Abstract
Receptor-like cytoplasmic kinases (RLCKs) are receptor kinases that lack extracellular ligand-binding domains and have emerged as a major class of signaling proteins that regulate plant cellular activities in response to biotic/abiotic stresses and endogenous extracellular signaling molecules. We have identified a rice RLCK (OsRLCK311) that was significantly higher in transgenic pSARK-IPT rice (Oryza sativa) that exhibited enhanced growth under saline conditions. Overexpression of OsRLCK311 full-length protein (RLCK311FL) and the C-terminus of OsRLCK311 (ΔN) in Arabidopsis confirmed its role in salinity tolerance, both in seedlings and mature plants. Protein interaction assays indicated that OsRLCK311 and ΔN interacted in-vivo with the plasma membrane AQP AtPIP2;1. The RLCK311-PIP2;1 binding led to alterations in the stomata response to ABA, which was characterized by more open stomata of transgenic plants. Moreover, OsRLCK311-ΔN effect in mediating enhanced plant growth under saline conditions was also observed in the perennial grass Brachypodium sylvaticum, confirming its role in both dicots and monocots species. Lastly, OsRLCK311 interacted with the rice OsPIP2;1. We suggest that the rice OsRLCK311 play a role in regulating the plant growth response under saline conditions via the regulation of the stomata response to stress. This role seems to be independent of the RLCK311 kinase activity, since the overexpression of the RLCK311 C-terminus (ΔN), which lacks the kinase full domain, has a similar phenotype to RLCK311FL.
Keywords: receptor-like cytoplasmic kinases, salinity tolerance, Oryza sativa, aquaporins
1. Introduction
Crop productivity is severely limited by soil salinity. Salinity affects major biosynthetic processes such as photosynthesis, protein synthesis and lipid metabolism [1]. Plant responses to salinity comprise two phases: (i) a fast response to the decrease in soil water potential (more negative) and (ii) a slower response to the toxic effects of Na+ ions in the leaves [2]. In general, these responses lead to a strong reduction in stomata conductance and growth retardation [3,4]. While stomata closure will lead to water conservation and enhanced survival to a severe stress, it will also result in a decreased photosynthesis and reduced biomass and yield production [3,5]. Thus, a precise balance between stomata opening, plant growth and active gas exchange needs to be maintained. Under mild stress, plant stress survival responses (i.e., stomata closure and decreased growth) are not necessarily beneficial agronomical traits [3]). The maintenance of a lesser degree of salinity-induced stomata closure might lead to enhanced gas exchange with improved plant biochemical activity and enhanced growth [5,6,7,8,9]. Thus, one might speculate that fine-tuning of the stomata response to salt stress can affect the threshold between water loss, active gas exchange and plant growth. While this phenomenon has been characterized [5,6,7,9], the molecular mechanism(s) regulating it is only beginning to emerge. In recent years, it has been shown that water channels play a role in the regulation of stomata pore opening and gas exchange, particularly under stress conditions [10,11]. The rose RsPIP2;1 was shown to contribute to plant growth strategies under water-deficit stress through alternating between survival (high RsPIP2;1 and low whole leaf transpiration) and biomass accumulation (low RsPIP2;1 and high whole leaf transpiration) [12] (In addition, reverse genetics in maize and Arabidopsis demonstrated that the plasma membrane intrinsic proteins 2 (PIP2) were needed for functional stomata closure under the ABA treatments [13,14]). In maize, ZmPIP2;5 overexpression and zmpip2;5 knockout mutant displayed a higher and lower ABA-dependent stomata closure than wild-type plants, respectively (Ding and Chaumont, 2020b). Interestingly, Arabidopsis atpip2;1 knockout mutant displayed a similar phenotype (i.e., stomata insensitivity to ABA) [13]. Furthermore, the role of AtPIP2;1 in ABA stomata closure was dependent on the AtOST1 protein kinase [13], emphasizing the involvement of kinase-mediated post-translational regulation of AtPIP2;1-dependent stomata closure. Proteomics-based studies showed that AtPIP2;1 could efficiently bind receptor-like kinases (RLK) and receptor-like cytoplasmic kinases (RLCKs), supporting the notion of kinase-mediated PIP2;1 post-translational regulation [15].
Receptor-like cytoplasmic kinases (RLCKs) in plants belong to the super family of receptor-like kinases (RLKs). These proteins are similar to RLKs in their kinase domain but lack an extracellular domain [16]. Based on expression analysis and functional studies, RLCKs have been proposed to play role(s) in diverse biological processes, including the response to biotic and abiotic stress [16]. For example, in rice, 376 RLCKs were identified and about 86 were shown to be responsive to abiotic stress [17]). Based on RLCK expression patterns and their involvement in aquaporin (AQP) channels’ post-translational regulation, we hypothesize that rice RLCKs could be involved in the regulation of the tolerance of rice to salinity stress, possibly through the regulation of stress-dependent stomata closure, plant growth and AQP activity. Here, we identified a single rice RLCK311 (putatively annotated as a serine/threonine-protein kinase BRI1-like 1 precursor) that was upregulated in the response of transgenic SARK:IPT rice [18] to salinity. We expressed RLCK311 in the dicot Arabidopsis thaliana and the monocot Brachypodium plants. Our results indicated that the RLCK311 can bind to PIP2;1 in vivo, deactivate PIP2;1-dependent stomata closure and contribute to enhanced plant growth under salinity conditions. Our results indicated that RLCK311-PIP2;1 binding and stomata reliant stress regulation was independent of RLCK311 kinase activity.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Homozygous transgenic T3 lines [18] expressing SARK::IPT and wild-type (WT) seeds of rice (Oryza sativa L. ssp. japonica cv. kitaake) were germinated on moist germination paper (25 × 38 cm; Anchor Paper Co., St. Paul, MN, USA) for 6 days at 30 °C in the dark. Seedlings were then transplanted into 4.5-L pots, filled with soil (Capay series, harvested in California rice fields, 38°32’23.93’’ N,121°48’30.81’’ W, shredded and steamed for 1.5 h to eradicate soil pathogens), with two plants per pot, and placed in water tubes. Plants were grown at 12 h/12 h day/night under an illumination of 1200 µmoL m−2s−1 at 30 °C⁄20 °C. Plants were fertilized with a solution 50% N:P:K (20:10:20) and 50% ammonium sulphate (total of 0.5 g of N) every 10 days until panicle initiation. Salinity treatments were applied at the panicle initiation stage; NaCl concentrations in the watering solution were increased progressively every 3 days (25, 50, 75 and 100 mM NaCl). A concentration of 100 mM NaCl was maintained for 40 days, until the end of the experiment. OsRLCK311 expression was measured from RNAseq data obtained from plants grown in the presence of 100 mM NaCl for 3 days (E. Blumwald, unpublished).
