Abstract
SARS-CoV-2 is causative of pandemic COVID-19. There is a sequence similarity between SARS-CoV-2 and SARS-CoV; however, SARS-CoV-2 RBDs (receptor-binding domain) binds 20-fold strongly with human angiotensin-converting enzyme 2 (hACE2) than SARS-CoV. The study aims to investigate protein–protein interactions (PPI) of hACE2 with SARS-CoV-2 RBD between wild and variants to detect the most influential interaction. Variants of hACE2 were retrieved from NCBI and subjected to determine the most pathogenic nsSNPs. Probability of PPIs determines the binding affinity of hACE2 genetic variants with RBD was investigated. Composition variations at the hACE2 and RBD were processed for PatchDock and refined by FireDock for the PPIs. Twelve nsSNPs were identified as the top pathogenic from SNPs (n = 7489) in hACE2 using eight bioinformatics tools. Eight RBD variants were complexed with 12 nSNPS of hACE2, and the global energy scores (Kcal/mol) were calculated and classified as very weak (–3.93 to −18.43), weak (–18.42 to −32.94), moderate (–32.94 to −47.44), strong (–47.44 to −61.95) and very strong (–61.95 to −76.46) zones. Seven composition variants in the very strong zone [G726R-G476S; R768W-V367F; Y252N-V483A; Y252N-V367F; G726R-V367F; N720D-V367F and N720D-F486L], and three in very weak [P263S-S383C; RBD-H378R; G726R-A348T] are significantly (p < 0.00001) varied for global energy score. Zonation of the five zones was established based on the scores to differentiate the effect of hACE2 and RBD variants on the binding affinity. Moreover, our findings support that the combination of hACE2 and RBD is key players for the risk of infection that should be done by further laboratory studies.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, ACE2, protein docking, in silico software, nsSNPs
Introduction
Server acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative of a global pandemic COVID-19, which originated late December 2019 in Wuhan, China (Huang et al., 2020). Based on genomic and phylogenetic analysis, SARS-CoV-2 is recognized to be one member of the coronaviridae family (Gorbalenya et al., 2020). The coronaviridae family are seven members, the common human coronaviruses termed as 229E, NL63, OC43 and HKU1 are causing mild symptoms, in contrast to SARS-CoV, which is causing severe symptoms (Li et al., 2020). Coronaviruses (CoVs) is named due to the crown-like shape of the spike protein that is present on the cell surface (Rabi et al., 2020). The genome is composed of non-segmented positive single-stranded RNA with length ranging between 26 and 32 Kb (Zheng, 2020), and the CoVs possess four structural proteins termed as spike (S) glycoprotein, envelope (E), membrane (M) and nucleocapsid (N) (Jiang et al., 2020). The Protein S is a huge protein with amino acid (1160-1542) length; however, it is known to specifically bind with the host cell. Spike (S) glycoprotein is divided into two functional subunits. S1 subunit has a role as host receptor, and the other is S2 subunit needed for membrane fusion (de Haan et al., 2006; Walls et al., 2020). The envelope (E) protein is a structural protein that determines the shape of the viral envelope, while nucleocapsid (N) protein binds with SARS-CoV-2 genome which gives the power of cycle replication within the host (Schoeman & Fielding, 2019). Human angiotensin-converting enzyme 2 (hACE2) is a part of the renin–angiotensin system (RAS), and it acts as multifunctional enzyme that has many protective roles in the human body such as inflammation response, and controller of body stability (Offringa et al., 2020). However, hACE2 has a strong interaction with S1 receptor of SARS-CoV, but not with SARS-CoV-2 (Hwang et al., 2006); moreover, there is study claiming that binding affinity is 10–20× higher of hACE2 with SARS-CoV-2 spike protein than the hACE2 with SARS-CoV (Wrapp et al., 2020). In this study, we aim to investigate the Protein–Protein Interactions (PPIs) of hACE2 with SARS-CoV-2 between wild and variants, and to detect the most impact on the interactions between them by using in silico tools. SARS-CoV-2 spike protein interacts with human hACE2 proteins irrespective of their combination of variations. Strong combinations of variants have been described.
Methods
SNPs data and protein sequence
hACE2 protein sequence was downloaded from NCBI databank [Accession Q9BYF1-1 UniProt database, https://www.uniprot.org] [UniProtKB ID: Q9BYF1 for hACE2]
Deleterious effect of coding nsSNPs of hACE2 receptor protein
For the purpose of detecting the most deleterious nsSNPs of hACE2 receptor protein, we used in silico tools, such as: SIFT (http://sift.jcvi.org/), Polyphen2 (http://genetics.bwh.harvard.edu/pp2), SNAP2 (https://www.rostlab.org/services/SNAP/), SNPs&Go (https://snps-and-go.biocomp.unibo.it/snps-and-go/), Provean (http://provean.jcvi.org/index.php), Panther (http://pantherdb.org) and Condel (http://bbglab.irbbarcelona.org/fannsdb/home).
SIFT for prediction of the effect of nsSNPs on protein function
Sorting Intolerant From Tolerant (SIFT) algorithm is predicting whether an amino acid substitution is affecting a protein function. The output is given as a SIFT score; a SIFT score ≤0.05 corresponds to tolerated nsSNPs, and a SIFT score ≥0.05 indicates a deleterious nsSNPs (Ng & Henikoff, 2003).
PolyPhen-2 for prediction of amino acid substation
Polymorphism phenotyping (PolyPhen-2) is a tool that predicts the possible impact of amino acid substitutions on the human protein structure and function using structural and comparative evolutionary considerations (Adzhubei et al., 2013).
PROVEAN for prediction of protein function influences
Protein Variation Effect Analyzer (PROVEAN) is a tool specifically to predict the functional effects of protein sequence variations, which includes single amino acid substitution or multiple amino acid substitutions and deletions or additions of frameshift mutations (Choi & Chan, 2015).
SNAP2 for prediction of the alteration of protein function
Screening for Non-Acceptable Polymorphisms 2 (SNAP2) and PolyPhen-2 has similar levels of performance. They are used to predict the change of a protein function, where, +100, strongly predicted to have an effect; −100, predicted to be neutral (Hecht et al., 2015).
