Abstract
A novel oligotrophic bacterium, designated strain CCA6, was isolated from leaf soil collected in Japan. Cells of the strain were found to be a Gram‐negative, non‐sporulating, motile, rod‐shaped bacterium. Strain CCA6 grew at 10–45°C (optimum 20°C) and pH 4.5–10.0 (optimum pH 5.0). The strain was capable of growth in poor‐nutrient (oligotrophic) medium, and growth was unaffected by high‐nutrient medium. The major fatty acid and predominant quinone system were C16:0 and ubiquinone‐8. Phylogenetic analysis based on 16S rRNA gene sequences indicated strain CCA6 presented as a member of the family Enterobacteriaceae. Multilocus sequence analysis (MLSA) based on fragments of the atpD, gyrB, infB, and rpoB gene sequences was performed to further identify strain CCA6. The MLSA showed clear branching of strain CCA6 with respect to Enterobacter type strains. The complete genome of strain CCA6 consisted of 4,476,585 bp with a G+C content of 54.3% and comprising 4,372 predicted coding sequences. The genome average nucleotide identity values between strain CCA6 and the closest related Enterobacter type strain were <88.02%. Based on its phenotypic, chemotaxonomic and phylogenetic features, strain CCA6 (=HUT 8142T =KCTC 62525T) can be considered as a novel species within the genus Enterobacter with the proposed name Enterobacter oligotrophica.
Keywords: average nucleotide identity value analysis, Enterobacter, genome sequence, multilocus sequence analysis, oligotroph, Voges–Proskauer test
In this study, we report a novel strain isolated from the leaf soil collected in Japan. Interesting features of this isolate were its growth unaffected by high‐nutrient medium and growth potential in poor‐nutrient (oligotrophic) medium. We performed physiological, chemotaxonomic, and phylogenetic analyses. Based on the results we propose the isolate represents a novel species within the genus Enterobacter.

1. INTRODUCTION
A variety of microorganisms are used in industrial fermentation for production of enzymes, medicines, and other organic compounds. Those microorganisms are generally grown in high‐nutrient medium containing large amounts of sugar, nitrogen, phosphorus, minerals, and other nutrients that are considered essential for their growth. Consequently, nutrient cost is an important factor when trying to achieve cost‐effective fermentation. This has led to the development of several related technologies, including bioengineering of microorganisms so as to enhance their productivity and yield (Min, Hwang, Lim, & Jung, 2017), use of agricultural byproducts as carbon and mineral sources (Thomsen, 2005), and production of chemicals such as bioemulsifiers (Banat, Satpute, Cameotra, Patil, & Nyayanit, 2014), biofuels (Ho, Ngo, & Guo, 2014), and biosurfactants (Banat et al., 2014) from renewable substrates.
Oligotrophs are organisms that grow under conditions of low levels of nutrients but grow more slowly at high levels (Kuznetsov, Dubinina, & Lapteva, 1979). Consequently, oligotrophs have not been applied for industrial use. We suggest that production costs could be reduced if oligotrophs could be used for industrial fermentation, and therefore screened for oligotrophs that are unaffected by a high‐nutrient condition. Here, we report the screening, isolation, and characterization of an oligotrophic bacterium from leaf soil, which is one kind of the compost and is accrued by fermenting the dry leaves. The isolate was named strain CCA6. This bacterium was capable of growth on poor‐nutrient medium, and its growth was unaffected by high‐nutrient mixtures. Moreover, physiological, chemotaxonomic, and phylogenetic analyses as well as average nucleotide identity (ANI) value analysis were performed to characterize strain CCA6. Based on the results of these analyses, we propose that strain CCA6 represents a novel species within the genus Enterobacter, for which the name E. oligotrophica sp. nov. is proposed.
2. MATERIALS AND METHODS
2.1. Bacterial isolation
Soil samples were collected from Higashi‐Hiroshima city in Hiroshima prefecture, Japan. A 1.5% agar (Nacalai tesque, Kyoto, Japan) plate (pH 7.2), which contained sulfates (>0.4%), calcium (>0.1%), iron (>0.01%), and a few fatty acids and/or other minerals at concentrations <0.01% was used for isolation. After 1 ml of a 10% (w/v) soil wash solution was inoculated onto a plate, the plate was incubated for 2 days at 37°C. Thereafter, a single colony was successively re‐streaked onto a new 1.5% agar plate at least three times to obtain a pure colony. The purified strain was then grown aerobically at 37°C in Nutrient Broth (Kyokuto, Tokyo, Japan) and preserved at −20°C as a suspension in Nutrient Broth supplemented with glycerol (30%, w/v).
2.2. Physiological characterization
Growth of strain CCA6 in Nutrient Broth was evaluated at various temperatures (4–50°C), pH (4.0–10.5), and NaCl concentrations (1–7%, w/v), and in the presence of selected antibiotics (ampicillin, chloramphenicol, and kanamycin). The OD600, which reflects cell growth, was measured by monitoring the difference between cellular and cell‐free turbidity values using an Eppendorf BioSpectrometer (Eppendorf, Hamburg, Germany). Carbon source utilization was assessed using API 20E (bioMérieux, Marcy‐l'Etoile, France) and API 50 CHE (bioMérieux) according to the manufacturer's instructions. Voges–Proskauer (VP) test was carried out using RapiD 20E (bioMérieux). Enzyme activities were evaluated using API ZYM (bioMérieux).
2.3. Chemotaxonomic analyses
The cellular fatty acid composition of strain CCA6 was determined using Sherlock Microbial Identification System Version 6.0 (MIDI, Newark, DE) with TSBA6 database (MIDI). Using the method of Bligh and Dyer (1959), lipids were extracted from lyophilized cells of strain CCA6 and loaded onto a Sep‐Pak Plus Silica cartridge (Waters, Milford, MA). The cartridge was then washed and the quinones were eluted. The quinones were quantified using an ACQUITY UPLC system (Waters) with an Eclipse Plus C18 column (Agilent technologies, Santa Clara, CA). The chromatographic conditions were as follows: mobile phase, methanol/isopropanol (3:1 v/v); flow rate, 0.5 ml/min; column oven temperature, 35°C. The quinone forms were identified as previously described (Tamaoka, Katayama‐Fujimura, & Kuraishi, 1983).