Seeds of wild-type and transgenic Arabidopsis thaliana (ecotype Columbia) were sterilized and then germinated in 1/2 Murashige and Skoog medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 0.5% sucrose. For plate-grown plants, seeds were incubated in medium containing 1% Phytagel ((Sigma-Aldrich, St. Louis, MO, USA)) for 10 days in a growth chamber at 23 ℃ under 100 µmoL m−2 s−1 light in a 16 h light/8 h dark regime. Seedlings were then transplanted to plates containing 1/2 Murashige and Skoog medium (MS) medium (control) or 1/2 MS medium supplemented with 75 mM NaCl (salt), and plates were incubated as described above in the same growth chamber. Photographs were taken 10 days after transplantation, and seedlings were sampled for fresh weight and malondialdehyde (MDA) measurements.
For soil-grown experiments, seeds of wild-type and transgenic Arabidopsis thaliana (Col-0) were grown in autoclaved Sunshine Mix 4 (SunGro, Sacramento, California). Plants were grown in a growth chamber as described above. Salt stress treatments were applied by irrigating with a solution containing 125mM NaCl at 30 days after seed germination. Photographs were taken at 7 days after salt stress was applied, and dry weight of the full rosettes were measured. Fully expanded leaves were sampled to determine the chlorophyll and MDA contents.
Seeds of wild-type Brachypodium sylvaticum and two T3 homozygous transgenic lines were germinated on moist germination paper for 5 days at 4 °C in the dark. Young seedlings in the paper rolls were then moved to the growth chamber under 100 µmoL m−2 s−1 light in a 16-h light/8 h-dark at 26 °C/20 °C. Twenty-day-old seedlings were transplanted to autoclaved Sunshine Mix 4 fertilized with Osmocote (ICL, Tel-Aviv, Israel) (N:P:K) (14:14:14) and kept well-watered with distilled water. Salt stress treatment was applied at 30 days after seed germination by irrigating with a 100mM NaCl solution for 7 days, and then increasing the salt concentration to 200 mM NaCl until harvest. Shoot dry weight was measured 2 weeks after the initiation of the stress treatment. Fully expanded leaves were sampled for qRT-PCR analysis. All plants were irrigated with DI water for another 25 days until the second harvest.
2.2. Protein Sequences Alignment
Protein sequences of RLCK311 full-length (FL) (LOC_Os11g06780) and RLCK311 C-terminal domain (ΔN) (Os11g0168600) were obtained from rice genome databases (http://rice.plantbiology.msu.edu/ and https://rapdb.dna.affrc.go.jp/). The protein sequences alignment was performed in the plasmid editor ApE.
2.3. Generation of Transgenic Plants
All the constructs were generated using the Gateway system (Invitrogen, Carlsbad, CA, USA).
For 35S::RLCK311-ΔN::3×Flag Arabidopsis thaliana transgenic plants, the ORFs of OsRLCK311-ΔN and OsRLCK311-FL were amplified from Oryza sativa cDNA and cloned into pDONR207 by BP reaction to generate pEN-ΔN and pEN-OsRLCK311FL. Meanwhile, 35S promoter was also amplified from pEarlyeGate101 [19] and cloned into pDONR P4P1R vector (Invitrogen). These two entry clones and pEN-R2-3xFlag-L3 [20] were recombined via multisite LR reaction into pB7m34GW [21], resulting in pB7m34GW-35S:ΔN /OsRLCK311FL:3XFlag.
For transgenic Brachypodium sylvaticum plants expressing pSARK:OsRLCK311300, the sequence of SARK promoter was amplified and cloned into pDONR P4P1R vector by BP reaction to generate pEN-pSARK. The pEN-ΔN and the pEN-pSARK were recombined via multisite LR reaction into pH7m24GW,3 [21], resulting in pH7m24GW-pSARK:ΔN.
For transgenic Brachypodium sylvaticum plants expressing pSARK::GUS(+), GUSplus gene was amplified from pCAMBIA1305.2 and cloned into pDONR207 by BP reaction to generate pEN-GUS+. Gateway sequence fragment was amplified from pH7m24GW and inserted into pZH2B (Kuroda et al., 2010) to accommodate two fragment multisite gateways in a 35S:Hyg vector for monocot transformation as pZH2B-GW. pEN-pSARK and pEN-GUS+ were recombined via multisite LR reaction into pZH2B-GW, resulting in pSARK:Gus(+).
2.4. Malondialdehyde Measurements
Young seedlings or fully expanded leaves were weighed and then ground in liquid N2. The tissue was homogenized in the presence of 10% trichloroacetic acid containing 0.25% thiobarbituric acid. The mixture was heated at 95 °C for 30 min, quickly cooled in ice, and then centrifuged at 10,000 xg for 10 min. The absorbance of the supernatant was measured at 532 nm, 600 nm and 450 nm (SynergyTM Mx Microplate Reader; BioTek, Winooski, VT, USA). The concentration was calculated according to the equation C (μmol/L) = 6.45 × (A532 − A600) − 0.56 × A450 [22].