SNPs&GO for the functional gene
SNPs&Go is a tool used to predict the damage of a Single Amino acid Polymorphism (SAPs) by processing the structure and the function of a protein. Scores larger than 0.5 are considered to relate to diseases (Capriotti et al., 2013).
PANTHER evolutionary tool linked with protein stability and its function
Protein ANalysis THrough Evolutionary Relationships (PANTHER) is a tool used to predict the damaging effect of a protein variant. If subPSEC score is ≥0.5, then nsSNPs are considered deleterious (Mi et al., 2012, 2019).
CONDEL for prediction of pathogenic nsSNPs
CONDEL is tool that only uses the negative control by taking deleterious scores; however, it is possible to provide the mutation impact on the biological activity of the protein (González-Pérez & López-Bigas, 2011).
Homology modeling and validation
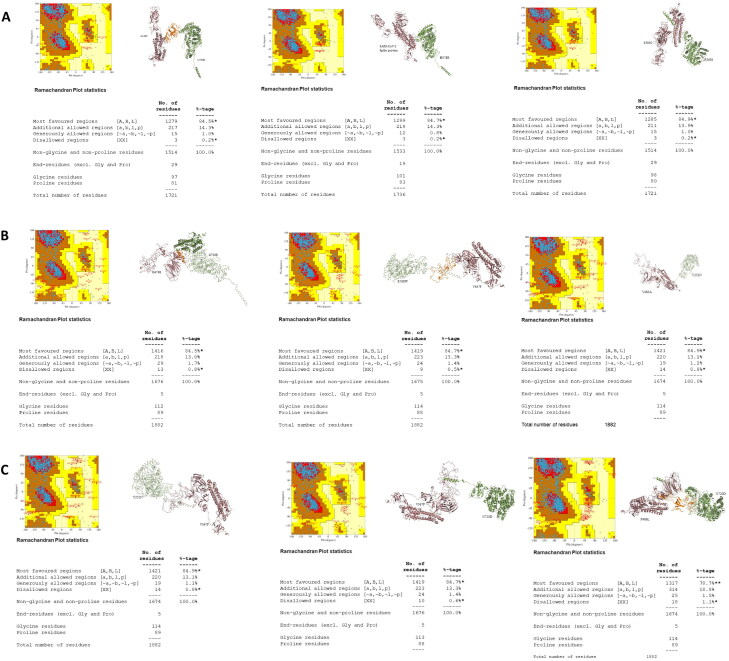
SARS-CoV-2 spike protein FASTA sequence (PDB ID: 6VSB) was retrieved from RCSB PDB databank (http://www.rcsb.org/) (Berman et al., 2002). hACE2 receptor protein and SARS-CoV-2 spike protein three-dimensional (3D) structures were created through SWISS-MODEL (https://swissmodel.expasy.org/) and Swiss-Pdb viewer software (Guex & Peitsch, 1997) as described earlier (Abdulazeez et al., 2019; AbdulAzeez & Borgio, 2016; Borgio et al., 2016), and structures then were carried on for validation through RAMPAGE and Ramachandran plot analysis (http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php) (Wang et al., 2016). Mutant structures of hACE2 receptor and SARS-CoV-2 spike protein were created using PyMol software (ver. 2.4, Schrödinger) (Seeliger & De Groot, 2010).
Multiple sequence alignment and phylogenetic analysis of SARS-CoV-2
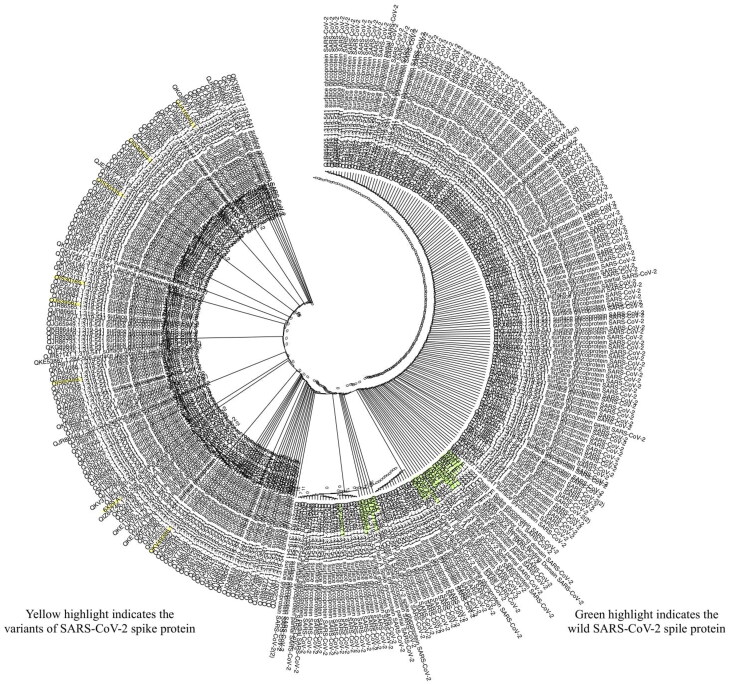
Multiple sequence alignment and phylogenetic analysis was conducted through open source software named Molecular Evolutionary Genetics Analysis (MEGA X), (ver. 10.1.8) (Stecher et al., 2020). We tested 100 wild and 316 mutants for RBD (receptor-binding domain) region that was obtained from blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi), by maximum-likelihood 8000 bootstrap (Boratyn et al., 2013).
Root-mean-square deviation (RMSD) prediction
Superimpose of RBD regions for SARS-CoV-2 spike protein (Arg319–Phe541) (Lan et al., 2020) was implemented to energy minimize by SuperPose (http://superpose.wishartlab.com/) (Maiti et al., 2004). For hACE2 receptor, it was implemented by SWISS-MODEL (https://swissmodel.expasy.org/) (Waterhouse et al., 2018).
Protein–protein interactions (PPIs)
Protein–protein interactions of hACE2 receptor and SARS-CoV-2 Spike protein were docked through PatcnhDock (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php). Then, results were refined using FireDock (http://bioinfo3d.cs.tau.ac.il/FireDock/php.php) (Mashiach et al., 2008; Schneidman-Duhovny et al., 2005).