2.4. Phylogenetic analysis based on 16S rRNA gene
After strain CCA6 was cultured aerobically for 6 hr at 37°C in Nutrient Broth, the cells were harvested by centrifugation, and their genomic DNA was extracted and purified using an illustra bacteria genomicPrep Mini Spin Kit (GE Healthcare, Chicago, IL) according to the manufacturer's instructions. The 16S rRNA gene was amplified using KOD plus DNA Polymerase (TOYOBO, Osaka, Japan) with the bacterial universal primers 27f (5′‐AGAGTTTGATCMTGGCTCAG‐3′; Lane, 1991) and 1391r (5′‐GACGGGCGGTGTGTRCA‐3′; Turner, Pryer, Miao, & Palmer, 1999). After purifying the amplified PCR product using a Wizard SV Gel and PCR Clean‐up System (Promega, Madison, WI), the purified product was cloned into pTA2 vector (TOYOBO) and sequenced. Sequence was then compared with reference sequences available in the GenBank/EMBL/DDBJ databases using BLAST. Multiple alignment and construction of a maximum‐likelihood tree were performed using MEGA‐X (Kumar, Stecher, Li, Knyaz, & Tamura, 2018) with Tamura and Nei model (1993).
2.5. Multilocus sequence analysis based on housekeeping genes
Multilocus sequence analysis (MLSA) was performed using the method of Brady et al. (2008), Brady, Cleenwerck, Venter, Coutinho, and De Vos (2013) with some modifications. A phylogenetic tree of concatenated sequences (2,637 bp), including partial sequences of four housekeeping genes [atpD (β subunit of ATP synthase; 642 bp), gyrB (DNA gyrase; 743 bp), infB (translation initiation factor 2; 615 bp), and rpoB (β subunit of RNA polymerase; 637 bp)] from strain CCA6, was also reconstructed using the maximum‐likelihood method with Tamura and Nei model (1993). The housekeeping genes of strain CCA6 and the related type strains are available in the GenBank/DDBL/EMBL databases.
2.6. Genome sequencing and ANI value analysis
The concentration and purity of the genomic DNA were measured using a NanoDrop ND‐1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and a Quant‐iT dsDNA BR assay kit (Invitrogen, Waltham, MA), respectively. After fragmenting the genomic DNA (20.9 μg) into approximately 20‐kb pieces using g‐TUBE (Covaris, Brighton, UK), the resultant fragments were ligated to SMRTbell sequencing adapters using a SMRTbell Template Prep Kit 1.0 (Pacific Biosciences, Menlo Park, CA), yielding SMRTbell libraries. The library size was measured using Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA). The SMRTbell libraries were then bound to polymerases and sequencing primers using a DNA/Polymerase binding kit P6 v2 (Pacific Biosciences), yielding the sequencing templates. The concentration of the sequencing templates was calculated using Binding Calculator v2.3.1.1 (Pacific Biosciences), after which the templates were bound to MagBeads using a MagBead kit (Pacific Biosciences) and loaded onto SMRT Cells 8Pac v3 (Pacific Biosciences). Sequencing was then performed using PacBio RS II (Pacific Biosciences).
The raw data included 100,771 reads with 330 coverage and were assembled de novo using SMRT Analysis v2.3.0 (Pacific Biosciences; Chin et al., 2013) to filter the subreads. Genome annotation was performed using CRITICA (Badger & Olsen, 1999) and Glimmer2 (Delcher, Harmon, Kasif, White, & Salzberg, 1999). The tRNA and rRNA genes were detected using tRNAScan‐SE (Lowe & Eddy, 1997) and BLASTN (Altschul, Gish, Miller, Myers, & Lipman, 1990), respectively. ANI values were calculated through pairwise genome comparison of whole‐genome sequences of strain CCA6 and its related Enterobacter type strains using the ANI algorithm (Goris et al., 2007) implemented within OrthoANIu tools (Yoon, Ha, Lim, Kwon, & Chun, 2017).
The genome properties of type strains of Enterobacter, Klebsiella, Kosakonia, Lelliottia, Pluralibacter, Pseudescherichia, Pseudomonas, and Raoultella species are presented in Table A1.
3. RESULTS AND DISCUSSION
3.1. Isolation of strain CCA6
To obtain oligtrophic microorganisms, filtrates were prepared from several soil samples and plated onto 1.5% agar (pH 7.2) without a carbon source or other medium components. After incubation for 2 days at 37°C, a single colony was obtained from the leaf soil filtrate. A purified colony was then obtained through standard dilution plating on the same plates and was named strain CCA6. Although high‐nutrient mixtures suppress the growth of some oligotrophic bacteria (Ohta, 2000; Ohta & Taniguchi, 1988), strain CCA6 showed a higher rate of growth, similar to that of Escherichia coli MG1655, when cultured in Nutrient Broth or LB media (Figure A1). By contrast, E. coli MG1655 did not grow on a 1.5% agar (pH 7.2). These results suggest we had successfully isolated the desired oligotroph.
3.2. Morphological and physiological characterization
Cells of strain CCA6 were Gram‐negative, motile, rod‐shaped and non‐sporulating. Colonies grown on Nutrient Broth plates were circular, smooth, glistening, light yellow, and 5.0 mm in diameter after incubation overnight at 37°C. When we examined the effect of culture temperature and pH, we found that the strain was capable of growing at temperatures between 10 and 45°C, but no growth was seen at 4 or 50°C (Figure A2a). The strain also grew effectively at pHs between 4.5 and 10.0, but growth rates were sharply lower at pHs below 4.0 or above 10.5 (Figure A2b). The strain was tolerant to 6% (w/v) NaCl (Figure A2c) and was resistant to ampicillin, but chloramphenicol and kanamycin inhibited its growth.
