Abstract
N6-methyladenosine (m6A)-mediated epitranscriptomic regulation is critical for various physiological processes. Genetic studies demonstrate that proper m6A-methylation is required for mouse brain development and function. Revealing landscapes of m6A-methylation in the cerebral cortex at different developmental stages will help to understand the biological meaning of epitranscriptomic regulation. Here, we depict the temporal-specific m6A-methylation status in mouse embryonic and postnatal cortices using methylated RNA immunoprecipitation (MeRIP) sequencing. We identified unique m6A binding motifs in stage-specific RNAs and found that more RNA transcripts are temporally methylated in embryonic cortices than in postnatal ones. Moreover, we found that cortical transcription factors and genes associated with neurological disorders are broadly as well specifically methylated at m6A sites. Our study highlights the importance of epitranscriptomic regulation in the developing cortex and provides a fundamental reference for future mechanistic examinations of m6A methylation-mediated gene expression regulation in normal brain development and neurological disorders.
Keywords: m6A, RNA methylation, cerebral cortex, neural development, neurological disorders
1. Introduction
Proper development of the mammalian cerebral cortex is crucial for its morphogenesis and function. In the mouse embryonic cortex, neural stem cells and neural progenitors proliferate and differentiate properly to set up the architectural foundation of the cortical size and shape [1,2]. In the postnatal cortex, migratory neurons form a multiple-layer structure and develop interaction networks within the cortex and with other regions in the central nervous system [3,4]. Temporal and spatial regulation of gene expression precisely drives developing progresses of the cerebral cortex throughout the embryonic and postnatal stages. Altered gene expression causes malformation in the cortex, and results in various neurological disorders, such as microcephaly, macrocephaly and autism [5,6,7].
Accumulating studies have demonstrated essential roles of genetic and epigenetic factors in controlling proper cortical development [8,9]. For instance, aberrance of DNA methylation and/or demethylation has been shown to alter transcription programs of neurogenic genes and lead to brain dysfunctions [10,11,12]. Notably, epitranscriptomic regulations in transcribed RNAs, in particular N6-methyladenosine (m6A) modification, have been reported to regulate RNA splicing, stability, translation rate and localization [13,14]. In particular, precise regulation of m6A-methylation is required for normal cortical development. Genetic deletion of m6A methyltransferase Mettl14 results in abnormal proliferation and differentiation of neural progenitor cells in the embryonic cortex [15,16,17]. Moreover, m6A-methylation is critical for maturation and function of postnatal and adult brains, as m6A-methylation has been shown to regulate dopamine transmission, sensory experience, learning and response to brain injury [18,19,20]. Altered m6A-methylation is also associated with neurological disorders, such as Alzheimer’s disease, glioblastoma and major depression [21,22,23,24]. While molecular mechanisms of m6A regulation are still unclear, understanding comprehensive profiles of RNA transcripts methylated by m6A will facilitate uncovering RNA methylation-mediated gene expression regulation and function.
In this study, to reveal m6A-methylation status in the developing cerebral cortex, we conduct methylated RNA immunoprecipitation (MeRIP) sequencing of RNAs collected from mouse embryonic and postnatal cortices. We identify unique m6A binding sites in temporally expressed RNAs and reveal specific m6A patterns carried by RNAs encoding transcription factors and disease-risk genes in the nervous system. Our results have provided strong evidence and useful reference of the importance of m6A-methylation in cortical development.
2. Materials and Methods
2.1. Animal Care
Wild-type C57BL/6 mice were purchased from Wushi company (Fujian, China). All animal experiments and euthanasia were approved (A2019009) and performed in accordance with the guidelines of Animal Care and Use, Committee of School of Medicine, Huaqiao University.
2.2. RNA Extraction and m6A-Methylated RNA Immunoprecipitation (MeRIP)
Mice were sacrificed by cervical dislocation and their cerebral cortex was removed and immediately digested with RNAisoTM Plus Reagent (TaKaRa, Dalian, China). Total RNA was extracted using the same reagent according to the manufacturer’s instructions. In general, embryonic-stage RNA was extracted from three E12.5–13 mouse embryos from one litter, and postnatal-stage RNA was extracted from three P14 mice from one litter. About 300 μg RNA was used for MeRIP, and all RNA samples were treated with DNase to remove genomic DNA.
The m6A-MeRIP reaction was performed according to a previous description [25]. Briefly, total RNA was chemically fragmented to 100–200 nucleotides (for m6A-seq). A portion of fragmented RNA was saved as input before immunoprecipitation (IP). The remaining fragmented RNA was incubated with an m6A-specific antibody (Synaptic Systems, cat. No. 202 003) at 4 °C for 2 h, followed by addition of Immobilized Recombinant Protein A (Repligen, cat. No. IPA300) for immunoprecipitation. After 2-h incubation at 4 °C, beads were extensively washed three times with IP Buffer (150 mM NaCl, 0.1% Igepal CA-630 and 10 mM Tris-HCl, pH 7.4). Immunoprecipitated RNA was recovered through elution with m6A nucleotide followed by ethanol precipitation. Due to the recognition feature of the m6A antibody, the detection of methylated sites might contain both m6A and m6A(m) [26]. Additionally, IgG (Sigma-Aldrich, cat. No. I8765) was used as a negative control of the immunoprecipitated reaction.
2.3. m6A Sequencing (m6A-seq) and Data Analysis
For high-throughput sequencing, purified RNA fragments from m6A-MeRIP were used for library construction with the NEBNext Ultra RNA library Prep kit from Illumina and were sequenced with Illumina HiSeq 4000. Library preparation and high-throughput sequencing were performed by Novogene (Beijing, China). The m6A-seq data (NCBI GEO: GSE141938) was analyzed with reference to the previous standardized pipeline [27]. Briefly, the m6A-seq data was cleaned by trimming the adapter using trim galore. The cleaned reads were aligned to the house mouse (Mus musculus) genome UCSC mm10 using Bowtie2 with default parameters and were then converted to a bam file by samtools. The PCR duplicates in the aligned data were removed by the MarkDuplicates function of Picard (version 1.95) with parameters, REMOVE_DUPLICATES = Ture. After filtering the duplicates, samtools was used to sort the bam file according to the coordinates. The m6A peaks were detected by MACS (Version 1.4.0rc 2) with parameters, -format = “BAM” -gsize = 282000000 -tsize = 101 -nomodel -shiftsize =150 -to-large False -w -S. The resulting m6A-seq data from m6A-antibody immunoprecipitation samples at E12.5–13 and P14 were normalized to their respective input RNA data before being used for the following analysis.
Hypergeometric Optimization of Motif EnRichment (HOMER) was used for m6A motif analysis. Briefly, we set the peak-harboring genome interval as detected data, while the peak-removed exon interval was set as background data. The detected data and background data were compared for motif analysis using HOMER with the following orders: -gc, -RNA, -len 6, -size given, -homer2, -p 2, -S 25, -dumpFasta. Each sample was independently measured, followed by intra-group comparison in E12.5–13 triplicates and P14 triplicates, respectively.
The integrative genomics viewer (IGV) tool was used for visualization of m6A peaks along the whole transcript [28]. Notably, we identified a methylation site as significant if the mean IGV value of peak was more than 3, which is approximately equal to log10 (500) (500 was the threshold of peak scores). Even if the IGV presentation showed a visible peak, the site was considered as non-methylated if the IGV value was less than 3. The heatmaps of the fold enrichment score for m6A peaks, the Volcano maps of transcript levels and the Venn analysis of targeted genes were all performed using the Omic Share tools, a free online platform for data analysis (http://www.omicshare.com/tools).