Statistical analysis
Cumulative score for various tools (SIFT, PolyPhen-2, PROVEAN, SNAP2, SNPs&GO, PANTHER, CONDEL and PhD-SNPs) was calculated by sum for the damaged nsSNPs as results from the eight in silico tools of predicting the effect of nsSNPs. The prediction of global energy scores (Jiang et al., 2018) was estimated by FireDock that applied Student’s t-test, and was calculated, and p < 0.05 is considered significant for the most impacting combinations of hACE2 receptor and SARS-CoV-2 spike protein.
Results
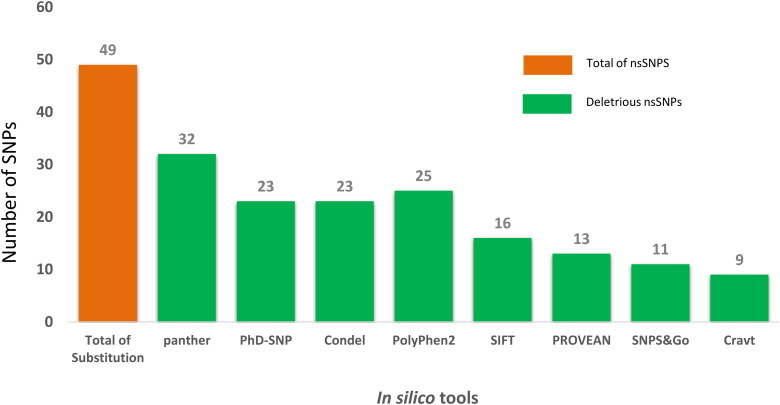
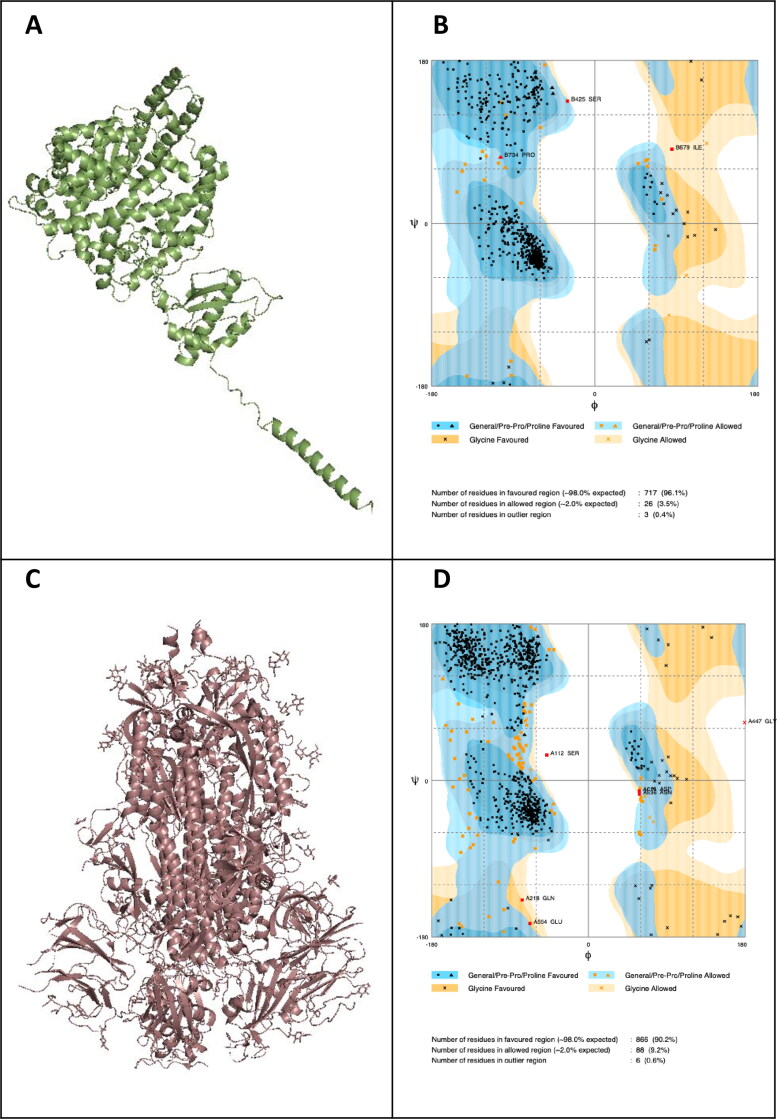
All nsSNPs were analyzed using eight in silico tools to identify the most deleterious nsSNPs, such as SIFT, PolyPhen-2, PROVEAN, SNAP2, SNPs&GO, PANTHER, CONDEL and PhD-SNPs. Nine out of the 48 nsSNPs among 7489 SNPs on hACE2 receptor extracted from NCBI, and were considered as the most pathogenic nsSNPs (Figure 1; Table 1). In addition to the nine pathogenic nsSNPs, three nsSNPs were included for further analysis (Table 1). The neutral results of nsSNPS in SNPs&Go 37 (77.1%), PROVEAN 35 (72.9%), SIFT 32 (66.6%), SNAP2 29 (60.4%), PhD-SNP 25 (52.1%), PolyPhen-2 23 (47.9%), Condel 20 (41.6%) and Panther 16 (33.3%) (Supplementary Table S1). The amino acid substitutions in hACE2 receptor: L595V, W459C, H378R, P263S and Y252N scored the highest cumulative scores (CS = 9), predicted by damaged by all the tested tools (Table 1). Three-dimensional structure of hACE2 receptor was constructed and validated (Figure 2). Maximum numbers of residues (99.5%) are falling in the allowed and favored regions, which indicated the constructed model is efficient for purpose of the protein docking (Figure 2(B)). Analysis of SARS-CoV-2 spike protein by MEGA X (Figures 3 and 4) identified eight variants such as A348T (QIS30335.1), S383C (6X29_A), A419S (QJA16640.1), G476S (Q1Q49882.1), V483A (QIS30165.1), F486L (QJS39567.1), A520S (QIS60489.1) and V367F (QK95522.1) were taken into consideration because of their high report in many countries (https://cov.lanl.gov/content/index) (Ou et al., 2020). Three-dimensional structure of SARS-CoV-2 spike protein was constructed and validated (Figure 2(D)). Maximum numbers of amino acid residues (99.37%) are in the favored and allowed regions; hence, the constructed model is competent for protein docking (Figure 2(D)).