Figure 2.
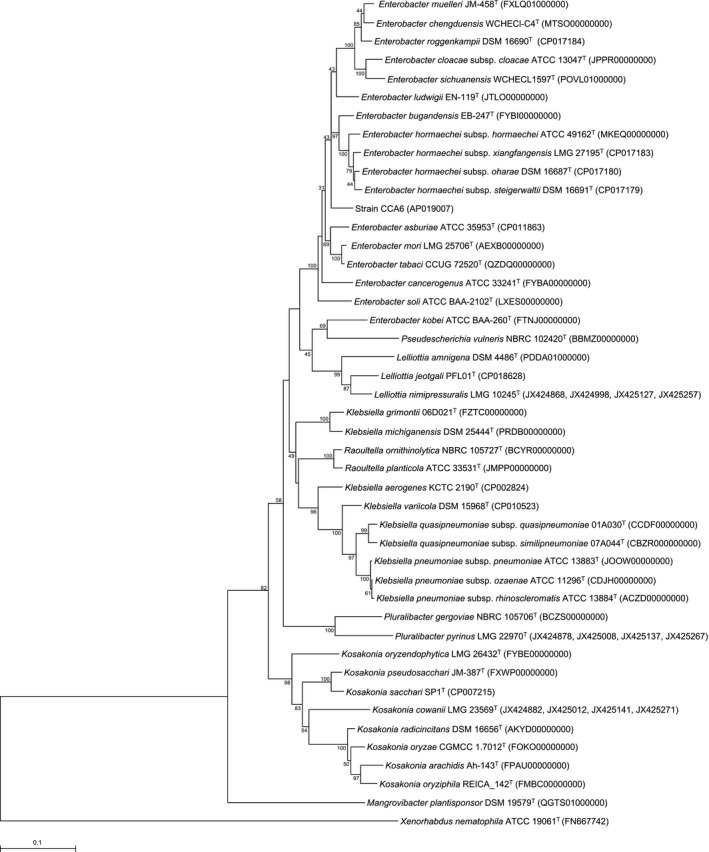
Phylogenetic tree reconstructed from analysis of the sequences of four housekeeping genes (atpD, gyrB, infB, and rpoB) and showing the relationships between strain CCA6 and the related type strains. The bar indicates a 0.1% nucleotide substitution rate. The tree was rooted using X. nematophila ATCC 19061T as the outgroup
Strain CCA6 showed a broad range of enzyme activities, including acid phosphatase, N‐acetyl‐β‐d‐glucosaminidase, alkaline phosphatase, cystine aminopeptidase, esterase lipase (C8), α‐galactosidase, β‐galactosidase, α‐glucosidase, β‐glucosidase, leucine aminopeptidase, α‐mannosidase, naphthol AS‐BI phosphate, trypsin, and valine aminopeptidase. By contrast, strain CCA6 did not exhibit α‐chymotrypsin, esterase (C4), α‐fucosidase, β‐glucuronidase, or lipase (C14) activity. These results suggest that strain CCA6 is capable of catabolizing a variety of different carbon sources. Culture with different carbon sources revealed that CCA6 was able to utilize the following compounds as a carbon source for growth: inulin, amygdalin, arbutin, esculin ferric citrate, 2‐nitrophenyl β‐d‐galactopyranoside, d‐cellobiose, d‐trehalose, d‐maltose, d‐lactose, d‐galactose, d‐glucose, l‐sorbose, d‐fructose, d‐mannose, l‐rhamnose, dulcitol, d‐sorbitol, d‐mannitol, N‐acetyl‐glucosamine, methyl‐β‐d‐xylopyranoside, d‐arabinose, l‐arabinose, d‐xylose, l‐xylose, l‐fucose, d‐lyxose, d‐ribose, adonitol, erythritol, glycerol, l‐arginine, l‐lysine, l‐ornithine, l‐tryptophane, 2‐keto gluconate, citrate, gluconate, pyruvate, and urea. By contrast, no growth occurred on glycogen, gelatin, starch, salicin, d‐melezitose, d‐raffinose, gentiobiose, d‐sucrose, d‐melibiose, d‐turanose, d‐tagatose, inositol, methyl‐α‐d‐glucopyranoside, methyl‐α‐d‐mannopyranoside, d‐fucose, d‐arabitol, l‐arabitol, xylitol, 5‐keto gluconate, or thiosulfate. Differences in phenotypic characteristics of strain CCA6 and its related type species are shown in Table 1.
Table 1.
