Abstract
The recent outbreak of SARS-CoV-2 disease, also known as COVID-19, has emerged as a pandemic. The unavailability of specific therapeutic drugs and vaccines urgently demands sincere efforts for drug discovery against COVID-19. The main protease (Mpro) of SARS-CoV-2 is a critical drug target as it plays an essential role in virus replication. Therefore for the identification of potential inhibitors of SARS-CoV-2 Mpro, we applied a structure-based virtual screening approach followed by molecular dynamics (MD) study. A library of 686 phytochemicals was subjected to virtual screening which resulted in 28 phytochemicals based on binding energy. These phytochemicals were further subjected to drug-likeness and toxicity analysis, which resulted in seven drug-like hits. Out of seven, five phytochemicals viz., Mpro-Dehydrtectol (-10.3 kcal/mol), Epsilon-viniferin (-8.6 kcal/mol), Peimisine (-8.6 kcal/mol), Gmelanone (-8.4 kcal/mol), and Isocolumbin (-8.4 kcal/mol) were non-toxic. Consequently, these phytochemicals are subjected to MD, post MD analysis, and MM/PBSA calculations. The results of 100 ns MD simulation, RMSF, SASA, Rg, and MM/PBSA show that Epsilon-viniferin (-29.240 kJ/mol), Mpro-Peimisine (-43.031 kJ/mol) and Gmelanone (-13.093 kJ/mol) form a stable complex with Mpro and could be used as potential inhibitors of SARS-CoV-2 Mpro. However, further investigation of these inhibitors against Mpro receptor of COVID-19 is needed to validate their candidacy for clinical trials.
Communicated by Ramaswamy H. Sarma
Keywords: Coronavirus, Mpro, COVID-19, molecular docking, molecular dynamic simulation
1. Introduction
SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2), which is known as Corona Virus Disease 2019 (COVID-19), was first identified in Wuhan, China, in December 2019 (Hui et al., 2020; Wang et al., 2020). The coronavirus spreads very rapidly from human-to-human by saliva droplets or nose discharge through coughs or sneezes of an infected person. It has globally infected over 11,113,345 individuals, with 526,764 deaths, dated July 3, 2020, as reported by WHO (World Health Organization, 2020). SARS-CoV-2 has been declared an international public health emergency and advocated rapid research efforts (Benvenuto et al., 2020; Ceraolo & Giorgi, 2020; Ren et al., 2020). There is no specific vaccine or effective drug or treatment for SARS-CoV-2 infection yet. The present condition puts the whole world under high pressure to produce a new vaccine or therapy against it (Huang et al., 2020).
For the control of viral infection, the inhibition of replication of viral genomic material is a well-known strategy (De Clercq, 2002). The reproduction of this virus in the host cell can be stopped by inhibiting the cleavage process. The viral genome codes for several non-structural proteins (NSP) including 3-chymotrypsin-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), and its helicase, papain-like protease (papLpro), the structural glycoprotein, and accessory proteins (Boopathi et al., 2020). Main protease (Mpro) has been reported as a potential drug target for SARS CoV-2 because it cuts the two replicate polyproteins required to mediate viral replication and transcription. Hence, Mpro can be used as a target for drug discovery to identify novel inhibitors of SARS-CoV-2. To discover a drug, the use of the computational technique for screening natural inhibitor is gaining attention among the researchers (Aanouz et al., 2020). Some natural compounds such as Baicalein, Luteolin, Quercetin, and Kaempferol are potential antiviral agents against many vital viruses, including Dengue, HIV, H5N1 influenza A virus, Coxsackie virus, CHIKV, and Japanese encephalitis virus (Habbu et al., 2009). So screening the natural compounds from medicinal plants is an excellent approach to find potential inhibitors against Mpro of SARS-CoV-2. Therefore, we selected the medicinal plants used in Chyawanprash for screening inhibitors of Mpro.
In the Ayurvedic system of India, many herbal formulations (i.e. Chyawanprash) are used for boosting the immune system. Similarly, many other plants are known to increase immunity, and their compounds can be used against the coronavirus. Chyawanprash is one of the most appreciated herbal preparations for boosting the immune system for all age groups (Parle & Bansal, 2006). It is formulated by processing around 40 such medicinal herbs and possesses multiple health benefits. Chyawanprash has been widely used since ancient times as a health supplement that acts as a medicine for enhancing immunity and longevity. Ministry of AYUSH reported Chyawanprash as an immunity enhancer and suggested 10 gm (1tsf) Chyawanprash every day in the morning to prevent the coronavirus infection. (https://www.mohfw.gov.in/pdf/ImmunityBoostingAYUSHAdvisory.pdf).
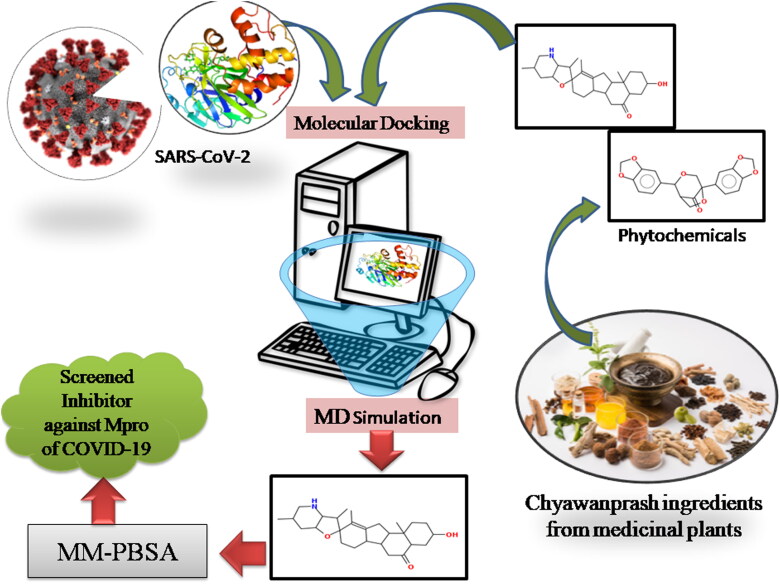
Thus, to find novel SARS-CoV-2 Mpro inhibitors, this study was focused on virtual screening of phytochemicals from medicinal plants used in the preparation of Chyawanprash, which have been reported for their antifungal, antibacterial, and antiviral activities. In light of these kinds of literature, we examined the potential natural compounds against SARS-CoV-2 by using bioinformatics tools. Therefore we used Main Protease (306 amino acids), essential for the functional polypeptide generation and virus survival in host cells, as a target in this Figure 1.
Figure 1.
Summary of work used in the present study to identify the phytochemical based inhibitors of SARS-CoV-2 Mpro.
2. Materials and methods
2.1. Construction of phytochemical library
The library of phytochemicals of medicinal plants used in Chyawanprash was constructed by text mining analysis using DLAD4U (Disease List Automatically Derived For you), PubTator, and Carrot2 servers. Text mining results showed that these medicinal plants have anti-inflammatory, anti-diabetic, anti-cancer, antimicrobial, antifungal, and potential antiviral activity. Hence, to screen out antiviral compounds against coronavirus, a library of 686 phytochemicals from 40 medicinal plants which are used in Chyawanprash was constructed by searching the scientific literature and by using the PubChem database (Supplementary Table 1).
Table 1.
