Abstract
Recent outbreak of COVID-19 pandemic caused by severe acute respiratory syndrome-Coronavirus-2 (SARS-CoV-2) has raised serious global concern for public health. The viral main 3-chymotrypsin-like cysteine protease (Mpro), known to control coronavirus replication and essential for viral life cycle, has been established as an essential drug discovery target for SARS-CoV-2. Herein, we employed computationally screening of Druglib database containing FDA approved drugs against active pocket of SARS-CoV-2 Mpro using MTiopen screen web server, yields a total of 1051 FDA approved drugs with docking energy >−7 kcal/mol. The top 10 screened potential compounds against SARS-CoV-2 Mpro were then studied by re-docking, binding affinity, intermolecular interaction, and complex stability via 100 ns all atoms molecular dynamics (MD) simulation followed by post-simulation analysis, including end point binding free energy, essential dynamics, and residual correlation analysis against native crystal structure ligand N3 inhibitor. Based on comparative molecular simulation and interaction profiling of the screened drugs with SARS-CoV-2 Mpro revealed R428 (−10.5 kcal/mol), Teniposide (−9.8 kcal/mol), VS-5584 (−9.4 kcal/mol), and Setileuton (−8.5 kcal/mol) with stronger stability and affinity than other drugs and N3 inhibitor; and hence, these drugs are advocated for further validation using in vitro enzyme inhibition and in vivo studies against SARS-CoV-2 infection.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, drug repurposing, structure-based virtual screening, molecular docking, molecular dynamics simulation
1. Introduction
Upper and lower respiratory infections caused by viruses are very common in temperate climates (Hing et al., 2006) and cause rhinitis, pharyngitis, sinusitis, bronchiolitis and pneumonia (Canducci et al., 2008; Jevšnik et al., 2012). Coronaviruses (CoVs) are a large group of RNA viruses with single-stranded RNA genomes that cause 30% of upper and lower respiratory tract infections in humans. However, in the last two decades, four human coronaviruses (HCoVs) have been identified, including severe acute respiratory syndrome (SARS)-CoV in 2003 (Fouchier et al., 2003), HCoV-NL63 in 2004 (van der Hoek et al., 2004), HCoV-HKU1 in 2005 (Vabret et al., 2006), and MERS-CoV in 2012 (de Groot et al., 2013). Recent COVID-19 pandemic caused by the fifth HCoVs considered as novel coronavirus (2019-nCoV), also known as SARS-CoV-2; suggested to emerged from Wuhan, Hubei Province of China (Giwa & Desai, 2020), which is already announced as pandemic by the WHO (Chen et al., 2020; Zhu et al., 2020). Generally, person infected with SARS-CoV-2 exhibits clinical symptoms, such as fever, cough, shortness of breath and dyspnea; in severe case, infection can cause pneumonia, kidney failure, SARS and even death (Wu et al., 2020). However, there is currently no specific therapeutic drug or vaccine has been approved for the treatment of HCoVs; hence, under present conditions, HCoVs outbreak is a potential danger to the human health and world economy (Kirtipal et al., 2020).
SARS-CoV-2 is a medium-sized, enveloped, positive-strand RNA virus (∼30 kb) of genus Betacoronavirus and characterized by a crown-shaped (corona) appearance in electron micrographs of negatively stained preparations (Chang et al., 2016; Kim et al., 2020). The viral genome deciphers several structural and non-structural proteins that assist virion in the infection to reproduce in a conserved linear arrangement (Kirtipal et al., 2020). Since, HCoVs, including SARS-CoV-2, encoded the polyprotein which further releases the functional polypeptides through proteolytic digestion by 33.8 kDa main protease (Mpro), also known as 3-chymotrypsin-like cysteine protease, 3CLpro (Kirtipal et al., 2020; Yang et al., 2003). Hence, because of functional importance of Mpro in the viral life cycle, viral Mpro has been suggested as potential target in the antiviral development against SARS-CoV-2 (Cui et al., 2019; Du Toit, 2020). Although, lack of therapeutic agents against SARS-CoV-2 posing a great difficulty to clinicians in disease management, but recent studies have highlighted the significance of existing drugs and their repurposing for disease management. For instance, some of noted recommendations are the synergistic use of antimalarial drugs, such as Chloroquine-Hydroxychloroquine (Colson et al., 2020) and Remdesivir-Favipiravir (Dong et al., 2020). Additionally, Huanzhu Lu has suggested neuraminidase inhibitors, Remdesivir, Peptide (EK1), abidol, RNA synthesis inhibitors (TDF and 3TC), anti-inflammatory drugs (hormones and other molecules), and Chinese traditional medicine (ShuFengJieDu Capsule and Lianhuaqingwen Capsule) as possible treatment against SARS-CoV-2 (Lu, 2020).