Differential characteristics of strain CCA6 and phylogenetically related species
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Carbon source utilization | ||||||||||
| d‐Sucrose | ‐ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| d‐Melibiose | ‐ | + | ++ | ++ | W | ++ | ++ | ++ | ++ | ++ |
| d‐Turanose | ‐ | + | + | ‐ | W | + | ‐ | + | W | + |
| l‐Rhamnose | ++ | + | ++ | ‐ | ++ | ++ | ++ | ++ | ++ | ++ |
| Inositol | ‐ | + | ++ | ++ | ‐ | W | W | + | ++ | ++ |
| Dulcitol | + | + | W | ‐ | ++ | ‐ | ‐ | + | + | + |
| d‐Sorbitol | ++ | ++ | ++ | ++ | W | ++ | ++ | ++ | ++ | ++ |
| Methyl‐α‐d‐glucopyranoside | ‐ | ++ | ++ | ++ | ++ | + | + | ++ | ++ | ++ |
| d‐Arabinose | ++ | + | ++ | ++ | ++ | ++ | ++ | + | ++ | + |
| l‐Fucose | ++ | + | + | ‐ | ++ | + | ++ | + | ‐ | W |
| d‐Lyxose | ++ | ++ | W | ++ | + | ++ | ++ | ++ | + | ++ |
| Adonitol | + | + | + | ‐ | W | W | ++ | + | ‐ | + |
| d‐Arabitol | ‐ | + | + | ‐ | W | W | ++ | + | ‐ | + |
| 2‐Keto gluconate | + | + | ++ | + | W | ‐ | + | + | W | ++ |
| Enzyme activity | ||||||||||
| Arginine dihydrolase | + | ++ | ++ | ++ | ++ | ++ | ++ | ‐ | ++ | ++ |
| Ornithine decarboxylase | + | ++ | ++ | ++ | ++ | ++ | ++ | ‐ | ++ | ++ |
| Lysine decarboxylase | + | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | W |
| Esculin hydrolysis | + | ++ | + | ++ | ‐ | W | W | + | W | + |
| Voges–Proskauer test | ‐ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
Strains: 1, strain CCA6; 2, E. asburiae ATCC 35953T; 3, E. cloacae subsp. cloacae ATCC13047T; 4, E. cloacae subsp. dissolvens ATCC 23373T; 5, E. hormaechei subsp. hormaechei ATCC 49162T; 6, E. hormaechei subsp. oharae DSM 16687T; 7, E. hormaechei subsp. steigerwaltii DSM 16691T; 8, E. hormaechei subsp. xiangfangensis LMG 27195T; 9, E. kobei ATCC BAA‐260T; 10, E. ludwigii EN‐119T. ++, strong positive; +, positive; W, weak positive; —, not detected.
Nearly all Enterobacter species produce acetoin as the end product of glucose metabolism, which yields a red complex in the VP test medium. The related Enterobacter type strains show a positive VP test; however, strain CCA6 was negative (Table 1).
3.3. Chemotaxonomic characterization
When strain CCA6 was cultured aerobically in Nutrient Broth, the major fatty acids were C16:0 and summed feature 8 (comprising C18:1ω6c and/or C18:1ω7c). The overall fatty acid profile of strain CCA6 was similar to that of E. hormaechei subsp. hormaechei ATCC 49162T (Table 2). Respiratory quinone analysis showed the presence of ubiquinone‐7 (4.2%), ubiquinone‐8 (87.2%), and menaquinone‐8 (8.6%).
Table 2.
Comparative fatty acid contents (%) of strain CCA6 and phylogenetically related reference strains
| Fatty acids | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Saturated fatty acids | ||||||
| C10 : 0 | 0.04 | ‐ | ‐ | ‐ | ‐ | ‐ |
| C11 : 0 | 0.11 | ‐ | ‐ | 0.08 | ‐ | ‐ |
| C12 : 0 | 3.7 | 3.03 | 2.47 | 2.50 | 2.26 | 3.96 |
| C13 : 0 | 1.4 | 0.72 | 0.61 | 1.18 | 0.37 | 0.90 |
| C14 : 0 | 5.6 | 6.46 | 6.74 | 8.39 | 9.73 | 6.18 |
| C16 : 0 | 22.7 | 27.89 | 29.27 | 21.75 | 30.16 | 25.71 |
| C17 : 0 | 4.0 | 3.73 | 3.43 | 4.94 | 2.02 | 3.18 |
| C18 : 0 | 0.3 | 0.47 | 0.53 | 0.38 | 0.47 | 0.39 |
| Branched‐chain fatty acids | ||||||
| iso‐C15 : 0 3‐OH | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| anteiso‐C19 : 0 | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| iso‐C19 : 0 | 0.2 | ‐ | 0.11 | ‐ | ‐ | ‐ |
| Unsaturated fatty acids | ||||||
| C15 : 1ω6c | 0.1 | ‐ | ‐ | ‐ | ‐ | ‐ |
| C15 : 1ω8c | 0.3 | ‐ | 0.09 | 0.14 | ‐ | ‐ |
| C16 : 1ω5c | ‐ | 0.21 | 0.21 | 0.22 | ‐ | 0.21 |
| C17 : 1ω8c | 0.4 | ‐ | ‐ | 0.44 | ‐ | ‐ |
| 11‐methyl‐C18 : 1ω7c | ‐ | 0.26 | ‐ | ‐ | ‐ | ‐ |
| C18 : 1ω5c | ‐ | ‐ | 0.18 | 0.19 | ‐ | ‐ |
| Hydroxy fatty acids | ||||||
| C15 : 0 3‐OH | ‐ | 0.16 | 0.15 | 0.36 | ‐ | 0.21 |
| Cyclopropane acids | ||||||
| cyclo‐C17 : 0 | 14 | 26.01 | 20.4 | 21.08 | 21.69 | 25.17 |
| cyclo‐C19 : 0ω8c | 3.3 | 7.04 | 6.10 | 4.35 | 5.79 | 5.99 |
| Summed feature | ||||||
| 1 | 1.3 | 0.93 | 0.64 | 1.46 | 0.34 | 0.74 |
| 2 | 8.1 | 6.14 | 6.85 | 6.21 | 6.79 | 6.79 |
| 3 | 12.6 | 3.51 | 6.50 | 5.71 | 4.49 | 4.9 |
| 8 | 21.3 | 13.44 | 14.27 | 20.62 | 15.88 | 15.68 |
Strains: 1, strain CCA6; 2, E. asburiae ATCC 35953T; 3, E. cloacae subsp. cloacae ATCC 13047T; 4, E. hormaechei subsp. hormaechei ATCC 49162T; 5, E. hormaechei subsp. xiangfangensis LMG 27195T; 6, E. ludwigii EN‐119T. Data from 2 to 6 are from Gu, Li, Yang and Huo (2014). —, not detected/not reported. Summed feature 1 consists of iso‐C15 : 1 H and/or C13 : 0 3‐OH; Summed feature 2 consists of iso‐C16:1 I and/or C14:0 3‐OH and/or C12:0 unidentified aldehyde or an unidentified fatty acid with an equivalent chain length of 10.928; Summed feature 3 consists of C16:1ω6c and/or C16:1ω7c; summed feature 8 consists of C18:1ω6c and/or C18:1ω7c.