The DAVID tool [29] was used for the Gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis by applying default parameters. Color intensity indicated the value of −log10 (p value), while the length of columns or the size of the circles indicated the gene counts. A full list of all selected terms of the biological process, cellular components, molecular functions and KEGG pathway category are provided in the Supplementary Materials.
2.4. Statistics
All data are shown as mean ± SEMT. One-way analysis of variance (ANOVA) with post hoc contrasts were used for statistical analysis. The results were considered significant at probability level less than 0.05.
3. Results
3.1. m6A-Methylation Profiles in Embryonic and Postnatal Cerebral Cortices
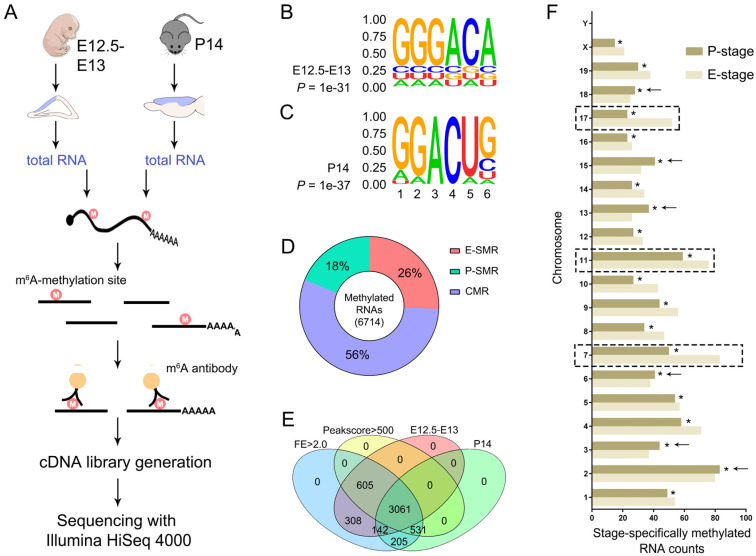
Cortical development is a continuous process of neural progenitor proliferation, differentiation and newborn neuron migration and maturation [1,3,30]. If m6A-methylation is essential for cortical development, we suspected that m6A-methylation profiles should reflect the dynamic process in developing cortices. Because mouse cortices at embryonic days 12.5 to 13 (E12.5–E13), termed here as embryonic stage (E-stage), and cortices at postnatal day 14 (P14), termed as postnatal stage (P-stage), consisted of more neural progenitors and mature neurons, respectively, we performed MeRIP sequencing (MeRIP-Seq) using cortical tissues from E12.5–E13 and P14 mouse brains (Figure 1A).
Figure 1.
Profiles of m6A-methylation in mouse embryonic and postnatal cerebral cortices. (A) Schematic presentation of methylated RNA immunoprecipitation (MeRIP) sequencing. Total RNA was extracted from the cerebral cortex (blue) of embryonic day 12.5–13 (E12.5–E13) and postnatal day 14 (P14) mice. (B,C) Sequence motifs identified within m6A peaks. The motif GGGACA and GGACUG were highly enriched in m6A peaks from E12.5–E13 and P14 samples, respectively. (D) The proportion of embryonic stage-specifically methylated RNAs (SMRs), termed as E-SMRs, postnatal stage-specifically methylated RNAs, named P-SMRs, and continuously methylated RNAs at both stages, termed as CMRs. (E) Identification of temporally-specifically methylated RNAs. The parameter value was limited as threshold. Peak score values > 500, fold enrichment > 2.0. p-values for all presenting RNAs were less than 0.001. (F) The counts of temporally-specifically methylated RNAs in each chromosome. The counting numbers for E-SMRs were significantly higher in chromosomes 7, 11 and 17 (dashed boxes), and for P-SMRs were higher in chromosomes 2, 3, 13, 15 and 18 (arrows). The asterisked bar in each group differ significantly from those unmarked (Tukey’s HSD (Honestly significant difference), p < 0.05).
We identified an average of 27,113,335 and 25,574,954 clean reads from E-stage and P-stage cortices, respectively. The coverage was 545 and 521 reads/1000 bp in E-stage and P-stage cortices, respectively [26,31]. Pearson correlation analysis showed related a co-efficiency approximately to 1 among each group of three replicates at each stage, indicating that MeRIP-Seq data displays high consistency and low variation (Supplementary Figure S1). Each read was frequently mapped to transcribed RNAs, and was clustered as a distinct peak, which was converged to about 100 nucleotides (nts) wide fragment with a summit in the center (Supplementary Figure S2A). After normalization to the input data, we detected 8355, 8488 and 8451 methylated RNAs from three E-stage cortices, and 8365, 8373, and 8151 methylated RNAs from three P-stage cortices (Supplementary Figures S2B,C). We identified 4733 and 4449 intersecting m6A-methylated RNAs with high-confidence in all three E-stage and P-stage replicates, respectively (500 value of sequencing peak-scores and 0.001 p-value were set as threshold limit) (Supplementary Figure S2B,C, Supplementary Table S1).
To identify m6A motif in temporal-specific cortical RNAs, we applied a sensitive and unbiased tool, Hypergeometric Optimization of Motif EnRichment (HOMER), for motif discovering. We detected the most confident motif, GGGACA, which was highly enriched in m6A peaks from triplicated E-stage cortices (Figure 1B). Motif variants, such as GGACUG and UGACU[GC], which shared a conserved sequence, GAC, with m6A in the middle, were also predicted. Similarly, we also detected the most confident motif, GGACUG, and predicted variants, UAGGAC and GACU[GA][AG], in m6A peaks from triplicated P-stage cortices (Figure 1C). These results indicate that m6A-methylation displays unique motifs in RNAs from embryonic and postnatal cortices.
3.2. None or Weak Correlations between m6A-Methylation Profile and Transcription Profile
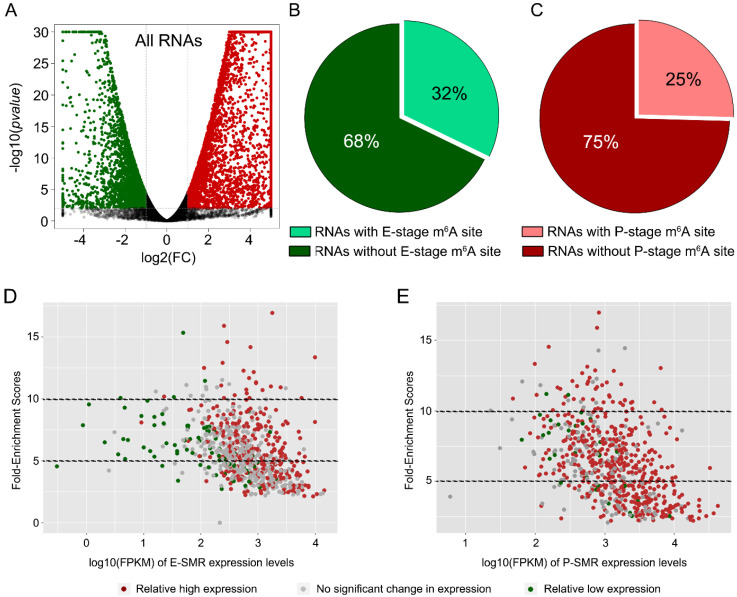
We analyzed RNA sequencing data of E- and P-stage cortices (NCBI GEO: GSE116056), and identified 2970 RNA transcripts from E-stage neural progenitors displaying higher expression levels than those from P-stage cortices, while 5416 transcripts showed higher expression from P-stage cortices than those from E-stage ones (Figure 2A). We detected 32% of 2970 RNAs and 25% of 5416 RNAs containing m6A-methylation sites in E-stage and P-stage cortices, respectively (Figure 2B,C). These data suggest that only parts of temporally-highly expressed RNAs are methylated at the m6A site. In addition, correlation analysis between levels of RNA transcription and m6A-methylation showed mixed distribution, suggesting none or weak correlations between two aspects (Figure 2D,E).