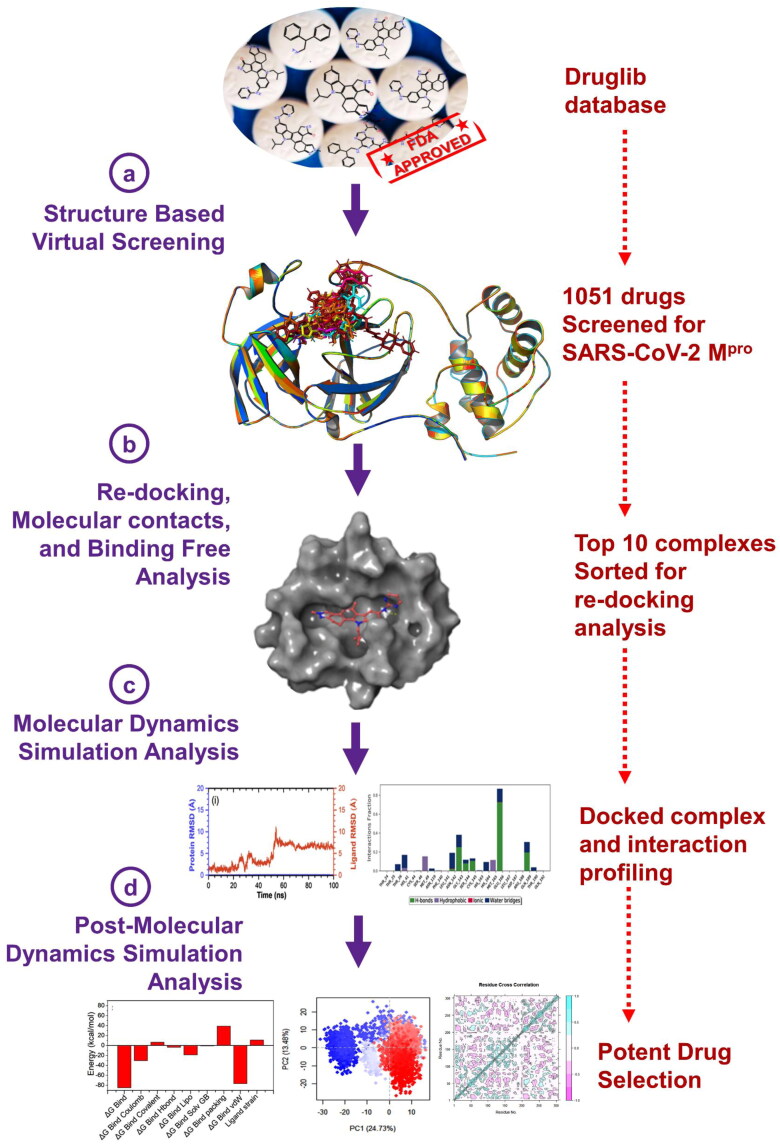
Provided with rapid surge in the information about the viral structure, mechanism and pathology, targeted drug discovery processes have been accelerated against SARS-CoV-2. However, considering the conventional timeframe for new drug development and alarming SARS-CoV-2 proliferation, drug repurposing has been established as better solution against SARS-CoV-2 as recommended by WHO (Kupferschmidt & Cohen, 2020). As limited research is conducted to screen the known FDA approved drugs against SARS-CoV-2 Mpro, therefore, we investigate the potential FDA approved drugs using structure-based virtual screening, all atoms molecular simulation, and post-simulation analysis against SARS-CoV-2 Mpro, as shown in Figure 1.
Figure 1.
Repurposing of FDA approved drugs from Druglib database via MTiOpenScreen virtual screening web server against SARS-CoV-2 Mpro compounds.
2. Materials and methods
2.1. Structure-based virtual screening
Structure-based virtual screening was performed against SARS-CoV-2 Mpro (PDB ID: 6LU7) (Jin et al., 2020) to identify the potential inhibitors from Druglib database-containing FDA approved drugs using MTiopen screen web server (Labbe et al., 2015). The viral protease contained three structural domains; domain I (Phe8-Tyr101), domain II (Lys102-Pro184), and domain III (Thr201-Val303), interconnected by loop of residues Phe185 to Ile200. The active pocket with catalytic dyad (Cys145 and His41) was defined in the cleft between domains I and II of SARS-CoV-2 Mpro (Jin et al., 2020). For virtual screening, SARS-CoV-2 Mpro was prepared under default parameters using Dock prep tool in Chimera-1.14 (Pettersen et al., 2004). Besides, residues (His41, Phe140, Gly143, Cys145, His163, His164, Glu166, Gln189, and Thr190) interacting with native ligand N3 inhibitor in the crystal structure of viral protease were considered as active residues for virtual screening against Druglib database via MTiOpenScreen virtual screening web server (Labbe et al., 2015).
2.2. Re-docking and free-binding energy calculations
Following, top 10 docked poses of protein-drug complexes were retrieved from virtual screening and re-docked again in the pocket covering same active residues using Chimera-1.14-AutoDock Vina plugin setup, as reported earlier (Bharadwaj, Lee, Dwivedi, Kang, et al., 2020; Bharadwaj, Lee, Dwivedi, Yadava, et al., 2020). Briefly, protein and screened drugs were minimized under default parameters in structure minimization tool in USCF Chimera-1.14 (Pettersen et al., 2004) and saved in PDB format. Following, these 3D structures were prepared with Dock prep tool in Chimera-1.14, where polar hydrogen atoms and charges were added. Later, molecular docking was conducted using AutoDock Vina (Trott & Olson, 2010), as plugin in USCF Chimera-1.14 under default parameters on the active residues covered by docking grid box of size 60 × 40 × 40 Å along the three (X, Y and Z) axes while center at −8.918, 17.918, and 62.905 Å region. Finally, top poses with the highest docking score and least RMSD (0 value by default) were selected for intermolecular interaction profiling using ligand-receptor interaction module in academic version of Maestro version 12.3 (Schrödinger Release 2020-1: Maestro, Schrödinger, LLC, New York, NY, 2020). Herein, various intermolecular interactions between ligands and active residues of protein, i.e. hydrogen bonding, hydrophobic, π-cation, π–π interaction, interaction, negative, positive, glycine, polar, and salt bridges formation, were calculated at cutoff radius of 4 Å under default conditions. Likewise, same docking procedure was also employed for the selected reference compound, i.e. Michael acceptor inhibitor or N3 inhibitor, which was reported as native ligand in the crystal structure of SARS-CoV-2 Mpro (Jin et al., 2020), to justify the docking methodology and for relative analysis against screened FDA approved drugs. Both 3D and 2D interaction images were rendered using academic version of Maestro version 12.3 package (Schrödinger Release 2020-1:Maestro, Schrödinger, LLC, New York, NY, 2020).
Furthermore, re-docked complexes were also analyzed for binding free energy using Prime molecular mechanics (MM)/generalized born surface area (GBSA) module in Schrodinger suite (Schrödinger Release 2019-2:Maestro, Schrödinger, LLC, New York, NY, 2019) and can be expressed using the following Equation (1):
| (1) |
where ΔGMMGBSA Bind depicted the binding free energy, ΔGComplex marked for the binding energy of docked receptor-ligand complex while ΔGLigand and ΔGReceptor showed the energy of receptor and ligand, respectively.