3.4. Phylogenetic affiliation of strain CCA6
The genus Enterobacter was first proposed by Hormaeche and Edwards (1960), and was classified as Gram‐negative, rod‐shaped, motile bacteria. To date, more than 18 Enterobacter species have been reported, and the Enterobacter cloacae complex has been rearranged in E. cloacae, E. asburiae, E. hormaechei, E. kobei, E. ludwigiii, and their subspecies based on whole‐genome DNA–DNA hybridizations and phenotypic characteristics (Mezzatesta, Gona, & Stefani, 2012).
To confirm the phylogenetic position of strain CCA6, the 16S rRNA gene sequence (1,294 bp) was determined. In the maximum‐likelihood tree based on almost complete sequences of the 16S rRNA gene, strain CCA6 fell inside the cluster comprising members of the genus Enterobacter and Kosakonia (Figure 1). The sequences of the following E. cloacae complex species showed similarity to that of strain CCA6: E. cloacae subsp. dissolvens LMG 2683T (98.3%), E. cloacae subsp. cloacae ATCC 13047T (98.0%), E. sichuanensis WCHECL1597T (97.8%), E. chengduensis WCHECl‐C4T (97.7%), K. oryzendophytica LMG 26432T (97.7%), E. ludwigii EN‐119T (97.6%), E. roggenkampii DSM 16690T (97.6%), and E. mori LMG 25706T (97.4%).
Figure 1.

Phylogenetic tree constructed from analysis of 16S rRNA gene sequences showing the relationships between strain CCA6 and the related type strains. The bar indicates a 0.02% nucleotide substitution rate. The tree was rooted using Xenorhabdus nematophila ATCC 19061T as the outgroup
According to Brady et al. (2008, 2013), MLSA is also useful for identification of bacterial species. The MLSA showed that strain CCA6 exhibited similarities of 96.0%, 96.0%, 96.0%, 95.9%, 95.9%, 95.8%, 95.8%, 95.7%, and 95.6% to its closest relatives, E. bugandensis EB‐247T, E. hormaechei subsp. xiangfangensis LMG 27195T, E. ludwigii EN‐119T, E. hormaechei subsp. hormaechei ATCC 49162T, E. hormaechei subsp. steigerwaltii DSM 16691T, E. asburiae ATCC 35953T, E. hormaechei subsp. oharae DSM 16687T, E. tabaci CCUG 72520T, and E. mori LMG 25706T, respectively. Moreover, strain CCA6 clusters on its own branch separately from other Enterobacter species. (Figure 2).
3.5. Genome properties and ANI values
The genome sequence of strain CCA6 was 4,476,585 bp. The G+C content was 54.3%, which fell within range of those of Enterobacter type strains (Table S1). Within the genomic DNA of strain CCA6, 4,372 predicted coding sequences were identified. In addition, 85 tRNA genes and 25 rRNA genes were detected.
To carry out a phylogenetic comparison of strain CCA6 and the related species in the family Enterobacteriaceae, ANI values were calculated (Table S1). The ANI values between strain CCA6 and the related type strains belonging to the genera Klebsiella, Kosakonia, Pluralibacter, Pseudescherichia, and Raoultella were all <79.70%. Moreover, the ANI values between strain CCA6 and the related Enterobacter type strains were in the range of 79.75–88.02%, which was clearly below the cutoff of 95–96% for prokaryotic species delineation as established by Richter and Rosselló‐Móra (2009).
4. CONCLUSION
We have isolated a Gram‐negative, non‐sporulating, rod‐shaped bacterium from leaf soil collected in Japan, which was designated strain CCA6. 16S rRNA gene sequence analysis revealed that strain CCA6 presented as a member of the family Enterobacteriaceae. (Figure 1). Moreover, MLSA based on partial sequences of the atpD, gyrB, infB, and rpoB gene showed clear separation between strain CCA6 and the related Enterobacter type strains (Figure 2). The ANI values between strain CCA6 and its closely related type strains were <88.02% (Table S1). Interesting features of strain CCA6 were its growth potential in oligotrophic medium and the fact that its growth was unaffected by high‐nutrient media. Strain CCA6 therefore has potential for utilization as a host bacterium for industrial fermentation of valuable compounds. Although the related Enterobacter type strains are capable of utilizing disaccharides such as d‐sucrose and d‐turanose, strain CCA6 did not catabolize those disaccharides (Table 1). When cellular fatty acids were compared between strain CCA6 and the related Enterobacter type strains, we found that fatty acids C16:0 and summed feature 8 occur in most members of the related Enterobacter type strains. By contrast, the ratio of C11:0, C15:1ω8c, C17:1ω8c, and iso‐C19:0 in strain CCA6 was significantly higher than in the close relatives, and the fatty acid C15:1ω6c was only detected in strain CCA6 (Table 2).
Based on its phylogenetic, phenotypic, and chemotaxonomic features, strain CCA6 can be considered as a novel species in the genus Enterobacter, which we propose to name E. oligotrophica sp. nov.
4.1. Description of E. oligotrophica sp. nov
Enterobacter oligotrophica (o.li.go.tro'phi.ca. Gr. adj. oligos little; Gr. adj. trophikos nursing, tending or feeding; N.L. fem. adj. oligotrophica eating little, referring to a bacterium living on low‐nutrient media).