2.5. Chlorophyll Measurements
Fully expanded leaves were ground in liquid N2, and chlorophyll was extracted in 80% acetone. Absorbance at 663 nm and 645 nm was measured (SynergyTM Mx Microplate Reader; BioTek, Winooski, VT, USA), and chlorophyll contents were calculated as described in [23].
2.6. Protein Interaction Assays
For RLCK311-ΔN, OsRLCK311-FL and AtPIP2;1 colocalization assays, the ORFs of RLCK311-ΔN and OsRLCK311-FL, excluding the stop codon, were amplified from rice leaf cDNA and cloned into pDONR207 by BP reaction. pEN-ΔN or pEN-OsRLCK311FL was recombined via LR reaction into the destination vector pEarleyGate 103 [19] for green fluorescent protein (GFP)fusion. The genes AtPIP2;1, OsPIP2;1 and OsBIN2 were fused with RFP of another destination vector pH7RWG2 using the same strategy as described above. The resulting constructs were introduced into Agrobacterium tumefaciens GV3101 and co-infiltrated into the abaxial surface of the leaves of 4-week-old Nicotiana benthamiana as described previously [24]. Fluorescence microscopy was performed two d after infiltration with an inverted Zeiss LSM 710 confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany) equipped with a 20× water immersion objective. The excitation wavelength/emission were as follows: GFP (488 nm/500–530 nm) and red fluorescent protein (RFP, 561 nm/600–660 nm).
For bimolecular fluorescence complementation (BiFC), the vectors pDEST-GWVYNE and pDEST-GWSCYCE from the Gateway-based BiFC vector systems [25] were employed to fuse OsRLCK311300 and OsRLCK311FL with the N-terminus of yellow fluorescent protein Venus (VenusN) and fused to AtPIP2;1, OsPIP2;1 and OsBIN2 with the C-terminus of super CFP (SCFPC) to obtain the constructs ΔN /OsRLCK311FL-VenusN and AtPIP2;1/OsPIP2;1/OsBIN2-SCFPC. All the constructs were introduced into Agrobacterium tumefaciens GV3101. Transient expression and fluorescence microscopy were performed at the same conditions as described for protein colocalization.
For co-immunoprecipitation and mass spectrometry analysis, three-week-old seedlings of wild-type and transgenic plants overexpressing OsRLCK311-ΔN /OsRLCK311-FL were ground in liquid N2, and incubated on an end-over-end rotator at 4 °C for 4 h with lysis buffer provided in the μMACS Isolation Kit (Miltenyl Biotec, Bergisch Gladbach, Germany), containing protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA) and 1 mM PMSF. The mixture was centrifuged at 10,000×g for 10 min at 4 °C, and the supernatant was used for co-immunoprecipitation (COIP). COIP was performed using anti-Flag magnetic beads according to the manufacturer protocol (Pierce™ Anti-DYKDDDDK Magnetic Agarose, Invitrogen, USA). LC-MS/MS analysis was performed at the Proteomics Core Facility of the University of California, Davis, as described previously [26]. Scaffold (version Scaffold 4; www.proteomesoftware.com) was used to validate tandem MS-based peptide and protein identification. The results were filtered with peptide thresholds (63% minimum) and protein thresholds (80% minimum), and two unique peptide required for identification. Although the filtration thresholds used in our study are somewhat low, they allowed for the identification of proteins forming complexes through binding both FL and ΔN-OsRLCK311. The putative interactions were further confirmed by BiFC assays (see above) and co-immunoprecipitation. Full interactors peptide and samples report is presented in Tables S3 and S4.
For co-immunoprecipitation by transient expression and western blots, OsRLCK31-ΔN-GFP/OsRLCK311FL-GFP was co-infiltrated with AtPIP2;1-RFP, OsPIP2;1-RFP or free RFP as described above. Samples displaying protein expression as measured by fluorescence microscopy (Figures S5 and S6) were used 2 daysafter infiltration. Leaf tissue was grinded in liquid N2, and lysis buffer (as described above) was added (2 mL/g fresh weight). Following incubation and centrifugation, 1 mL of the supernatants was purified through anti-GFP magnetic beads from the μMACS GFP Isolation Kit (Miltenyl Biotec, Bergisch Gladbach, Germany) and applied for immunoblot analysis using anti-GFP (NB600-308SS, Novud Biologicals, Centennial, CO, USA) and anti-RFP antibody (NBP2-25157SS, Novus biologicals, Colorado, USA) with a 1:1000 or 1:500 dilution, respectively.
2.7. GUS Staining
Seeds of wild-type Brachypodium sylvaticum and two transgenic lines were germinated on moist germination paper for 5 days at 4 °C in the dark. Young seedlings were then moved from the paper rolls to the growth chamber and grown at 26 °C/20 °C (day/night) 100 µmol m−2 s−1 light in 16h/8h day/night regime. Paper rolls with 20 d-old seedlings were exposed to 200mM NaCl for 24 h and stained using standard GUS protocols [27].
2.8. Quantification of Stomata Aperture
The epidermis of fully expanded leaves was carefully peeled from the abaxial surface and immediately incubated in MES-KCl buffer (5 mM KCl/10 mM Mes/ 50 µM CaCl2, pH 6.15) under 100 µmol m−2 s−1 light for 2 h. After 2 h, 50 µM ABA in DMSO was added to the MES-KCl buffer. Epidermis peels and buffers were mixed by gentle shaking for 20 min and kept under 100 µmol m−2 s−1 light all the time. Following incubation, stomata were observed using a fluorescence digital microscope (Olympus, Tokyo, Japan) and stomata aperture was analyzed by Image J.