Binding Energy and compound identification number of screened phytochemicals.
| S.no | Name of plants | Phytochemicals | Docking energy (kcal/mol) | Compound Identification number |
|---|---|---|---|---|
| 1. | Mpro (6W63) | (X77) | −8.4 | |
| 2. | Phyllanthus emblica | Emblicanin B | −15.9 | CID 119058017 |
| 3. | Vitis vinifera | Vitilagin | −13.3 | CID 131752915 |
| 4. | Terminalia chebula | Chebulanin | −12.9 | CID 102004757 |
| 5. | Phyllanthus emblica | Punigluconin | −12.8 | CID 44631480 |
| 6. | Mesua ferrea | Mesuabixanthone A | −11.7 | CID 102149342 |
| 7. | Mesua ferrea | Mesuaferrol | −11.2 | CID 101994076 |
| 8. | Phyllanthus amarus | Rutin | −10.8 | CID 73568 |
| 9. | Stereospermum suaveolens | Dehydrotectol | −10.2 | CID 3037329 |
| 10. | Phyllanthus emblica | Phyllanemblinin B | −9.5 | CID 10941235 |
| 11. | Phyllanthus amarus | Furosin | −9.4 | CID 10416810 |
| 12. | Tinospora cordifolia | Tinocordiside | −9.2 | CID 102504931 |
| 13. | Mesua ferrea | Mesuaferrone B | −9.1 | CID 90472563 |
| 14. | Terminalia chebula | Casuarin | −9.1 | CID 442673 |
| 15. | Solanum indicum | Solanine | −9.1 | CID 262400 |
| 16. | Mesua ferrea | Mesuaferrone A | −8.9 | CID 101324837 |
| 17. | Vitis vinifera | Proanthocyanidins | −8.9 | CID 21881649 |
| 18. | Leptadenia reticulate | Luteolin | −8.8 | CID 5280637 |
| 19. | Tribulus terrestris | Neohecogenin | −8.6 | CID 90473944 |
| 20. | Tinospora cordifolia | Tinosporin | −8.8 | CID 442068 |
| 21. | Lilium polyphyllum | Peimisine | −8.6 | CID 161294 |
| 22. | Vitis vinifera | epsilon-viniferin | −8.6 | CID 5281728 |
| 23. | Phyllanthus emblica | Verbascoside | −8.5 | CID 5281800 |
| 24. | Terminalia chebula | Terchebulin | −8.5 | CID 131675995 |
| 25. | Leptadenia reticulate | Apigenin | −8.5 | CID 5280443 |
| 26. | Martynia annua | Cyaniding −3-galactoside | −8.5 | CID 44256700 |
| 27. | Tinospora cordifolia | Isocolumbin | −8.4 | CID 24721165 |
| 28. | Gmelina arborea | Gmelanone | −8.4 | CID 21722946 |
| 29. | Terminalia chebula | Chebulinic acid | −8.4 | CID 72284 |
2.2. Protein and ligand preparation
The three-dimensional crystal structure of the Mpro (PDB ID 6W63) receptor was retrieved from the Protein Data Bank (https://www.rcsb.org). For the screening, the protein structure was prepared by using the software MGL Tools and PyMOL. The protein preparation involved in two steps. Firstly, protein was prepared by removing all water molecules using PyMOL software. Further hydrogen atoms were added into the receptor by using MGL Tools (Trott & Olson, 2010). The structure of the protein was saved in pdbqt format for further analysis. The 3 D structure of each phytochemical and reference molecule X77 was downloaded from PubChem [https://pubchem.ncbi.nlm.nih.gov] in SDF format and then converted into PDB format using Open Babel GUI (O'Boyle et al., 2011).
2.3. Molecular docking and visualization
Before the docking, the center of mass of co-crystallized ligand with receptor was calculated by using the "centerofmass" command in PyMOL software to retrieve x y and z coordinates of the reference compound. The coordinates of the centerofmass of X77 were X= −23.315, Y = 13.893, and Z= −29.698, which was used for docking with grid box size 30 × 30 × 30 Å. The same grid configuration was used for virtual screening of all phytochemicals using Mpro protein using PyRx open software with AutoDockVina keeping ligand molecules flexible throughout the process. After docking, the 2 D interactions of the protein-ligand complexes, including hydrogen bonds and the bond lengths, were analyzed and depicted by using Ligplot + v.1.4.5 software.
2.4. Drug likeness and toxicity prediction
Drug-likeness and toxicity prediction of the screened compounds were predicted by DruLiTo and OSIRIS software (Mabkhot et al., 2016). DruLiTo indicates various properties such as drug-likeness, water solubility (LogS) etc. of a compound. OSIRIS open-source software was used to predict drug toxicity and risk properties like mutagenicity, tumorigenicity, irritation, reproductive development and drug score. All screened ligands were evaluated for their drug-like nature under Lipinski's rules of five; 'RO5 (Lipinski, 2000).
2.5. Molecular dynamics (MD) simulation
The obtained docked complexes were subjected to MD simulation using GROMACS 5.0.7 to examine the insight behavior of the protein-ligand complexes (Pronk et al., 2013). The CHARMM 36 force field was applied to build the topology of ligand and protein (Vanommeslaeghe et al., 2009). The dynamic behavior of all protein-ligand complexes was observed in the presence of explicit water molecules after solvation with the TIP3P water model in an orthorhombic periodic boundary box. To prevent interaction of the protein-ligand complexes with the distance between the complex and the box wall was kept 10 Å and then add ions for neutralized the system. The energy of the prepared systems was minimized for 40,000 steps. The equilibrated system was then subjected to 10 ps simulation in NPT ensemble with 300 K temperature, constant pressure of 1 atm, and a time step of 2fs. Further, MD simulation was performed at a constant temperature of 300 K and a constant of 1 atm pressure using two fs for a 100 ns time scale. After successful completion of 100 ns MD simulation, the Root mean square deviation (RMSD), Root mean square fluctuation (RMSF), Radius of Gyration (Rg), SASA, and H-bond were calculated. This was done to quantify the strength of the interaction between protein-ligand complexes. We also computed the non-bonded interaction energy between protein and ligands with the same parameter as MD simulation.
2.6. Binding free energies
The binding free energy of the complexes was determined by the molecular mechanics' method to analyze complex stability. Therefore, the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) (Shukla & Singh, 2019) was used for calculating the binding free energy of protein-ligand complexes. The whole process was as summarized in the below equation:
Finally, the MM-G/PBSA value of the protein-ligand complex was calculated by summing the gas-phase electrostatic energy (Eele), van der Waals (EvdW), polar (Gpolar), and nonpolar (Gnonpolar) components.
3. Results
3.1. Virtual screening
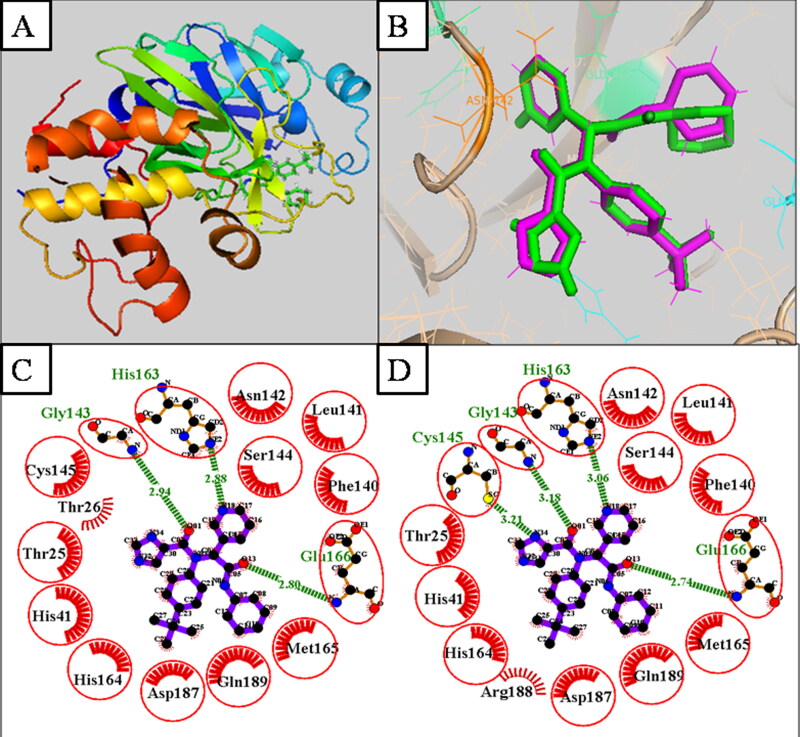
Before the virtual screening, docking of reference compound X77, the co-crystallized ligand of Mpro (Figure 2A), was validated by the docking protocol. The result revealed that the docked reference X77 (Green) was completely superimposed with a co-crystallized X77 (Purple) (Figure 2B) and the RMSD of the superimposed structure is 0.62 Å. The 2 D interaction of active site residue of experimental reference (X77) viz., Ser46, Met49, Pro52, Tyr54, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His163,Thr25, Thr26, Leu27, His41, Cys44, Thr45, His164, Met165, Glu166, Leu167, Pro168, His172, Asp187, Arg188, and Gln189 similar to docked reference (X77) of Mpro (Figure 2C and D).
Figure 2.
(A.) 3 D structure of Mpro (6W63) with the co-crystalized ligand. (B.) Superimposition of docked (X77) (Green color) and Experimental reference (X77), (Purple color) by 3 D structure. (C.)Docked reference (X77) of Mpro. (D.) Experimental reference (X77) of Mpro.