Cells are aerobic, Gram‐negative, non‐sporulating, and rod‐shaped (1.0–2.0 μm × 4.0–5.0 μm). Colonies are circular, smooth, glistening, light yellow, and grow to 5.0 mm in diameter on Nutrient Broth plates after incubation for 24 hr at 37°C. Growth is observed in poor‐nutrient medium, and growth is unaffected by high‐nutrient medium. The VP test is negative. The major cellular fatty acids are C16:0 and sums of C16 : 1ω6c and/or C16 : 1ω7c or C18 : 1ω6c and/or C18 : 1ω7c. The predominant quinone system is ubiquinone‐8. Growth is observed in Nutrient Broth at 10–45°C and pH 4.5–10.0, with optimal growth at 20°C and pH 5.0. Growth occurs in the presence of 0–6% (w/v) NaCl as well as ampicillin. Strain CCA6 is positive for lysine decarboxylase. No growth occurs on d‐sucrose, d‐melibiose, d‐turanose, d‐tagatose, inositol, or methyl‐α‐d‐glucopyranoside. Strain CCA6 is clearly separated from the related Enterobacter type strains by MLSA based on partial sequences of the atpD, gyrB, infB, and rpoB gene. The genome size of the type strain is 4,476,585 bp, which has a G+C content of 54.3%.
CONFLICT OF INTERESTS
None declared.
AUTHOR CONTRIBUTIONS
HA an ZK designed, carried out the experiments, and wrote the manuscript. AM revised the manuscript.
ETHICS STATEMENT
None required.
Supporting information
ACKNOWLEDGMENTS
We are grateful to all members of the Bio‐conversion Research Group at our Institute [Research Institute for Sustainable Chemistry, National Institute of Advanced Industrial Science and Technology (AIST)] for their technical assistance and valuable discussion. This work was supported by JSPS KAKENHI Grant Number 16K18683.
APPENDIX 1.
1.1.
Figure A1.
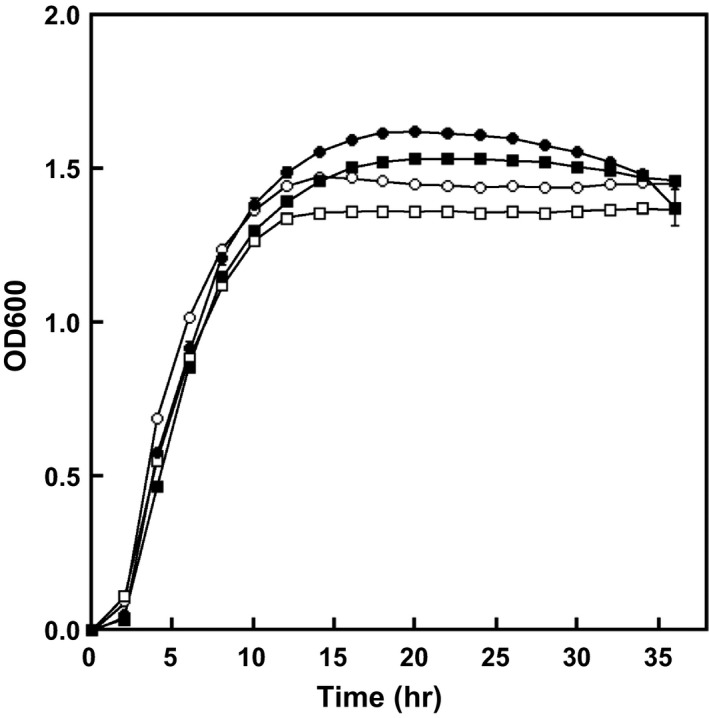
Growth rates at 30°C of strain CCA6 and E. coli MG1655. The results for strain CCA6 and E. coli MG1655 are shown as filled and open symbols, respectively. The media are indicated as follows: circles, Nutrient Broth (pH 7.0) and squares, LB media (pH 7.0). The OD600 was measured using a Bio Microplate Reader HiTS (Scinics, Tokyo, Japan). Experiments were performed in triplicate
Figure A2.
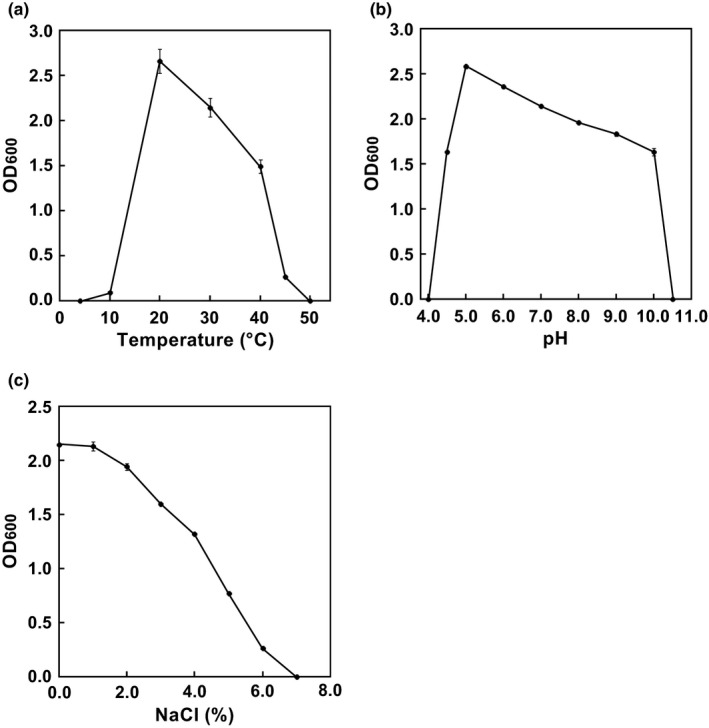
Effects of culture conditions on growth of strain CCA6. (a) Effects of culture temperature. Cells were cultured in Nutrient Broth (pH 7.0). (b) Effects of culture pH. Cells were cultured in Nutrient Broth at 30°C. (c) Effect of NaCl concentration. Cells were cultured in Nutrient Broth (pH 7.0) at 30°C. Error bars indicate SE (n = 3)
Table A1.