Figure 2.
None-correlation between fold enrichment of MeRIP sequencing and reads of transcriptome sequencing. (A) The Volcano map of transcription profiles of all RNAs in P-stage compared to E-stage. The transcriptome data were downloaded and used from (NCIB GEO: GSE116056). (B) The proportion of down-regulated RNAs with or without an E-stage m6A site. (C) The proportion of up-regulated RNAs with or without a P-stage m6A site. (D,E) The correlation analysis of embryonic stage-specifically methylated RNAs (E-SMRs) or postnatal stage-specifically methylated RNAs (P-SMRs) between their fold enrichment score values of m6A-methylation levels and the FPKM reads of their expression levels.
To comprehensively evaluate the correlation between the transcribed change of RNAs and the position of m6A, the amounts of m6A sites were calculated according to their distribution region in RNAs with different changes in their transcription level (Supplementary Figure S3). We found that the up-regulated RNAs in postnatal stage harbor more postnatal-special m6A sites than embryonic-special m6A sites in RNA regions of 5′ untranslated region (5′UTR), coding sequence (CDS) and 3′UTR (Supplementary Figure S3A). On the contrary, both down-regulated and none-changed RNAs had more embryonic-special m6A sites than postnatal-special m6A sites in targeted regions (Supplementary Figure S3B,C). Additionally, the amounts of common m6A sites (detected in both embryonic and postnatal stage) were generally higher than of E-special and P-special m6A sites in targeted regions of both changed and none-changed RNAs (Supplementary Figure S3). Our analysis indicates that the transcription changes of RNAs show partial correlation with temporal-specificity of m6A rather than the position of m6A.
3.3. Temporally Methylated RNAs
We next analyzed distributions of m6A summits among transcribed RNAs residing in different chromosomes. We observed that there are, overall, slightly higher m6A summits in RNAs from E-stage cortices than those from P-stage, suggesting a higher global methylation level in embryonic cortices (Supplementary Figure S4A). Moreover, we counted numbers of m6A-methylated RNAs in each chromosome and found that more m6A-methylated RNAs were detected in E-stage cortices than in P-stage ones in most chromosomes (Supplementary Figure S4B). These results further suggest that m6A-methylation appears to be more active in embryonic cortices than in postnatal ones.
We further compared RNAs that were temporally and continuously methylated either in E-stage cortices or in P-stage ones. We identified 26% embryonic stage-specifically methylated RNAs (SMRs), termed as E-SMRs, 18% postnatal stage specifically methylated RNAs, named P-SMRs, and 56% continuously methylated RNAs, termed as CMRs, at both stages (Figure 1D). Notably, there were more E-SMRs than P-SMRs (about 1.5 times). These results suggest that the majority of RNAs are continuously methylated in the developing cortex, and more RNAs are temporally methylated in embryonic cortices than in postnatal ones.
To exclude an effect of input signals, we further identified significantly methylated RNAs by analyzing the fold enrichment (FE) score, which is calculated as peak-reads of immunoprecipitation samples versus those of relative input samples. We detected 3061 CMRs, 605 E-SMRs and 531 P-SMRs (p < 0.001, peak score > 500, FEs > 2.0) (Figure 1E). Moreover, similar to all methylated RNAs, the number of E-SMRs was more than that of P-SMRs, and E-SMRs showed significant enrichment in chromosomes 7, 11 and 17 (Figure 1F). These data suggest that many RNAs from different chromosomes are temporally modified by m6A-methylation in the developing cortices.
3.4. Patterns of m6A-Methylation within Transcribed RNAs
To examine the distribution pattern of m6A binding sites within transcribed RNAs, we first analyzed the number of m6A binding sites in them. In E-stage cortices, one single m6A peak was detected in 58% RNAs, two m6A peaks in 26% RNAs, three m6A peaks in 10% RNAs, and four or more m6A peaks in 6% RNAs (Supplementary Figure S4C). In addition, one single, two, three and four or more m6A peaks were observed in 61%, 25%, 9% and 5% RNAs transcribed in P-stage cortices, respectively (Supplementary Figure S4D). These results indicate that a majority of cortical RNAs contains one or two m6A binding sites.
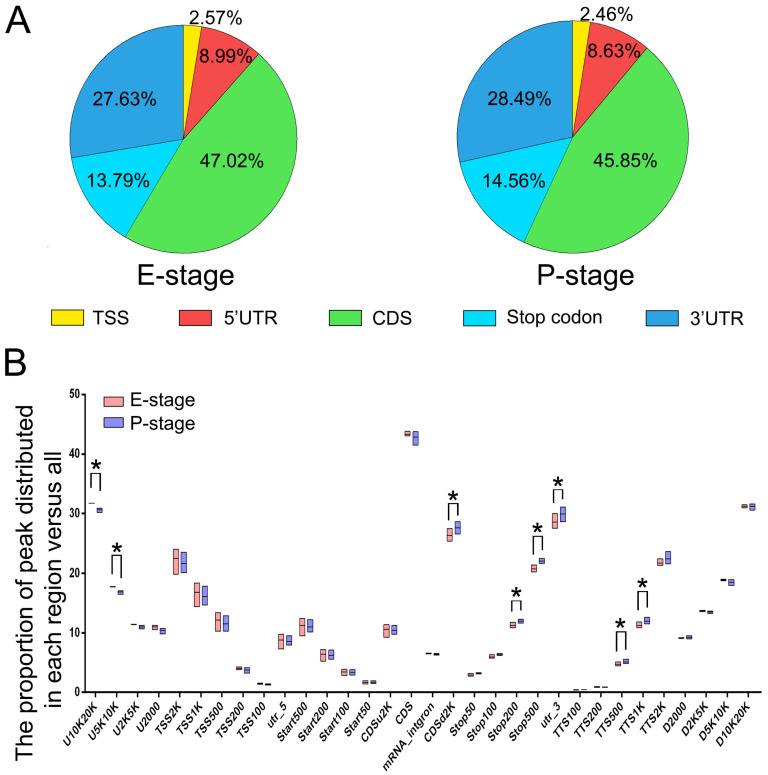
We then determined the distribution position of m6A binding sites in RNAs. We observed that m6A binding sites resided in coding sequences (CDS) in 47.02% and 45.85% RNAs from E-stage and P-stage cortices, respectively, and in untranslated regions (UTRs) in 36.62% and 37.12% RNAs from E-stage and P-stage cortices, respectively (Figure 3A). Interestingly, m6A binding sites were more abundant in the 3′UTR than in the 5′UTR in both stages (Figure 3A). Moreover, m6A binding sites were also detected in regions of the stop codon in many RNAs. To further investigate the distribution patterns of m6A mapping to genomic region, we calculated the numbers of m6A peaks mapped to specific genomic regions versus numbers of all detected m6A peaks (Figure 3B). Significant differences of m6A distributions mapping to genomic regions from E-stage and P-stage cortices were mostly located in the upstream region (5–10 kb), downstream of CDS (2 kb), near stop codon (NSC, 200–500 bp), 3′UTR and transcriptional termination site (TTS, 500–1000 bp) (Figure 3B). Additionally, we also calculated the percentage of m6A summits in different regions of mRNAs versus the numbers of all detected summits and found an enrichment of m6A summits in the region of near stop codon (NSC) and 3′UTR regions (Supplementary Figure S5). Collectively, these data indicate variations of m6A-methylated sites in transcribed RNAs, beside the CDS, with a high enrichment in the NSC and 3′UTR regions.
