokinins in leaf senescence [7, 51–53].
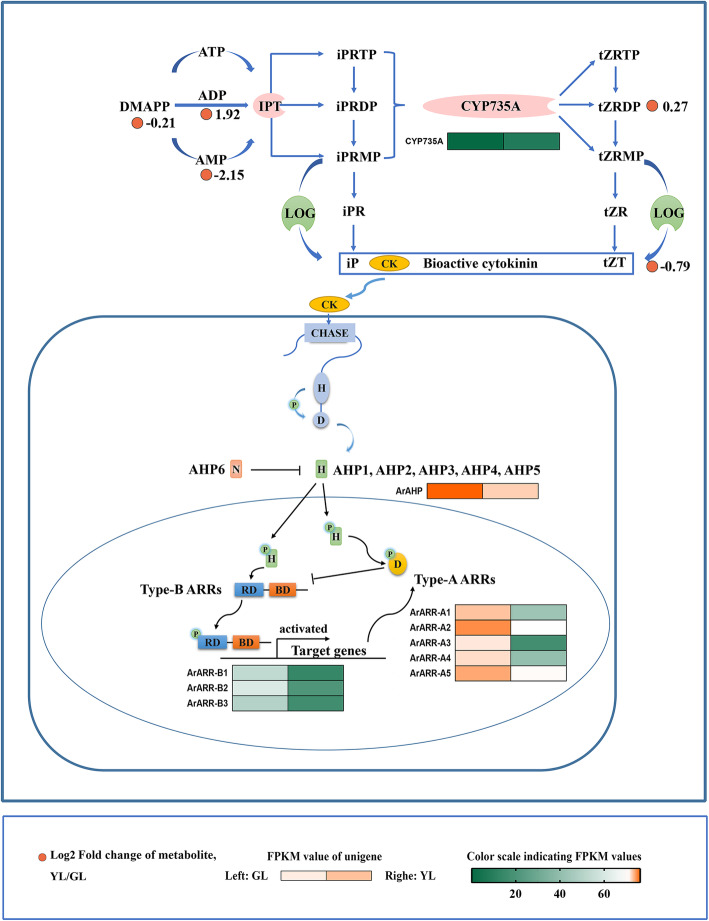
In this study, the content of cytokinins in senescing leaves was far above that of non-senescing leaves, and genes related to various aspects of CK homeostasis were collected (Fig. 4). Cytokinin signaling is based on two-component system (TCS) that is achieved by the continuous transfer of phosphate groups between major components. Our results showedthat all differentially expressed genes in this signal transduction pathway were down-regulated in senescing leaves (Fig. 4).
Fig. 4.
Biosynthetic and signal transduction pathways of cytokinins. DMAPP, dimethylallyl diphosphate; ATP, adenosine triphosphate; ADP, adenosine diphosphate; AMP, adenosine monophosphate; IPT, isopentenyl transferase; iPRTP, isopentenyl adenosine-5′-triphosphate; iPRDP, isopentenyl adenosine-5′-diphosphate; iPRMP, isopentenyl adenosine-5′-monophosphate; CYP735A, cytokinin trans-hydroxylase; tZRTP, trans-Zeatin riboside triphosphate; tZRDP, trans-Zeatin riboside diphosphate; tZRMP, trans-Zeatin riboside monophosphate; LOG, cytokinin-activating enzyme LONELY GUY; iPR, isopentenyl adenosine; tZR, trans-zeatin riboside; iP, isopentenyl adenine; tZT, trans-zeatin; AHP, histidine-containing phosphotransfer peotein; Type-A ARRs, type-A response regulators; Type-B ARRs, type-B response regulators. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, completely yellow; ~ 95% chlorophyll loss. The colored cell represents the expression profiles of differentially expressed genes (DEGs). The heatmaps were generated using the Rpackage and the color bar at the lower right. Orange dots show the fold change of the content of compounds in non-senescing and senescing leaves
Auxin
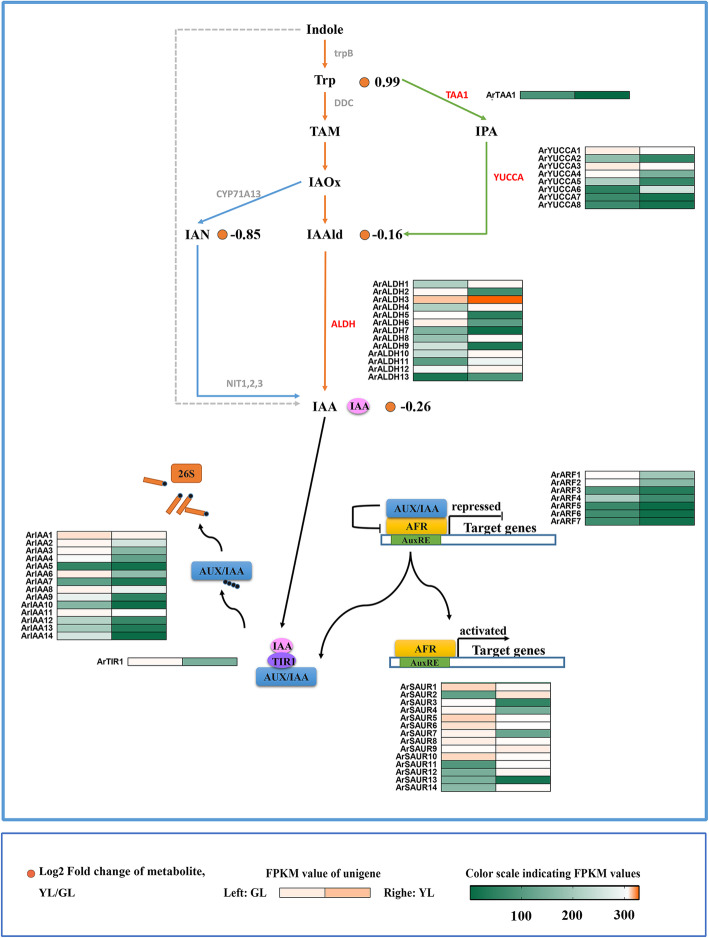
Early study revealed that exogenous application of auxin was inversely related to leaf senescence, and that the endogenous auxin levels of many plants decreased with advancing age [54]. Plants achieve the synthesis of tryptophan to auxin through three pathways, named after the intermediate products, these are include the indole-3-pyruvate pathway, the tryptamine pathway, and the indole-3-acetonitrile pathway [55]. At present, these two of three synthetic pathways have not been fully investigated, and key genes controlling the relevant synthetic steps have yet to be discovered. Many of these steps are still potential models based on existing experiments.
In senescing leaves of red maple, the tryptophan concentration is higher than in non-senescing leaves. Indole-3-acetaldehyde (IAAld), indole-3-acetonitrile (IAN), and indole-3-acetic acid (IAA) have a higher content in non-senescing leaves (Fig. 5). A total of 22 key differential expression genes were obtained in the auxin synthesis pathway, including the YUCCA, ALDH, and TAA1 family genes. There are seven YUCCA gene expression levels in senescing leaves that are significantly down-regulated (indole-3-pyruvate pathway). The indole-3-pyruvate pathway is the first completely elucidated auxin biosynthetic pathway, and the most fundamental and important auxin synthesis pathway in plants [56, 57].
Fig. 5.
Biosynthetic and signal transduction pathways of auxin. trpB, tryptophan synthase beta chain; Trp, tryptophan; DDC, tryptophan decarboxylase; TAA, tryptophan aminotransferase; TAM, tryptamine; IPA, indole-3-pyruvate; CYP71A13, indoleacetaldoxime dehydratase; IAOx, indole 3-acetaldoxime; YUCCA, indole-3-pyruvate monooxygenase; IAAld, Indole-3-acetaldehyde; IAN, Indole-3-acetonitrile; ALDH, aldehyde dehydrogenase; NIT, nitrilase; IAA, Indole-3-acetate; AUX/IAA, auxin-responsive protein IAA; ARF, auxin response factor; TIR1, transport inhibitor response 1; SAUR, small auxin up-regulated RNA. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, completely yellow; almost 95% chlorophyll loss. The colored cells represent the expression profiles of differentially expressed genes (DEGs). Heatmaps were generated by Rpackage and the color bar at the lower right. Orange dots show the fold change of the content of compounds in non-senescing and senescing leaves
Many studies have shown that YUCCA inactivation in this pathway reduces the synthesis of auxin in plants, whereas overexpression of the YUCCA gene leads to the overproduction of auxin, which causes developmental defects [58]. Therefore, YUCCA is a rate-limiting factor for auxin synthesis, and its gene expression pattern plays a crucial role in auxin synthesis. In this study, the expression of ArYUCCA genes in senescing leaves was down-regulated, suggesting its key role in auxin synthesis. Through differential gene screening, 35 differentially expressed genes, including TIR1, IAA, ARF, and SAUR family genes, were found in the auxin signal transduction pathway. The analysis of each family gene expression pattern revealed that the TIR1, IAA, and ARF gene families were up-regulated in non-senescing leaves, while the SAUR gene family had both up−/down-regulated genes in non-senescing leaves (Fig. 5).