Figure 1.
Predicting the effect of nsSNPs using various in silico tools.
Table 1.
The list of selected pathogenic nsSNPs out of 7489 SNPs.
| SNP | Coordinate | Substitution | Sift |
Polyphen-2 |
Panther | Provean |
Snap2 |
Snps&Go |
Phd-SNP |
Condel |
CS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | S | P | S | P | P | S | P | S | P | S | P | S | P | Label | ||||
| rs148036434 | 15589801 | L595V | 1 | 0 | 1 | 1 | 1 | 1 | –2.56 | 1 | 16 | 1 | 0.69 | 1 | 0.727 | 1 | D | 9 |
| rs11798104 | 15593854 | W459C | 1 | 0 | 1 | 1 | 1 | 1 | –12.36 | 1 | 68 | 1 | 0.809 | 1 | 0.907 | 1 | D | 9 |
| rs142984500 | 15596376 | H378R | 1 | 0 | 1 | 1 | 1 | 1 | –7.93 | 1 | 70 | 1 | 0.879 | 1 | 0.906 | 1 | D | 9 |
| rs200745906 | 15605891 | P263S | 1 | 0 | 1 | 1 | 1 | 1 | –7.33 | 1 | 42 | 1 | 0.871 | 1 | 0.768 | 1 | D | 9 |
| rs371464495 | 15605924 | Y252N | 1 | 0.002 | 1 | 1 | 1 | 1 | –6.84 | 1 | 82 | 1 | 0.815 | 1 | 0.931 | 1 | D | 9 |
| rs375352455 | 15589896 | S563L | 1 | 0.001 | 1 | 1 | 1 | 1 | –5.78 | 1 | 48 | 1 | 0.716 | 1 | 0.862 | 1 | D | 8 |
| rs139980377 | 15582280 | G726R | 1 | 0.004 | 1 | 1 | 1 | 1 | –2.82 | 1 | 79 | 0 | 0.206 | 1 | 0.778 | 1 | D | 8 |
| rs140016715 | 15582154 | R768W | 1 | 0 | 1 | 1 | 1 | 1 | –2.82 | 1 | 62 | 1 | 0.517 | 1 | 0.853 | 1 | D | 8 |
| rs267606408 | 15603648 | P284S | 0 | 0.053 | 1 | 0.991 | 1 | 1 | –7.65 | 0 | 0 | 1 | 0.838 | 1 | 0.864 | 1 | D | 7 |
| rs41303171 | 15582298 | N720D | 0 | 0.092 | 0 | 0.006 | 1 | 0 | –1.19 | 0 | –32 | 0 | 0.139 | 1 | 0.5 | 1 | D | 3# |
| rs143936283 | 15599428 | E329G | 0 | 0.121 | 0 | 0.027 | 0 | 0 | –2.21 | 1 | 5 | 0 | 0.108 | 0 | 0.343 | 1 | D | 2*# |
| rs146676783| | 15618926 | E37K | 0 | 0.16 | 1 | 0.712 | 1 | 0 | –1.86 | 0 | –35 | 0 | 0.175 | 0 | 0.449 | 0 | N | 2* |
CS: Cumulative Score; P: Prediction; S: Score. * These two amino acid substitutions were selected as they are reported as contact residues of RBD–hACE2 (Lan et al., 2020). # SNP was observed in the Saudi population and was added for the regional interest. The values in P: 0 indicates neutral (N); 1 indicated damaged (D).
Figure 2.
Three-dimensional structures and their validations of A&B: hACE2, and C&D: SARS-CoV-2 spike protein.
Figure 3.
Annotation of SARS-CoV-2 variants in the multiple sequence alignment of the spike protein sequence and reported variants. Arrow indicates the amino acid substitutions.
Figure 4.
Phylogenetic analysis of SARS-CoV-2 spike protein was accomplished through maximum-likehood 8000 bootstrap. The list of sequences used to construct phylogeny are listed in the Supplementary Data 1.
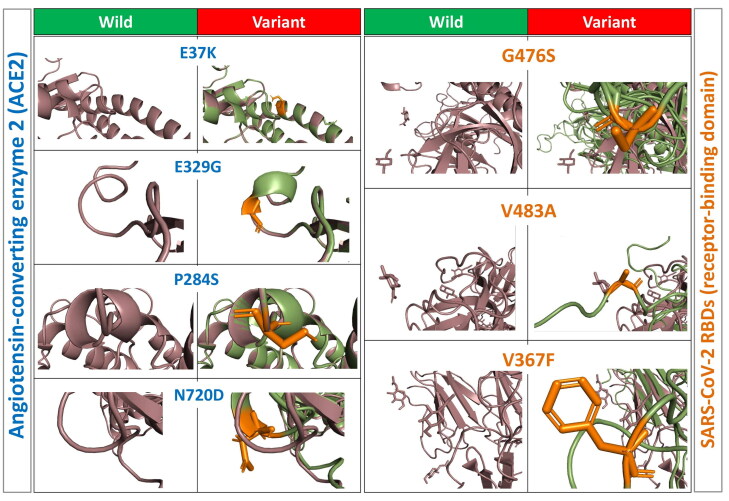
Each point of amino acid variations was generated in the native protein model structure of hACE2 receptor, and SARS-CoV-2 spike protein separately using PyMol software (Figure 5). RMSD of hACE2 receptor and SARS-CoV-2 spike protein variants structures was calculated after the energy minimization (Table 2). The protein–protein interactions of wild and variants for both hACE2 receptor and SARS-CoV-2 protein were analyzed using PatchDock, and the top 300 predictions we refined using FireDock (Figure 6). The global energy score (kcal/mol) was considered the final score to predict the interaction between the hACE2 receptor and SARS-CoV-2 spike protein (Figure 7(A)). The global energy score indicates the stability of the interaction, which means less binding energy is giving higher binding affinity, and higher binding energy is giving less binding affinity (Jiang et al., 2018). In order to identify the variations in the binding affinity, the global energy score (kcal/mol) was scaled in range of five zones for all composition variations by the following formula:
Figure 5.