Figure 3.
Patterns of m6A-methylation within transcribed RNAs. (A) The proportion of m6A peaks distributed among five cortical RNA regions at E- or P-stage. (B) The proportion of m6A peaks distributed in the genome. U: upstream; D: downstream; TSS: translational start site; TTS: translational termination site; UTR: untranslated region; start: translation start codon; stop: translation stop codon; CDS: coding sequence; K: kilobase. The U2000 and D2000 refer to 2000 bp upstream of genes and 2000 bp downstream of genes, respectively. CDSu2k and CDSd2k refer to 2KB upstream and downstream of CDS, respectively. Tss100, TTS100, Stop100 and Start100 refer to the range of 100bp centered on TSS, TTS, start codon and stop codon. There might be overlapping parts among some of the regions above. The asterisked bar in each group differ significantly from those unmarked (Tukey’s HSD, p < 0.05).
3.5. Functional Association of m6A-Methylation with Transcribed RNAs
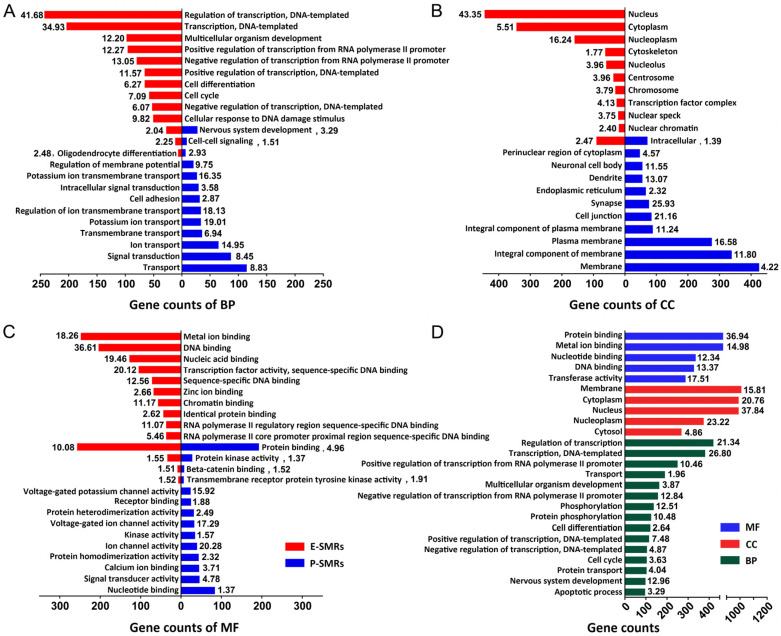
We found that m6A binding sites show enrichment in the NSC and 3′UTR regions, and within these regions, more m6A binding sites are detected in RNAs from P-stage than those from E-stage cortices (Figure 3B). Therefore, we focused the remaining analyses on RNAs with enriched m6A binding sites in the NSC and 3′UTR regions. We first performed a gene ontology (GO) analysis on E-SMRs and P-SMRs (Figure 4A, Supplementary Table S2). The top categories of biological process (BP) for E-SMRs were mainly involved in regulation of transcription, cell differentiation and cell cycle, while they were associated with ion transport, signal transduction and cell adhesion for P-SMRs (Figure 4A). Interestingly, three common categories of nervous system development, cell-cell signaling and oligodendrocyte differentiation were identified in both E-SMRs and P-SMRs (Figure 4A).
Figure 4.
Functional association of m6A-methylation with transcribed RNAs. (A–C) Gene ontology (GO) analysis on E-SMRs and P-SMRs in categories of biological process (BP), cellular component (CC) and molecular function (MF). (D) GO analysis on continuously methylated RNAs (CMRs). The –log10(p-Value) of each category is shown by the respective box.
Moreover, we analyzed cellular component (CC) and found that enriched categories for E-SMRs are associated with DNA-replication and RNA-synthesis, and those for P-SMRs are enriched in cell membrane construction and assembly of specific neuronal cell bodies, dendrites and synapses (Figure 4B). In addition, molecular function (MF) analyses showed that categories for E-SMRs are generally enriched in DNA and ion binding, and those for P-SMRs are involved in channel activities (Figure 4C). GO analyses for CMRs showed enriched categories, mainly in fundamental biological processes, such as transport, membrane and protein binding (Figure 4D). Collectively, our results indicate that temporal-specific m6A-methylation is highly associated with E-SMRs that function in transcription and cell cycle regulations, and with P-SMRs that play a role in regulating differentiation and signal transduction, which is consistent with major biological events occurring in neural progenitors and mature neurons.
3.6. Status of m6A-Methylation in Genes Involved in Major Signaling Pathways
Because cortical development is precisely modulated by several signaling pathways, we examined whether genes in major pathways are regulated by m6A-methylation. We first analyzed E-SMRs with m6A binding sites in the NSC and 3′UTR and identified 27 enriched pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (Supplementary Table S2). In particular, genes in the PI3K-Akt signaling pathway, hippo signaling pathway, p53 signaling pathway and hedgehog signaling pathway displayed significant m6A-methylation (Supplementary Figure S6).