3. Results
3.1. Overexpression of OsRLCK311 and Its C Terminus Domain (ΔN) in Arabidopsis Conferred Salt Tolerance
RLCKs, which lack extracellular ligand binding domains, are associated with signalling and many cellular processes [16], and a number of RLCKs were shown to be associated with the response(s) of rice to abiotic stress [17]. OsRLCK311, a gene encoding a Receptor-Like Cytoplasmic Kinase (RLCK), one of these proteins, was highly expressed in transgenic rice plants overexpressing IPT, a gene encoding ISOPENTENYL TRANSFERASE (regulating cytokinin synthesis), driven by PSARK, a stress-induced promoter. The expression of PSARK:IPT resulted in delayed stress-induced senescence and increased abiotic stress tolerance of the transgenic rice plants ([18]; Figure S1). OsRLCK311, annotated as a brassinosteroid-insensitive like precursor (BRIL) in the rice genome database (http://rice.plantbiology.msu.edu/), comprises 389 amino acids. It has been also annotated as a shorter version with 91 amino acids in a different rice genome database (https://rapdb.dna.affrc.go.jp/). Protein sequence alignment revealed that the shorter version is similar to the C-terminus domain (ΔN) of the full-length protein (FL) (Figure S2).
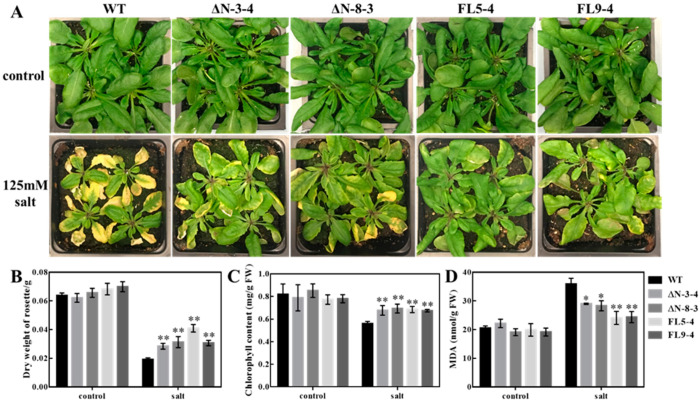
In order to evaluate the roles of OsRLCK311 in salinity stress responses, the full-length OsRLCK311 (FL) and the OsRLCK311 C-terminus (ΔN) were overexpressed and transgenic lines were generated in Arabidopsis (Figure S3). When grown under salinity conditions, FL- and ΔN-overexpressing Arabidopsis plants displayed enhanced growth, higher dry weight and lower MDA contents than wild-type plants (Figure 1 and Figure S4).
Figure 1.
Salt stress tolerance of wild-type (WT) and RLCK311 over-expressing lines grown in soil. (A) Growth of wild-type (WT) and different RLCK311 over-expressing lines under normal condition (control) or salt stress (treated with 125mM NaCl for 7 days). (B) Dry weight of each rosette, (C) chlorophyll content and (D) malondialdehyde (MDA) content of wild-type (WT) and all RLCK311 over-expressing plants. Values represent means ± SE (n = 12). Asterisks represent significant differences between transgenic lines and wild-type within a treatment by the Dunnet test (*, p < 0.05 and **, p < 0.01).
3.2. OsRLCK311 Binds to Aquaporin AtPIP2;1
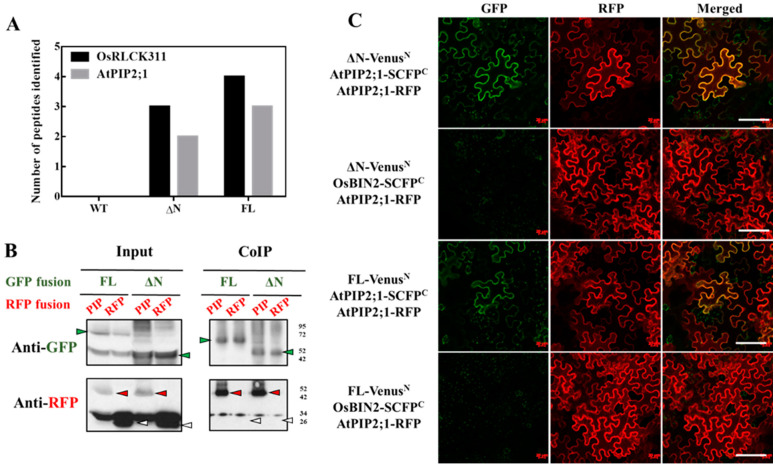
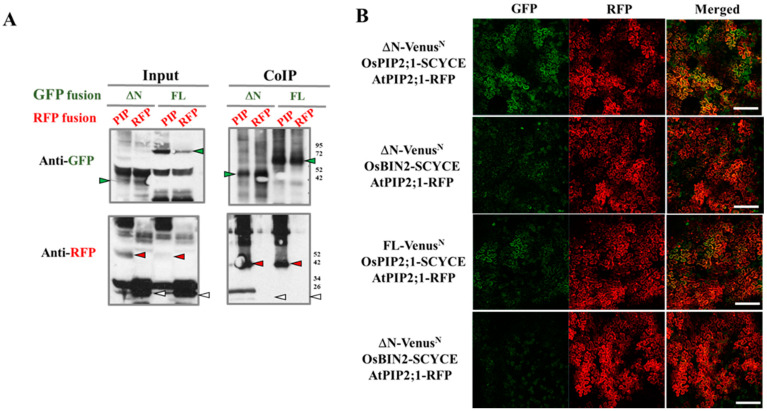
To reveal possible mechanisms mediating OsRLCK311 function(s) in the response to salinity, proteins interacting with OsRLCK311-FL and its C-terminus (ΔN) were identified by immunoprecipitation and subsequent mass spectrometry. Immunoprecipitated ΔN/FL–3×Flag and its interacting proteins were obtained from total protein extracts from 3-week-old seedlings of wild-type and transgenic plants overexpressing OsRLCK311FL/ΔN. Protein extracts from wild-type plants were used as a negative control to exclude the proteins non-specifically binding to the anti-Flag magnetic beads. A number of candidate proteins were identified (Table S2), among them Aquaporin AtPIP2;1 (Figure 2A). Co-immunoprecipitation by OsRLCK311ΔN/FL transient expression of tobacco leaves and western blots confirmed the interaction between OsRLCK311ΔN/FL and AtPIP2;1 (Figure 2). Full-length OsRLCK311-FL fused with GFP (FL-GFP) or OsRLCK311- ΔN fused with GFP (ΔN-GFP) were co-infiltrated with AtPIP2;1-RFP, and leaves displaying high GFP and RFP abundance were used for immunoprecipitation (Figure S5). AtPIP2;1-RFP, but not free RFP, was immunoprecipitated by ΔN/FL-OsRLCK311-GFP (Figure 2B). BiFC was used to confirm the interaction. The transient expression of fusion proteins ΔN/FL-VenusN and AtPIP2;1-SCFPC in N. benthamiana resulted in BiFC fluorescence, which co-localized with the plasma membrane marker AtPIP2;1-RFP (Figure 2C). Similar results were obtained when OsRLCK311 and Os OsPIP2;1 were transiently expressed in tobacco leaves. CoIP and western blots showed that OsPIP2;1-RFP was immunoprecipitated by ΔN/FL-GFP (Figure 3 and Figure S6), and that the interaction occurred at the plasma membrane.