Jasmonic acid
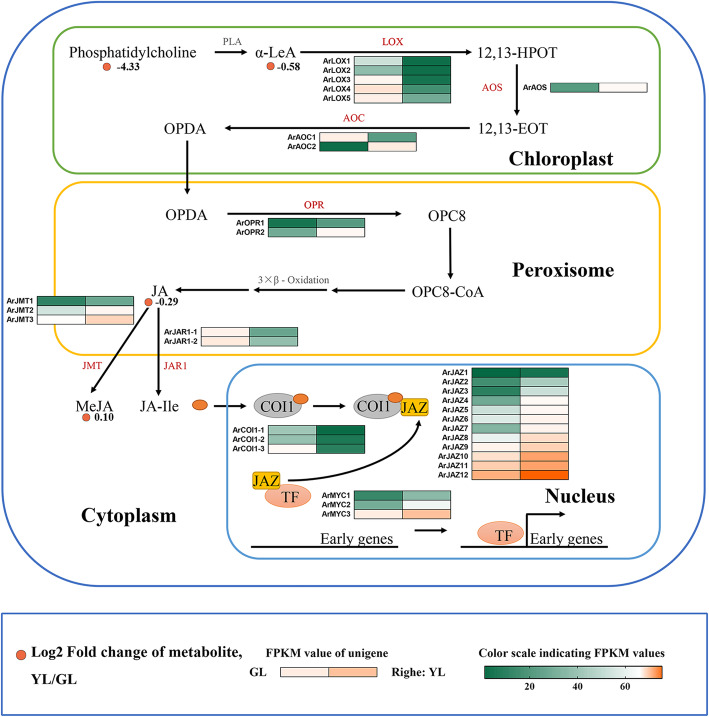
Jasmonic acid and its derivatives are an important class of plant hormone that is required for plant growth and development and assists with enduring stress and completing the life cycle. The first documented function of jasmonic acid was the promotion of senescence in isolated oat leaves [59, 60]. Therefore, we investigated the metabolite content and the expression patterns of DEGs in jasmonic acid biosynthesis and signal transduction related pathways of red maple (Fig. 6). The content of α-Linolenic acid and methyl jasmonate in non-senescing leaves was higher than that in senescing leaves, and the content of phosphatidylcholine and jasmonic acid was higher in the latter.
Fig. 6.
Biosynthetic and signal transduction pathways of jasmonic acid. LA, phospholipase A1; α-LeA, alpha-Linolenic acid; LOX, lipoxygenase; 12,13-HPOT, 12,13-hydroperoxylinoleic acid; AOS, allene oxide synthase; 12,13-EOT, 12,13-epoxyoctadecatrienoic acid; AOC, allene oxide cyclase; OPDA, oxophytodienoic acid; OPR, oxophytodienoic acid reductase; JA, jasmonic acid; JMT, jasmonic acid carboxyl methyltransferase; JAR1, jasmonic acid resistant 1; MeJA, Methyl jasmonate; JA-Ile, Jasmonoyl-L-isoleucine; COI1, coronatine-insensitive protein 1; JAZ, jasmonate ZIM domain-containing protein. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, completely yellow; ~ 95% chlorophyll loss. Colored cells represent the heatmaps of differentially expressed genes (DEGs). With the heatmaps, the redder the color, the higher the DEGs expression, and the greener the color, the lower the DEGs expression. Orange dots show the fold change the content of the compounds in non-senescing and senescing leaves
The expression of related genes showed a clear pattern, and the gene expression profiles of the same gene family, except for allene oxide cyclase (AOC), were consistent. Among them, one ArAOC gene and five lipoxygenase (LOX) genes were down-regulated in senescing leaves. Seven biosynthesis genes were up-regulated in senescing leaves, including allene oxide synthase (AOS), allene oxide cyclase (AOC), oxophytodienoate reductase (OPR), and jasmonate O-methyltransferase (JAT), which catalyzed a series of reactions in the JA-biosynthetic pathway. Jasmonate-resistant 1 (JAR1), coronatine insensitive 1 (COI1), jasmonate ZIM (JAZ) proteins, and the transcription factor MYC2 are important constituent elements of the JA-signaling pathway. The first two are down-regulated in senescing leaves, whereas the latter two are up-regulated.
Salicylic acid
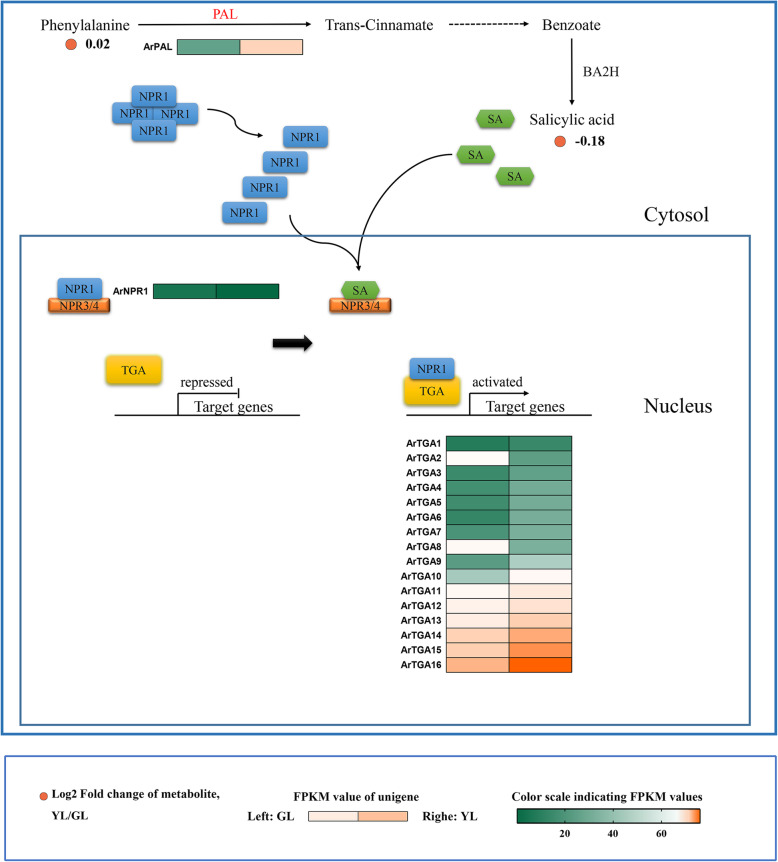
Salicylic acid (SA) is a phenolic plant hormone that can regulate seed germination, cell growth, respiration, stomatal closure, stress response, nitrogen fixation, and the seed setting rate of various plants [61]. The role of salicylic acid in leaf senescence has also been recognized in recent years [62]. Therefore, we investigated the metabolite content and the expression of related genes in the salicylic acid SA synthesis, and the SA-signaling pathway in red maple (Fig. 7). The phenylalanine pathway was the first discovered salicylic acid synthesis pathway. We detected that the phenylalanine content was almost equivalent in both senescing and non-senescing leaves. However, the content of salicylic acid in non-senescing leaves of red maple is higher, although the expression of the core enzyme phenylalanine ammonia lyase (PAL) gene in the senescing leaves is much higher than that of non-senescing leaves. Interestingly, this result was different from those of other studies. In Arabidopsis, the content of endogenous salicylic acid increased with leaf senescence [63].
Fig. 7.
Biosynthetic and signal transduction pathways of salicylic acid. PAL, phenylalanine ammonia-lyase; BA2H, benzoate; NADPH: oxygen oxidoreductase (2-hydroxylating); NPR, nonexpressorofpathogenesis-relatedgenes1; TGA, transcription factor TGA. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, completely yellow; ~ 95% chlorophyll loss. Colored cells represent the expression profiles of differentially expressed genes (DEGs). Heatmaps were generated using the Rpackage and the color bar at the lower right. Orange dots show the fold change of the content of compounds in non-senescing and senescing leaves
Studies have shown that salicylic acid signal transduction pathways are primarily NPR1-dependent pathways [64]. Following the depolymerization of NPR1 into a reactive monomer and transfer to the nucleus, it can interact directly with some members of the transcription factor TAG family to induce the expression of downstream disease resistance genes. The expression levels of ArNPR1 gene in senescing leaves and non-senescing leaves were similar; however, 15 members of the 16 TGA genes were up regulated during senescence. Therefore, we hypothesized that the role of SA in the leaf senescence of red maple was dependent on the efficient expression of related genes in signal transduction pathways.