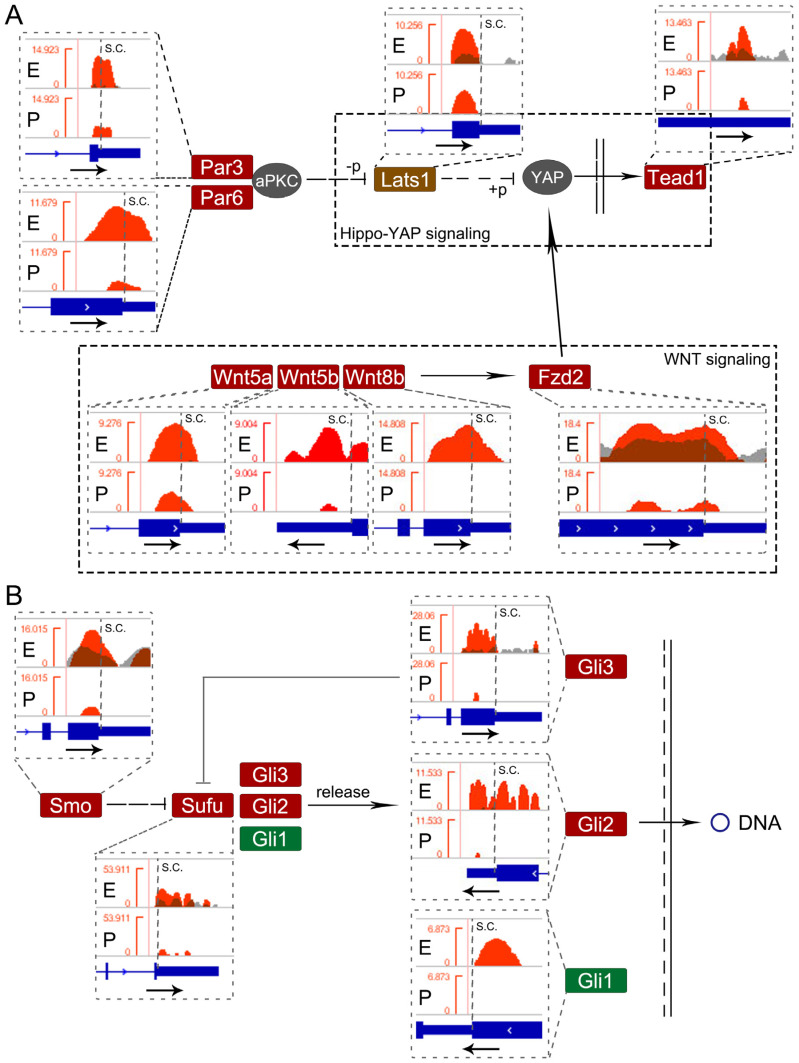
Among these pathways, hippo and hedgehog signaling pathways have been shown to play crucial roles in cortical development [32,33]. Hence, we investigated m6A-methylation status in major genes among E-SMRs in these two pathways. Studies have shown that Par3/Par6/aPKC complex, Lats1, YAP and Tead1 are associated with the hippo-YAP signaling pathway [34,35]. We analyzed their m6A-methylation patterns using the Integrative Genomics Viewer (IGV) (Figure 5). We detected one m6A binding site near stop codon of Par3 and Par6 transcripts (Figure 5A). Moreover, Lats1 displayed one m6A binding site near the stop codon, and Tead1 had one m6A site in the 3′UTR. In addition, studies have shown association of the Wnt and hippo-YAP signaling pathways [36]. We found that Wnt5a/b, Wnt8b and Fzd2 are methylated near their stop codons with one or two m6A sites. Furthermore, we detected one or more m6A sites in the NSC or 3′UTR region in Smo, Sufu, and Gli1/2/3 in the hedgehog signaling pathway (Figure 5B and Supplementary Figure S7). Collectively, our data indicate that m6A-methylation appears to be a common event in genes which are involved in major signaling pathways regulating cortical development.
Figure 5.
Status of m6A-methylation in genes in the hippo-YAP pathway and hedgehog pathway. (A) Integrative Genomics Viewer (IGV) analysis of four E-SMRs (Par3, Par6, Lats1 and Tead1) in the hippo-YAP pathway. Four Wnt-related E-SMRs (Wnt5a/b, Wnt8b and Fzd2) participated in the hippo-YAP pathway. (B) IGV analysis of five E-SMRs (Smo, Sufu, Gli1/2/3) in the hedgehog pathway.
Furthermore, we examined the m6A-methylation status in P-SMRs with m6A binding sites in the NSC and 3′UTR using the KEGG analysis. We observed that neural signaling transmissions, such as neuroactive ligand-receptor interaction pathways and multi-chemical synapses, are specifically enriched in P-SMRs (Supplementary Figure S6 and Table S2). In detail, we detected 20 P-SMRs involved in the glutamatergic synapse pathway, 16 in the cholinergic synapse pathway, 13 in the GABGergic synapse pathway, 12 in the serotonergic synapse pathway, and 10 in the dopaminergic synapse pathway (Supplementary Tables S1 and S2). We also noticed that a majority of P-SMRs involved in the above pathways encode receptors located in the postsynaptic membrane; in particular, P-SMRs involved in the glutamatergic synapse pathway are coordinated with calcium ion (Ca2+) and the calcium signaling pathway (Supplementary Figure S8). In addition, these targeted genes had multiple m6A sites along their transcripts (Supplementary Table S3), suggesting an important role of m6A-methylation in gene functional regulation. These analyses indicate that m6A methylated genes in P-stage cortices are mostly involved in signaling transmission of postsynaptic membrane reception.
3.7. Cortical-Specific Transcription Factors Methylated at the m6A Sites
Because transcription factors play crucial roles in cortical development and function [37], we next focused on analyzing m6A-harboring status in RNAs encoding cortical-specific transcription factors with m6A binding sites near their stop codon and the 3′UTR.
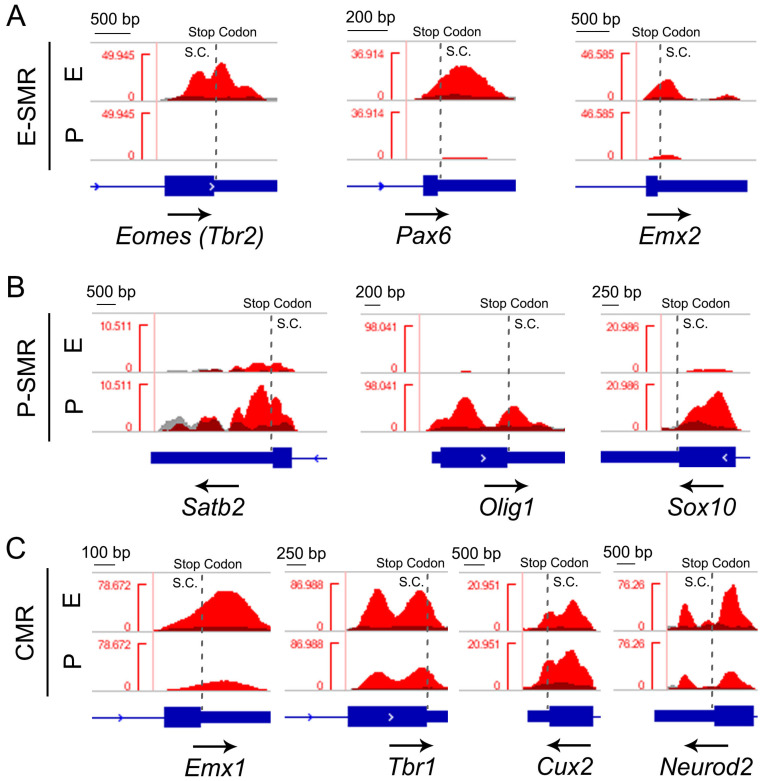
Among 930 E-stage-specifically methylated RNAs, we detected 60 genes functioning as transcription factors such as PAX6, EMX2 and TBR2. Pax6 and Tbr2 harbored one m6A site near the stop codon, and Emx2 contained one m6A site near stop codon and one in the 3′UTR, illustrated by the IGV (Figure 6A). Moreover, we detected 17 transcription factors among 794 P-SMRs (Supplementary Table S4). For instance, Satb2 and Sox10 harbored one m6A site near the stop codon, and Olig1 had two m6A sites near the stop codon (Figure 6B).
Figure 6.
Status of m6A-methylation in cortical transcription factors. (A) Integrative Genomics Viewer (IGV) analysis of three E-SMRs that function as transcription factors (Pax6, Emx2 and Tbr2). (B) IGV analysis of three P-SMRs (Satb2 Olig1 and Sox10). (C) IGV analysis of four CMRs encoding transcription factors.