2.3. Explicit molecular dynamics simulation
The selected docked poses for each protein-ligand complexes were further subjected to 100 nanoseconds (ns) molecular dynamics (MD) simulation under Linux environment on HP Z280 workstation using academic version of Maestro-Desmond version 5.6 module of Schrödinger-Maestro version 11.8 suite (Schrödinger Release 2018-4, Schrödinger, LLC, New York, NY, 2018) (Bowers et al., 2006). Briefly, protein was prepared using protein preparation wizard module of Schrödinger suite under default parameters followed by generation of orthorhombic grid box (10 × 10 × 10Å buffer) as simulation box using system building tool. The complete prepared system was immersed in Monte Carlo equilibrated periodic transferable intermolecular potential with 4 points (TIP4P) water bath and neutralized with suitable number of sodium counter ions; salt and ions placement were omitted at 20 Å resolution around ligand. Following, 0.002 ps time steps for anisotropic diagonal position scaling were selected to maintain constant pressure for MD simulation process. Moreover, temperature was gradually raised from 100 to 300 K coupled with 20 ps NPT reassembly at 1 atm pressure. Following, density of the complete simulation system was preserved at 1 g/cm3 and MD calculation was conducted under default force field, i.e. optimized potentials for liquid simulations (OPLS)-2005, parameters in academic version of Desmond version 5.6-Maestro suite version 11.8. Finally, MD simulation was performed for each selected docked complex under similar conditions for 100 ns interval. Subsequently, MD simulation trajectory for each system was investigated using simulation interaction diagram tool of the Desmond version 5.6 module in Schrödinger-Maestro version 11.8 package (Bharadwaj, Rao, et al., 2020; Dwivedi et al., 2020).
2.4. Post-molecular dynamics simulation analysis
2.4.1. Molecular mechanics generalized born surface area calculations
The end-point binding free energy in terms of MM/GBSA was calculated for each simulated complex of SARS-CoV-2 Mpro with screened FDA approved drugs and reference ligand using Prime MMGBSA module of the MM/GBSA protocol in Schrödinger suite as mentioned in Section 2.2. Herein, MM/GBSA calculations were computed under OPLS-2005 force field with default parameters on the extracted snapshots from 100 ns MD simulation trajectory of each system, where complexes were refined by deletion of explicit TIP4P water molecules and counter ions, as reported earlier (Mena-Ulecia et al., 2015).
2.4.2. Essential dynamics and dynamic cross-correlation matrix (DCCM) profiling
Essential dynamics aided in the elucidation of correlated fluctuations in protein structure that are essentially required for the protein function. Hence, generated MD trajectory for each complex was considered for essential dynamics via principle components analysis (PCA) using Bio3d package (Grant et al., 2006). Moreover, residual displacement in the docked protein structure during the simulation interval was also scrutinized for residue correlation coefficient by dynamic cross-correlation analysis in Bio3d package (Grant et al., 2006). Both essential dynamics and DCCM analysis were conducted on all the Carbon-alpha (Cα) atoms of each complex extracted as 5000 frames from 100 ns MD trajectory and later superimposed to the initial docked pose to reduce the root mean square variances among the equivalent residues in the protein structure. These calculations were computed for each simulation trajectory under R environment (Team, 2013) with Bio3d package for simulation trajectory analysis (Grant et al., 2006).
3. Results and discussion
3.1. Virtual screening, re-docking and MM/GBSA analysis
Virtual screening is the use of high-performance computing to screen the large-small molecule databases for the identification of potential ligands against a specified drug target (Bharadwaj, Rao, et al., 2020; Dwivedi et al., 2016). MTiOpenScreen webserver predicts the potential drug molecules based on the minimum binding energy and correspondingly generates three significant binding poses against each screened molecule (Labbe et al., 2015). In this study, structure-based virtual screening of SARS-CoV-2 Mpro against Druglib database yields a total of 1051 FDA approved drugs with significant docking score between −10.3 and −7.5 kcal/mol (Table S1). Following, top 10 docked drugs were selected for re-docking analysis in AutoDock Vina (Figure 2) revealed improved binding score, as mentioned in Table 1, and occupied the same site on in the protease active pocket (Figure S1). Interestingly, all the selected drug molecules showed binding affinity greater than −8 kcal/mol against N3 inhibitor (7.6 kcal/mol), which is considerably significant for SARS-CoV-2 Mpro docked ligands, as reported earlier ( Bharadwaj, Lee, Dwivedi, Yadava, et al., 2020; Bharadwaj, Lee, Dwivedi, Kang, et al., 2020; Elmezayen et al., 2020).
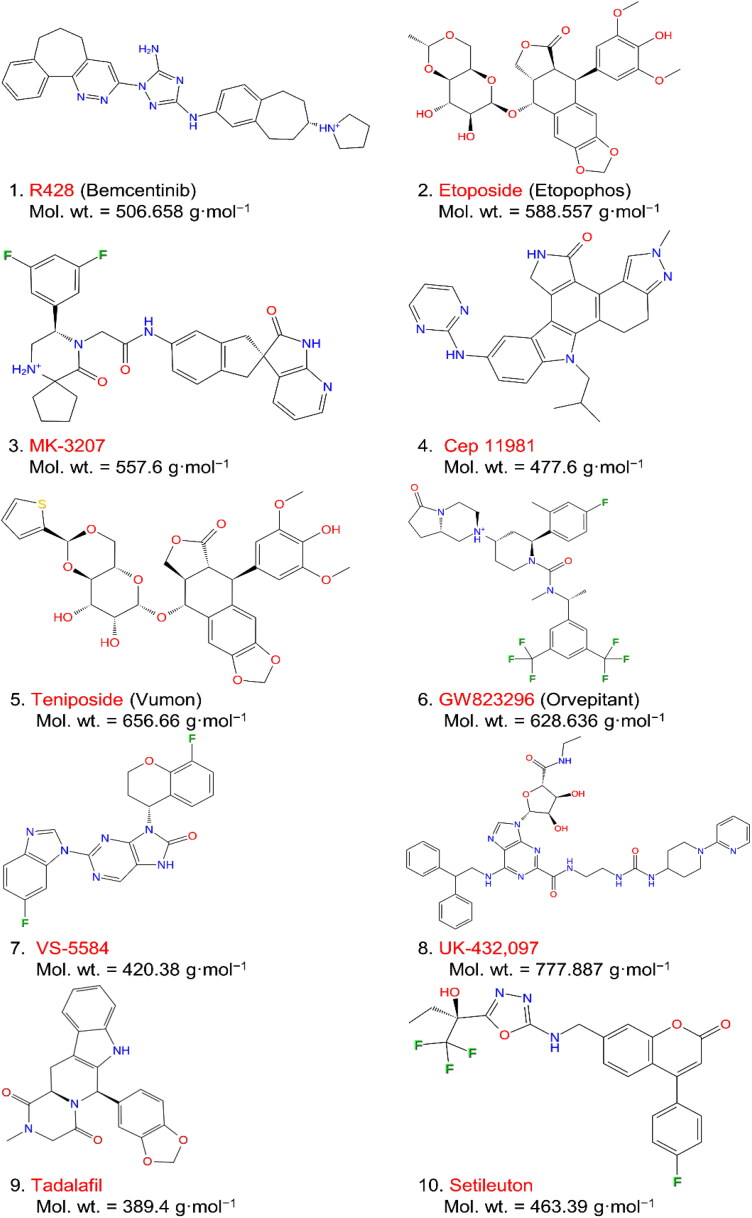
Figure 2.