Brassinosteroid
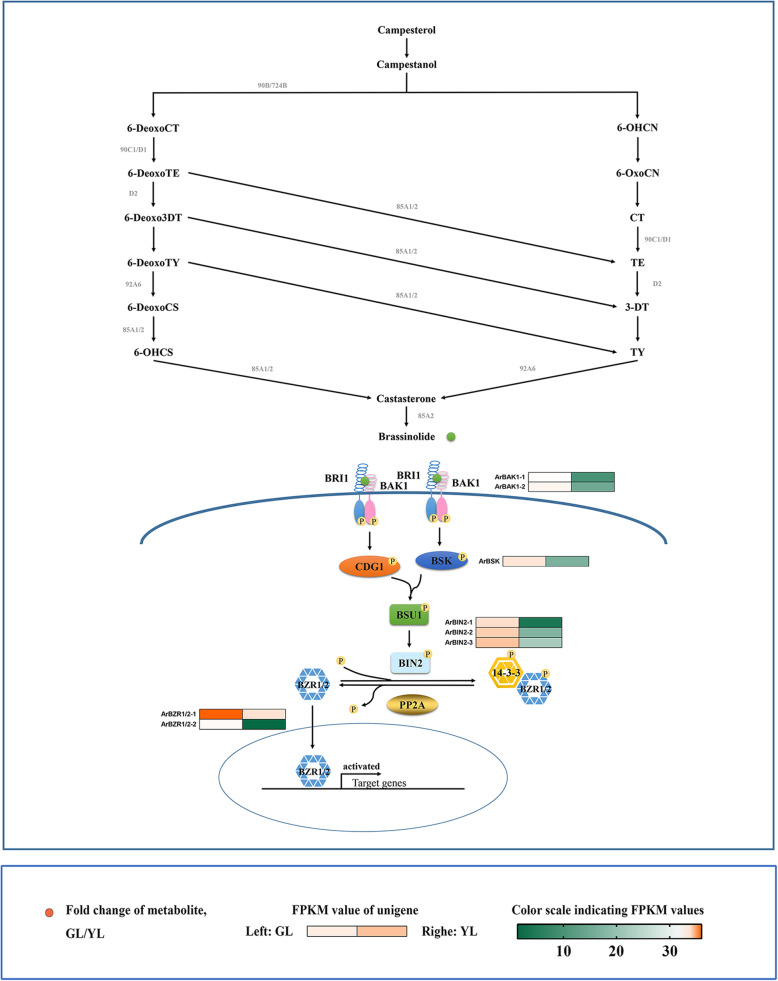
The plant hormone brassinosteroid (BR) play a role in a variety of developmental processes including leaf senescence. Some Arabidopsis mutants of BR biosynthesis (e.g., det2,) [65], or with the loss of a signal transduction pathway (e.g., BRI1) have delayed leaf senescence phenotypes and associated aging characteristics [66–69]. However, this phytohormone was not detected in our metabolome test results. Previous studies have shown that the content of brassinosteroid in plant tissues varies greatly [70]. Pollen, immature seeds, and fruits have the highest BR content. Young growing tissue contains higher BR levels; however, mature leaves have lower BR concentrations [71].
The transmission of the BR signal from BRI1 located on the membrane to the transcription factor BZR1 in the nucleus is accomplished through a series of protein phosphorylation and dephosphorylation reactions. This series of phosphatases or phosphokinases, including BAK1, BSK, BIN2, and BZR1/2, are down-regulated in senescing leaves (Fig. 8).
Fig. 8.
Biosynthetic and signal transduction pathways of brassinosteroids. 90B/724B, steroid 22-alpha-hydroxylase; 6-DeoxoCT, 6-Deoxocathasterone; 6-OHCN, 6alpha-Hydroxycampestanol; 6-DeoxoTE, 6-Deoxoteasterone; 6-OxoCN, 6-Oxocampestanol; 6-Deoxo3DT, 3-Dehydro-6-deoxoteasterone; CT, cathasterone; 6-DeoxoTY, 6-Deoxotyphasterol; TE, teasterone; 6-DeoxoCS, 6-Deoxocastasterone; 3-DT, 3-Dehydroteasterone; 6-OHCS, 6alpha-Hydroxy-castasterone; TY, Typhasterol; 85A1/2, brassinosteroid-6-oxidase 1; 92A6, typhasterol/6-deoxotyphasterol 2alpha-hydroxylase; 85A2, brassinosteroid-6-oxidase 2; BRI1, protein brassinosteroid insensitive 1; BAK1, brassinosteroid insensitive 1-associated receptor kinase 1; BSK, BR-signaling kinase; BSU1, serine/threonine-protein phosphatase; BIN2, brassinosteroid insensitive 2; BZR1/2, brassinosteroid resistant 1/2; PP2A, protein phosphatase type 2A. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, completely yellow; ~ 95% chlorophyll loss. Colored cells represent the expression profiles of differentially expressed genes (DEGs). Heatmaps were generated with the Rpackage and the color bar at the lower right. Orange dots show the fold change of the content of compounds in non-senescing and senescing leaves
Gibberellin
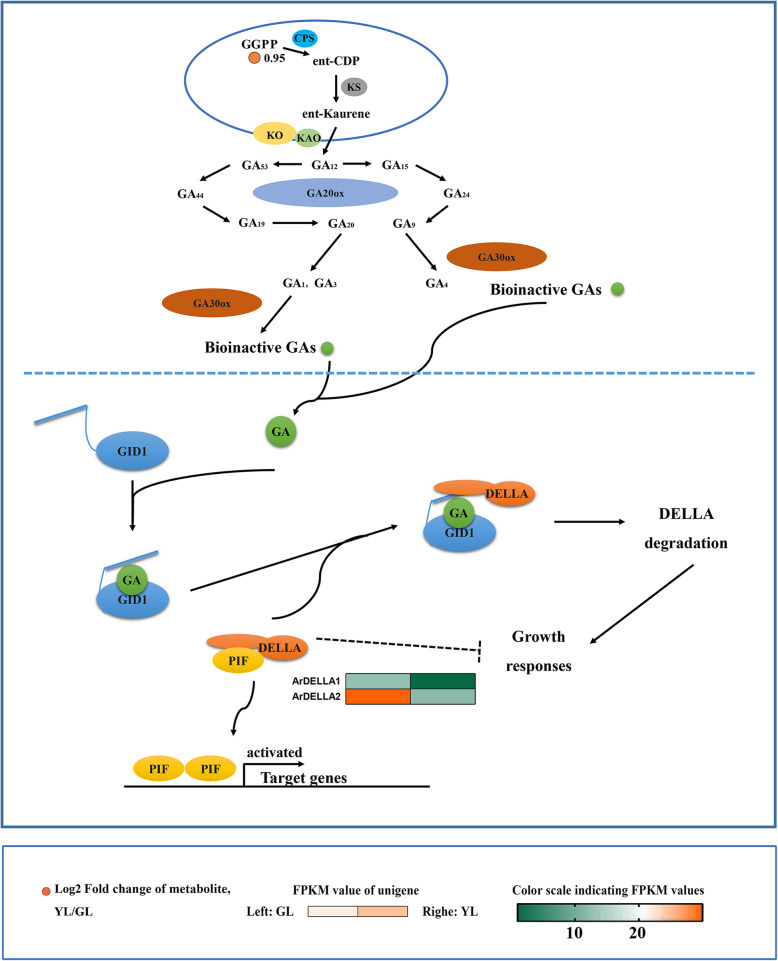
Gibberellin (GAs) are a class of phytohormones that belongs to the biguanide compounds, which play an important role in the entire life cycle of plants. The precursor of GA biosynthesis in higher plants is geranylgeranyldiphosphate (GGPP), which is synthesized from isopentenyl pyrophosphate. The impeding effect of gibberellin on leaf senescence has been discussed in many previous studies [72]. Senesent leaves have double the amout of GGPP than non-senescent (GL/YL = 0.5; Fig. 9). Of all the genes involved in the gibberellin signaling, only two differentially expressed genes (ArDELLA1 and ArDELLA2) were found, all of which were highly expressed in non-senescing leaves.
Fig. 9.
Biosynthetic and signal transduction pathways of gibberellins. GGPP, geranylgeranyldiphosphate; CPS, ent-copalyl diphosphate synthase; ent-CDP, ent-Copalyl diphosphate; KS, ent-kaurene synthase; KO, ent-Kaurene oxidase; KAO, ent-Kaurenoicacid oxidase; GA, Gibberellin; GA20ox, GA20-oxidase; GA30ox, GA30-oxidase; GID1, Gibberellin insensitive dwarf 1. GL, non-senescing leaves, fully expanded green leaves; YL, senescing leaves, completely yellow; ~ 95% chlorophyll loss. Colored cells represent the expression profiles of differentially expressed genes (DEGs). Heatmaps were generated using the Rpackage and the color bar at the lower right. Orange dots show the fold change of the content of compounds in non-senescing and senescing leaves
Discussion
The promotion of leaf senescence in red maple
Ethylene is a central regulator of growth and physiology in plants. From seed germination to organ senescence, and from sex determination to cell elongation, ethylene plays critical roles [73]. In some species, ethylene has been shown to be effective in the regulation of leaf senescence [74]. In this study, we observed a positive correlation between the levels of three ACC oxidases (ArACO3, ArACO6, and ArACO7) and ethylene in red maple (Fig. 10). The expression level of three ArACOs in senescing leaves was higher than in non-senescent and consistent with the level of ethylene. Early studies have demonstrated that ACC oxidase catalyzed the final step in ethylene biosynthesis, and under conditions of high ethylene production, ACO is often the rate-limiting step in biosynthesis [75, 76]. A study indicated that leaf senescence, as assessed by color change from green to yellow, was clearly delayed by 10–14 days in transgenic ACC oxidase antisense tomato plants when compared with wild-type plants [77]. ET signal transduction is performed according to the “linear” approach. It starts with a receptor and end with transcription factors (TFs). ETR (ethylene response) and CTR1 (Raf-like CONSTITUTIVE TRIPLE-RESPONSE1) are the negative regulatory factors of this pathway. For this study, two CTR1 genes (ArCTR1–1 and ArCTR1–2) and six ETR genes (ArETR1 and ArETR3/4/5/6/7) were negatively correlated with ethylene in red maple. Analyses of the etr mutants in Arabidopsis and rice, respectively, did indicate that etr leaves (ethylene insensitivity) had greater longevity than did wild-type leaves [78, 79]. Similarly, in Arabidopsis thaliana the ctr1 mutation was analyzed in detail to show that the mutation causes a delay in functional leaf senescence [80]. These results suggested that the ArACOs, ArETRs, and ArCTRs were involved in the leaf senescence affected by ethylene in red maple.