Figure 2.
Rice RLCK (OsRLCK311) interacts with an aquaporin protein AtPIP2;1 in vivo. (A) AtPIP2;1 interacts with both ΔN and full-length (FL) through co-immunoprecipitation and identified by MS. (B) Co-immunoprecipitation of ΔN or FL with AtPIP2;1 by transient expression in Nicotiana benthamiana, identified by western blots. The proteins extracted from tobacco leaves infiltrated with both ΔN-GFP/FL-GFP (green arrows) and AtPIP2;1-RFP (red arrows) were co-immunoprecipitated (IP) with anti-GFP antibodies and blotted with anti-GFP or anti-red fluorescent protein (RFP) antibodies. Free RFP is marked in white arrows. ‘’Input’’ stands for protein extracts before immunoprepicitaton. (C) Bimolecular fluorescence complementation (BiFC) analysis of the interaction between ΔN or FL with AtPIP2;1 by transient expression in Nicotiana benthamiana. VenusN was fused at the C-terminus of ΔN and FL, while SCFPC was fused to the C-terminus of AtPIP2;1 and OsBIN2. Infiltration with OsBIN2-SCFPC were used as negative controls. AtPIP2;1-mcherry was used as a plasma membrane marker. Bar = 200 μM.
Figure 3.
OsRLCK311 interacts with an aquaporin protein OsPIP2;1 in vivo. (A) Co-immunoprecipitation of ΔN or FL with OsPIP2;1 by transient expression in Nicotiana benthamiana and then identified by western blot. The proteins extracted from tobacco leaves infiltrated with both ΔN-GFP/FL-GFP (green arrows) and OsPIP2;1-RFP (red arrows) were co-immunoprecipitated (IP) with anti-GFP antibody and blotted with anti-GFP or anti-RFP antibody. Free RFP is marked in white arrows. ‘’Input’’ stands for protein extracts before immunoprepicitaton. (B) BiFC analysis of the interaction between ΔN or FL with OsPIP2;1 by transient expression in Nicotiana benthamiana. VenusN was fused at the C-terminus of ΔN and FL, while SCFPC was fused to the C-terminus of OsPIP2;1 and OsBIN2. Infiltration with OsBIN2-SCFPC were used as negative controls. AtPIP2;1-mcherry was used as a plasma membrane marker. Bar = 200 μM.
3.3. OsRLCK311 Negatively Regulates Stomata Response to ABA
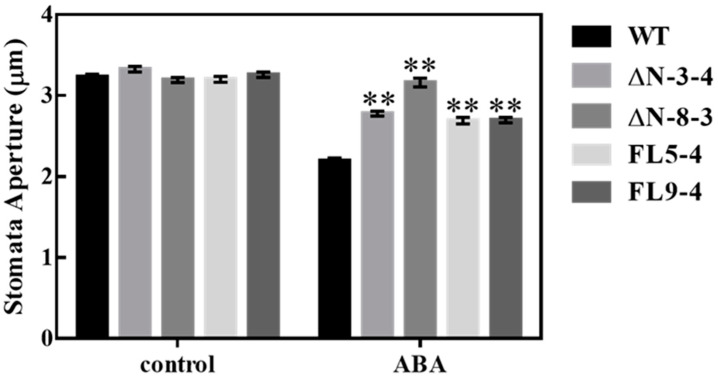
AtPIP2;1 knockout resulted in the loss of ABA-induced stomata closure [13]. In order to assess whether the OsRLCK311-AtPIP2;1 interaction led to the deactivation of AtPIP2;1-mediated stomata response to ABA, stomatal aperture was measured in epidermal peels of wild-type and transgenic plants over-expressing ΔN/FL. Over-expression of ΔN/FL significantly suppressed ABA-mediated stomatal closure (Figure 4). Notably, the expression of both OsRLCK311-FL and OsRLCK311- ΔN suppressed ABA-mediated stomatal closure, suggesting that the action of OsRLCK311 was independent of its kinase activity.
Figure 4.
Epidermal strips of wild-type (WT) and all RLCK311 over-expressing lines were first incubated in stomatal opening buffer under the light and then treated with 0 or 50 μM ABA for 20 min, and stomatal pore area was microscopically assessed. Values plots of > 700 measurements (stomata). Asterisks represent significant differences between transgenic lines and wild-type within a treatment by the Dunnet test (*, p < 0.05 and **, p < 0.01).