3D structures of wild and variants of hACE2 receptor and SARS-CoV-2 spike protein.
Table 2.
RMSD calculations for hACE2 and SARS-CoV-2 spike protein variants.
| hACE2 receptor variants |
SARS-CoV-2 spike protein variants |
||||
|---|---|---|---|---|---|
| nsSNPs | Residue Change | RMSD | Number | Residue Change | RMSD |
| rs148036434 | L595V | 0.06 | QIS30335 | A348T | 1.488 |
| rs11798104 | W459C | 0.06 | 6X29_A | S383C | 1.488 |
| rs142984500 | H378R | 0.06 | QJA16640 | A419S | 1.488 |
| rs143936283 | E329G | 0.06 | QIQ49882 | G476S | 3.2 |
| rs267606408 | P284S | 0.06 | QIS30425 | G476S | 3.2 |
| rs200745906 | P263S | 0.06 | QIS30165 | V483A | 3.2 |
| rs371464495 | Y252N | 0.06 | QJS39567 | F486L | 3.2 |
| rs146676783 | E37K | 0.06 | QIS60489 | A520S | 1.488 |
| rs375352455 | S563L | 0.06 | QK95522.1 | V367F | 1.488 |
| rs139980377 | G726R | 0.05 | |||
| rs140016715 | R768W | 0.06 | |||
| rs41303171 | N720D | 1.514 | |||
Figure 6.
Significant composition variations of hACE2 receptor and SARS-CoV-2 spike protein. A: Very weak Composition variation. B & C: Very strong composition variations.
Figure 7.
A: Heat map of Global energy score for composition variations of hACE2 and SARS-CoV-2 spike protein-generated Heatmapper (http://www.heatmapper.ca/pairwise/) with matrix of Displaying data as-is. and their significant impact; B: graphical representations of all composition variations that reflect the degree of binding to SARS-CoV-2.
Z = Length of the zone
A1 = Lowest global energy score (kcal/mol)
A2 = Highest global energy score (kcal/mol)
Global energy score in different zones: Very weak (–3.93 to −18.43), Weak (–18.42 to −32.94), Moderate (–32.94 to −47.44), Strong (–47.44 to −61.95) and Very strong (–61.95 to −76.46) (Figure 6). Seven composition variants in the very strong zone [G726R-G476S; R768W-V367F; Y252N-V483A; Y252N-V367F; G736R-V367F, N720D-V367F and N720D-F486L], and three composition variants in the safe zone [P263S-S383C; Spike protein-H378R and G726R-A348T] (Figures 6 and 7(B)). Both zones are significantly (p < 0.00001) varied for global energy score (Kcal/mol).
Discussion
The information about the variations in SARS-CoV-2 spike protein and hACE2 receptor protein and their interaction in the infection scale have not been studied properly, while the reason remains unknown. In this study, eight in silico tools (i.e. SIFT, PolyPhen-2, Provean, SNAP2, SNPs&GO, PANTHER, CONDEL and PhD-SNPs) were used to determine the most deleterious hACE2 receptor variants. Simultaneously, multiple sequence alignment and phylogenetic analysis allowed us to reveal the newly reported SARS-CoV-2 spike protein variants from the protein sequence retrieved from RCSB PDB protein databank. Homology modeled hACE2 receptor, and SARS-CoV-2 spike proteins were validated and used to generate the mutated protein for protein–protein interaction studies using PatchDock. The estimated PPIs binding affinity of various hACE2 receptor and SARS-CoV-2 spike protein combinations (Figure 7(A)) were refined from FireDock to attain the global energy score (kcal/mol) to calculate the overall view about the binding affinity degree of wild and hACE2 receptor variants with SARS-CoV-2 spike protein. Moreover, in order to understand what is the meaning of these scores, we categorized the results into five zones (Figure 7(B)). One notable finding is that irrespective of the variants in hACE2 receptor and SARS-CoV-2 spike protein, the binding established from weak to strong depends on the combinations of variants (Figure 7(A)). The second finding is that we categorized the risk of infection into five zones, such as very weak, weak, moderate, strong and very strong due to the spectrum of binding affinity among variants composition of hACE2 receptor and SARS-CoV-2 protein. The present observations are the first to study about the interactions between wild and variants of SARS-CoV-2 spike protein and pathogenic variants of hACE2 receptor. The hACE2 receptor variant [E329G] showed a strong binding affinity that is present at three zones, very strong [E329G-V483A, E329G-G476S], strong [E329G-A419S, E329G-A348T] and moderate [E329G-S383C, E329G-F486L]. Cyro-electron microscopy study for hACE2 receptor with and without the RBD has discovered that hACE2 receptor [E329] variant would get weaken due to the variant R426-N439 in spike (Wu et al., 2020), and this clearly is in line with our observations on the variants in spike that can influence the binding affinity. Study by Amin et al (2020) reported hACE2 receptor [E329] variant with SARS-CoV spike protein with high contribution for binding energy. Recently, a study proved that wild hACE2 receptor and SARS-CoV-2 spike protein (–35.33 Kcal/mol) are having a stable interaction and are present at the moderate zone similar to the present results (Veeramachaneni et al., 2020). Previous study reported that SARS-CoV-2 spike protein [G476S] variant has a significant very strong point and a strong direct binding affinity with hACE2 receptor, and similarly, the G476S variant showed strong binding [G726R] of spike and falling in the very strong zone (Korber et al., 2020). Another study reported three [A348T; G467S and V483A] SARS-CoV-2 spike protein variants in RBD region from 579 whole genome of SARS-CoV-2 isolated from US COVID-19 patients (Kaushal et al., 2020), and all these three variants are analyzed for their impact on the binding affinity. The analysis revealed that these three variants are present at the very weak zone [G726R-A348T] with low binding affinity (–3.93 Kcal/mol), at the strong zone [P284S-A348T; E329G-A348T; L595V-A348T; H378R-A348T and P263S-A348T, −48.72,–48.72, −48.72 and −48.72 Kcal/mol, respectively], weak zone [ACE2-A348T (–31.24 Kcal/mol) and N720D-A348T (–24.98 Kcal/mol)]; furthermore, [G476S] variant of spike protein is present at all of the three zones (very strong, strong and weak) and [G726] variant of spike protein is present at very strong [G726R-G476S] with binding affinity (–76.46 Kcal/mol), strong [ACE2-G476S; E37K-G476S; P284S-G476S; E329G-G476S; L595V-G476S; W459C-G476S; S563L-G476S; R768W-G476S and Y252N-G476S] with binding affinity (–64.5, −64.5, −64.5, −64.5, −64.5, −64.5, −64.5, −55.96 and −55.87 Kcal/mol, respectively) and weak [H378R-G476S; N720D-G476S and P263S-G476S] with binding affinity (–31.47, −23.58 and −22.67 Kcal/mol, respectively). [V483A] variant of spike protein is present at very strong [Y252N-V483A; E37K-V483A; hACE2-V483A; E329G-V483A; L595V-V483A; H378R-V483A; P263S-V483A and S563L-V483A] with binding affinity (–72.09, −66.33, −65.57, −65.57, −65.57, −65.57, −65.57 and −65.57 Kcal/mol, respectively) and weak [R768W-V483A and W459C-V483A] with binding affinity (–30.47 and −29.17 Kcal/mol) (Figure 7(B)). The spectrum of binding observed among the four variants of spike reported from the USA gave a clue on the spectrum of severity among COVID-19 patients; however, these computational observations need to be confirmed (Kaushal et al., 2020).