Fig. 10.
Gene regulatory network of leaf senescence with integrated data from red maple. Squares represent hormone-related metabolites and triangles represent regulatory genes. Two colored lines represent two different regulatory relationships between the regulatory genes and plant hormone-related metabolites; red refers to a positive regulatory relationship, blue refers to a negative regulatory relationship. CK, Cytokinins; JA, Jasmonic acid; SA, Salicylic acid; GA, Gibberellin; BR, Brassinosteroid; ABA, Abscisic Acid
It has been widely proven that ABA has a significant role in promoting leaf senescence [81]. Endogenous ABA levels can induce the expression of a variety of genes in various plants. Besides transcription factors and other signaling molecules, the products of these genes are functional genes such as LEA proteins, detoxification enzymes, and enzymes for osmoprotectant synthesis [82]. During the Arabidopsis senescence, most of the genes involved in the synthesis of abscisic acid were up-regulated [83–86]. In many circumstances, changes in product accumulation were consistent with gene expression. Interestingly, this presented the opposite results to our study. The ABA response has since been shown to be strictly controlled by intracellular signal transduction pathways, and many likely signaling intermediates correlated with ABA responses have also been identified by molecular and biochemical studies. Recent progress toward understanding early ABA signal transduction has led to the construction of a PYL–PP2C–SnRK2 signal transduction model [87]. A co-regulation network of ABA metabolic pathways in red maple showed that seven members of three gene families had correlations with ABA (Fig. 10). There was a negative correlation between the expression levels of ArPP2C1/2/3 and ABA. ArPYL2 and ArSnRK2–7/8/9 were shown to be positively correlated with ABA levels. Numerous genetic evidences suggested that phosphatases 2C (PP2C) were negative ABA signaling regulators. Zhang et al. noted that the SAG113 gene in Arabidopsis encoded a protein phosphatase 2C, and the role of SAG113 protein phosphatase 2C was to inhibit stomatal closure during leaf senescence. This allowed increased oxygen to enter the leaves to speed up the respiration rate of the mesophyll cells, and the respiration rates of aging cells tended to accelerate, while simultaneously promoting the rapid evaporation of the leaf water until it was depleted [81, 88]. Our results indicated that four ArPP2C genes were highly expressed in senescing leaves, which suggested an important role for these genes in the leaf senescence of red maple. Previous research has shown that PYR/PYLs are ABA receptors functioning at the apex of a negative regulatory pathway that controls ABA signaling by inhibiting PP2Cs. Consequently, we speculated that in the senescing leaves of red maple, ArPYL2 might interact with ArPP2C1/2/3 and inhibit phosphatase activity, allowing ArSnRK2–7/8/9 activation and the phosphorylation of target proteins, to then regulate leaf senescence.
The inhibition of leaf senescence in red maple
Previous research suggests that cytokinins are one of the most crucial phytohormones in the regulation of plant growth and development [89, 90]. Cytokinins are likely the most commonly studied hormone that regulates leaf senescence. The application of exogenous cytokinin on detached plant leaves, can retard leaf senescence [89, 91]. The overexpression of components in the cytokinin signaling pathway may also delay plant leaf senescence [92]. In red maple, the content of cytokinin in non-senescing leaves is high. It has been suggested that cytokinin may inhibit leaf senescence in red maple. Correlation analysis showed that the content of cytokinins in red maple was positively correlated with ArARR-A1/3/4 (Fig. 10). It is known that the expression of type-A ARR genes is rapidly and specifically induced by cytokinins [93, 94]. This type of gene is considered to be a marker gene in the cytokinin signaling pathway. The overexpression of A-type ARR genes will lead to an earlier flowering time, longer roots, increased lateral roots, premature senescence, and reduced cytokinin sensitivity in transgenic plants. ARR16 in Arabidopsis may be involved in regulating the senescence process of its leaves [95]. In conclusion, our results suggested that ArARR-A1/3/4 in red maple may be involved in regulating the senescence process of leaves through the cytokinin signaling pathway.
Auxin plays an important role in all parts and stages of plant growth and development [96]. Early studies have shown that the external application of auxin had a negative correlation with leaf senescence, which suggests a role of auxin on the delay of leaf senescence [97]. In most cases, the application of auxin can delay both chlorophyll degradation and protein degradation in leaf disks for many plants, while the level of endogenous auxin were decreased in leaves that began/were in the process of senescing [54]. In this study, non-senescing red maple leaves were shown to contain elevated auxin levels in contrast to senescing leaves. Correlation analysis revealed that the auxin content of red maple was positively correlated with ArYUCCA2/3/5/8 (Fig. 10). Previous studies have shown that yuc6-1d, a YUCCA6 activation mutant, had a high level of free IAA and displayed typical high-auxin phenotypes. The transgenic plants showed a significant life cycle extension and exhibited a significant inhibitory effect on dark- and hormone-induced senescence [98]. In addition, the expression of ArSAUR14 was observed to increase during red maple senescence, which exhibited a negative regulatory relationship with auxin content. Recently, several molecular genetic studies found that the small auxin-up RNA (SAUR) gene, a member of the auxin-responsive gene family, played a critical role in plant growth by regulating the accumulation of the auxin phytohormone [R]. Transgenic rice plants overexpressing SAUR39 showed that this gene was expressed at higher levels in senescing leaves, unlike auxin biosynthesis. The transgenic plants had a lower auxin level and reduced polar auxin transport, as well as the down-regulation of some putative auxin biosynthesis and transporter genes [99]. Overall, the results suggested that ArYUCCA2/3/5/8 and ArSAUR14 in red maple may regulate leaf senescence through auxin metabolism and signaling.
Two plant hormones in red maple that are contrary to previous studies
Jasmonates (JAs) and their oxylipin derivatives are well recognized phytohormones that regulate plant growth and plant adaptations to biotic stresses, as well as abiotic stresses [100]. The first confirmed biological function of JA was found as a senescence-promoting substance to promote the senescence of detached oat leaves [60]. He et al. (2002) showed that the JA content in senescing leaves was four-fold higher than that in non-senescing leaves [101]. However, our test results indicated that the JA content in the non-senescing leaves of red maple was higher than in senescing leaves. The detection results of JA downstream products revealed that the content of the MeJA in senescing leaves was moderately higher than in non-senescing leaves (GL/YL = 0.9). It is possible that JA generated additional MeJA to participate in the anti-stress reaction. The results of correlation analysis showed that the ArJAZ1/4/6/9/11/12, ArMYC1, and JA content were negatively correlated (Fig. 10). Previous studies have shown that JAZs can inhibit JA-induced leaf senescence by interacting with a series of transcription factors or signal transduction proteins such as MYC2 [102–105]. Therefore, the high expression of ArJAZs transcripts in senescing leaves of red maple may inhibit leaf senescence induced by JA.
Although SA has been well studied as a signaling molecule in plant disease defense responses, its role in leaf senescence has been recognized until recently. In Arabidopsis, the content of endogenous SA increased with the leaf senescence process [106], and the leaves of Arabidopsis mutants showed early or late senescence with increased or decreased SA content [107–112], respectively. We have shown that although the levels of phenylalanine dehydrogenase in senescing leaves is high, the content of salicylic acidis higher in non-senescent than senescent leaves. In many plants, salicylic acid is synthesized via the phenylalanine pathway [113]. Under the action of phenylalanine dehydrogenase, phenylalanine produces cinnamic acid, and then benzoic acid, and finally salicylic acid. However, cinnamic acid is the precursor of many polyphenols, such as flavonoids [114]. Therefore, we speculated that cinnamic acid in red maple is more involved in plant metabolism as a precursor of this type of substance. Correlation analysis revealed that there was a positive correlation between the ArNPR1 content and SA (Fig. 10). Previous studies confirmed that NPR1 plays an important role in leaf senescence. npr1, which is a mutant of SA signal receptor loss in Arabidopsis, exhibited the phenomenon of delaying leaf senescence and also changed expression of some Senescence-Associated Gene (SAGs) [106]. Consequently, we speculated that the lower accumulation of SA in senescing leaves might be related to the lower expression of the ArNPR1 genes. Of course, this conjecture needs to be verified by further experiments in the future.