After validation of the docking protocol, all 686 phytochemicals were docked on the active site of Mpro by using the default parameter of the active site. Docking results reveal that 28 phytochemicals out of 686 phytochemicals have binding energy from −15.9 kcal/mol to −8.4 Kcal/mol, which is lower or equivalent to binding energy the reference (X77) (-8.4 kcal/mol) (Table 1). Therefore, 28 phytochemicals were screened as potential inhibitors. These phytochemicals belong to following plants; Phyllanthus emblica, Vitis vinifera, Terminalia chebula, Mesua ferrea, Phyllanthus amarus, Stereospermum suaveolens, Tinospora cordifolia, Solanum indicum, Tribulus terrestris, Lilium polyphyllum, Leptadenia reticulate, Gmelina arborea, and Martynia annua. Screened phytochemicals of medicinal plants, their compound identification number, and binding energy are given in Table 1. These 28-screened phytochemicals were used for further analysis to find the potential inhibitors against the Mpro enzyme.
3.2. Drug Likeness analysis
After the screening of docking, all twenty-eight phytochemicals were applied for drug-likeness prediction, in which only seven phytochemicals viz. peimisine, apigenin, dehydrotectol, tinosporin, isocolumbin, epsilon-viniferin, and gmelanone showed better pharmacokinetics. As per RO5, all seven phytochemicals showed Log p ≤ 5, molecular weight ≤ 400, number of (HBA) hydrogen bond acceptors ≤ 10, and the number of (HBD) hydrogen bond donors ≤ five and Lipinski rule of violation were 0 as compared to the reference, and they successfully passed in RO5 evaluation. The compounds, which showed better pharmacokinetics and satisfied the fundamental RO5 accepted as drug-like molecules. As per RO5, the drug-likeness result of screened phytochemicals has been given in Table 2.
Table 2.
The physicochemical properties of screened phytochemicals by DruLiTo.
| S.no | Name of phytochemicals | Molecular weight |
LogP | HBA | HBD | Lipinski Rule Violation |
Drug likeness alert |
|---|---|---|---|---|---|---|---|
| 1. | Reference (X77) | 430.04 | 3.634 | 7 | 4 | 0 | Accepted |
| 2. | Dehydrotectol | 423.98 | 3.95 | 4 | 0 | 0 | Accepted |
| 3. | Tinosporin | 352.97 | 1.251 | 7 | 1 | 0 | Accepted |
| 4 | Peimisine | 388.0 | 2.624 | 4 | 2 | 0 | Accepted |
| 5. | Epsilon-viniferin | 437.01 | 2.728 | 6 | 5 | 0 | Accepted |
| 6. | Apigenin | 263.0 | −1.043 | 5 | 0 | 0 | Accepted |
| 7. | Isocolumbin | 336.98 | 2.293 | 6 | 1 | 0 | Accepted |
| 8. | Gmelanone | 351.98 | 0.548 | 7 | 0 | 0 | Accepted |
3.3. Toxicological properties analysis
Since the toxic potential can significantly impact the discovery and development of drugs, the toxicological properties were analyzed by the OSIRIS server. The various toxicity properties, such as mutagenicity, carcinogenicity, and irritation, showed mostly acceptable parameters (Table 3).
Table 3.
Toxicity prediction of the screened phytochemicals of Chyawanprash by OSIRIS.
| S.No | Name of phytochemicals | Mutagenic | Tumorigenic | irritant | Water Solubility (LogS) |
Drug score |
|---|---|---|---|---|---|---|
| 1. | Reference (X77) | No risk | No risk | No risk | −4.74 | 0.31 |
| 2. | Peimisine | No risk | No risk | No risk | −4.91 | 0.57 |
| 3. | Apigenin | Medium risk | No risk | Medium risk | −2.41 | 0.3 |
| 4. | Dehydrotectol | No risk | No risk | No risk | −5.49 | 0.12 |
| 5. | Tinosporin | No risk | High risk | High risk | −3.19 | 0.27 |
| 6. | Isocolumbin | No risk | No risk | No risk | −3.51 | 0.45 |
| 7. | Epsilon-viniferin | No risk | No risk | No risk | −4.88 | 0.17 |
| 8. | Gmelanone | No risk | No risk | No risk | −4.29 | 0.61 |
The drug score was between the ranges of 0 to 1. All screened phytochemicals drug scores were less than 1; they indicate the excellent possibility of being drug candidates. The toxicity test showed that the reference molecule, X77, and five phytochemicals, Peimisine, Isocolumbin, Epsilon-viniferin, Dehydrotectol, and Gmelanone, had no risk of toxicity, and the remaining two compounds were toxic. Apigenin showed medium risk with mutagenic, while tinosporin showed high risk with tumorigenic and irritation. Log S refers to the water solubility of the phytochemicals that ideally ranges between −6.5 and 0.5. Apigenin showed a higher LogS value (-2.40), but it showed medium risk mutagenicity and irritation. Dehydrotectol showed a minimum LogS value (-5.49) as compared to other complexes. All the phytochemicals were acceptable range. Finally, five phytochemicals, Peimisine, Isocolumbin, Epsilon-viniferin, Dehydrotectol, and Gmelanone show no risk with mutagenicity, carcinogenicity, irritation, and the value of LogS and drug scores are in the acceptable range.
3.4. 2 D Visualization of the docked complex
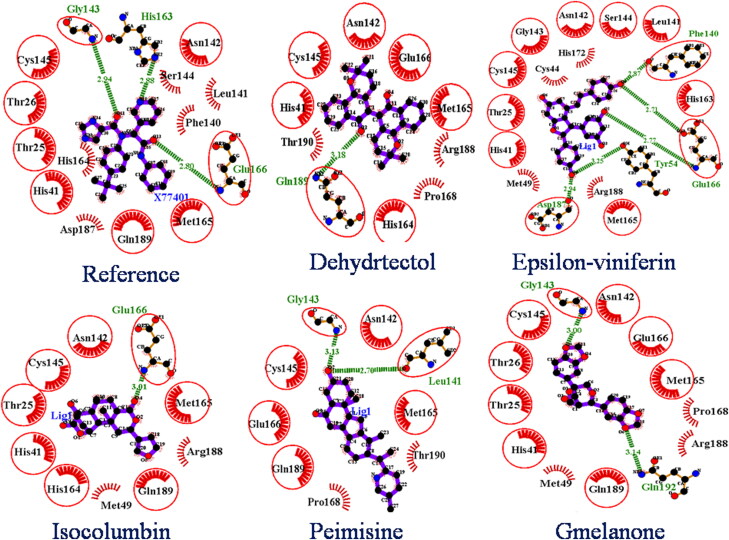
From the molecular docking, drug-likeness, and toxicity prediction, Peimisine, Gmelanone, and Isocolumbin were found as potential inhibitors of SARS-CoV-2 Mpro. The 2 D interactions of the reference molecule and all screened phytochemicals were analyzed by using LigPlot + v.1.4.5 software and are shown in Figure 3. The reference molecule X77 showed interaction with several residues. X77 forms three hydrogen bonds with Gly143, His163, and Glu166 of 3.24 Å, 3.09 Å, and 2.75 Å respectively, and twelve hydrophobic bonds with His164, Met165, Asp187, Arg188, Gln189 Thr25, His41, Phe140, Leu141, Asn142, Ser144, Cys145, and yields the binding energy −8.4 kcal/mol by docking (Figure 3).
Figure 3.
2 D interaction of reference compounds (X77) and screened phytochemicals. In all phytochemicals, dotted green line indicates the hydrogen bond, red sparked arc represents hydrophobic interaction, and red circle and red ellipses represent common protein residue with reference.
Mpro-Dehydrtectol shows the highest binding affinity (-10.3 kcal/mol) compared to other ligands against Mpro. The Dehydrtectol showed interaction with several residues and formed one hydrogen bond with residue Gln189 of 3.18 Å and also exhibits hydrophobic interaction with Cys145, Glu166, Met165, Thr190, Pro168, Asn142, His164, Arg188, His41.