Wild hACE2 showed higher binding affinity with spike variants [V483A and G476S] (ACE2-V483A −65.57 Kcal/mol and ACE2-V367F −53.46 Kcal/mol) indicating that the wild hACE2 also binding acutely with variant of the spike protein and in the strong zone, but not in the very strong; however, previous study reported same variants with low binding affinity (Nelson-Sathi et al., 2020). Previous observations by Ou et al. (2020) on [V367F and V483A] variants of spike divided the variants into two groups, similar and higher binding affinity, one variant [V367F] was considered with the higher binding affinity and another [V483A] was considered with the similar binding affinity with wild hACE2; however, we have observed that [V483A] has relatively higher affinity with most of the hACE2 receptor variants and are present at the very strong zone (Ou et al., 2020).
Many genomic studies are working on detecting the type of SARS-CoV-2 mutation (Hassan et al., 2020; Kaushal et al., 2020). There are many studies on the variants of spike protein (Hassan et al., 2020), claiming the (S) protein is mainly causing the severity of the COVID-19 condition (Zhang et al., 2020), and mutation has a big impact on the severity of the condition (Hassan et al., 2020; Othman et al., 2020); however, the present observation has given an insight on the combination of variants on the interaction between spike and receptor. The [F486] of SARS-CoV-2 spike protein forms a strong binding affinity with [Y83] of hACE2 receptor, and this study observed [N720D] of hACE2 receptor and [F486L] of SARS-CoV-2 spike protein as significant composition variation located at the very strong zone (Wang et al., 2020). Zou et al. (2020) revealed six out of eight SARS-CoV-2 variants with high binding affinity with hACE2, one of them was [F486], which showed similar results and in the very strong zone with the present observation. The reported SARS-CoV-2 spike protein variant [D614G] has got attention is (Korber et al., 2020; Ou et al., 2020); however this is not in the RBD [Arg319-Phe541] (Lan et al., 2020), and hence, this has not been included in the study. Variants of hACE2 receptor were reported to be associated with severity of COVID-19 based on the status of hypertension (Gomez et al., 2020). Variants of hACE2 receptor will have stronger binding with wild SARS-CoV-2 spike protein and variants. However, the severity depends on the variant type, missense variants in hACE2 [T27R; F28W; L79W; G326E; G352Y; D30E; K31E; Y41N; Q41E; L45E; Y83F; N330K; L353H] inhibit the interaction with SARS-CoV-2 spike protein and contribute to the genetic risk in COVID-19 patients (MacGowan & Barton, 2020), in line with the present observation, suggesting that variants of hACE2 receptor cause susceptibility that will eventually lead to the risk of COVID-19 severity (MacGowan & Barton, 2020). Jia et al.’s (2020) study were the reports on SARS-CoV-2 variants with low binding affinity with hACE2 receptor, but that results were not comparable for this study due to mismatch in selection of variants. Moreover, this study found that SARS-CoV-2 protein variants are causing a high or low binding affinity with hACE2 receptor, depending on the variants. The present observations are computational that have its notable limitation and further laboratory work will provide the confirmation of all different zones and their phenotypic association among COVID-19 patients. Signaling pathways and intricate protein–protein interaction networks are governed by proteins for cellular life (Paladino et al., 2020; Serapian & Colombo, 2020; Serapian & van der Kamp, 2019); furthermore, the enhanced spike-ACE2 interaction increases chances on the entrance into the host cells (Serapian et al., 2020; Spinello et al., 2020), and hence, the present observations on the strong and very strong protein–protein interaction will be a base for future clinical and wet lab studies on selected variations on spike and ACE2.
Conclusion
The proposed risk of infection scale in COVID-19 patients depends on the composition variations of hACE2 receptor and SARS-CoV-2 spike protein is one of a kind to use as reference for further computational and laboratory studies to correlate the spectrum of severity.
The principle cause of COVID-19 disease severity has been categorized into five zones (very weak, weak, moderate, strong and very strong). The wild hACE2 receptor and SARS-CoV-2 spike protein are binding strongly to each other.
Supplementary Material
Acknowledgements
The authors thank the Dean, Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia, for her continuous support and encouragement. Authors thank Mr. Ranilo, M. Tumbaga and Mr. Horace T. Pacifico for their assistance.