A role of brassinosteroids and GA in leaf senescence of red maple
Brassinosteroids are a unique class of plant hormones that are essential for normal plant growth [115, 116]. However, to date, the roles of these substances in plant leaf senescence have not been studied in depth. At present, it is known that the seedling leaves of mung bean [117] and cucumber cotyledons [118] will appear to prematurely senesce when treated with epibrassinolide (epiBR). In Arabidopsis, different BR mutants (e.g., det2, bri1) showed the phenotype of accelerating or delaying leaf senescence [67–69]. In our results, the content of BRs in red maple was not detected, which may have been because the content of BRs varies significantly between different organisms. Previous studies revealed that the BRs content in pollen, immature seeds, and fruits was highest, followed by young growing tissues, whereas the BRS concentration in mature leaves was lowest [119, 120]. No differentially expressed genes were identified in the BRs synthesis pathway of red maple, and the genes of the signal transduction pathway were also highly expressed in non-senescing leaves without suggesting that there may be no significant accumulation of brassinosteroid during the senescence process.
Gibberellin is a type of plant hormone that belongs to diterpenoids, which plays an critical role over the entire life cycle of plants [121]. Based on the treatment of gibberellin and related research results, gibberellin was reported to maintain chlorophyll levels and RNA synthesis; thus, delaying post mitotic leaf senescence [R]. The spraying of exogenous GA (3) on Paris polyphylla can obviously inhibit the senescence of aerial components [122], and GA (4) can delay the senescence of Alstroemeria hybrida leaves [123, 124]. According to our results, no gibberellin was detected in red maple, and no differentially expressed genes were observed in the gibberellin synthesis pathway. Although the content of the initial GGPP metabolite in senescing leaves was high, the change of GA content during the senescing process of red maple could not be predicted. This is because the GGPP in plants can form GA, ent-kaurene [125], Chl-phytol (Chl (Phy)) [126–129], phytoene, and carotenoid [130] according to different branching pathways. The manipulation of any branching pathway using GGPP as the precursor, will also significantly affect other branching paths. For example, the up- or down-regulation of phytoene synthetase will lead to the increase or decrease of carotenoid synthesis and changes in GA levels [131]. Therefore, the effect of GA on leaf senescence of red maple requires further study.
Conclusion
Ethylene and ABA are likely to play a positive role in the leaf senescence of Acer rubrum. The key factors involved in the ethylene regulation of leaf senescence may include members of the ArACO, ArETR, and ArCTR gene families. In the ABA signaling pathway, the PYL-PP2C-SnRK2 component plays a critical role in regulating ABA production. Cytokinins and auxins are likely to play negative roles in the regulation of leaf senescence of red maple. A-type nuclear response regulators are very likely to be key regulators of cytokinin in red maple to delay leaf senescence. It is suggested that future research on the mechanisms of auxin delaying leaf senescence of this species might focus on genes such as YUCCA. The higher accumulation of JA and SA in non-senescing leaves of red maple was contrary to the research results of others in that the content of endogenous JA and SA in other species increased with leaf senescence.
Based on the analytical results of the changes in the content of related metabolites of the JA synthesis pathway, we speculated that JA in the senescing leaves of red maple was involved a period of rapid accumulation. We speculated further, that the precursor of SA, cinnamic acid, was more involved with other secondary metabolites, such as flavonoids, during aging, which may have been the reason that SA was not further accumulated. We did not detect BRs and GAs in red maple. Based on the analysis of the expression of differential genes in the BRs signaling pathway, we speculated that it was unlikely that the content of this substance increased during the senescence of red maple leaves. Due to the complexity of the downstream branch pathway of the initial metabolite of the GA metabolic pathway, GGPP, we still cannot speculate on the possible accumulation of GAs. In the future, we will further elucidate the mode and mechanism of action of the above hormones in the regulation of senescing red maple leaves through additional experimental evidence.
Supplementary information
Additional file 1: Table S1. Differentially expressed plant hormone-related metabolites candidates in red maple.
Additional file 2: Table S2. Differentially expressed gene (DEG) candidates involved in plant hormone metabolism in red maple.
Additional file 3: Table S3. Correlation matrix of metabolites (plant hormone-related) and gene expression levels.
Additional file 4: Table S4. Classification of macromolecules in senescing leaves compared with non-senescing leaves in red maple. “Total” refers to the total number of metabolites changed from senescent to non-senescent leaves.
Additional file 5: Table S5. Functional categories of genes correlated with the anabolism of macromolecules differentially expressed in senescing leaves compared with non-senescing leaves in red maple.
Acknowledgments
Thanks to all the members of the garden plant and landscape engineering team for their contributions.
Abbreviations
- UHPLC-QE-MS
Liquid chromatograph Q extractive mass spectrometry
- ET
Ethylene
- ABA
Abscisic acid
- JA
Jasmonic acid
- SA
Salicylic acid
- BR
Brassinosteroid
- CK
Cytokinin
- IAA
Auxin
- GA
Gibberellin
- Non-senescing leaves
Fully expanded green leaves
- senescing leaves
100% yellowing leaf surface, ~ 95% chlorophyll loss
- IDA
Information-dependent acquisition
- ESI +
Electrospray ionization-positive ion mode
- ESI–
Electrospray ionization-negative ion mode
- NGS
Next-generation sequencing
- SMRT
Single-molecule real-time sequencing
- Iso-Seq
Isoform Sequencing protocol
- FPKM
Fragments per kilobase of transcript per million mapped reads
- DEGs
Differentially expressed genes
- NCBI
National Center for Biotechnology Information
- GL
Non-senescing leaves, fully expanded green leaves
- YL
Senescing leaves, 100% yellowing of the leaf surface; ~ 95% chlorophyll loss
- mtnK
5-methylthioribose kinase
- mtnA
Methylthioribose-1-phosphate isomerase
- MTN
5′-methylthioadenosine nucleosidase
- mtnB
Methylthioribulose-1-phosphate dehydratase
- speE
Spermidine synthase
- mtnC
2,3-diketo-5-methylthiopentyl-1-phosphate enolase-phosphatase
- speD
S-adenosylmethionine decarboxylase proenzyme
- mtnD
1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase
- metK
S-adenosylmethionine synthetase
- TAT
Tyrosine aminotransferase
- ACS
1-aminocyclopropane-1-carboxylate synthase
- ACO
Aminocyclopropanecarboxylate oxidase
- ETR
Ethylene receptor
- CTR
Constitutive
- EIN2
Ethylene insensitive 2
- EIN3
Ethylene insensitive 3
- ERF1/2
Ethylene-responsive transcription factor 1/2
- TF
Transcription factor
- ZEP
Zeaxanthin epoxidase
- NSY
Neoxanthin synthase
- VDE
Violaxanthin de-epoxidase
- ABA
Xanthoxin dehydrogenase
- NCED
9-cis-epoxycarotenoid dioxygenase
- AAO
Abscisic-aldehyde oxidase
- PYL
Pyrabactin-resistance 1-like
- PP2C
Protein phosphatase type 2C
- SnRK
Serine/threonine-protein kinase
- ABF
Abscisic acid responsive element binding factor
- DMAPP
Dimethylallyl diphosphate
- ATP
Adenosine triphosphate
- ADP
Adenosine diphosphate
- AMP
Adenosine monophosphate
- IPT
Isopentenyl transferase
- iPRTP
Isopentenyl adenosine-5′-triphosphate
- iPRDP
Isopentenyl adenosine-5′-diphosphate
- iPRMP
Isopentenyl adenosine-5′-monophosphate