Mpro-Epsilon-vinifein shows the binding affinity (-8.6 kcal/mol) as compared to other ligands against Mpro. The Epsilon-vinifein formed five hydrogen bonds with residue Phe140, Glu166, Asp187, and Tyr45 of 2.87 Å, 2.71 Å, 2.77 Å, 2.94 Å, and 3.25 Å and exhibited thirteen hydrophobic interactions with Cys145, Gly143, Asn142, Ser144, Leu141, His163, Arg188, Met149, Thr25, Cys44, His172, His41, and Met165.
Mpro-Peimisine shows the highest binding affinity (-8.6 kcal/mol) as compared to other ligands against Mpro. The Peimisine showed interaction with several residues and formed two hydrogen bonds with residue Gly143 and Leu141 of 3.13 Å and 2.76 Å and also exhibits hydrophobic interaction with Cys145, Glu166, Met165, Thr190, Pro168, and Gln189. Mpro-Gmelanone showed a binding affinity of −8.4 Kcal/mol with the Mpro complex. It formed two hydrogen bonds with Gly143, Gln192 with length 3.00 Å, 3.14 Å, and yielded 12 hydrophobic interactions with His 41, Thr26, Thr25, Asn142, Glu166, Met49, Cys145, Met165, Pro168, Asp187, and Arg188, of Mpro receptor. While, binding affinity of Mpro-Isocolumbin was showed −8.4 Kcal/mol from AutoDockVina, which is similar to the reference molecule. It formed one hydrogen bond with Glu166 of 3.01 Å and formed nine hydrophobic interactions with His41, His164, Thr25, Asn142, Met49, Cys145, Met165, Asp187, and Arg188, of Mpro receptor. From the findings, we observed that most of the screened phytochemicals show common interaction and were involved in H-bond interaction and hydrophobic interaction with the same residue, as shown in Figure 3.
3.5. Molecular dynamic simulation
All biological systems can be represented as dynamic networks of molecular interactions, while molecular docking represents only a single glimpse of protein-ligand interaction. Therefore, to sample the various conformations that these complexes might acquire in the solvated state, we simulated the dynamics behavior of all the five complexes (Mpro-Dehydrtectol, Mpro-Epsilon-vinifein, Mpro-Peimisine, Mpro-Gmelanone, and Mpro-Isocolumbin) and reference (Mpro-X77) using TIP3P water models for 100 ns each. We restricted our simulation runs to 100 ns because all the systems included in this study, Mpro protein, native reference (Mpro-X77), and docked complexes viz Mpro-Epsilon-vinifein, Mpro-Peimisine, and Mpro-Gmelanone achieved stabilization within 5–10 ns of the production runs except Mpro-Dehydrtectol and Mpro-Isocolumbin. Mpro-Dehydrtectol and Mpro-Isocolumbin were unstable after 25 ns. The dynamic behaviour of three-screened protein-ligand complexes were analyzed by RMSD, RG, RMSF, SASA, H-bond, and interaction energy are given in Table 4.
Table 4.
The average values of RMSD, RMSF, Rg, SASA, number of H-bond, and Interaction energy for protein-ligand complexes.
| S.no | Phytochemicals | MD simulation |
Post MD simulation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Average Rmsd (nm) |
Average Rmsf (nm) |
Average Rg (nm) |
Average Sasa (nm2) |
H-bond (Å) |
Interaction Energy (kJ/mol) |
|||||
| Average Energy |
Error Estimate |
Rmsd | Total- Drift |
|||||||
| 1. | Native protein(Mpro) | 0.18 ± 0.03 | 0.09 ± 0.02 | 1.88 ± 0.05 | – | – | – | – | – | – |
| 2. | Mpro-reference complex | 0.17 ± 0.02 | 0.13 ± 0.04 | 1.88 ± 0.08 | 149.29 ± 2.17 | 4 | −137.52 | 9.2 | 22.13 | 52.17 |
| 3. | Mpro--Epsilon-vinifein complex | 0.17 ± 0.02 | 0.10 ± 0.03 | 1.89 ± 0.05 | 151.88 ± 2.09 | 6 | −106.305 | 4.3 | 19.27 | 8.81 |
| 4. | Mpro-Peimisine complex | 0.20 ± 0.03 | 0.10 ± 0.03 | 1.89 ± 0.07 | 149.93 ± 2.01 | 3 | −92.94 | 8.3 | 22.06 | 52.44 |
| 5. | Mpro-Gmelanone complex | 0.22 ± 0.03 | 0.13 ± 0.04 | 1.90 ± 0.06 | 156.07 ± 3.49 | 3 | −99.98 | 6.5 | 19.24 | −39.89 |
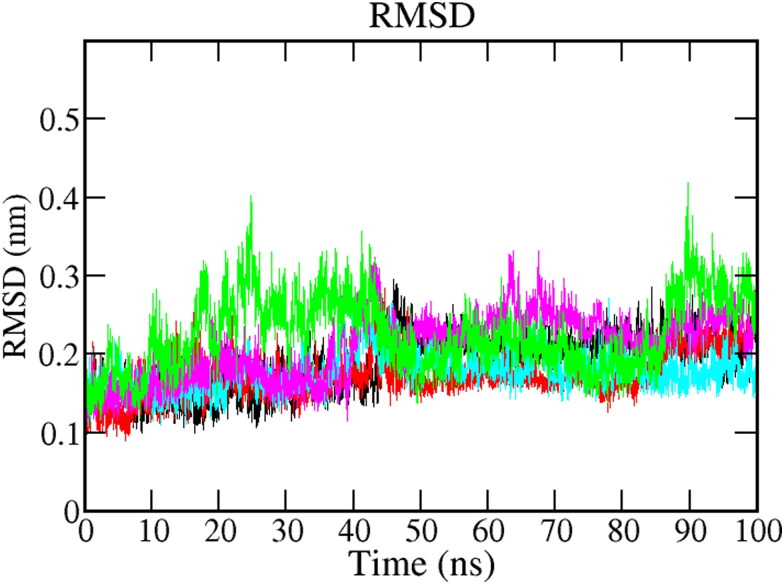
3.5.1. Root mean square deviation (RMSD)
All protein-ligand complexes' configuration changes were studied in terms of Root mean square deviation (RMSD) during the 100 ns MD simulation. We calculated coordinates of the protein backbone and ligand from the coordinates in the initial docked pose by analyzing RMSD. Figure 4 shows the RMSD plot for native protein and all protein-ligand complexes. The RMSD trajectory of protein and all the complexes attained the equilibration in initial ∼5-10 ns of simulation and produced stable trajectories. The native protein showed stability in 100 ns simulation with an average RMSD 0.18 nm (Black). In the case of the bound system, all complexes are stable in the 100 ns simulation trajectory period. The average value of RMSD has calculated 0.17 nm for Mpro-Epsilon-vinifein (cyan), 0.20 nm for Mpro-Peimisine complex (magenta), and 0.22 nm for Mpro-Gmelanone (green), respectively as compared to the reference (X77), 0.17 nm (red) Table 4. The reference complex, Mpro-X77 and Mpro-Epsilon-vinifein complex, showed the same RMSD value, which confirmed the stability of the complexes. In conclusion, the RMSD fluctuation analysis suggests that the MD trajectories were overall stable and in are acceptable range during the simulation period for the entire studied complex.
Figure 4.
RMSD plot of Mpro protein and screened protein-ligand complexes for 100 ns MD Simulation of protein (Black) protein-reference (X77) (Red), Mpro-Epsilon-vinifein (Cyan), Mpro-Peimisine (Magenta), and Mpro-Gmelanone (Green).