Funding Statement
The project is funded by Deanship of Scientific Research (Covid19-2020-007-IRMC), Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- AbdulAzeez, S., & Borgio, J. F. (2016). In-silico computing of the most deleterious nsSNPs in HBA1 gene. PLoS One, 11(1), e0147702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulazeez, S., Sultana, S., Almandil, N. B., Almohazey, D., Bency, B. J., & Borgio, J. F. (2019). The rs61742690 (S783N) single nucleotide polymorphism is a suitable target for disrupting BCL11A-mediated foetal-to-adult globin switching. PLoS One, 14(2), 1–18. 10.1371/journal.pone.0212492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei, I., Jordan, D. M., & Sunyaev, S. R. (2013). Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols, 76(1), 7–20. 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, M., Sorour, M. K., & Kasry, A. (2020). Comparing the binding interactions in the receptor binding domains of SARS-CoV-2 and SARS-CoV. The Journal of Physical Chemistry Letters, 11(12), 4897–4900. 10.1021/acs.jpclett.0c01064 [DOI] [PubMed] [Google Scholar]
- Berman, H. M., Battistuz, T., Bhat, T. N., Bluhm, W. F., Bourne, P. E., Burkhardt, K., Feng, Z., Gilliland, G. L., Iype, L., Jain, S., Fagan, P., Marvin, J., Padilla, D., Ravichandran, V., Schneider, B., Thanki, N., Weissig, H., Westbrook, J. D., & Zardecki, C. (2002). The protein data bank. Acta Crystallographica. Section D, Biological Crystallography, 58(Pt 6 No 1), 899–907. 10.1107/s0907444902003451 [DOI] [PubMed] [Google Scholar]
- Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., & Ma, N. (2013). BLAST: A more efficient report with usability improvements. Nucleic Acids Research, 41(W1), W29–W33. 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgio, J. F., Al-Madan, M. S., & AbdulAzeez, S. (2016). Mutation near the binding interfaces at α-hemoglobin stabilizing protein is highly pathogenic. American Journal of Translational Research, 8(10), 4224–4232. PMID: 27830006 [PMC free article] [PubMed] [Google Scholar]
- Capriotti, E., Calabrese, R., Fariselli, P., Martelli, P. L., Altman, R. B., & Casadio, R. (2013). WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics, 14(Suppl 3), S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y., & Chan, A. P. (2015). PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics (Oxford, England)), 31(16), 2745–2747. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan, C. A. M., Te Lintelo, E., Li, Z., Raaben, M., Wurdinger, T., Bosch, B. J., & Rottier, P. J. M. (2006). Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. Journal of Virology, 80(22), 10909–10918. 10.1128/JVI.00950-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, J., Albaiceta, G. M., Garcia-Clemente, M., Lopez-Larrea, C., Amado, L., & Hermida, T. (2020). Angiotensin-converting enzyme (ACE1, ACE2) gene variants are associated with COVID19 severity depending on the hypertension status. MedRxiv, 1–24. 10.1101/2020.06.11.20128033 [DOI] [Google Scholar]
- González-Pérez, A., & López-Bigas, N. (2011). Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. American Journal of Human Genetics, 88(4), 440–449. 10.1016/j.ajhg.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., & Gulyaeva, A. A. (2020). The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 5, 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis, 18(15), 2714–2723. 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Hassan, S. S., Choudhury, P. P., & Roy, B. (2020). SARS-CoV2 envelope protein: Non-synonymous mutations and its consequences. Genomics, 112(6), 3890–3892. 10.1016/j.ygeno.2020.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan, D. S. S., Choudhury, P. P., Roy, B., & Jana, S. S. (2020). Missense mutations in SARS-CoV2 genomes from Indian patients. SSRN Electron Journal, 1–8. 10.31219/osf.io/2wm8h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, M., Bromberg, Y., & Rost, B. (2015). Better prediction of functional effects for sequence variants. BMC Genomics, 16(S8), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England), 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W. C., Lin, Y., Santelli, E., Sui, J., Jaroszewski, L., Stec, B., Farzan, M., Marasco, W. A., & Liddington, R. C. (2006). Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. The Journal of Biological Chemistry, 281(45), 34610–34616. 10.1074/jbc.M603275200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., Shen, G., Zhang, Y., Huang, K.-S., Ho, H.-Y., Hor, W.-S., Yang, C.-H., Li, C., & Wang, W.-L. (2020). Analysis of the mutation dynamics of SARS-CoV-2 reveals the spread history and emergence of RBD mutant with lower ACE2 binding affinity. BioRxiv, 1–17. 10.1101/2020.04.09.034942 [DOI] [Google Scholar]
- Jiang, M., Niu, C., Cao, J., Ni, D., & Chu, Z. (2018). In silico-prediction of protein–protein interactions network about MAPKs and PP2Cs reveals a novel docking site variants in Brachypodium distachyon. Scientific Reports, 8(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal, N., Gupta, Y., Goyal, M., Khaiboullina, S. F., Baranwal, M., & Verma, S. C. (2020). Mutational frequencies of SARS-CoV-2 genome during the beginning months of the outbreak in USA. Pathogens, 9(7), 516–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, B., Fischer, W., Gnanakaran, S. G., Yoon, H., Theiler, J., Abfalterer, W., Foley, B., Giorgi, E. E., Bhattacharya, T., Parker, M. D., Partridge, D. G., Evans, C. M., Freeman, T. M., de Silva, T. I., LaBranche, C. C., & Montefiori, D. C. (2020). Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. BioRxiv, 1–33. 10.1101/2020.04.29.069054 [DOI] [Google Scholar]
- Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., & Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581(7807), 215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Li, X., Zai, J., Zhao, Q., Nie, Q., Li, Y., Foley, B. T., & Chaillon, A. (2020). Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. Journal of Medical Virology, 92(6), 602–611. 10.1002/jmv.25731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGowan, S. A., & Barton, G. J. (2020). Missense variants in ACE2 are predicted to encourage and inhibit interaction with SARS-CoV-2 Spike and contribute to genetic risk in COVID-19. BioRxiv, 1–38. 10.1101/2020.05.03.074781 [DOI] [Google Scholar]