- CYP735A
Cytokinin trans-hydroxylase
- tZRTP
Trans-Zeatin riboside triphosphate
- tZRDP
Trans-Zeatin riboside diphosphate
- tZRMP
Trans-Zeatin riboside monophosphate
- LOG
Cytokinin-activating enzyme LONELY GUY
- iPR
Isopentenyl adenosine
- tZR
Trans-zeatin riboside
- iP
Isopentenyl adenine
- tZT
Trans-zeatin
- AHP
Histidine-containing phosphotransfer peotein
- Type-A ARRs
Type-A response regulators
- Type-B ARRs
Type-B response regulators
- trpB
Tryptophan synthase beta chain
- Trp
Tryptophan
- DDC
Tryptophan decarboxylase
- TAA
Tryptophan aminotransferase
- TAM
Tryptamine
- IPA
Indole-3-pyruvate
- CYP71A13
Indoleacetaldoxime dehydratase
- IAOx
Indole 3-acetaldoxime
- YUCCA
Indole-3-pyruvate monooxygenase
- IAAld
Indole-3-acetaldehyde
- IAN
Indole-3-acetonitrile
- ALDH
Aldehyde dehydrogenase
- NIT
Nitrilase
- IAA
Indole-3-acetate
- AUX/IAA
Auxin-responsive protein IAA
- ARF
Auxin response factor
- TIR1
Transport inhibitor response 1
- SAUR
Small auxin up-regulated RNA
- LA
Phospholipase A1
- α-LeA
Alpha-Linolenic acid
- LOX
Lipoxygenase
- 12,13-HPOT
12,13-hydroperoxylinoleic acid
- AOS
Allene oxide synthase
- 12,13-EOT
12,13-epoxyoctadecatrienoic acid
- AOC
Allene oxide cyclase
- OPDA
Oxophytodienoic acid
- OPR
Oxophytodienoic acid reductase
- JMT
Jasmonic acid carboxyl methyltransferase
- JAR1
Jasmonic acid resistant 1
- MeJA
Methyl jasmonate
- JA-Ile
Jasmonoyl-L-isoleucine
- COI1
Coronatine-insensitive protein 1
- JAZ
Jasmonate ZIM domain-containing protein
- PAL
Phenylalanine ammonia-lyase
- BA2H
Benzoate
- NADPH
Oxygen oxidoreductase (2-hydroxylating)
- NPR
Nonexpressorofpathogenesis-relatedgenes1
- TGA
Transcription factor TGA
- 90B/724B
Steroid 22-alpha-hydroxylase
- 6-DeoxoCT
6-Deoxocathasterone
- 6-OHCN
6alpha-Hydroxycampestanol
- 6-DeoxoTE
6-Deoxoteasterone
- 6-OxoCN
6-Oxocampestanol
- 6-Deoxo3DT
3-Dehydro-6-deoxoteasterone
- CT
Cathasterone
- 6-DeoxoTY
6-Deoxotyphasterol
- TE
Teasterone
- 6-DeoxoCS
6-Deoxocastasterone
- 3-DT
3-Dehydroteasterone
- 6-OHCS
6alpha-Hydroxy-castasterone
- TY
Typhasterol
- 85A1/2
Brassinosteroid-6-oxidase 1
- 92A6
Typhasterol/6-deoxotyphasterol 2alpha-hydroxylase
- 85A2
Brassinosteroid-6-oxidase 2
- BRI1
Protein brassinosteroid insensitive 1
- BAK1
Brassinosteroid insensitive 1-associated receptor kinase 1
- BSK
BR-signaling kinase
- BSU1
Serine/threonine-protein phosphatase
- BIN2
Brassinosteroid insensitive 2
- BZR1/2
Brassinosteroid resistant 1/2
- PP2A
Protein phosphatase type 2A
- GGPP
Geranylgeranyldiphosphate
- CPS
Ent-copalyl diphosphate synthase
- ent-CDP
Ent-Copalyl diphosphate
- KS
Ent-kaurene synthase
- KO
Ent-Kaurene oxidase
- KAO
Ent-Kaurenoicacid oxidase
- GA
Gibberellin
- GA20ox
GA20-oxidase
- GA30ox
GA30-oxidase
- GID1
Gibberellin insensitive dwarf 1
Authors’ contributions
CZ: Conceptualization, Writing- Original draft preparation, Writing- Reviewing and Editing. LXY: Methodology, Writing- Reviewing and Editing. GJL: Validation. XY: Investigation. RJ: Supervision, Funding acquisition. All authors have read and approved the manuscript.
Funding
This work was supported by the Anhui Provincial Natural Science Foundation [grant numbers 1908085QC113]; the Anhui Provincial Natural Science Foundation [grant number1708085MC57]; and the National Natural Science Foundation of China [grant number31600543]. All these fundings play roles in the design of the study and collection, analysis, and writing.
Availability of data and materials
All the sequencing data were submitted to NCBI Sequence Read Archive database under accession number PRJNA531583.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chen Zhu, Email: chenzhu0621@126.com.
Lu Xiaoyu, Email: maggie890407@126.com.
Gao Junlan, Email: 372595137@qq.com.
Xuan Yun, Email: 120090133@qq.com.
Ren Jie, Email: renjieaaas@sina.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02628-5.
References
- 1.Lim PO, Kim HJ, Gil Nam H. Leaf senescence. Annu Rev Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 2.Lers A. 6 Environmental regulation of leaf senescence. Annu Plant Rev Senescence ProcessPlants. 2007;26:108. [Google Scholar]
- 3.Khanna-Chopra R. Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma. 2012;249(3):469–481. doi: 10.1007/s00709-011-0308-z. [DOI] [PubMed] [Google Scholar]
- 4.Guo Y, Gan S. Leaf senescence: signals, execution, and regulation. Curr Top Dev Biol. 2005;71:83–112. doi: 10.1016/S0070-2153(05)71003-6. [DOI] [PubMed] [Google Scholar]
- 5.Davies PJ. The plant hormones: their nature, occurrence, and functions. Dordrecht: Springer Netherlands; 2010.
- 6.Jibran R, Hunter DA, Dijkwel PP. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol Biol. 2013;82(6):547–561. doi: 10.1007/s11103-013-0043-2. [DOI] [PubMed] [Google Scholar]
- 7.Gan S, Amasino RM. Cytokinins in plant senescence: from spray and pray to clone and play. Bioessays. 2010;18(7):557–565. [Google Scholar]
- 8.Li Z, Peng J, Wen X, Guo H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell. 2013;25(9):3311–3328. doi: 10.1105/tpc.113.113340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Y, Adachi Y, Ye W, Hayashi M, Nakamura Y, Kinoshita T, Mori IC, Murata Y. Difference in abscisic acid perception mechanisms between closure induction and opening inhibition of stomata. Plant Physiol. 2013;163(2):600–610. doi: 10.1104/pp.113.223826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinbothe C, Springer A, Samol I, Reinbothe S. Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009;276(17):4666–4681. doi: 10.1111/j.1742-4658.2009.07193.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhang K, Halitschke R, Yin C, Liu C-J, Gan S-S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci. 2013;110(36):14807–14812. doi: 10.1073/pnas.1302702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Zhao Z, Si J, Di C, Han J, An L. Brassinosteroids alleviate chilling-induced oxidative damage by enhancing antioxidant defense system in suspension cultured cells of Chorispora bungeana. Plant Growth Regul. 2009;59(3):207–214. [Google Scholar]
- 13.Zwack PJ, Rashotte AM. Cytokinin inhibition of leaf senescence. Plant Signal Behav. 2013;8(7):e24737. doi: 10.4161/psb.24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartmann A, Senning M, Hedden P, Sonnewald U, Sonnewald S. Reactivation of meristem activity and sprout growth in potato tubers require both cytokinin and gibberellin. Plant Physiol. 2011;155(2):776–796. doi: 10.1104/pp.110.168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazan K, Manners JM. Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci. 2009;14(7):373–382. doi: 10.1016/j.tplants.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Noodén LD. Senescence and aging in plants. Leopold: Academic Press; 1988.
- 17.Van Bragt J. Biochemistry and physiology of plant hormones. Heidelberg: Springer-Verlag; 1979.
- 18.Thomas H. Senescence, ageing and death of the whole plant. New Phytol. 2013;197(3):696–711. doi: 10.1111/nph.12047. [DOI] [PubMed] [Google Scholar]
- 19.Zari MP. Biomimetic design for climate change adaptation and mitigation. Archit Sci Rev. 2010;53(2):172–183. [Google Scholar]
- 20.Engels C, Kirkby E, White P. Mineral nutrition, yield and source–sink relationships. San Diego: Academic Press; 2012.
- 21.Ogata K. A systematic study of the genus Acer. Bull Tokyo Univ Forests. 1967;63:89–206. [Google Scholar]
- 22.de Jong PC. Flowering and sex expression in Acer L. A biosystematic study. Wageningen: Veenman; 1976.
- 23.Wu Z, Raven P, Hong D. Flora of China. Vol. 11: Oxalidaceae through Aceraceae. Beijing: Science Press and St. Louis: Missouri Botanical Garden Press; 2008. [Google Scholar]
- 24.van Gelderen DM, De Jong PC, Oterdoom HJ. Maples of the world. Portland: Timber Press; 1994.
- 25.Weakley A. Flora of the Southern and Mid-Atlantic States. UNC Herbarium, North Carolina Botanical Garden; 2012.