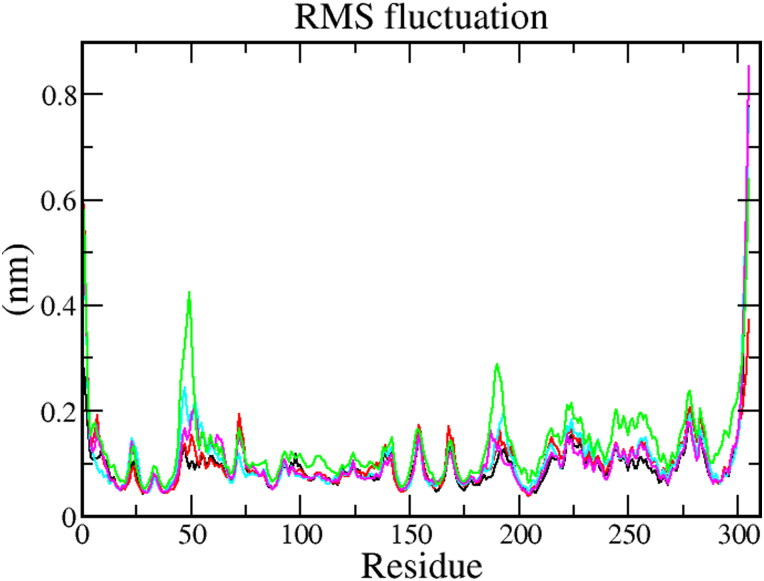
3.5.2. Root mean square fluctuation (RMSF)
RMSF was used to calculate the average fluctuation of each amino acid of the protein. RMSF plot (Figure 5) shows that the secondary conformations of Mpro remain stable during the simulation of 100 ns. The average RMSF value for Mpro protein 0.09 ± 0.02 nm, Mpro-X77complex 0.13 ± 0.04 nm, Mpro-Epsilon-vinifein 0.10 ± 0.03 nm, Mpro-Peimisine complex 0.10 ± 0.03 nm, and Mpro-Gmelanone complex 0.13 ± 0.04 nm respectively (Table 4). The fluctuation during all protein-ligand interaction was below 0.2 nm, which is perfectly acceptable. In conclusion, it indicates that RMSF of all complexes is significantly similar as compared to reference resulted in less fluctuation indicating good stability.
Figure 5.
The RMSF values of the Mpro protein and screened protein-ligand complexes for 100 ns MD Simulation of protein (Black) protein-reference(X77) (Red), Mpro-Epsilon-vinifein (Cyan), Mpro-Peimisine (Magenta), and Mpro-Gmelanone (Green).
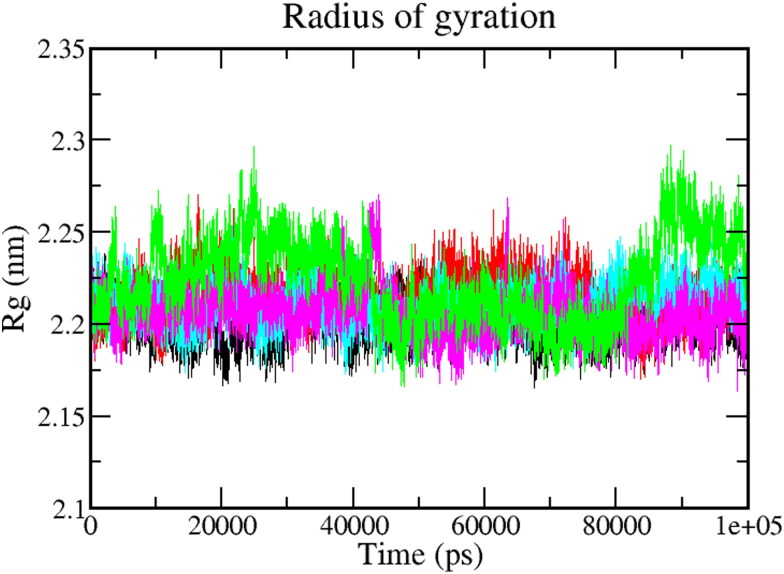
3.5.3. The radius of gyration (Rg)
Rg is used to determine whether the complexes are stably folded or unfolded during the MD simulation. The folding and unfolding of protein structure upon binding of the ligands can be measured by the Radius of gyration (Rg). In conclusion, higher Rg shows less compactness (more unfolded) of the protein-ligand complex. The average Rg value of native protein Mpro was found to be around 1.88 ± 0.05 (Black). Furthermore, the average Rg values of Mpro-Epsilon-vinifein, Mpro-Peimisine, Mpro-Gmelanone, complexes were 1.89 ± 0.05 nm, 1.89 ± 0.07 nm, and 1.90 ± 0.06 nm respectively, which was significantly similar as compared to reference1.88 ± 0.08 nm molecules (Figure 6). As stated earlier, if protein will likely maintain a relatively steady value of Rg throughout the MD simulation, it can be regarded as stably folded. If the value of Rg changes over time, it would be considered unfolded. As a result, each complex exhibited relatively similar behavior of compactness and consistent amounts of Rg as compared to the native protein and reference (Table 4).
Figure 6.
Radius of Gyration plot depicting the changes observed in the conformational behavior of the Mpro protein and all protein-ligand complexes; 100 ns MD Simulation of protein (Black) protein-reference(X77) (Red), Mpro-Epsilon-vinifein (Cyan), Mpro-Peimisine (Magenta), and Mpro-Gmelanone (Green).
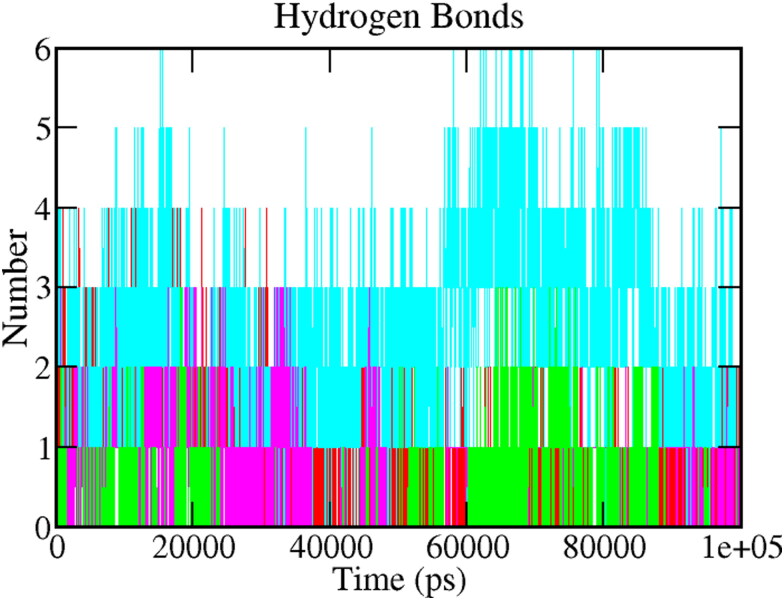
3.5.4. Hydrogen bonds
Hydrogen bonds are very specific interactions between receptor and ligand, which play a crucial role in the stabilization of the protein-ligand complex. It is also responsible for drug specificity, metabolization, and adsorption in drug design. The hydrogen bonds between each ligand-protein complex were also examined. Figure 7 depicts the total number of hydrogen bonds present in the complexes, which were calculated after the 100 ns simulation period. Analyzing the reference complex, around five hydrogen bonds (red) were observed in the complex. In comparison, Mpro-Epsilon-vinifein was found to establish six (cyan) hydrogen bonds; three hydrogen bonds were observed in the Mpro-Peimisine (Magenta) and Mpro-Gmelanone complex (blue), respectively. Through the above detail H-bond analysis, we can conclude that both the compounds were bound to the Mpro as effectively and tightly as reference X77.
Figure 7.
A plot showing hydrogen bonds. The color code for all panels is as follows protein-reference(X77) (Red), Mpro-Epsilon-vinifein (Cyan), Mpro-Peimisine (Magenta), and Mpro-Gmelanone (Green).
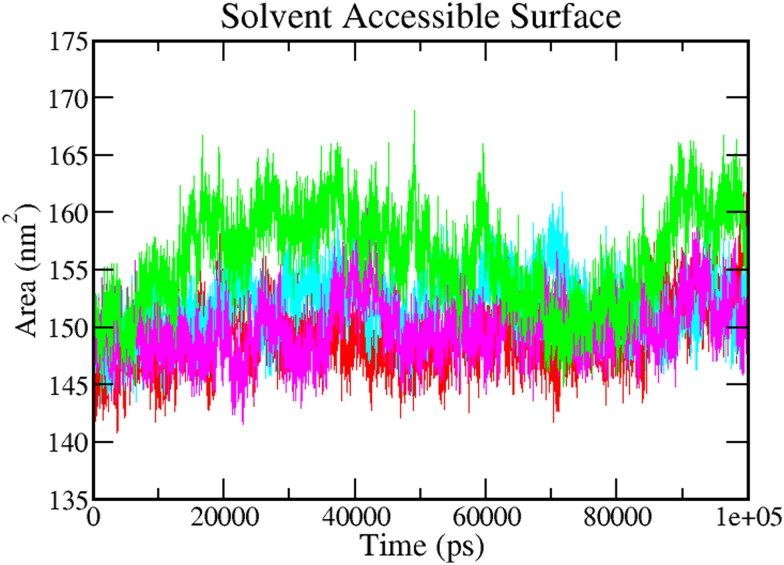
3.5.5. Solvent accessible surface area (SASA)
The calculation of SASA of the protein-ligand complex helps to predict the extent of the conformational changes, which can be accessible by the water solvent. Hence, SASA was carried out to analyze interactions between complex and solvent during the 100 ns MD simulation. The figure shows the plot of SASA value vs. time for all the protein-ligand complexes (Figure 8). The average SASA values for Mpro-Epsilon-vinifein, Mpro-Peimisine, Mpro-Gmelanone were 151.88 ± 2.17 nm2, 149.93 ± 2.01 nm2, and 156.07 ± 2.01 nm2, respectively. The average value was significantly better than the reference (Red), i.e. 149.29 ± 2.17 nm2. The average value of SASA is shown in Table 4. These calculations show that all the complexes have a significantly similar value of SASA as the reference complex.