3.4. Salinity-Induced Expression of OsRLCK311-ΔN Conferred Salt Tolerance in the Perennial Grass Brachypodium sylvaticum
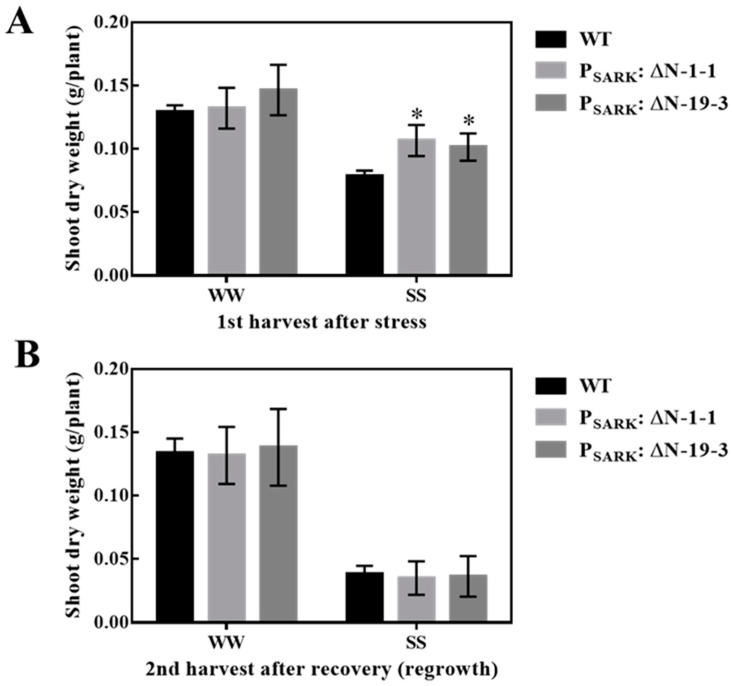
OsRLCK311-ΔN, driven by the stress-inducible promoter SARK, was introduced into the perennial grass Brachypodium sylvaticum (Figure S7). Transgenic PSARK::RLCK311-ΔN B. sylvaticum plants displayed enhanced growth under salt stress (Figure 5A). The shoot dry weight of transgenic plants was higher than that of wild-type plants at the first harvest after stress (Figure 5A). Since B. sylvaticum is a perennial grass, the re-growth capacity was assessed by a 2nd harvest following recovery under well-watered conditions. No significant difference was observed between genotypes, indicating no negative effect on perenniality (Figure 5B).
Figure 5.
Salt stress tolerance of wild-type (WT) and transgenic Brachypodium sylvaticum plants expressing pSARK: ΔN. (A) Shoot dry weight of WT and two transgenic lines (SARK: ΔN-1-1 and SARK: ΔN-19-3) grown under well-watered (WW) and salt stress (SS) conditions during the first harvest. Plants were harvested for the first time at around 45 days old, right after being treated with 100mM NaCl solution for 1 week, and then 200mM NaCl solution for 1 week continuously. (B) Shoot dry weight of wild-type (WT) and two transgenic lines during the second harvest. The second harvest was performed at 25 days after the first harvest, during which all the plants were well irrigated with distilled water. Values are means and SE (n= 12 for SARK:ΔN and 48 for WT). Asterisks represent significant differences between transgenic lines and WT within a treatment by the Dunnet test (p < 0.05).
4. Discussion
RLCKs have been shown to be important players in plant receptor kinase-mediated signalling [16]. OsRLCK311 expression was correlated with the enhanced stress tolerance of transgenic rice plants, brought about delayed stress-induced senescence [18], and the overexpression of OsRLCK311 in Arabidopsis plants resulted in enhanced growth under saline conditions (Figure 1). These results agreed with observations showing that soybean RLCKs negatively regulated ABA signalling and contributed to increased abiotic stress tolerance in Arabidopsis transgenic plants expressing GsRLCKs [28,29].
OsRLCK311 interacted with AtPIP2;1 (Figure 2). Aquaporins have been shown to regulate the stomata aperture under stress-like conditions [10,11,13,14] and to regulate the trade-off between plant survival and plant growth (i.e., water conservation versus growth/gas exchange) [12]. Plants overexpressing OsRLCK311 displayed an altered response to ABA, leading to increased stomata opening, possibly due to the deactivation of PIP2;1 [13]. RLCK311 also possibly interacted with a GRF 14-3-3 protein (Figure S8). 14-3-3 proteins have been shown to interact with water channels [30], and more specifically, with Arabidopsis AtPIP2;1, to regulate oscillations in water homeostasis [31]. Our results suggest that RLCK311 forms a complex with PIP2;1 and a 14-3-3 protein, regulating ABA-dependent stomata response. Moreover, phospholipase D-alpha 1, which is involved in ABA-dependent stomata regulation [32], was also identified as potential interactor with RLCK311 (Table S2), supporting the notion of a role of RLCK311 in the response of stomata to ABA. Interestingly, the transgenic plants overexpressing OsRLCK311-ΔN also displayed enhanced salinity tolerance (Figure 1). Moreover, the contribution of OsRLCK311-ΔN to salinity tolerance was also observed in the perennial grass Brachypodium sylvaticum (Figure 5). Since in-silico analysis and autophosphorylation assays showed no active kinase domain in OsRLCK-ΔN (not shown), our results suggest that the role(s) of OsRLCK311 in stress tolerance was independent of this kinase activity. Research is needed to assess the possibility that OsRLCK311 may function as a scaffold protein, bringing together cytoplasmic signalling components affecting aquaporin function(s).
Depending on the type and severity of the stress, the overall environmental conditions, and the plant species, plant response(s) to stress include, avoidance, escape or tolerance strategies [33]. For example, an avoidance strategy, which includes decreased stomata conductance and plant growth, could be beneficial for plant survival under severe stress, but not necessarily as a response to a mild stress [3] where the maintenance of an active gas exchange could contribute to plant growth and metabolism [9,34,35,36]. Moreover, under low vapour pressure conditions (e.g., high humidity) where transpiration flow is relatively low, open stomata would allow continuous CO2 diffusion with minimum water loss. OsRLCK311 could contribute to determining a stomata threshold response to environment conditions via its interactions with stomata response proteins. Although OsRLCK311 binding to PIP2;1 led to adjustment in stomata closure, similar interactions in other tissues (e.g., roots, mesophyll, xylem parenchyma) cannot be excluded. In rice, OsPIP2;1 was shown to regulate root hydraulic conductivity [37] while Arabidopsis AtPIP2;1 was shown to regulate leaf hydraulic conductivity via xylem parenchyma [38]. In addition, since OsRLCK311 has been identified in transgenic plants with modified cytokinin production, it would be interesting to further test the link between OsRLCK311-mediated stress tolerance and cytokinin response.