In addition, we analyzed 2562 CMRs, identified 71 transcription factors expressed in both E- and P-stage cortices, and found 54 of them harbored one m6A site in the NSCs or 3′UTR region (Supplementary Table S4). For example, Emx1 and Tbr1 had one m6A site, and Cux2 and Neurod2 displayed two m6A sites (Figure 6C).
Besides the NSC and 3′UTR region, there were 569 RNAs harboring m6A sites at the 5′UTR and 1939 RNAs in the CDS region. Among them, 13 of 194 E-SMRs, 9 of 141 P-SMRs and 10 of 234 CMRs were RNAs encoding transcription factors harboring m6A sites in the 5′UTR region, while 51 E-SMRs, 12 P-SMRs and 46 CMRs were transcription factors with m6A sites in the CDS region (Supplementary Table S4). Similarly, more than one m6A site was detected from the targeted transcripts (Supplementary Table S3).
Taken together, our data indicate that cortical transcription factors are broadly methylated at the m6A site.
3.8. Detection of m6A-Methylated RNAs Related to Neurological Disorders
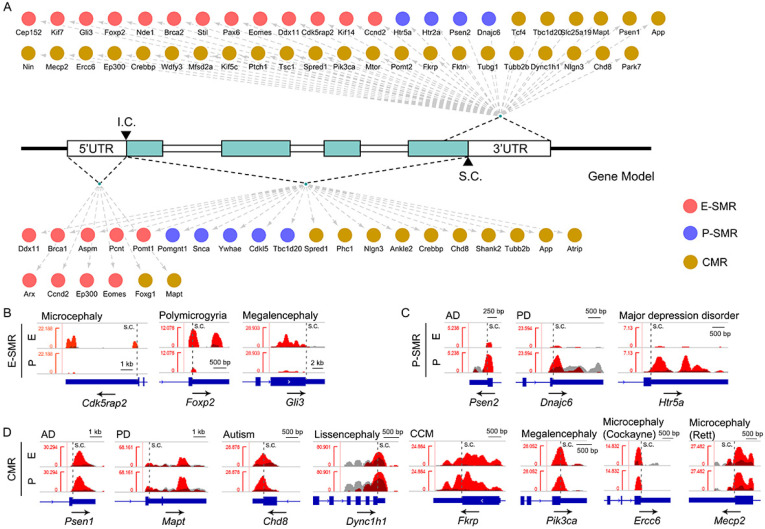
Previous studies have identified pathogenic genes associated with neurological disorders [38], so we next examined m6A-methylation patterns of these genes. We identified 45 disease-risk genes modified by m6A-methylation that is restricted to the NSC or the 3′UTR region. In total, 13 E-SMRs were reported to be associated with neurodevelopmental diseases, such as microcephaly, polymicrogyria and megalencephaly (Supplementary Table S5). We observed one single m6A site near the stop codon in 7 microcephaly-related E-SMRs, including Brca2, Cep152, Ddx11, Nde1, Kif14, Stil and Cdk5rap2, 3 polymicrogyria-related E-SMRs (Eomes/Tbr2, Pax6 and Foxp2), and 3 megalencephaly-related E-SMRs (Gli3, Ccnd2 and Kif7) (Figure 7A, Supplementary Tables S5 and S6). Moreover, Cdk5rap2 and Foxp2 contained two m6A sites near the stop codon and in the 3′UTR region (Figure 7B).
Figure 7.
Status of m6A-methylation sites of genes in neurological disorders. (A) Identification of methylated genes and their specifically methylated patterns in RNA transcripts. The red dots stand for E-SMRs, the blue dots stand for P-SMRs, and the dark yellow dots stand for CMRs. (B) IGV analysis of three representative E-SMRs that act as pathogenetic genes (Cdk5rap2, Foxp2 and Gli3) associated with neurological disorders. (C) IGV analysis of three representative P-SMRs (Psen2, Dnajc6 and Htr5a). (D) IGV analysis of eight representative CMRs (Psen1, Mapt, Chd8, Dync1h1, Fkrp, Pik3ca, Ercc6 and Mecp2) encoding neurological disease-risk genes.
Additionally, we detected 4 P-SMRs involved in Alzheimer’s disease (Psen2), Parkinson’s disease (Dnajc6) and major depression disorder (Htr2a and Htr5a) (Supplementary Table S5). Htr5a contained two m6A sites located near stop codon and in the 3′UTR, and the remaining 3 P-SMRs harbored one m6A site near stop codon (Figure 7C, Supplementary Table S6).
Moreover, we identified 28 CMRs associated with neurological disorders (Supplementary Table S6). Among them, Mapt, Nlgn3 and Mtor contained two m6A sites in the NSC or 3′UTR, Nin displayed three sites, and the rest of the CMRs contained one m6A site in the NSC or 3′UTR (Figure 7D, Supplementary Table S6). Our analyses indicate that the targeted RNAs associated with neurological diseases are modified by m6A-methylation with a variation of m6A binding sites.
Furthermore, we found that disease-risk RNA transcripts also display m6A-methylation sites in the 5′UTR and CDS. For example, 2 CMRs, Mapt and Foxg1, had one m6A-methylation site in the 5′UTR, microcephaly-related genes, Brca1 and Ddx11, had one m6A site, and Aspm and Pcnt harbored four or more m6A sites within the CDS (Figure 7A, Supplementary Table S5). The data suggest that m6A-methylation also modulates pathogenic RNAs via the 5′UTR and CDS region.
4. Discussion
Development of the cerebral cortex is precisely controlled by both genetic and epigenetic regulations. Epitranscriptomic regulation mediated by m6A is a prominent RNA modification and plays a crucial role in various biological processes. In this study, we uncovered the m6A-methylation status in mouse embryonic and postnatal cortices using MeRIP sequencing and identified distinct m6A binding motifs. While m6A-methylation appears to be a common phenomenon in transcribed RNAs associated with transcription regulation and disease conditions, it displays temporal and functional specificity. Our study highlights a fundamental reference of epitranscriptomic regulation in the developing cortex.
Besides the cerebral cortex, m6A-methylation occurs in the whole brain, including the cerebellum, suggesting a common epitranscriptomic modification in the central nervous system [39,40,41]. Dynamic m6A modification is reflected not only in region-specificity between the cerebellum and cerebral cortex [39], but also in temporal-specificity. Through comparing m6A-methylation between embryonic and postnatal cortices, we have found a generally higher methylation status in embryonic cortices than in postnatal cortices, which may reflect a more methylated gene expression regulation in embryonic neural progenitors and differentiated neurons [15]. Moreover, m6A-methylation displays distinct motifs in RNAs from embryonic and postnatal cortices. Despite this difference, the core sequence of m6A motifs (GGAC) in cortices at each stage is in accordance with previous studies of the mouse cerebellum and cortex [39]. Our finding suggests that temporal methylation of m6A is likely achieved by binding to specific sites in transcribed RNAs specifically functioning in neural progenitors and mature neurons. Finally, while m6A-methylation occurs in the coding sequence in an RNA, it appears more frequently near the stop codon and in the 3′ UTRs. Our finding is consistent with previous reports of m6A binding sites [40,42].