List of screened top 10 FDA approved drugs against SARS-CoV-2 Mpro with 2D structural formula and molecular weight.
Table 1.
List of virtual screened drug molecules from Druglib database with re-docking score and type of interaction in the active pocket of SARS-CoV-2 Mpro.
| S.no. | Drugs | Docking energy (kcal/mol) | H-bond | π–π stacking |
Hydrophobic | Polar | Negative | positive | Glycine |
|---|---|---|---|---|---|---|---|---|---|
| 1. | R428 | −10.5 | Phe140, Gly143, Glu166, Thr190 |
His41 | Leu27, Val42, Met49, Phe140, Leu141, Cys145, Met165, Leu167, Pro168, Ala191 |
Thr25, Thr26, His41, Asn142, Ser144, His163, His164, His172, Gln189, Thr190, Gln192 | Glu166 | Arg188 | Gly143 |
| 2. | Etoposide | −10.3 | Leu141, His163, Thr190 |
His41 | Leu27, Met49, Tyr54 Phe140, Leu141, Cys145, Met165, Leu167, Pro168, Ala191 |
His41, Asn142, Ser144, His163, His172, Gln189, Thr190, Gln192 | Glu166, Asp187 |
Arg188 | Gly143 |
| 3. | MK-3207 | −9.9 | Gln189 | -- | Leu27, Val42, Met49, Phe140, Leu141, Cys145, Met165, Pro168 |
Thr25, Thr26, His41, Asn142, Ser144, His163, His164, His172, Gln189, Thr190, Gln192 | Glu166 | Arg188 | Gly143 |
| 4. | CEPT-11981 | −9.9 | -- | -- | Leu27, Met49, Tyr54, Phe140, Lleu141, Cys145, Met165, Leu167, Pro168 | Thr24, Thr25, Thr26, His41, Asn142, Ser144, His163, His164, His172, Gln189, Thr190, Gln192 |
Glu166, Asp187 | Arg188 | Gly143 |
| 5. | Teniposide | −9.8 | Thr26, His41, Glu166 | -- | Leu27, Met49, Pro52, Tyr54, Cys145, Met165, Leu167, Pro168 | Thr24, Thr25, Thr26, His41, Asn142, Ser144, His163, His164, Gln189, Thr190, Gln192 | Glu166, Asp187 |
Arg188 | Gly143 |
| 6. | Orvepitant | −9.6 | Glu166, Gln192 | -- | Leu27, Met49, Phe140 Leu141, Cys145, Met165, Leu167, Pro168, Ala191 |
Thr25, Thr26, His41, Thr45, Ser46, Asn142, Ser144, His163, His164, His172, Gln189, Thr190, Gln192 |
Glu166 | Arg188 | Gly143 |
| 7. | VS-5584 | −9.4 | Gly143, Glu166 | -- | Leu27, Met49, Phe140, Leu141, Cys145, Met165, Leu167, Pro168 | His41, Asn142, Ser144, His163, His164, His172, Gln189, Thr190, Gln192 | Glu166, Asp187 |
Arg188 | Gly143 |
| 8. | UK-432097 | −9.3 | Leu141, Gly143, Ser144, Glu166 | -- | Leu27, Met49, Phe140, Leu141, Cys145, Met165, Leu167, Pro168, Val171, Ala191 | Thr25, His41, Ser139, Asn142, Ser144, His163, His164, His172, Gln189, Thr190, Gln192 | Glu166 | Lys137, Arg188 |
Gly138, Gly143, Gly170 |
| 9. | Tadalafil | −9.2 | Gly143 | His41 | Met49, Tyr54, Phe140, Leu141, Cys145, Met165 |
His41, Asn142, Ser144, His163, His164, His172, Gln189 |
Glu166, Asp187 |
Arg188 | Gly143 |
| 10. | Setileuton | −8.5 | -- | -- | Leu27, Met49, Leu141, Cys145, Met165, Leu167, Pro168, Ala191 | Thr25, Thr26, His41, Asn142, Ser144, His163, His164, Gln189, Thr190 Gln192 |
Glu166, Asp187 |
Arg188 | Gly143 |
| 11. | N3 inhibitor |
−7.6 | Cys145, Glu166, Gln189(2) | – | Leu27, Met49, Tyr54 | Thr24, Thr25, Thr26, His41, Thr45, Ser46, Asn142, Ser144, His163, His164, His172, Gln189, Thr190, Gln192 | Glu166, Asp187 | Arg188 | Gly143 |
3.2. Intermolecular interaction and MM/GBSA analysis
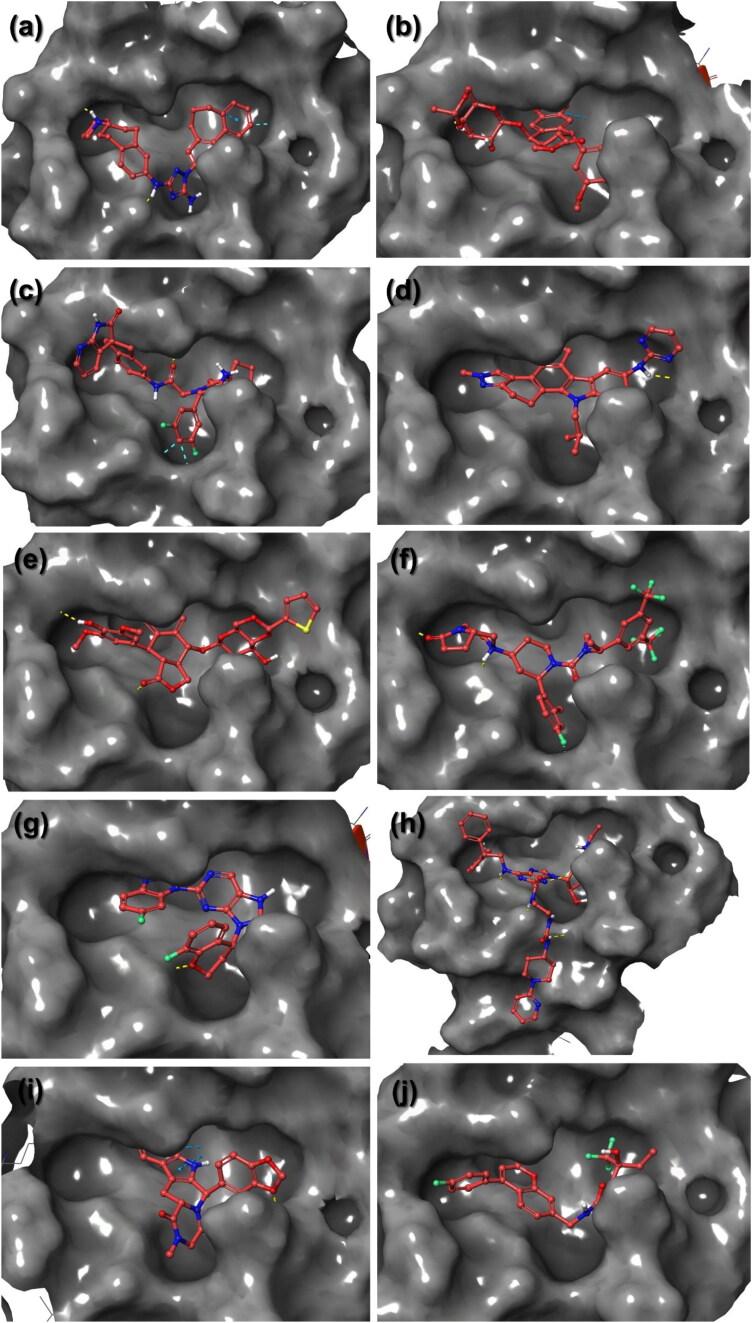