- Maiti, R., Van Domselaar, G. H., Zhang, H., & Wishart, D. S. (2004). SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Research, 32, 590–594. 10.1093/nar/gkh477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiach, E., Schneidman-Duhovny, D., Andrusier, N., Nussinov, R., & Wolfson, H. J. (2008). FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Research, 36, 229–232. 10.1093/nar/gkn186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H., Muruganujan, A., Ebert, D., Huang, X., & Thomas, P. D. (2019). PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Research, 47(D1), D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H., Muruganujan, A., & Thomas, P. D. (2012). PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Research, 41(D1), D377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi, S., Umasankar, P. K., Easwaran, S., Nair, R. R., Joseph, I., Nori, S. R. C., Philip, J. S., Prasad, R., Navyasree, K. V., Ramesh, S. T., Pillai, H., Ghosh, S., Santosh Kumar, T. R., & Radhakrishna Pillai, M. (2020). Structural and functional implications of spike protein mutational landscape in SARS-CoV-2. BioRxiv, 1–11. 10.1101/2020.05.02.071811 [DOI] [Google Scholar]
- Ng, P. C., & Henikoff, S. (2003). SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Research, 31(13), 3812–3814. 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offringa, A., Montijn, R., Singh, S., Paul, M., Pinto, Y. M., & Pinto-Sietsma, * S.-J. (2020). The mechanistic overview of SARS-CoV-2 using angiotensin-converting enzyme 2 to enter the cell for replication: Possible treatment options related to the renin–angiotensin system. European Heart Journal - Cardiovasc Pharmacotherapy, 6(5), 1–9. 10.1093/ehjcvp/pvaa053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman, H., Bouslama, Z., Brandenburg, J.-T., da Rocha, J., Hamdi, Y., Ghedira, K., Srairi-Abid, N., & Hazelhurst, S. (2020). Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: Similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochemical and Biophysical Research Communications, 527(3), 702–708. 10.1016/j.bbrc.2020.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, J., Zhou, Z., Dai, R., Zhang, J., Lan, W., Zhao, S., Wu, J., Seto, D., Cui, L., Zhang, G., & Zhang, Q. (2020). Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. BioRxiv, 1–24. [Google Scholar]
- Paladino, A., Woodford, M. R., Backe, S. J., Sager, R. A., Kancherla, P., Daneshvar, M. A., Chen, V. Z., Bourboulia, D., Ahanin, E. F., Prodromou, C., Bergamaschi, G., Strada, A., Cretich, M., Gori, A., Veronesi, M., Bandiera, T., Vanna, R., Bratslavsky, G., Serapian, S. A., Mollapour, M., & Colombo, G. (2020). Chemical perturbation of oncogenic protein folding: From the prediction of locally unstable structures to the design of disruptors of Hsp90-client interactions. Chemistry (Weinheim an Der Bergstrasse, Germany), 26(43), 9459–9465. 10.1002/chem.202000615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabi, F. A., Al Zoubi, M. S., Al-Nasser, A. D., Kasasbeh, G. A., & Salameh, D. M. (2020). SARS-CoV-2 and coronavirus disease 2019: What we know so far. Pathogens, 9(3), 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny, D., Inbar, Y., Nussinov, R., & Wolfson, H. J. (2005). PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Research, 33, 363–367. 10.1093/nar/gki481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman, D., & Fielding, B. C. (2019). Coronavirus envelope protein: Current knowledge. Virology Journal, 16(1), 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger, D., & De Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Journal of Computer-Aided Molecular Design, 24(5), 417–422. 10.1007/s10822-010-9352-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serapian, S. A., & Colombo, G. (2020). Designing molecular spanners to throw in the protein networks. Chemistry–A European Journal, 26(21), 4656–4670. 10.1002/chem.201904523 [DOI] [PubMed] [Google Scholar]
- Serapian, S. A., & van der Kamp, M. W. (2019). Unpicking the cause of stereoselectivity in actinorhodin ketoreductase variants with atomistic simulations. ACS Catalysis, 9(3), 2381–2394. [Google Scholar]
- Serapian, S. A., Marchetti, F., Triveri, A., Morra, G., Meli, M., Moroni, E., Sautto, G. A., Rasola, A., & Colombo, G. (2020). The answer lies in the energy: How simple atomistic molecular dynamics simulations may hold the key to epitope prediction on the fully glycosylated SARS-CoV-2 spike protein. The Journal of Physical Chemistry Letters, 11(19), 8084–8093. 10.1021/acs.jpclett.0c02341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S., Hillyer, C., & Du, L. (2020). Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends in Immunology, 41, 335–359. 10.1016/j.it.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinello, A., Saltalamacchia, A., & Magistrato, A. (2020). Is the rigidity of SARS-CoV-2 spike receptor-binding motif the hallmark for its enhanced infectivity? Insights from all-atoms simulations. The Journal of Physical Chemistry Letters, 11(12), 4785–4790. 10.1021/acs.jpclett.0c01148 [DOI] [PubMed] [Google Scholar]
- Stecher, G., Tamura, K., & Kumar, S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Molecular Biology and Evolution, 37(4), 1237–1239. 10.1093/molbev/msz312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramachaneni, G. K., Thunuguntla, V. B. S. C., Bobbillapati, J., & Bondili, J. S. (2020). Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. Journal of Biomolecular Structure and Dynamics, 0, 1–11. 10.1080/07391102.2020.1773318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Xia, M., Chen, J., Deng, F., Yuan, R., Zhang, X., & Shen, F. (2016). Data set for phylogenetic tree and RAMPAGE Ramachandran plot analysis of SODs in Gossypium raimondii and G. arboreum. Data in Brief, 9, 345–348. 10.1016/j.dib.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Zhang, Y., Wu, L., Niu, S., Song, C., Zhang, Z., Lu, G., Qiao, C., Hu, Y., Yuen, K.-Y., Wang, Q., Zhou, H., Yan, J., & Qi, J. (2020). Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell, 181(4), 894–904.e9. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F. T., de Beer, T. A. P., Rempfer, C., Bordoli, L., Lepore, R., & Schwede, T. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), W296–303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., & Wang, Y. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 3, 1–8. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Jackson, C. B., Mou, H., Ojha, A., Rangarajan, E. S., Izard, T., Farzan, M., & Choe, H. (2020). The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. BioRxiv, 1–25. 10.1101/2020.06.12.148726 [DOI] [Google Scholar]
- Zheng, J. (2020). SARS-coV-2: An emerging coronavirus that causes a global threat. International Journal of Biological Sciences, 16(10), 1678–1685. 10.7150/ijbs.45053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J., Yin, J., Fang, L., Yang, M., Wang, T., & Wu, W. (2020). Computational prediction of mutational effects on the SARS-CoV-2 binding by relative free energy calculations. ChemRxiv, 0–19. 10.1021/acs.jcim.0c00679 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.