- 26.Bi W, Gao Y, Shen J, He C, Liu H, Peng Y, Zhang C, Xiao P. Traditional uses, phytochemistry, and pharmacology of the genus Acer (maple): a review. J Ethnopharmacol. 2016;189:31–60. doi: 10.1016/j.jep.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Ball DW. The chemical composition of maple syrup. J Chem Educ. 2007;84(10):1647. [Google Scholar]
- 28.González-Sarrías A, Li L, Seeram NP. Anticancer effects of maple syrup phenolics and extracts on proliferation, apoptosis, and cell cycle arrest of human colon cells. J Funct Foods. 2012;4(1):185–196. doi: 10.1002/ptr.3677. [DOI] [PubMed] [Google Scholar]
- 29.Perkins TD, van den Berg AK. Maple syrup—production, composition, chemistry, and sensory characteristics. Adv Food Nutr Res. 2009;56:101–143. doi: 10.1016/S1043-4526(08)00604-9. [DOI] [PubMed] [Google Scholar]
- 30.Legault J, Girard-Lalancette K, Grenon C, Dussault C, Pichette A. Antioxidant activity, inhibition of nitric oxide overproduction, and in vitro antiproliferative effect of maple sap and syrup from Acer saccharum. J Med Food. 2010;13(2):460–468. doi: 10.1089/jmf.2009.0029. [DOI] [PubMed] [Google Scholar]
- 31.Park KH, Yoon KH, Yin J, Le TT, Ahn HS, Yoon SH, Lee MW. Antioxidative and anti-inflammatory activities of galloyl derivatives and antidiabetic activities of Acer ginnala. Evid Based Complement Alternat Med. 2017;2017:6945912. doi: 10.1155/2017/6945912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sulpice R, Pyl E-T, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC. Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci. 2009;106(25):10348–10353. doi: 10.1073/pnas.0903478106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukushima A, Kusano M, Redestig H, Arita M, Saito K. Integrated omics approaches in plant systems biology. Curr Opin Chem Biol. 2009;13(5–6):532–538. doi: 10.1016/j.cbpa.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 34.Zu Castell W, Ernst D. Experimental ‘omics’ data in tree research: facing complexity. Trees. 2012;26(6):1723–1735. [Google Scholar]
- 35.Batushansky A, Kirma M, Grillich N, Toubiana D, Pham PA, Balbo I, Fromm H, Galili G, Fernie AR, Fait A. Combined transcriptomics and metabolomics of Arabidopsis thaliana seedlings exposed to exogenous GABA suggest its role in plants is predominantly metabolic. Mol Plant. 2014;7(6):1065–1068. doi: 10.1093/mp/ssu017. [DOI] [PubMed] [Google Scholar]
- 36.Jin J, Zhang H, Zhang J, Liu P, Chen X, Li Z, Xu Y, Lu P, Cao P. Integrated transcriptomics and metabolomics analysis to characterize cold stress responses in Nicotiana tabacum. BMC Genomics. 2017;18(1):496. doi: 10.1186/s12864-017-3871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fasano C, Diretto G, Aversano R, D'Agostino N, Di Matteo A, Frusciante L, Giuliano G, Carputo D. Transcriptome and metabolome of synthetic Solanum autotetraploids reveal key genomic stress events following polyploidization. New Phytol. 2016;210(4):1382–1394. doi: 10.1111/nph.13878. [DOI] [PubMed] [Google Scholar]
- 38.Xue Q, Fan H, Yao F, Cao X, Liu M, Sun J, Liu Y. Transcriptomics and targeted metabolomics profilings for elucidation of pigmentation in Lonicera japonica flowers at different developmental stages. Industrial Crops and Products. 2019;145:111981.
- 39.Tsaballa A, Sarrou E, Xanthopoulou A, Tsaliki E, Kissoudis C, Karagiannis E, Michailidis M, Martens S, Sperdouli E, Hilioti Z. Comprehensive approaches reveal key transcripts and metabolites highlighting metabolic diversity among three oriental tobacco varieties. Ind Crop Prod. 2020;143:111933. [Google Scholar]
- 40.Zhou S, Chen J, Lai Y, Yin G, Chen P, Pennerman KK, Yan H, Wu B, Zhang H, Yi X. Integrative analysis of metabolome and transcriptome reveals anthocyanins biosynthesis regulation in grass species Pennisetum purpureum. Ind Crop Prod. 2019;138:111470. [Google Scholar]
- 41.Guo J, Wu Y, Wang G, Wang T, Cao F. Integrated analysis of the transcriptome and metabolome in young and mature leaves of Ginkgo biloba L. Ind Crop Prod. 2020;143:111906. [Google Scholar]
- 42.Wang J, Zhang T, Shen X, Liu J, Zhao D, Sun Y, Wang L, Liu Y, Gong X, Liu Y. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics. 2016;12(7):116. [Google Scholar]
- 43.Forsberg EM, Huan T, Rinehart D, Benton HP, Warth B, Hilmers B, Siuzdak G. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS online. Nat Protoc. 2018;13(4):633. doi: 10.1038/nprot.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z, Lu X, Xuan Y, Tang F, Wang J, Shi D, Fu S, Ren J. Transcriptome analysis based on a combination of sequencing platforms provides insights into leaf pigmentation in Acer rubrum. BMC Plant Biol. 2019;19(1):240. doi: 10.1186/s12870-019-1850-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60(12):3311–3336. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- 46.Abeles FB, Morgan PW, Saltveit Jr ME. Ethylene in plant biology. New York: Academic Press; 1992.
- 47.Adams DO, Yang SF. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc Natl Acad Sci. 1979;76(1):170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta N, Gupta S, Kumar A. Exogenous cytokinin application increases cell membrane and chlorophyll stability in wheat (Triticum aestivum L.) Cereal Res Commun. 2000;28(3):287–291. [Google Scholar]
- 49.Osborne DJ. Effect of kinetin on protein & nucleic acid metabolism in xanthium leaves during senescence. Plant Physiol. 1962;37(5):595–602. doi: 10.1104/pp.37.5.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staden JV, Cook EL, Noodén LD. Cytokinins and Senescence. Leopold: Academic Press; 1988.
- 51.Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270(5244):1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- 52.Humbeck K. Senescence in plants. J Plant Growth Regul. 2014;33(1):1–3.
- 53.Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci. 2007;104(49):19631–19636. doi: 10.1073/pnas.0709453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gan S. The hormonal regulation of senescence: Springer Netherlands; 2010.
- 55.Yunde Z. Auxin Biosynthesis and Its Role in Plant Development. Annu Rev Plant Biol. 2010;61(1):49–64. [DOI] [PMC free article] [PubMed]
- 56.Normanly J. Auxin metabolism. Physiol Plant. 1997;100(3):431–442. [Google Scholar]
- 57.Taiz L, Zeiger E. Plant physiology. Q Rev Biol. 2006;167(4):161–168. [Google Scholar]
- 58.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20(13):1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ueda J, Kato J, Yamane H, Takahashi N. Inhibitory effect of methyl jasmonate and its related compounds on kinetin-induced retardation of oat leaf senescence. Physiol Plant. 1981;52(2):305–309. [Google Scholar]
- 60.Ueda J, Kato J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.) Plant Physiol. 1980;66(2):246–249. doi: 10.1104/pp.66.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vlot AC, Dempsey DMA, Klessig DF: Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206. [DOI] [PubMed]
- 62.Rivas-San Vicente M, Plasencia J. Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot. 2011;62(10):3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- 63.Yusuf M, Hayat S, Alyemeni MN, Fariduddin Q, Ahmad A. Salicylic Acid: Physiological Roles in Plants. Dordrecht: Springer Netherlands; 2013.