The screened best poses for all the 10 drugs molecules docked with SARS-CoV-2 Mpro were analyzed for intermolecular interaction which assisted in the formation of stable docked complex. All the selected molecules showed significant placement in the binding pocket of SARS-CoV-2 Mpro (Figure 3) and shared some common residues for intermolecular interaction (Table 1, Figure S2). Since, hydrogen bonding in the docked complex marked for stability of the ligand with receptor, a maximum and a minimum of four and one hydrogen bonds, respectively were observed in the respective docked complexes; in addition to other intermolecular interactions (Table 1, Figure S2). Based on the number of hydrogen bond formation, selected drugs can be placed in the descending order; R428, UK-432097, Etoposide, Teniposide, Orvepitant, Vs5584, MK-3207, Tadalafil, CEP-11981, and Setileuton, as potent inhibitor of SARS-CoV-2 Mpro. Moreover, all the drugs showed polar and hydrophobic interactions with the catalytic dyad residues His41 and Cys145, respectively, except R428 and Etoposide also formed π–π stacking with His41 while Teniposide exhibited hydrogen bond formation with Cys145 residue. Moreover, these re-docked poses were also studied in comparison to the best pose of SARS-CoV-2 Mpro-N3 inhibitor for intermolecular interactions profiling which showed occupancy of similar residues (Table 1, Figure S3). It is important to mention that SARS-CoV-2 Mpro-N3 inhibitor complex showed intermolecular molecular interaction via formation of four hydrogen bonds with residues Cys145, Glu166, and Gln189(2) along with other intermolecular interactions (Table 1, Figures S2 and S3). Interestingly, in SARS-CoV-2 Mpro-FDA approved drug complexes, a maximum of four hydrogen bonds were exhibited by R428 and UK-432097 in the active pocket of viral protease (Table1, Figures S2). Additionally, both screened FDA drugs and N3 inhibitor showed substantial hydrophobic, polar, negative, positive, and glycine interactions with common residues in the active pocket of SARS-CoV-2 Mpro (Table 1, Figures S2 and S3). Moreover, the screened drugs also depicted interactions with the same residues as documented for the native ligand N3 inhibitor, i.e. Phe140, Gly143, Ser144, His163, and Glu166, with SARS-CoV-2 Mpro (Table 1, Figures S2 and S3). These active residues have been previously reported for strong non-covalent contacts formation with the substrate-binding cleft located between domains I and II of SARS-CoV-2 Mpro crystal structure (Zhang et al., 2020). However, additional residue contacts were observed between the screened drugs and viral protease Mpro (Table 1, Figures S2 and S3). These drug molecules, except R428, MK-3207, Orvepitant, and UK-432097, also interacted with Asp187 residue which was suggested to enhance the catalytic efficiency of viral Mpro (Zhao et al., 2008). The molecular contacts of screened drugs with the essential residues in the protease structure suggested the inhibition of SARS-CoV-2 Mpro proteolytic function, which is essentially required for the viral replication and pathogenesis. Hence, comparative molecular docking analysis of screened FDA approved drugs against N3 inhibitor suggested the potential of selected drugs to inhibit SARS-CoV-2 Mpro by formation of hydrogen and non-covalent interaction with its catalytic dyad and substrate binding residues.
Figure 3.
3D docked poses of screened drugs; (a) R428, (b) Etoposide, (c) MK-3207, (d) CEP-11981, (e) Teniposide, (f) Orvepitant, (g) VS-5584, (h) UK-432097, (i) Tadalafil, and (j) Setileuton, in the active pocket of SARS-CoV-2 Mpro.