Acknowledgments
This research was funded by grants from United States Department of Energy Number #DE-SC0008797 and the Will W. Lester Endowment of University of California.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/10/1383/s1, Figure S1: Rice expressing PSARK::IPT showing salinity stress tolerance, Figure S2: Alignment of RLCK311 full length protein FL (LOC_Os11g0678) and RLCK311 C terminal ΔN (Os11g0168600) from the two rice genome databases, Figure S3: Expression of OsRLCK311 (ΔN or FL) in transgenic Arabidopsis plants. qRT-PCR analysis was conducted by using 30-day-old plants grown under normal conditions, Figure S4: Salt stress tolerance of 20 days-old Arabidopsis wild type (WT) and RLCK311 over-expressing lines grown in MS plates with and without 75 mM NaCl for 10 days, Figure S5: Infiltrated leaves taken for blots shown in Figure 2(B) The entire blots of Figure 2, Figure S6: Infiltrated leaves taken for blots shown in Figure 3(B). The entire blots of Figure 3. Figure S7: Gene expression driven by a stress and maturation induced promoter (SARK) promoter. Figure S8:MS identification of GRF9 and GRF10 interacting with both RCLK311-ΔN and RCLK311-FL, Table S1: List of the primers used in the experiments, Table S2: List of candidates RLCK311 interactors, Table S3: list of all MS interactors samples, Table S4: list of all peptide interactors.
Author Contributions
Conceptualization, N.S.; Data curation, N.S. and F.W.; Formal analysis, N.S., F.W., H.T., G.D. and Z.P.; Funding acquisition, E.B.; Investigation, F.W., H.T., Y.Z., L.Z. and M.d.M.R.W.; Writing—original draft, N.S. and F.W.; Writing—review & editing, N.S. and E.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Parida A.K., Das A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005;60:324–349. doi: 10.1016/j.ecoenv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Munns R., Tester M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 3.Skirycz A., Vandenbroucke K., Clauw P., Maleux K., De Meyer B., Dhondt S., Pucci A., Gonzalez N., Hoeberichts F., Tognetti V.B., et al. Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 2011;29:212–214. doi: 10.1038/nbt.1800. [DOI] [PubMed] [Google Scholar]
- 4.Isayenkov S.V., Maathuis F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019;10:80. doi: 10.3389/fpls.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang Q., Roche D., Monaco T.A., Hole D.G. Stomatal Conductance is a Key Parameter to Assess Limitations to Photosynthesis and Growth Potential in Barley Genotypes. Plant Biol. 2006;8:515–521. doi: 10.1055/s-2006-923964. [DOI] [PubMed] [Google Scholar]
- 6.James R.A., Von Caemmerer S., Condon A.T., Zwart A.B., Munns R. Genetic variation in tolerance to the osmotic stress componentof salinity stress in durum wheat. Funct. Plant Biol. 2008;35:111–123. doi: 10.1071/FP07234. [DOI] [PubMed] [Google Scholar]
- 7.Rahnama A., James R.A., Poustini K., Munns R. Stomatal conductance as a screen for osmotic stress tolerance in durum wheat growing in saline soil. Funct. Plant Biol. 2010;37:255–263. doi: 10.1071/FP09148. [DOI] [Google Scholar]
- 8.Singh A., Jha S.K., Bagri J., Pandey G.K. ABA Inducible Rice Protein Phosphatase 2C Confers ABA Insensitivity and Abiotic Stress Tolerance in Arabidopsis. PLoS ONE. 2015;10:e0125168. doi: 10.1371/journal.pone.0125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiani-Pouya A., Rasouli F., Rabbi B., Falakboland Z., Yong M., Chen Z.-H., Zhou M., Shabala S. Stomatal traits as a determinant of superior salinity tolerance in wild barley. J. Plant Physiol. 2020;245:153108. doi: 10.1016/j.jplph.2019.153108. [DOI] [PubMed] [Google Scholar]
- 10.Maurel C., Verdoucq L., Rodrigues O. Aquaporins and plant transpiration. Plantcell Environ. 2016;39:2580–2587. doi: 10.1111/pce.12814. [DOI] [PubMed] [Google Scholar]
- 11.Ding L., Chaumont F. Are Aquaporins Expressed in Stomatal Complexes Promising Targets to Enhance Stomatal Dynamics? Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J.-G., Feng M., Chen W., Zhou X., Lu J., Wang Y., Li Y., Jiang C.-Z., Gan S.-S., Ma N., et al. In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat. Plants. 2019;5:290–299. doi: 10.1038/s41477-019-0376-1. [DOI] [PubMed] [Google Scholar]
- 13.Grondin A., Rodrigues O., Verdoucq L., Merlot S., Leonhardt N., Maurel C. Aquaporins Contribute to ABA-Triggered Stomatal Closure through OST1-Mediated Phosphorylation. Plant Cell. 2015;27:1945–1954. doi: 10.1105/tpc.15.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L., Chaumont F. Aquaporin mediating stomatal closure is associated with water conservation under mild water deficit. bioRxiv. 2020 doi: 10.1101/2020.04.15.042234. [DOI] [Google Scholar]
- 15.Bellati J., Champeyroux C., Hem S., Rofidal V., Krouk G., Maurel C., Santoni V. Novel Aquaporin Regulatory Mechanisms Revealed by Interactomics. Mol. Cell. Proteom. 2016;15:3473–3487. doi: 10.1074/mcp.M116.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X., Zhou J.-M. Receptor-Like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase–Mediated Signaling. Annu. Rev. Plant Biol. 2018;69:267–299. doi: 10.1146/annurev-arplant-042817-040540. [DOI] [PubMed] [Google Scholar]