- 64.Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21(9):2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujioka S, Li J, Choi Y-H, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9(11):1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z-Y, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410(6826):380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 67.Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9(5):701–713. [Google Scholar]
- 68.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90(5):929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 69.Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123(4):1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S. Organ-specific expression of Brassinosteroid-biosynthetic genes and distribution of endogenous Brassinosteroids in Arabidopsis. Plant Physiol. 2003;131(1):287–297. doi: 10.1104/pp.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montoya T, Nomura T, Yokota T, Farrar K, Harrison K, Jones JGD, Kaneta T, Kamiya Y, Szekeres M, Bishop GJ. Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J. 2010;42(2):262–269. doi: 10.1111/j.1365-313X.2005.02376.x. [DOI] [PubMed] [Google Scholar]
- 72.Li J, Yu K, Wei J, Ma Q, Wang B, Yu D. Gibberellin retards chlorophyll degradation during senescence of Paris polyphylla. Biol Plant. 2010;54(2):395–399. [Google Scholar]
- 73.Light KM, Wisniewski JA, Vinyard WA, Kieber-Emmons MT. Perception of the plant hormone ethylene: known-knowns and known-unknowns. JBIC J Biol Inorg Chem. 2016;21(5–6):715–728. doi: 10.1007/s00775-016-1378-3. [DOI] [PubMed] [Google Scholar]
- 74.Iqbal N, Khan NA, Ferrante A, Trivellini A, Francini A, Khan M. Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front Plant Sci. 2017;8:475. doi: 10.3389/fpls.2017.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Giovannoni J. Molecular biology of fruit maturation and ripening. Annu Rev Plant Biol. 2001;52(1):725–749. doi: 10.1146/annurev.arplant.52.1.725. [DOI] [PubMed] [Google Scholar]
- 76.Giovannoni JJ. 12 Fruit ripening and its manipulation. Ann Plant Rev Senescence Process Plants. 2008;26:278. [Google Scholar]
- 77.John I, Drake R, Farrell A, Cooper W, Lee P, Horton P, Grierson D. Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: molecular and physiological analysis. Plant J. 1995;7(3):483–490. [Google Scholar]
- 78.Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262(5133):539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 79.Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim Y-S, Penfold CA, Jenkins D. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 2011;23(3):873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72(3):427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 81.Zhang K, Xia X, Zhang Y, Gan SS. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 2012;69(4):667–678. doi: 10.1111/j.1365-313X.2011.04821.x. [DOI] [PubMed] [Google Scholar]
- 82.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 83.Oh SA, Lee SY, Chung IK, Lee C-H, Nam HG. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol. 1996;30(4):739–754. doi: 10.1007/BF00019008. [DOI] [PubMed] [Google Scholar]
- 84.Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol. 1998;37(3):455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- 85.He Y, Tang W, Swain JD, Green AL, Jack TP, Gan S. Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 2001;126(2):707–716. doi: 10.1104/pp.126.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42(4):567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 87.Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24(16):1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang K, Gan S-S. An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 2012;158(2):961–969. doi: 10.1104/pp.111.190876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gan S, Amasino RM. Cytokinins in plant senescence: from spray and pray to clone and play. Bioessays. 1996;18(7):557–565. [Google Scholar]
- 90.Zwack PJ, Rashotte AM. Interactions between cytokinin signalling and abiotic stress responses. J Exp Bot. 2015;66(16):4863–4871. doi: 10.1093/jxb/erv172. [DOI] [PubMed] [Google Scholar]
- 91.Richmond AE, Lang A. Effect of kinetin on protein content and survival of detached Xanthium leaves. Science. 1957;125(3249):650–651. [Google Scholar]
- 92.Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413(6854):383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 93.Brandstatter I. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell. 1998;10(6):1009–1019. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proceedings of the National Academy of Sciences. 1998;95(5):2691–96. [DOI] [PMC free article] [PubMed]
- 95.Ren B, Liang Y, Deng Y, Chen Q, Zhang J, Yang X, Zuo J. Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 2009;19(10):1178–1190. doi: 10.1038/cr.2009.88. [DOI] [PubMed] [Google Scholar]
- 96.Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot. 2013;64(9):2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nooden LD. Abscisic acid, auxin, and other regulations of senescence. Senescence Aging Plants. 1988;3(3):329–368. [Google Scholar]
- 98.Kim JI, Murphy AS, Baek D, Lee S-W, Yun D-J, Bressan RA, Narasimhan ML. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot. 2011;62(11):3981–3992. doi: 10.1093/jxb/err094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kant S, Bi Y-M, Zhu T, Rothstein SJ. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009;151(2):691–701. doi: 10.1104/pp.109.143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang H, Liu B, Liu L, Song S. Jasmonate action in plant growth and development. J Exp Bot. 2017;68(6):1349–1359. doi: 10.1093/jxb/erw495. [DOI] [PubMed] [Google Scholar]
- 101.He Y, Fukushige H, Hildebrand DF, Gan S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002;128(3):876–884. doi: 10.1104/pp.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Uji Y, Akimitsu K, Gomi K. Identification of OsMYC2-regulated senescence-associated genes in rice. Planta. 2017;245(6):1241–1246. doi: 10.1007/s00425-017-2697-5. [DOI] [PubMed] [Google Scholar]
- 103.Qi T, Wang J, Huang H, Liu B, Gao H, Liu Y, Song S, Xie D. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell. 2015;27(6):1634–1649. doi: 10.1105/tpc.15.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song S, Huang H, Wang J, Liu B, Qi T, Xie D. MYC5 is involved in jasmonate-regulated plant growth, leaf senescence and defense responses. Plant Cell Physiol. 2017;58(10):1752–1763. doi: 10.1093/pcp/pcx112. [DOI] [PubMed] [Google Scholar]
- 105.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21(1):131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morris K, Mackerness SAH, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 2000;23(5):677–685. doi: 10.1046/j.1365-313x.2000.00836.x. [DOI] [PubMed] [Google Scholar]
- 107.Barth C, Moeder W, Klessig DF, Conklin PL. The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1. Plant Physiol. 2004;134(4):1784–1792. doi: 10.1104/pp.103.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. Salicylic acid–independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell. 2006;18(4):1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 1996;8(11):2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nawrath C, Métraux J-P. Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11(8):1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roetschi A, Si-Ammour A, Belbahri L, Mauch F, Mauch-Mani B. Characterization of an Arabidopsis–Phytophthora pathosystem: resistance requires a functional PAD2 gene and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 2001;28(3):293–305. doi: 10.1046/j.1365-313x.2001.01148.x. [DOI] [PubMed] [Google Scholar]
- 112.Mishina TE, Zeier J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 2006;141(4):1666–1675. doi: 10.1104/pp.106.081257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee H-I, León J, Raskin I. Biosynthesis and metabolism of salicylic acid. Proc Natl Acad Sci. 1995;92(10):4076–4079. doi: 10.1073/pnas.92.10.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau J-P, Vuylsteke M. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell. 2004;16(10):2749–2771. doi: 10.1105/tpc.104.023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li J, Chory J. Brassinosteroid actions in plants. J Exp Bot. 1999;50(332):275–282. [Google Scholar]
- 116.Krishna P, Prasad BD, Rahman T. Brassinosteroid action in plant abiotic stress tolerance. New York: Humana Press; 2017. [DOI] [PubMed]
- 117.He Y, Xu R, Zhao Y. Enhancement of senescence by epibrassinolide in leaves of mung bean seedling. Acta Phytophysiol Sinica. 1996;22(1):58–62. [Google Scholar]
- 118.Ding W, Zhao Y. Effect of epi-BR on activity of peroxidase and soluble protein content of cucumber cotyledon. Acta Phytophysiol Sinica. 1995;21(3):259–264. [Google Scholar]
- 119.Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S. Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;126(2):770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Montoya T, Nomura T, Yokota T, Farrar K, Harrison K, Jones JG, Kaneta T, Kamiya Y, Szekeres M, Bishop GJ. Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J. 2005;42(2):262–269. doi: 10.1111/j.1365-313X.2005.02376.x. [DOI] [PubMed] [Google Scholar]
- 121.Ya-li N, Qian Z, Xiao-han Z, Qiu-shi A, Shui-shan S. Research progress on the role and regulation mechanism of gibberellin singal in response to abiotic stress. Biotechnol Bull. 2015;31(10):31–37. [Google Scholar]
- 122.Yu K, Wei J, Ma Q, Yu D, Li J. Senescence of aerial parts is impeded by exogenous gibberellic acid in herbaceous perennial Paris polyphylla. J Plant Physiol. 2009;166(8):819–830. doi: 10.1016/j.jplph.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 123.Ranwala AP, Miller WB. Preventive mechanisms of gibberellin4+ 7 and light on low-temperature-induced leaf senescence in Lilium cv. Stargazer. Postharvest Biol Technol. 2000;19(1):85–92. [Google Scholar]
- 124.Kappers I, Jordi W, Tsesmetzis N, Maas F, Van der Plas L. GA 4 does not require conversion into GA 1 to delay senescence of Alstroemeria hybrida leaves. J Plant Growth Regul. 1998;17(2):89–93. [Google Scholar]
- 125.Su P, Tong Y, Cheng Q, Hu Y, Zhang M, Yang J, Teng Z, Gao W, Huang L. Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway. Sci Rep. 2016;6:23057. doi: 10.1038/srep23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Soll J, Schultz G, Rüdiger W, Benz J. Hydrogenation of geranylgeraniol: two pathways exist in spinach chloroplasts. Plant Physiol. 1983;71(4):849–854. doi: 10.1104/pp.71.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schoefs B, Bertrand M. The formation of chlorophyll from chlorophyllide in leaves containing proplastids is a four-step process. FEBS Lett. 2000;486(3):243–246. doi: 10.1016/s0014-5793(00)02309-7. [DOI] [PubMed] [Google Scholar]
- 128.Tanaka R, Oster U, Kruse E, Rüdiger W, Grimm B. Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 1999;120(3):695–704. doi: 10.1104/pp.120.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou Y, Gong Z, Yang Z, Yuan Y, Zhu J, Wang M, Yuan F, Wu S, Wang Z, Yi C. Mutation of the light-induced yellow leaf 1 gene, which encodes a geranylgeranyl reductase, affects chlorophyll biosynthesis and light sensitivity in rice. PLoS One. 2013;8(9):e75299. doi: 10.1371/journal.pone.0075299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kato S, Takaichi S, Ishikawa T, Asahina M, Takahashi S, Shinomura T. Identification and functional analysis of the geranylgeranyl pyrophosphate synthase gene (crtE) and phytoene synthase gene (crtB) for carotenoid biosynthesis in Euglena gracilis. BMC Plant Biol. 2016;16(1):4. doi: 10.1186/s12870-015-0698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fray RG, Wallace A, Fraser PD, Valero D, Hedden P, Bramley PM, Grierson D. Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J. 1995;8(5):693–701. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Differentially expressed plant hormone-related metabolites candidates in red maple.
Additional file 2: Table S2. Differentially expressed gene (DEG) candidates involved in plant hormone metabolism in red maple.
Additional file 3: Table S3. Correlation matrix of metabolites (plant hormone-related) and gene expression levels.
Additional file 4: Table S4. Classification of macromolecules in senescing leaves compared with non-senescing leaves in red maple. “Total” refers to the total number of metabolites changed from senescent to non-senescent leaves.
Additional file 5: Table S5. Functional categories of genes correlated with the anabolism of macromolecules differentially expressed in senescing leaves compared with non-senescing leaves in red maple.
Data Availability Statement
All the sequencing data were submitted to NCBI Sequence Read Archive database under accession number PRJNA531583.