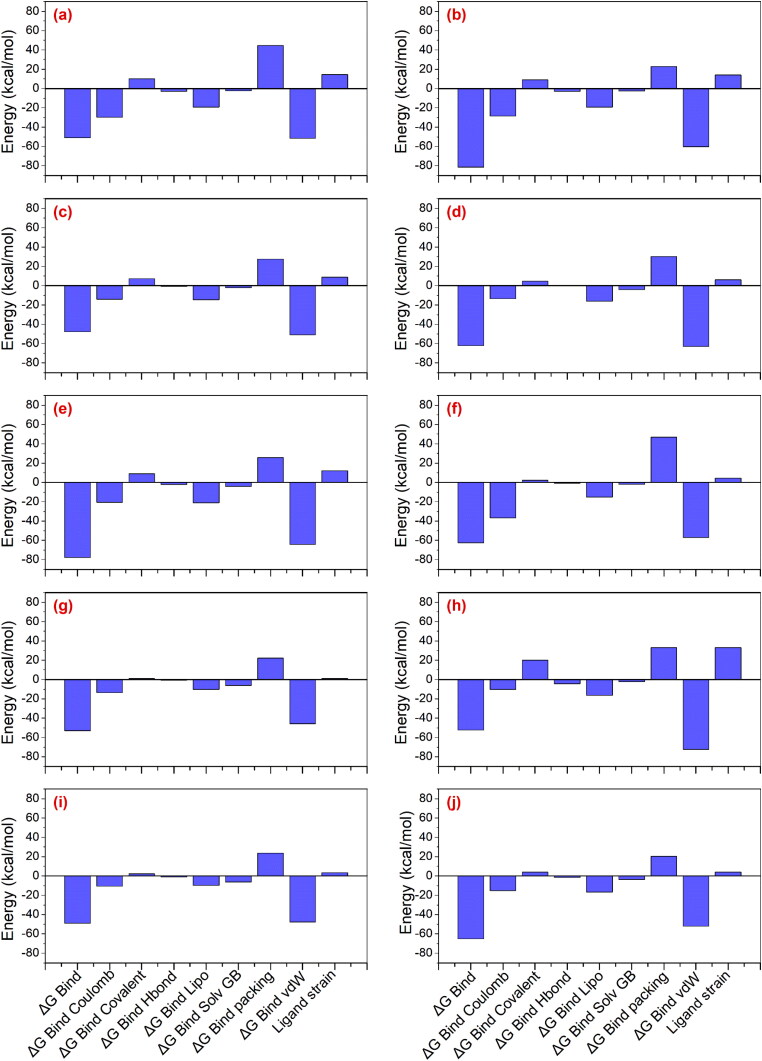
To further established the role of intermolecular interactions between screened drugs and SARS-CoV-2 Mpro, binding free energy along with contributing energy components were assessed using Prime MM/GBSA method for each docked complex (Table S2, Figures 4 and S4). These results suggested the maximum contribution of ΔGBind Coulomb and ΔGBind vdW in the stability of the respective complexes while ΔGBind Covalent and ΔGBind Packing contribute in the destabilization of the docked respective complexes. Remarkably, SARS-CoV-2 Mpro-Etoposide, SARS-CoV-2 Mpro-Cept-11981, SARS-CoV-2 Mpro-Teniposide, SARS-CoV-2 Mpro-Orvepitant, and SARS-CoV-2 Mpro-Setileuton docked complexes exhibited relatively higher binding free energy (>−60 kcal/mol) against other docked drug complexes and N3 Inhibitor complex (−84.75 kcal/mol). These observations further support the potential of screened drugs for the inhibition of SARS-CoV-2 Mpro.
Figure 4.
Total molecular mechanics generalized born surface area (MM/GBSA) binding free energy (kcal/mol) values calculated for screened drugs, i.e. (a) R428, (b) Etoposide, (c) MK-3207, (d) CEP-11981, (e) Teniposide, (f) Orvepitant, (g) Vs-5584, (h) UK-432097, (i) Tadalafil, and (j) Setileuton, docked with SARS-CoV-2 Mpro.
3.3. Molecular dynamics simulation analysis
MD simulation is a widely accepted computational method in drug discovery in order to understand the physical interactions, such as structure-function relationships, intramolecular/intermolecular interactions, and other structural properties, at the atomic-level for the biological macromolecules (Bharadwaj, Lee, Dwivedi, Yadava, et al., 2020; Bharadwaj, Lee, Dwivedi, Kang, et al., 2020). Thereof, the selected SARS-CoV-2 Mpro-screened drug complexes were monitored for the docked complex stability under 100 ns MD simulations conducted in Desmond suite. The produced MD trajectory for each docked complex were scrutinized in terms of last snapshot of simulated docked complex, root mean square deviation (RMSD), root mean square-fluctuations (RMSF), and protein-ligand contacts mapping with respect to 100 ns interval, as reported earlier ( Bharadwaj et al., 2019).
The last snapshot from each simulated complex trajectory was analyzed for the occupancy in the active pocket and formation of molecular contacts with viral protease by comparison to N3 inhibitor as reference ligand (Figures 5 and S5). Interestingly, all the drugs were relatively noted within the catalytic pocket of SARS-CoV-2 Mpro and observed for formation of substantial interactions, including hydrogen bonds and other intermolecular interactions against N3 inhibitor (Figures 5, S5 and S6). Besides, Table S3 exhibits the intermolecular interaction profiling for extracted last snap shot from the respective 100 ns simulation trajectories and supports considerable stability of drugs R428, MK3207, CEPT-11981, Teniposide, VS-5584, and Setileuton with viral protease based on formation of hydrogen bonds with essential residues against N3 inhibitor, as noticed in the respective initial docked poses (Table 1). Hence, these observations indicate the considerable stability of these protein-drug complexes against other docked drugs and reference ligand N3 inhibitor with SARS-CoV-2 Mpro.
Figure 5.