We also found that m6A-methylated RNAs display distinct functions in embryonic and postnatal cortices. While genes specifically methylated in embryonic cortices function in cell differentiation, DNA repair and cell cycle, those in postnatal cortices play roles in transport and synapse assembly. Genes methylated at both stages function mostly in chromatin modification, nucleus and membrane structures, which represent fundamental cellular functions. Similarly, m6A-modified RNAs in the cerebellum are functionally enriched in cell cycle and DNA repair at early developing stages and changed to signal transduction and synaptic plasticity at later developing stages [41]. These studies and ours reveal that RNA methylation is in parallel with both cortical and cerebellar developmental progresses, with gene expression patterns that control these steps. An interesting question is that whether RNA methylation is a default event that occurs in dynamically expressed genes or an internal drive that participates temporal gene expression regulation. Future work will be to decipher RNA methylation-mediated gene expression phenomenon.
Genetic studies of depleting either m6A methyltransferases, such as METTL3 and METTL14 or demethylase FTO (fat mass and obesity associated protein), have demonstrated their important roles in controlling cortical development and regulating brain cognitive functions [16,43,44]. Our study has provided a strong reference supporting the cause of those phenotypes. For example, PAX6 and TBR2 control proliferation of neural progenitors, and TBR1 and SATB2 regulate migration and maturation of postmitotic neurons [45,46,47]. We found that the RNAs encoding these transcription factors are harboring m6A sites, even though the position and number of methylation sites are different among them. Moreover, Par3 and Par6 are involved in the hippo-YAP signaling pathway and function in controlling polarity of cortical neural progenitors [48,49]. Our study has demonstrated m6A-methylation in these and other key genes in the hippo pathway. Furthermore, we found that major genes associated with neurological disorders also are methylated at the m6A site. Even though genetic mutations in these genes are associated with disorders, such as microcephaly-related genes, it is unclear why m6A-methylation seems to be a common event in them and how it may modulate proper expression and translation of these disease genes.
5. Conclusions
Our study has uncovered an image of m6A-methylation in the mouse developing cortex and revealed the basic rules of m6A-methylation in different genes highly expressed in the cortex. Our finding has provided an informative reference for future mechanistic examinations of m6A methylation-mediated gene expression regulation in normal brain development and neurological disorders.
Acknowledgments
This work was supported by the Scientific Research Funds of Huaqiao University (Grant No. Z16Y0023 (L.B.Z.) and No. Z16Y0017 (T.S.)), the National Natural Science Foundation of Fujian Province, China (Grant No. 2018J05058), and the National Natural Science Foundation of China (Grant No. 81701132 and Grant No. 31771141).
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/10/1139/s1, Figure S1: The Pearson correlation of read counts, Figure S2: The status of m6A summits and sequencing replications, Figure S3: The correlation between the up-/down-regulation of RNAs and the positions of m6A, Figure S4: The status and distribution of temporally methylated RNAs counted in each chromosome, Figure S5: The distribution of m6A summits in relative position of a gene, Figure S6: The collection of the most impacted KEGG terms, Figure S7: E-SMRs (NSC and 3’ UTR restricted) involved in the hippo signaling pathway and Hedgehog signaling pathway, Figure S8: P-SMRs (NSC and 3’ UTR restricted) regulate multi chemical signals transport via relative synapse pathway, Figure S9: Three examples for status of m6A-methylation in genes from triplicated samples Table S1: The collection of all detected methylation RNAs in E12.5-E13 and P14 cortices, respectively, Table S2: The GO and KEGG analysis of E-SMRs, P-SMRs and CMRs (NSC and 3’ UTR restricted), Table S3: The numbers of m6A site in targeted genes shown in Figure 4 and Figure 5, Table S4: Genes encoding transcript factors expressing in cortex, temporal- and spatial-specifically methylated, Table S5: Pathogenic genes of nervous-systamic disorders with temporal- and spatial-specific methylation form, Table S6: The IGV performance of all risk genes of brain disorders with temporal-specific methylation in NSC and 3’ UTR region.
Author Contributions
T.S. and L.Z. conceived and designed the study. L.Z. performed major experiments and analyzed the data. K.D. and J.W. assisted the experimental performance. Y.N. performed the transcriptome analysis. L.Z., T.L. and T.S. interpreted the data and wrote the paper, and all authors provided feedback. All authors have read and agreed to the published version of the manuscript.
Funding
The National Natural Science Foundation of China (Grant No. 81701132 and No. 31771141); the National Natural Science Foundation of Fujian Province, China (Grant No. 2018J05058); the Scientific Research Funds of Huaqiao University (Grant No. Z16Y0023 (L.B.Z.) and No. Z16Y0017 (T.S.)).
Conflicts of Interest
The authors declare that there is no conflict of interest associated with this study.
Data Availability
The raw RNA-seq data (GSE141938) presented in this study can be downloaded from NCBI’s GEO database.
References
- 1.Sun T., Hevner R.F. Growth and folding of the mammalian cerebral cortex: From molecules to malformations. Nat. Rev. Neurosci. 2014;15:217–232. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franco S.J., Muller U. Shaping our minds: Stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ismail F.Y., Fatemi A., Johnston M.V. Cerebral plasticity: Windows of opportunity in the developing brain. Eur. J. Paediatr. Neurol. 2017;21:23–48. doi: 10.1016/j.ejpn.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Christopherson K.S., Ullian E.M., Stokes C.C., Mullowney C.E., Hell J.W., Agah A., Lawler J., Mosher D.F., Bornstein P., Barres B.A. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 5.Parikshak N.N., Luo R., Zhang A., Won H., Lowe J.K., Chandran V., Horvath S., Geschwind D.H. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozpedek W., Pytel D., Poplawski T., Walczak A., Gradzik K., Wawrzynkiewicz A., Wojtczak R., Mucha B., Diehl J.A., Majsterek I. Inhibition of the PERK-dependent Unfolded Protein Response signaling pathway involved in the pathogenesis of Alzheimer’s disease. Curr. Alzheimer Res. 2019;16:209–218. doi: 10.2174/1567205016666190228121157. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Mubarak T., Chen Y., Lee T., Pollock A., Sun T. Counter-Balance Between Gli3 and miR-7 Is Required for Proper Morphogenesis and Size Control of the Mouse Brain. Front. Cell. Neurosci. 2018;12:259. doi: 10.3389/fncel.2018.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu X.L., Wang Y., Shen Q. Epigenetic control on cell fate choice in neural stem cells. Protein Cell. 2012;3:278–290. doi: 10.1007/s13238-012-2916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satterlee J.S., Basanta-Sanchez M., Blanco S., Li J.B., Meyer K., Pollock J., Sadri-Vakili G., Rybak-Wolf A. Novel RNA modifications in the nervous system: Form and function. J. Neurosci. 2014;34:15170–15177. doi: 10.1523/JNEUROSCI.3236-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong X., Gong Z., Zhang L., Sun X., Ou Z., Xu B., Huang J., Long D., He X., Lin X., et al. JAK2/STAT3 signaling mediates IL-6-inhibited neurogenesis of neural stem cells through DNA demethylation/methylation. Brain Behav. Immun. 2019;79:159–173. doi: 10.1016/j.bbi.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 11.Fan G., Martinowich K., Chin M.H., He F., Fouse S.D., Hutnick L., Hattori D., Ge W., Shen Y., Wu H., et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 12.Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y.E. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roignant J.Y., Soller M. m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Widagdo J., Anggono V. The m6A-epitranscriptomic signature in neurobiology: From neurodevelopment to brain plasticity. J. Neurochem. 2018;147:137–152. doi: 10.1111/jnc.14481. [DOI] [PubMed] [Google Scholar]