- 17.Vij S., Giri J., Dansana P.K., Kapoor S., Tyagi A.K. The Receptor-Like Cytoplasmic Kinase (OsRLCK) Gene Family in Rice: Organization, Phylogenetic Relationship, and Expression during Development and Stress. Mol. Plant. 2008;1:732–750. doi: 10.1093/mp/ssn047. [DOI] [PubMed] [Google Scholar]
- 18.Peleg Z., Reguera M., Tumimbang E., Walia H., Blumwald E. Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol. J. 2011;9:747–758. doi: 10.1111/j.1467-7652.2010.00584.x. [DOI] [PubMed] [Google Scholar]
- 19.Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 20.Van Leene J., Witters E., Inzé D., De Jaeger G. Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 2008;13:517–520. doi: 10.1016/j.tplants.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Karimi M., De Meyer B., Hilson P. Modular cloning in plant cells. Trends Plant Sci. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Sade N., Umnajkitikorn K., Wilhelmi M.D.M.R., Wright M., Wang S., Blumwald E. Delaying chloroplast turnover increases water-deficit stress tolerance through the enhancement of nitrogen assimilation in rice. J. Exp. Bot. 2017;69:867–878. doi: 10.1093/jxb/erx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porra R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002;73:149–156. doi: 10.1023/A:1020470224740. [DOI] [PubMed] [Google Scholar]
- 24.Li X. Infiltration of Nicotiana benthamiana Protocol for Transient Expression via Agrobacterium. Bio-Protoc. 2011;1 doi: 10.21769/BioProtoc.95. [DOI] [Google Scholar]
- 25.Gehl C., Waadt R., Kudla J., Mendel R.R., Hänsch R. New GATEWAY vectors for High Throughput Analyses of Protein–Protein Interactions by Bimolecular Fluorescence Complementation. Mol. Plant. 2009;2:1051–1058. doi: 10.1093/mp/ssp040. [DOI] [PubMed] [Google Scholar]
- 26.Roston R.L., Ruppel N.J., Damoc C., Phinney B.S., Inoue K. The Significance of Protein Maturation by Plastidic Type I Signal Peptidase 1 for Thylakoid Development in Arabidopsis Chloroplasts. Plant Physiol. 2010;152:1297–1308. doi: 10.1104/pp.109.151977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jefferson R.A., Kavanagh T.A., Bevan M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L., Ji W., Zhu Y., Gao P., Li Y., Cai H., Bai X., Guo D. GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 2010;61:2519–2533. doi: 10.1093/jxb/erq084. [DOI] [PubMed] [Google Scholar]
- 29.Sun X., Sun M., Luo X., Ding X., Ji W., Cai H., Bai X., Liu X., Zhu Y. A Glycine soja ABA-responsive receptor-like cytoplasmic kinase, GsRLCK, positively controls plant tolerance to salt and drought stresses. Planta. 2013;237:1527–1545. doi: 10.1007/s00425-013-1864-6. [DOI] [PubMed] [Google Scholar]
- 30.Moeller H.B., Slengerik-Hansen J., Aroankins T., Assentoft M., Macaulay N., Moestrup S.K., Bhalla V., A Fenton R. Regulation of the Water Channel Aquaporin-2 via 14-3-3θ and -ζ. J. Biol. Chem. 2015;291:2469–2484. doi: 10.1074/jbc.M115.691121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prado K., Cotelle V., Li G., Bellati J., Tang N., Tournaire-Roux C., Martinière A., Santoni V., Maurel C. Oscillating Aquaporin Phosphorylation and 14-3-3 Proteins Mediate the Circadian Regulation of Leaf Hydraulics. Plant Cell. 2019;31:417–429. doi: 10.1105/tpc.18.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacob T., Ritchie S., Assmann S.M., Gilroy S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA. 1999;96:12192–12197. doi: 10.1073/pnas.96.21.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu S., Ramegowda V., Kumar A., Pereira A. Plant adaptation to drought stress. F1000Research. 2016;5:1554. doi: 10.12688/f1000research.7678.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sade N., Gebretsadik M., Seligmann R., Schwartz A., Wallach R., Moshelion M. The Role of Tobacco Aquaporin1 in Improving Water Use Efficiency, Hydraulic Conductivity, and Yield Production Under Salt Stress. Plant Physiol. 2009;152:245–254. doi: 10.1104/pp.109.145854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou L., Wang C., Liu R., Han Q., Vandeleur R.K., Du J., Tyerman S.D., Shou H. Constitutive overexpression of soybean plasma membrane intrinsic protein GmPIP1;6 confers salt tolerance. BMC Plant Biol. 2014;14:181. doi: 10.1186/1471-2229-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohamed I.A.A., Shalby N., Bai C., Qin M., Agami R.A., Jie K., Wang B., Zhou G. Stomatal and Photosynthetic Traits Are Associated with Investigating Sodium Chloride Tolerance of Brassica napus L. Cultivars. Plants. 2020;9:62. doi: 10.3390/plants9010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding L., Uehlein N., Kaldenhoff R., Guo S., Zhu Y., Kai L. Aquaporin PIP2;1 affects water transport and root growth in rice (Oryza sativa L.) Plant Physiol. Biochem. 2019;139:152–160. doi: 10.1016/j.plaphy.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 38.Prado K., Boursiac Y., Tournaire-Roux C., Monneuse J.-M., Postaire O., Da Ines O., Schäffner A.R., Hem S., Santoni V., Maurel C. Regulation of Arabidopsis Leaf Hydraulics Involves Light-Dependent Phosphorylation of Aquaporins in Veins. Plant Cell. 2013;25:1029–1039. doi: 10.1105/tpc.112.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.