Genome properties of type strains of Enterobacter, Klebsiella, Kosakonia, Lelliottia, Pluralibacter, Pseudescherichia, Pseudomonas, and Raoultella species used in this study
| Strains | Size (bp) | G+C (mol%) | Protein | Accession no. |
|---|---|---|---|---|
| Enterobacter | ||||
| E. asburiae ATCC 35953T | 4,713,742 | 55.4 | 4,436 | CP011863 |
| E. bugandensis EB‐247T | 4,971,744 | 56.0 | 4,344 | FYBI00000000 |
| E. cancerogenus ATCC 33241T | 4,879,939 | 55.6 | 4,521 | FYBA00000000 |
| E. chengduensis WCHECl‐C4T | 5,138,130 | 55.7 | 4,745 | MTSO00000000 |
| E. cloacae subsp. cloacae ATCC 13047T | 5,551,574 | 54.6 | 5,393 | JPPR00000000 |
| E. hormaechei subsp. hormaechei ATCC 49162T | 4,890,213 | 55.2 | 4,522 | MKEQ00000000 |
| E. hormaechei subsp. oharae DSM 16687T | 4,724,316 | 55.6 | 4,390 | CP017180 |
| E. hormaechei subsp. steigerwaltii DSM 16691T | 4,782,480 | 55.6 | 4,424 | CP017179 |
| E. hormaechei subsp. xiangfangensis LMG 27195T | 4,661,849 | 55.3 | 4,306 | CP017183 |
| E. kobei ATCC BAA‐260T | 4,700,329 | 55.5 | 4,424 | FTNJ00000000 |
| E. ludwigii EN‐119T | 4,952,770 | 54.6 | 4,459 | JTLO00000000 |
| E. mori LMG 25706T | 4,953,765 | 55.3 | 4,496 | AEXB00000000 |
| E. muelleri JM‐458T | 4,695,678 | 55.9 | 4,423 | FXLQ01000000 |
| E. roggenkampii DSM 16690T | 4,748,414 | 56.0 | 4,451 | CP017184 |
| E. sichuanensis WCHECL1597T | 4,897,201 | 55.2 | 4,634 | POVL01000000 |
| E. soli ATCC BAA‐2102T | 4,960,767 | 53.8 | 4,571 | LXES00000000 |
| E. tabaci CCUG 72520T | 4,927,887 | 55.5 | 4,627 | QZDQ00000000 |
| Klebsiella | ||||
| K. aerogenes KCTC 2190T | 5,280,350 | 54.8 | 4,912 | CP002824 |
| K. grimontii 06D021T | 6,168,876 | 55.4 | 5,986 | FZTC00000000 |
| K. michiganensis DSM 25444T | 6,193,009 | 56.0 | 5,732 | PRDB00000000 |
| K. pneumoniae subsp. ozaenae ATCC 11296T | 4,925,250 | 57.2 | 4,458 | CDJH00000000 |
| K. pneumoniae subsp. pneumoniae ATCC 13883T | 5,544,684 | 57.0 | 5,205 | JOOW00000000 |
| K. pneumoniae subsp. rhinoscleromatis ATCC 13884T | 5,280,675 | 56.9 | 5,671 | ACZD00000000 |
| K. quasipneumoniae subsp. quasipneumoniae 01A030T | 5,465,736 | 58.0 | 5,287 | CCDF00000000 |
| K. quasipneumoniae subsp. similipneumoniae 07A044T | 5,109,717 | 58.2 | 4,927 | CBZR000000000 |
| K. variicola DSM 15968T | 5,521,203 | 57.6 | 5,200 | CP010523 |
| Kosakonia | ||||
| K. arachidis Ah‐143T | 5,135,597 | 52.5 | 4,861 | FPAU00000000 |
| K. oryzae CGMCC 1.7012T | 5,380,462 | 54.0 | 4,980 | FOKO00000000 |
| K. oryzendophytica LMG 26432T | 4,878,776 | 53.7 | 4,459 | FYBE00000000 |
| K. oryziphila REICA_142T | 4,814,900 | 52.7 | 4,667 | FMBC00000000 |
| K. pseudosacchari JM‐387T | 4,956,546 | 53.9 | 4,638 | FXWP00000000 |
| K. radicincitans DSM 16656T | 5,817,639 | 53.7 | 5,660 | AKYD00000000 |
| K. sacchari SP1T | 4,902,027 | 53.7 | 4,545 | CP007215 |
| Lelliottia | ||||
| L. amnigena DSM 4486T | 4,370,208 | 52.9 | 4,070 | PDDA01000000 |
| L. jeotgali PFL01T | 4,603,334 | 54.2 | 4,243 | CP018628 |
| Pluralibacter | ||||
| P. gergoviae NBRC 105706T | 5,662,775 | 58.6 | 5,176 | BCZS00000000 |
| Pseudescherichia | ||||
| P. vulneris NBRC 102420T | 4,374,581 | 56.4 | 4,196 | BBMZ00000000 |
| Raoultella | ||||
| R. ornithinolytica NBRC 105727T | 5,533,930 | 55.7 | 5,099 | BCYR00000000 |
| R. planticola ATCC 33531T | 5,668,028 | 55.8 | 5,237 | JMPP00000000 |
| Xenorhabdus | ||||
| X. nematophila ATCC 19061T | 4,587,917 | 44.3 | 3,754 | FN667742 |
Data are from the GenBank/EMBL/DDBJ databases.
Akita H, Matsushika A, Kimura Z‐I. Enterobacter oligotrophica sp. nov., a novel oligotroph isolated from leaf soil. MicrobiologyOpen. 2019;8:e843 10.1002/mbo3.843
DATA ACCESSIBILITY
The 16S rRNA gene sequence of strain CCA6 is available in the GenBank/EMBL/DDBJ databases under accession number LC368255. The complete genome sequence of strain CCA6 has been deposited in the DDBJ/EMBL/GenBank under accession number AP019007. The type strain is CCA6T and was deposited in two international strain collection institutes with the following accession numbers: HUT 8142T = KCTC 62525T.