Figure 8.
Solvent accessible surface area (SASA). The color code for all panels is as follows protein-reference(X77) (Red), Mpro-Epsilon-vinifein (Cyan), Mpro-Peimisine (Magenta), and Mpro-Gmelanone (Green).
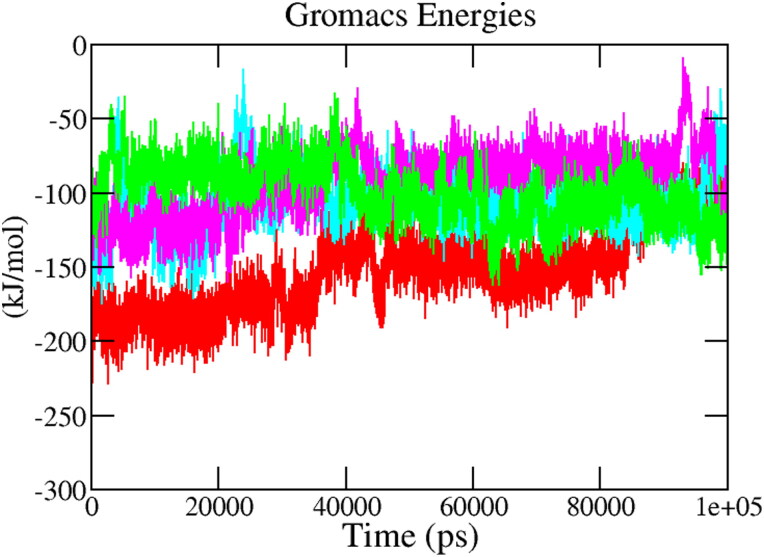
3.5.6. Interaction energy
The interaction energy reflects the strength of the receptor-ligand complex. To validate the binding energy of molecular docking studies, a detailed analysis was performed regarding the calculation of the free interaction energies associated with the binding of hit compounds with the structure of Mpro using the Parrinello–Rahman parameter implemented in GROMACS. Figure 9 displays the average short-range Lennard–Jones energy plot of protein-ligand complexes Mpro-X77(Red), Mpro-Epsilon-vinifein, Mpro-Peimisine, Mpro-Gmelanone for 100 ns of MD Simulation. The average interaction energy of all complexes was observed in the acceptable range of −100 kJ/mol to −200 kJ/mol during the 100 ns simulation period (Table 4). The reference complex, Mpro-X77, shows that the average interaction energy calculated was −186.73 kJ mol-1, hit phytochemicals, i.e. Epsilon-vinifein, Peimisine, and Gmelanone showed similar and comparable interaction energy as the reference compound. Among the complexes, the Mpro- Epsilon-vinifein showed the best average interaction energy, i.e. −106.30 kJ/mol, Mpro-Peimisine had the interaction energy of −92.94 kJ/mol. In comparison, Mpro-Gmelanone had interaction energy of −99.98 kJ/mol. These interaction energy calculations validated the molecular docking results and indicated stable complex formation with Mpro and potential drug candidates against COVID-19.
Figure 9.
Interaction energy. The color code for all panels is as follows protein-reference(X77) (Red), Mpro-Epsilon-vinifein (Cyan), Mpro-Peimisine (Magenta), and Mpro-Gmelanone (Green).
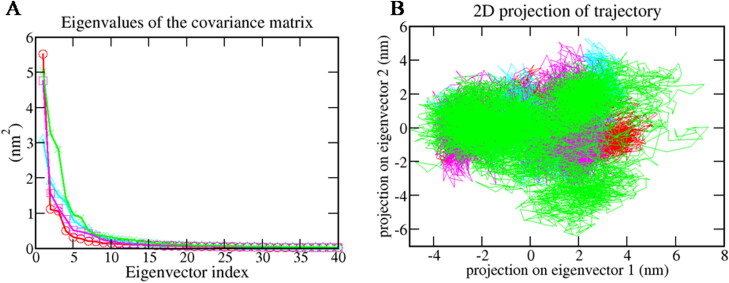
3.6.7. Principal component analysis (PCA)
PCA was carried out to investigate the significant motions during ligand binding. In this study, the matrix diagonalization was used to calculate the eigenvectors and eigenvalues. For this study, the first 40 eigenvectors were selected to calculate concerted motions. Figure 10A represents the eigenvalues that were obtained from the diagonalization of the covariance matrix of atomic fluctuations in decreasing order versus the corresponding eigenvector for Mpro-X77, Mpro-Epsilon-vinifein (Cyan), Mpro-Peimisine (Magenta), and Mpro-Gmelanone (Green).
Figure 10.
Principal component analysis (A.) Plots of eigenvalues vs. first 40 eigenvectors. (B) First two eigenvector describing the protein motion in phase space for all the complexes. The color code for all panels is as follows protein-reference(X77) (Red), Mpro-Epsilon-vinifein (Cyan), Mpro-Peimisine (Magenta), and Mpro-Gmelanone (Green).
It was observed that out of the 40 eigenvectors, the first ten eigenvectors accounted for 84.79% for Mpro-X77(Red), 86.13% for Mpro-Epsilon-vinifein (Cyan), 88.73% of total motions for Mpro-Peimisine (Magenta), and 86.95% for Mpro-Gmelanone (Green) (Figure 10a). All the studied complexes showed very fewer motions as compared with the reference compound. Therefore, we concluded that all complexes had very fewer motions from the PCA analysis and formed a stable complex with Mpro. The 2 D projection plot generation in PCA is another way to achieve the dynamics of complexes. Figure 10b shows the 2 D projection of the trajectories in the phase space for the first two principal components, PC1 and PC2 for Mpro-X77, Mpro-Epsilon-vinifein, Mpro-Peimisine, and Mpro-Gmelanone complexes, respectively. The complex, which occupied less phase space, showed a stable cluster while the complex that occupied more space showed a less-stable cluster. From the figure, the Mpro-Peimisine(Magenta) and Mpro-Gmelanone (green) were highly stable as they occupied less space in the phase space, and the cluster was well defined as compared to Mpro- X77 (Red), and Mpro-Gmelanone, (green) complexes.