3D poses extracted at the end of 100 ns MD simulation for the screened drugs, i.e. (a) R428, (b) Etoposide, (c) MK-3207, (d) CEP-11981, (e) Teniposide, (f) Orvepitant, (g) VS-5584, (h) UK-432097, (i) Tadalafil, and (j) Setileuton, docked with SARS-CoV-2 Mpro where protein surface is generated based on the nature of residue property and ligand color was computed based on the nature of atom in the structure.
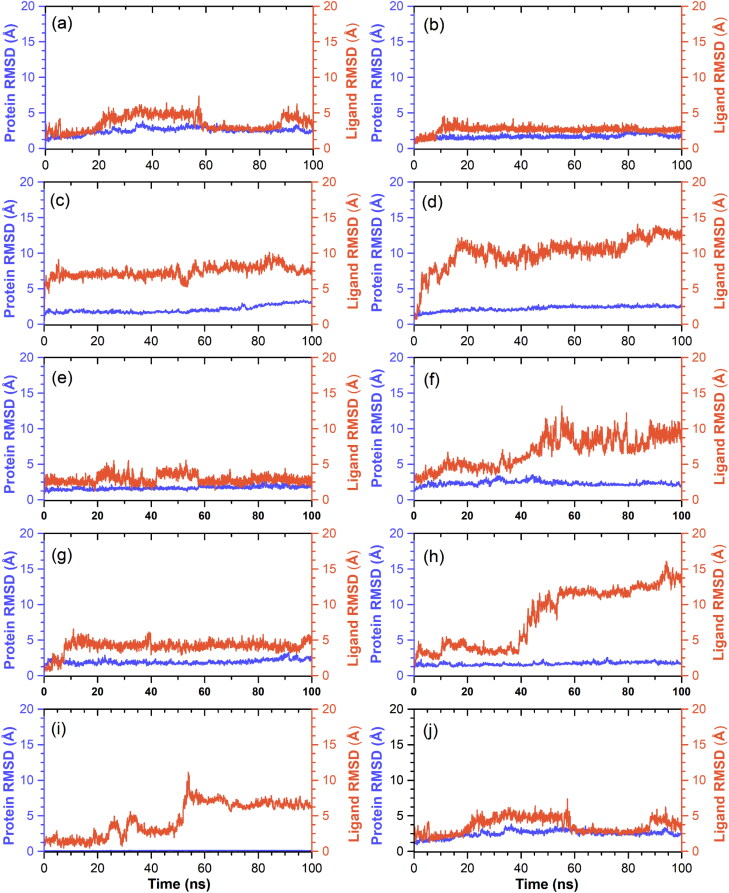
Figure 6.
RMSD values for alpha carbon atoms (violet color curves) of SARS-CoV-2 Mpro and selected ligands (red curves), viz. (a) R428, (b) Etoposide, (c) MK-3207, (d) CEP-11981, (e) Teniposide, (f) Orvepitant, (g) VS-5584, (h) UK-432097, (i) Tadalafil, and (j) Setileuton, were plotted with respect to 100 ns simulation time.
Moreover, an average change in the displacement of Cα atoms (RMSD) of viral protease and protein fit drug molecule as ligand in reference to their corresponding initial frames were analyzed for each SARS-CoV-2 Mpro-drug complex by comparison to SARS-CoV-2 Mpro-N3 inhibitor complex (Figures 6 and S7). Remarkably, Cα atoms in each docked complex showed acceptable deviation (<2.5 Å) during 100 ns simulation interval (Figures 6 and S7). These variations in Cα atoms were further supported by RMSF of respective receptor in docked complexes, where residues showed deviations <2.5 Å except in N- and C-terminal of the viral protease (<4.5 Å) (Figures S8 and S9). As N- and C-terminal of the protein structure does not contribute substantially to the activity of the viral protease, these residual fluctuations in each complex are considered as acceptable during the molecular dynamic’s simulation. Likewise, RMSD values for protein fit drugs were extracted and analyzed from 100 ns simulation interval (Figure 6). Although, occasional fluctuations <6.0 Å were recorded for some of protein fit drugs, i.e. R428, Etoposide, MK-3207, Teniposide, VS-5584, Tadalafil, and Setileuton, but these docked ligands in the active pocket of SARS-CoV-2 Mpro exhibited equilibrium within 40–60 ns and remained till end of 100 ns simulation interval. Thus, final RMSD values for ligands R428 (<3.2 Å), Etoposide (<2.6 Å), MK-3207 (<8.0 Å), CEP-11981 (<13.0 Å), Teniposide (<2.7 Å), Orvepitant (<10.0 Å), VS-5584 (<4.8 Å), UK-432097 (<12.8 Å), Tadalafil (<6.0 Å), and Setileuton (<3.2 Å) were noted at the end of 100 ns simulation interval (Figure 6). However, N3 inhibitor docked with viral protease initially observed with considerable RMSD values followed by higher deviations (<18.0 Å) after 80 ns till end of MD simulation (Figures S7), suggested that N3 inhibitor required more than 100 ns interval to achieve the equilibrium with viral protease. Besides, RMSF analysis for all the screened drug atoms showed acceptable variations (<3.5 Å) except in the regions with heavy atoms (<10.5 Å) by comparison to N3 inhibitor (Figures S10 and S11).
Figure 7.
Protein-ligand interactions mapping recorded for the screened drugs; (a) R428, (b) Etoposide, (c) MK-3207, (d) CEP-11981, (e) Teniposide, (f) Orvepitant, (g) VS-5584, (h) UK-432097, (i) Tadalafil, and (j) Setileuton, with SARS-CoV-2 Mpro were extracted from 100 ns MD simulations.
Figure 9.
Principal component analysis of MD simulation trajectories for SARS-CoV-2 Mpro docked with (a) R428, (b) Etoposide, (c) MK-3207, (d) CEP-11981, (e) Teniposide, (f) Orvepitant, (g) VS-5584, (h) UK-432097, (i) Tadalafil, and (j) Setileuton. Herein, continuous color changes from blue to white to red exhibit periodic jumps between structural conformations extracted from 100 ns simulation trajectories.