REFERENCES
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Badger, J. H. , & Olsen, G. J. (1999). CRITICA: Coding region identification tool invoking comparative analysis. Molecular Biology and Evolution, 16, 512–524. 10.1093/oxfordjournals.molbev.a026133 [DOI] [PubMed] [Google Scholar]
- Banat, I. M. , Satpute, S. K. , Cameotra, S. S. , Patil, R. , & Nyayanit, N. V. (2014). Cost effective technologies and renewable substrates for biosurfactants’ production. Frontiers in Microbiology., 5, 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh, E. G. , & Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology, 37, 911–917. 10.1139/y59-099 [DOI] [PubMed] [Google Scholar]
- Brady, C. , Cleenwerck, I. , Venter, S. , Coutinho, T. , & De Vos, P. (2013). Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter . Systematic and Applied Microbiology., 36, 309–319. 10.1016/j.syapm.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Brady, C. , Cleenwerck, I. , Venter, S. , Vancanneyt, M. , Swings, J. , & Coutinho, T. (2008). Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Systematic and Applied Microbiology, 31, 447–460. 10.1016/j.syapm.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Chin, C. S. , Alexander, D. H. , Marks, P. , Klammer, A. A. , Drake, J. , Heiner, C. , … Korlach, J. (2013). Nonhybrid, finished microbial genome assemblies from long‐read SMRT sequencing data. Naure Methods, 10, 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Delcher, A. L. , Harmon, D. , Kasif, S. , White, O. , & Salzberg, S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Research, 27, 4636–4641. 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris, J. , Konstantinidis, K. T. , Klappenbach, J. A. , Coenye, T. , Vandamme, P. , & Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole‐genome sequence similarities. International Journal of Systematic and Evolutionary Microbiology, 57, 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Gu, C. T. , Li, C. Y. , Yang, L. J. , & Huo, G. C (2014). Enterobacter xiangfangensis sp. nov., isolated from Chinese traditional sourdough, and reclassification of Enterobacter sacchari Zhu et al. 2013 as Kosakonia sacchari comb. nov. International Journal of Systematic and Evolutionary Microbiology, 64, 2650–2656. [DOI] [PubMed] [Google Scholar]
- Ho, D. P. , Ngo, H. H. , & Guo, W. (2014). A mini review on renewable sources for biofuel. Bioresource Technology, 169, 742–749. 10.1016/j.biortech.2014.07.022 [DOI] [PubMed] [Google Scholar]
- Hormaeche, E. , & Edwards, P. R. (1960). A proposed genus Enterobacter . International Bulletin of Bacteriological Nomenclature and Taxonomy, 10, 71–74. [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov, S. I. , Dubinina, G. A. , & Lapteva, N. A. (1979). Biology of oligotrophic bacteria. Annual Review of Microbiology, 33, 377–387. 10.1146/annurev.mi.33.100179.002113 [DOI] [PubMed] [Google Scholar]
- Lane, D. J. (1991). 16S/23S rRNA sequencing In Stackebrandt E. & Goodfellow M. (Eds.), Nucleic acid techniques in bacterial systematics (pp. 115–175). New York, NY: John Wiley and Sons. [Google Scholar]
- Lowe, T. M. , & Eddy, S. R. (1997). tRNAscan‐SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research, 25, 955–964. 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzatesta, M. L. , Gona, F. , & Stefani, S. (2012). Enterobacter cloacae complex: Clinical impact and emerging antibiotic resistance. Future Microbiology, 7, 887–902. 10.2217/fmb.12.61 [DOI] [PubMed] [Google Scholar]
- Min, B. E. , Hwang, H. G. , Lim, H. G. , & Jung, G. Y. (2017). Optimization of industrial microorganisms: Recent advances in synthetic dynamic regulators. Journal of Industrial Microbiology & Biotechnology, 44, 89–98. 10.1007/s10295-016-1867-y [DOI] [PubMed] [Google Scholar]
- Ohta, H. (2000). Growth characteristics of Agromonas oligotrophica on ferulic acid. Microbes and Environments, 15, 133–142. 10.1264/jsme2.2000.133 [DOI] [Google Scholar]
- Ohta, H. , & Taniguchi, S. (1988). Growth characteristics of the soil oligotrophic bacterium Agromonas oligotrophica JCM 1494 on diluted nutrient broth. The Journal of General and Applied Microbiology, 34, 349–353. 10.2323/jgam.34.349 [DOI] [Google Scholar]
- Richter, M. , & Rosselló‐Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proceedings of the National Academy of Sciences of the United States of America, 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaoka, J. , Katayama‐Fujimura, Y. , & Kuraishi, H. (1983). Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. Journal of Applied Bacteriology, 54, 31–36. 10.1111/j.1365-2672.1983.tb01297.x [DOI] [Google Scholar]
- Tamura, K. , & Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10, 512–526. [DOI] [PubMed] [Google Scholar]
- Thomsen, M. H. (2005). Complex media from processing of agricultural crops for microbial fermentation. Applied Microbiology and Biotechnology, 68, 598–606. 10.1007/s00253-005-0056-0 [DOI] [PubMed] [Google Scholar]
- Turner, S. , Pryer, K. M. , Miao, V. P. , & Palmer, J. D. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. Journal of Eukaryotic Microbiology, 46, 327–338. 10.1111/j.1550-7408.1999.tb04612.x [DOI] [PubMed] [Google Scholar]
- Yoon, S. H. , Ha, S. M. , Lim, J. , Kwon, S. , & Chun, J. (2017). A large‐scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek, 110, 1281–1286. 10.1007/s10482-017-0844-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequence of strain CCA6 is available in the GenBank/EMBL/DDBJ databases under accession number LC368255. The complete genome sequence of strain CCA6 has been deposited in the DDBJ/EMBL/GenBank under accession number AP019007. The type strain is CCA6T and was deposited in two international strain collection institutes with the following accession numbers: HUT 8142T = KCTC 62525T.