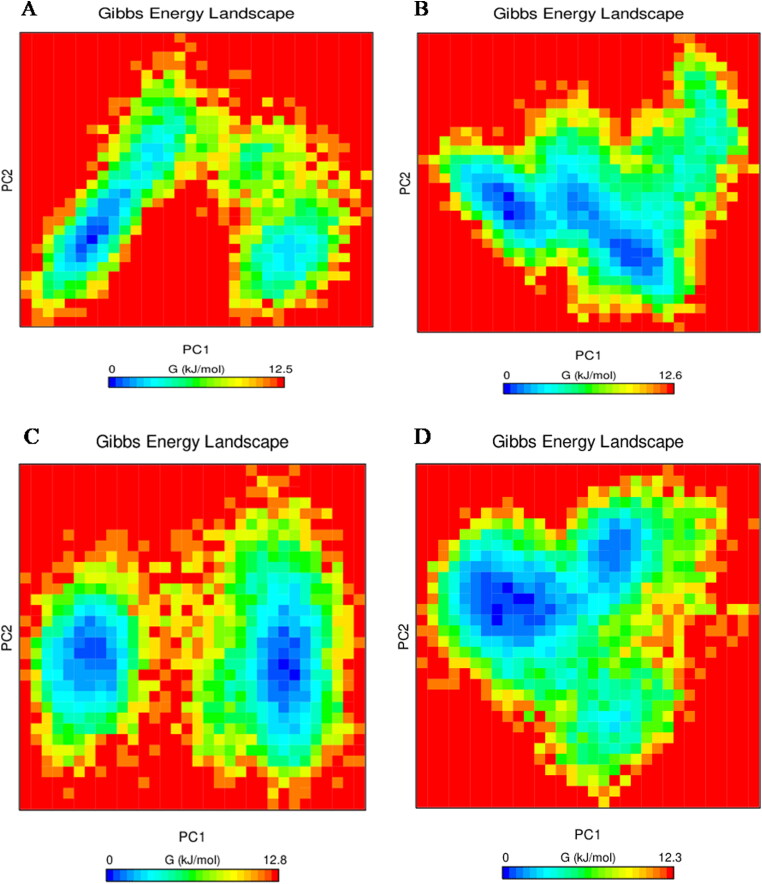
The Gibb's free energy plot for PC1 and PC2 was also calculated and is shown in Figure 11. The plot shows Gibbs free energy value ranging from 0 to 12.5 for Mpro-X77, for Mpro-Epsilon-vinifein Gibbs energy is 0 to 12.6 kJ/mol, Mpro-Peimisine Gibbs energy is 0 to 12.8 kJ/mol, and for Mpro-Gmelanone Gibbs energy is 0 to 12.3 kJ/mol, respectively. Thus screened compounds shows lower energy as compared to the reference with Mpro, which indicates that these complexes follow more favorable transitions energetically from one conformation to another as compared to the reference.
Figure 11.
Gibbs free energy plots (A.) protein-reference(X77), (B.) Mpro-Epsilon-vinifein (C.)Mpro-Peimisine (D) Mpro-Gmelanone.
3.6.8. Protein-ligand binding by MM/PBSA methods
The simulated complex's binding free energy was calculated to revalidate the inhibitor affinity of the Mpro-ligand complexes predicted by the docking simulation studies. The calculations of binding free energies were performed using the last 20 ns of MD trajectories. The summation of the nonpolar energies, polar, and non- bonded interaction energies (electrostatic interaction and Vander Waals), was calculated for all the complexes using the MM-PBSA methods and are shown in Table 5.
Table 5.
Free binding energy calculations of each Mpro and ligand complexes.
| Phytochemicals | Complexes Binding energy (kJ/mol) | van der Waal energy (EvdW) (kJ/mol) |
Electrostattic energy (Eelec) (kJ/mol) |
Polar solvation energy (DG polar) (kJ/mol) |
SASA energy (kJ/mol) |
|---|---|---|---|---|---|
| Mpro-Reference | -29.240 ± 76.632 | -34.067 ± 53.784 | -1.712 ± 3.838 | 10.997 ± 31.210 | -4.458 ± 7.095 |
| Mpro-Epsilon-vinifein | -29.009 ± 72.000 | -29.131 ± 47.694 | -15.576 ± 26.281 | 19.849 ± 106.630 | -4.150 ± 7.000 |
| Mpro-Peimisine | -43.031 ± 40.523 | -79.250 ± 43.186 | -4.320 ± 5.908 | 51.229 ± 16.128 | -10.690 ± 5.420 |
| Mpro-Gmelanone | -13.093 ± 35.373 | -43.559 ± 24.250 | -10.573 ± 9.241 | 47.521 ± 41.183 | -6.482 ± 3.433 |
Table 5 shows the binding energy of Mpro reference (-29.240 kJ/mol), Mpro-Peimisine (-43.031 kJ/mol), Mpro-Epsilon-vinifein (-29.009 kJ/mol), and Mpro Gmelanone (-13.093 kJ/mol), respectively. The van der Waal energy of phytochemicals, when compared with the reference, the Mpro-Epsilon-vinifein (-29.131), had less binding affinity while Mpro-Peimisine (-79.250) Gmelanone (-43.559) showed a strong binding affinity. The electrostatic energy show significant moderate values in the case of Peimisine, Gmelanone, and Epsilon-vinifein. The polar solvation and SASA energy showed moderate effects on binding energy in each complex. The binding energy showed complexes' stability and it suggests two complexes (Peimisine and Epsilon-vinifein) could be used as potential inhibitors against Mpro of SARS- CoV-2.
4. Discussion
The outbreak of COVID-19 opens up new horizons for the exploration of synthetic and natural small compounds to identify potential leads that could be used to design plausible antiviral drugs. The recently documented crystallographic structure of Mpro of SARS-CoV-2 could be a promising target for structural based drug discovery . Nevertheless, identifying and creating new effective medicines is a time consuming and expensive operation(Van Norman, 2016). Hence, the use of computational techniques has accelerated the efforts of drug discovery (Hirono, 2002). Plant-derived natural products play a significant role in discovering and producing drug candidates by being the lead molecule (Joseph et al., 2017). Ancient Indian scriptures, including Rig-Veda, Atherveda, and Charka Sanhita, demonstrated great benefits of plants for treating various human ailments (Kumar et al., 2018). Plants are an excellent natural source of high-value alkaloids, flavonoids, lignans, polyketides, alkanes, alkenes, alkynes, aromatics, peptides, terpenoid, phenols, coumarins, and steroids. In the current time of drug discovery, plants' enormous medicinal properties allow the researchers to exclusively use them for the discovery of drug-like natural molecules (Jee et al., 2018; Kumar et al., 2018; Panchangam et al., 2016).
In this study, we selected medicinal plants used in the preparation of Chyawanprash and constructed a phytochemical library of 686 phytochemicals. After that, we conducted virtual screening targeting the Mpro against SARS-CoV-2. The binding energy of screened 28 phytochemicals out of 686 is within the range of −15.8 Kcal/mol to −8.4Kcal/mol. To analyze physicochemical properties and the drug-like nature of compounds, we analyzed Drug-likeness parameters. According to the rule that most "drug-like" molecules have Log p ≤ 5, molecular weight ≤ 400, number of hydrogen bond acceptors ≤ 10, and number of hydrogen bond donors' ≤ 5. The seven-screened phytochemicals were accepted as a drug by the FAF Drug 3 server, and our study shows screened phytochemicals fulfill all the enlisted criteria of the drug-likeness rule. To determine biological activity and toxicity of screened phytochemicals, we analyzed the OSIRIS prediction of the screened phytochemicals under different criteria; mutagenicity, carcinogenicity, irritation, among others, through correlation rules established by the software in which five phytochemicals were screened against SARS-CoV-2. The comparison of 2 D interaction of all hit phytochemicals revealed stable interaction with one or both the catalytic dyad (His-41 and Cys-145) in Mpro of SARS-CoV-2. The Mpro-Peimisine did not retain the interaction with His-41 but its interaction with substrate binding site amino acid residue Cys-145, Thr-24 Gly-143, Glu-166, Met-165, Thr-190, Pro-168, Gln-189, and Leu-141. The residue Glu-166 in Mpro is considered to be involved in the development of its functional dimeric form. The potential of Epsilon-vinifein to bind to a catalytic group along with a substrate-binding region containing Glu-166 residues inhibits the dimerization of Mpro as Glu-166 residues of one monomer/promoter cannot interfere with N-finger residues of other monomers.
After that, to obtain a more in-depth insight into the structural changes of protein-ligand complexes, we conducted MD simulations. All complexes, including reference, showed relatively similar and consistent stability throughout 100 ns. The results of MMPBSA indicate that Mpro-Peimisine complex (-43.031 kJ/mol), Mpro-Epsilon-vinifein complex (-29.009 kJ/mol), and Mpro-Gmelanone complex (-13.093 kJ/mol) bind to the Mpro receptor of SARS-CoV-2 efficiently. In our study, we found Peimisine (Lilium polyphyllum), Epsilon-vinifein (Vitis vinifera) Gmelanone (Gmelina arborea) have better stability to Mpro receptor of SARS-CoV-2. In recent study, Peimisine was reported as potential active substances in Mongolian medicine Agsirga that can block the binding of ACE2 receptor with spike protein of SARS-CoV-2(Cheng et al., 2020). From the various study Tinospora cordifolia, reported as used for fever and to boost the immune system managing diabetes and makes our respiratory system stronger (Singh et al., 2003). Vitis vinifera and Gmelina arborea has anti-inflammatory and antiviral properties. It means that these three phytochemicals could be potent inhibitors against Mpro of SARS-CoV-2. The results suggest that all these compounds could be potential drug candidates against SARS-CoV-2. The study may pave the way to develop effective medications and preventive measures against SARS-CoV-2 in the future.