- 15.Boles N.C., Temple S. Epimetronomics: m6A Marks the Tempo of Corticogenesis. Neuron. 2017;96:718–720. doi: 10.1016/j.neuron.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L., et al. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell. 2017;171:877–889. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M., Zhao X., Wang W., Shi H., Pan Q., Lu Z., Perez S.P., Suganthan R., He C., Bjørås M., et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19:69. doi: 10.1186/s13059-018-1436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng Y.L., Wang X., An R., Cassin J., Vissers C., Liu Y., Liu Y., Xu T., Wang X., Wong S.Z., et al. Epitranscriptomic m(6)A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron. 2018;97:313–325.e6. doi: 10.1016/j.neuron.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Mao J., Wang X., Lin Y., Hou G., Zhu J., Xie B. Genome-wide screening of altered m6A-tagged transcript profiles in the hippocampus after traumatic brain injury in mice. Epigenomics. 2019;11:805–819. doi: 10.2217/epi-2019-0002. [DOI] [PubMed] [Google Scholar]
- 20.Hess M.E., Hess S., Meyer K.D., Verhagen L.A., Koch L., Bronneke H.S., Dietrich M.O., Jordan S.D., Saletore Y., Elemento O., et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013;16:1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 21.McTaggart J.S., Lee S., Iberl M., Church C., Cox R.D. Ashcroft FM. FTO is expressed in neurones throughout the brain and its expression is unaltered by fasting. PLoS ONE. 2011;6:e27968. doi: 10.1371/journal.pone.0027968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X., Yu C., Guo M., Zheng X., Ali S., Huang H., Zhang L., Wang S., Huang Y., Qie S., et al. Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem. Neurosci. 2019;10:2355–2363. doi: 10.1021/acschemneuro.8b00657. [DOI] [PubMed] [Google Scholar]
- 23.Du T., Rao S., Wu L., Ye N., Liu Z., Hu H., Xiu J., Shen Y., Xu Q. An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect. Disord. 2015;183:279–286. doi: 10.1016/j.jad.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bogler O., et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominissini D., Moshitch-Moshkovitz S., Amariglio N., Rechavi G. Transcriptome-Wide Mapping of N(6)-Methyladenosine by m(6)A-Seq. Methods Enzymol. 2015;560:131–147. doi: 10.1016/bs.mie.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Mclntyre A.B.R., Gokhale N.S., Cerchietti L., Jaffrey S.R., Horner S.M., Mason C.E. Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci. Rep. 2020;10:6590. doi: 10.1038/s41598-020-63355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 28.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Buchsbaum I.Y., Cappello S. Neuronal migration in the CNS during development and disease: Insights from in vivo and in vitro models. Development. 2019;146 doi: 10.1242/dev.163766. [DOI] [PubMed] [Google Scholar]
- 31.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. Topology of the human and mouse m6A RNA methylations revealed by m6A seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 32.Kandilya D., Maskomani S., Shyamasundar S., Tambyah P.A., Shiao Yng C., Lee R.C.H., Hande M.P., Mallilankaraman K., Chu J.J.H., Dheen S.T. Zika virus alters DNA methylation status of genes involved in Hippo signaling pathway in human neural progenitor cells. Epigenomics. 2019;11:1143–1161. doi: 10.2217/epi-2018-0180. [DOI] [PubMed] [Google Scholar]
- 33.Yabut O.R., Pleasure S.J. Sonic Hedgehog Signaling Rises to the Surface: Emerging Roles in Neocortical Development. Brain Plast. 2018;3:119–128. doi: 10.3233/BPL-180064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyu J., Kim H.R., Yamamoto V., Choi S.H., Wei Z., Joo C.K., Lu W. Protein phosphatase 4 and Smek complex negatively regulate Par3 and promote neuronal differentiation of neural stem/progenitor cells. Cell Rep. 2013;5:593–600. doi: 10.1016/j.celrep.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel S.H., Camargo F.D., Yimlamai D. Hippo Signaling in the Liver Regulates Organ Size, Cell Fate, and Carcinogenesis. Gastroenterology. 2017;152:533–545. doi: 10.1053/j.gastro.2016.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y.A., Lu C.Y., Cheng T.Y., Pan S.H., Chen H.F., Chang N.S. WW Domain-Containing Proteins YAP and TAZ in the Hippo Pathway as Key Regulators in Stemness Maintenance, Tissue Homeostasis, and Tumorigenesis. Front. Oncol. 2019;9:60. doi: 10.3389/fonc.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray P.A., Fu H., Luo P., Zhao Q., Yu J., Ferrari A., Tenzen T., Yuk D.I., Tsung E.F., Cai Z., et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 38.Mirzaa G.M., Paciorkowski A.R. Introduction: Brain malformations. Am. J. Med. Genet. C Semin. Med. Genet. 2014;166:117–123. doi: 10.1002/ajmg.c.31404. [DOI] [PubMed] [Google Scholar]
- 39.Chang M., Lv H., Zhang W., Ma C., He X., Zhao S., Zhang Z.W., Zeng Y.X., Song S., Niu Y., et al. Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017;7:170166. doi: 10.1098/rsob.170166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma C., Chang M., Lv H., Zhang Z.W., Zhang W., He X., Wu G., Zhao S., Zhang Y., Wang D., et al. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19:68. doi: 10.1186/s13059-018-1435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H., et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao X., Shin Y.H., Li M., Wang F., Tong Q., Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS ONE. 2010;5:e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du K., Zhang L., Lee T., Sun T. m(6)A RNA Methylation Controls Neural Development and Is Involved in Human Diseases. Mol. Neurobiol. 2018;56:1596–1606. doi: 10.1007/s12035-018-1138-1. [DOI] [PubMed] [Google Scholar]
- 45.Englund C., Fink A., Lau C., Pham D., Daza R.A., Bulfone A., Kowalczyk T., Hevner R.F. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Britanova O., de Juan Romero C., Cheung A., Kwan K.Y., Schwark M., Gyorgy A., Vogel T., Akopov S., Mitkovski M., Agoston D., et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 47.Kolk S.M., Whitman M.C., Yun M.E., Shete P., Donoghue M.J. A unique subpopulation of Tbr1-expressing deep layer neurons in the developing cerebral cortex. Mol. Cell. Neurosci. 2006;32:200–214. doi: 10.1016/j.mcn.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 48.Liu W.A., Chen S., Li Z., Lee C.H., Mirzaa G., Dobyns W.B., Ross M.E., Zhang J., Shi S.H. PARD3 dysfunction in conjunction with dynamic HIPPO signaling drives cortical enlargement with massive heterotopia. Genes Dev. 2018;32:763–780. doi: 10.1101/gad.313171.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou P.J., Xue W., Peng J., Wang Y., Wei L., Yang Z., Zhu H.H., Fang Y.X., Gao W.Q. Elevated expression of Par3 promotes prostate cancer metastasis by forming a Par3/aPKC/KIBRA complex and inactivating the hippo pathway. J. Exp. Clin. Cancer Res. 2017;36:139. doi: 10.1186/s13046-017-0609-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq data (GSE141938) presented in this study can be downloaded from NCBI’s GEO database.