5. Conclusion
This study aimed to identify novel inhibitors against the main protease of SARS-CoV-2. Herein, molecular docking and MD simulation were successfully performed to discover novel inhibitors of Mpro based on the natural compounds. A set of 686 phytochemicals from 40 medicinal plants were screened by the Molecular docking method. Finally, the relative stability of three-hit phytochemicals was validated by MD simulation and MMPBSA calculation. All complexes displayed structural stability during the 100 ns MD simulation period. From this study, three screened phytochemicals Peimisine, Gmelanone, and Epsilon-vinifein, were obtained, which showed promising high affinities against SARS-CoV-2. Thus, this study's outcome shows that the screened phytochemicals may be potential drug candidates against Mpro for SARS-CoV-2 and can be exploited to develop better antiviral candidates against COVID-19.
Supplementary Material
Acknowledgements
The authors are thankful to the Head Department of Botany, Kumaun University, Nainital, for providing the facility, space, and resources for this work. The Authors also acknowledge Rashtriya Uchchattar Shiksha Abhiyan (RUSA), Ministry of Human Resource Development, Government of India, to provide Computational infrastructure to establish the Bioinformatics Centre in Kumaun University, S. S. J Campus, Almora.
Glossary
Abbreviations
- COVID-19
Coronavirus disease 2019
- MD
Molecular dynamic
- Mpro
Main protease
- WHO
World health organization
- PDB
Protein Data Bank
- RMSD
Root Mean Square Deviation
- SASA
Solvent Accessible Surface Area; Rg: Radius of gyration
- RMSF
Root Mean Square Fluctuation
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus-2
Disclosure statement
The authors declare that there is no competing interest in this work.
Author contributions
Priyanka Sharma, Sushma Tamta, and Subhash Chandra designed the protocol, conducted experiments, collected data, and prepared the manuscript. Priyanka Sharma and Tushar Joshi help to analyze MD and post-MD simulation. Shalini Mathpal contributed to the construction and analysis of Ligplots. Hemlata Pundir and Tanuja Joshi collaborated in data collection for pharmacokinetic evaluation in the present study. Dr. Subhash Chandra guided in conducting the experiment and reviewing of the manuscript.
Reference
- Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., & Bouachrine, M. (2020). Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto, D., Giovanetti, M., Ciccozzi, A., Spoto, S., Angeletti, S., & Ciccozzi, M. (2020). The 2019-new coronavirus epidemic: Evidence for virus evolution. Journal of Medical Virology, 92(4), 455–459. 10.1002/jmv.25688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics., 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceraolo, C., & Giorgi, F. M. (2020). Genomic variance of the 2019-nCoV coronavirus. Journal of Medical Virology, 92(5), 522–528. 10.1002/jmv.25700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Tang, Y., Bao, B., & Zhang, P. (2020). Exploring the Active Compounds of Traditional Mongolian Medicine Agsirga in Intervention of Novel Coronavirus (2019-nCoV) Based on HPLC-Q-Exactive-MS/MS and Molecular Docking Method. ChemRxiv. [DOI] [PMC free article] [PubMed]
- De Clercq, E. (2002). Strategies in the design of antiviral drugs. Nature Reviews. Drug Discovery, 1(1), 13–25. 10.1038/nrd703 [DOI] [PubMed] [Google Scholar]
- Habbu, P. V., Mahadevan, K. M., Shastry, R. A., & Manjunatha, H. (2009). Antimicrobial activity of flavanoid sulphates and other fractions of Argyreia speciosa (Burm.f) Boj. Indian Journal of Experimental Biology, 47(2), 121–128. [PubMed] [Google Scholar]
- Hirono, S. (2002). An introduction to the computer-aided structure-based drug design–applications of bioinformatics to drug discovery, Rinsho byori. The Japanese Journal of Clinical Pathology, 50(1), 45–51. []. [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan. Lancet (London, England), 395(10223), 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, D. S., I Azhar, E., Madani, T. A., Ntoumi, F., Kock, R., Dar, O., Ippolito, G., Mchugh, T. D., Memish, Z. A., Drosten, C., Zumla, A., & Petersen, E. (2020). The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. International Journal of Infectious Diseases: IJID: Official Publication of the International Society for Infectious Diseases, 91, 264–266. 10.1016/j.ijid.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee, B., Kumar, S., Yadav, R., Singh, Y., Kumar, A., & Sharma, M. (2018). Ursolic acid and carvacrol may be potential inhibitors of dormancy protein small heat shock protein16.3 of Mycobacterium tuberculosis. Journal of Biomolecular Structure & Dynamics, 36(13), 3434–3443. 10.1080/07391102.2017.1389305 [DOI] [PubMed] [Google Scholar]
- Joseph, J., Bhaskaran, R., Kaliraj, M., Muthuswamy, M., & Suresh, A. (2017). Molecular Docking of Phytoligands to the viral protein receptor P. monodon Rab7. Bioinformation, 13(4), 116–121. 10.6026/97320630013116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Kumar, R., Sharma, M., Kumar, U., Gajula, M. N. P., & Singh, K. P. (2018). Uttarakhand medicinal plants database (UMPDB): A platform for exploring genomic. chemical, and traditional knowledge. Data, 3(1), 7. [Google Scholar]
- Lipinski, C. A. (2000). Drug-like properties and the causes of poor solubility and poor permeability. Journal of Pharmacological and Toxicological Methods, 44(1), 235–249. [DOI] [PubMed] [Google Scholar]
- Mabkhot, Y., Alatibi, F., El-Sayed, N., Al-Showiman, S., Kheder, N., Wadood, A., Rauf, A., Bawazeer, S., & Hadda, T. (2016). Antimicrobial Activity of Some Novel Armed Thiophene Derivatives and Petra/Osiris/Molinspiration (POM) Analyses. Molecules, 21(2), 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. J Cheminform, 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchangam, S. S., Vahedi, M., Megha, M. J., Kumar, A., Raithatha, K., Suravajhala, R., & Reddy, P. (2016). Saffron‘omics’: The challenges of integrating omic technologies. Avicenna Journal of Phytomedicine, 6(6), 604–620. [PMC free article] [PubMed] [Google Scholar]
- Parle, M., & Bansal, N. (2006). Traditional medicinal formulation, Chyawanprash—A review. Indian Journal Traditional Knowledge, 5, 484–488. [Google Scholar]
- Pronk, S., Páll, S., Schulz, R., Larsson, P., Bjelkmar, P., Apostolov, R., Shirts, M. R., Smith, J. C., Kasson, P. M., van der Spoel, D., Hess, B., & Lindahl, E. (2013). GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics (Oxford, England)), 29(7), 845–854. 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, L.-L., Wang, Y.-M., Wu, Z.-Q., Xiang, Z.-C., Guo, L., Xu, T., Jiang, Y.-Z., Xiong, Y., Li, Y.-J., Li, X.-W., Li, H., Fan, G.-H., Gu, X.-Y., Xiao, Y., Gao, H., Xu, J.-Y., Yang, F., Wang, X.-M., Wu, C., … Wang, J.-W. (2020). Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chinese Medical Journal, 133(9), 1015–1024. 10.1097/CM9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, R., & Singh, TR. (2019). Virtual Screening, Pharmacokinetics, Molecular dynamics and binding free energy analysis for small natural molecules against Cyclin-dependent kinase 5 for Alzheimer's disease. Journal of Biomolecular Structure and Dynamics., 1, 22. [DOI] [PubMed] [Google Scholar]
- Singh, J., Sinha, K., Sharma, A., Mishra, N. P., & Khanuja, SP. (2003). Traditional uses of Tinospora cordifolia (Guduchi). Journal of Medicinal Aromatic Plant Science, 25, 748–751. [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman, G. (2016). Drugs, Devices, and the FDA: Part 1: An Overview of Approval Processes for Drugs. JACC. Basic to Translational Science, 1(3), 170–179. 10.1016/j.jacbts.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe, K., Hatcher, E., Acharya, C., Kundu, S., Zhong, S., & Shim, J. (2009). CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. Journal of Computational Chemistry., 31(4), 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., & Wu, G. (2020). Detection of SARS-CoV-2 in Different Types of Clinical Specimens. Jama, 323,1843–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2020). Coronavirus disease (COVID-19) pandemic 2020. World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.