Furthermore, protein-ligand intermolecular interactions for the screened drugs against N3 inhibitor were also extracted during the course of 100 ns MD simulation, including hydrogen bonds, hydrophobic contacts, ionic interactions, and water bridge formations, which reflect the binding strength and permanency of docked ligands with viral protease (Figures 7 and S12). Interestingly, various residues were recorded for the substantial interactions with the screened drugs by comparison to N3 inhibitor during 100 ns simulation (Figures 7 and S12). For instance, screened drugs showed one or more than one type of interaction with active residues, viz. catalytic dyad (His41 and Cys145), substrate binding residues (Phe140, Leu141, Gly143, Ser144, His163, Met165, and Glu166), and other essential substrate-binding residues (Thr24, Thr25, Met49, Phe140, Asn142, His163, Met165, Asp187, and Gln189) (Figure 7), as defined in the crystal structure of SARS-CoV-2 Mpro with N3 inhibitor (Jin et al., 2020). Remarkably, these residues were also logged in the respective docked complexes of the protease with screened drugs (Table 1), further support the stability of selected drugs with active pocket of SARS-CoV-2 Mpro. Also, protein-ligand interactions profile extracted at 30% of simulation interval for each MD trajectory demonstrates the highest number of non-covalent interactions for all the docked drugs, except Orvepitant, with catalytic dyad residues and other essential residues in the active pocket of SARS-CoV-2 Mpro against N3 inhibitor (Figures S13 and S14).
Conclusively, only drugs R428, Teniposide, VS-5584, and Setileuton against N3 inhibitor docked with SARS-CoV-2 Mpro were noted for significant molecular contacts via hydrogen bonding with viral protease in the extracted last pose from MD trajectories, acceptable variations values, and substantial interactions with essential residues of viral protease. Hence, based on MD simulation analysis, R428, Teniposide, VS-5584, and Setileuton drugs are concluded as potential inhibitors of SARS-CoV-2 Mpro than other drug molecules and N3 inhibitor.
3.4. Post-molecular dynamics simulation analysis
3.4.1. Binding free energy calculations
To compute the influence of MD simulation on the binding free energy of ligands with the active pocket of SARS-CoV-2 Mpro, snapshots collected from respective MD simulations were analyzed for binding affinity using Prime MM/GBSA method (Figure 8). Total ΔGBind and individual energy components values computed on the respective snapshots from simulation trajectories are given Table S4. All the respective snapshots for SARS-CoV-2 Mpro-FDA approved drugs exhibited no considerable change in the net binding energy, except reduction in binding affinity was noted for Etoposide, Orvepitant, Setileuton, and N3 inhibitor docked with viral protease (Figures 8 and S15). Additionally, analysis of individual components contributing in the total binding free energy of the respective complexes, i.e. ΔGBind Coulomb, ΔGBind Covalent, ΔGBind Hbond, ΔGBind Lipo (hydrophobic interactions), ΔGBind Solv GB (Generalized Born electrostatic solvation energy), ΔGBind packing (π–π interactions), and ΔGBind vdW (van der Waals energy) revealed significant contribution of ΔGBind Coulomb and ΔGBind vdW energy in the respective complex stability (Table S4, Figures 8 and S15), supports the favorable enthalpy formation in the respective complexes. Similar results for the energy components ΔGBind Coulomb and ΔGBind vdW contributing in the docked complexes were reported earlier ( Bharadwaj et al., 2019). Moreover, no significant contribution of ligand strain energy was observed in all the complexes, indicates the considerable affinity of screened drugs by comparison to N3 inhibitor with the active pocket of SARS-CoV-2 Mpro.
Figure 8.
End point binding free energy (kcal/mol) values calculated for snapshots extracted for SARS-CoV-2 Mpro docked with screened drugs, i.e. (a) R428, (b) Etoposide, (c) MK-3207, (d) CEP-11981, (e) Teniposide, (f) Orvepitant, (g) VS-5584, (h) UK-432097, (i) Tadalafil, and (j) Setileuton, from respective 100 ns MD simulation trajectories.
3.4.2. Essential dynamics and dynamic cross-correlation matrix analysis
Essential dynamics were performed on the MD trajectories to collect the major Eigen values, also known as principal component analysis (PCA) via a covariance-matrix-based mathematical technique to elucidate the protein domain dynamics and residual displacements. Herein, PCA components were extracted for SARS-CoV-2 Mpro docked with screened drugs compounds, R428, Etoposide, MK-3207, CEP-11981, Teniposide, Orvepitant, VS-5584, UK-432097, Tadalafil, and Setileuton, as shown in Figures S16; exhibit the percentage of variance (%) (Eigen fraction) for mean square positional fluctuations in the covariance matrix as a function of 20 Eigen modes. In all SARS-CoV-2 Mpro-drugs complexes, each system demonstrated a rapid drop in Eigen fraction against N3 inhibitor corresponds to the early three Eigen modes (Figures S16 and S17). These values suggest a major level of conformational mobility caused by docked ligand in active pocket of viral protease. However, successive elbow point and no notable change in variations of the Eigen fraction were detected following 4th Eigen value (Figures S16 and S17). These results intimated that substantial flexibility exist in SARS-CoV-2 Mpro docked with selected compounds during initial phase of MD simulation that diminished with respect to time. Also, slow decrement in proportional contribution of the Eigen modes advised the additional localized fluctuations in SARS-CoV-2 Mpro docked with each ligand to obtain the favorable stability. Hence, these variations in each complex were advised to play a key role in the stability of respective docked complexes.
Furthermore, first two Eigen vectors for SARS-CoV-2 Mpro docked with each compound that were obtained from respective MD trajectory as cluster groups, indicated compact and cluster motions for SARS-CoV-2 Mpro during 100 ns simulation, except in SARS-CoV-2 Mpro-Etoposide complex and SARS-CoV-2 Mpro-MK-3207 complex (Figures 9 and S17). Also, the generated plots suggested the variations in cluster distribution in each conformation during MD simulation via color gradient change from blue to red stands for periodic jumps among the various conformational poses of docked viral protease. In conclusion, a restricted correlated fluctuations and motions of the viral protease in all the studied systems represent the rigidity and stability of respective docked complexes, except in SARS-CoV-2 Mpro-Etoposide complex and SARS-CoV-2 Mpro-MK-3207 complex, during MD simulation.
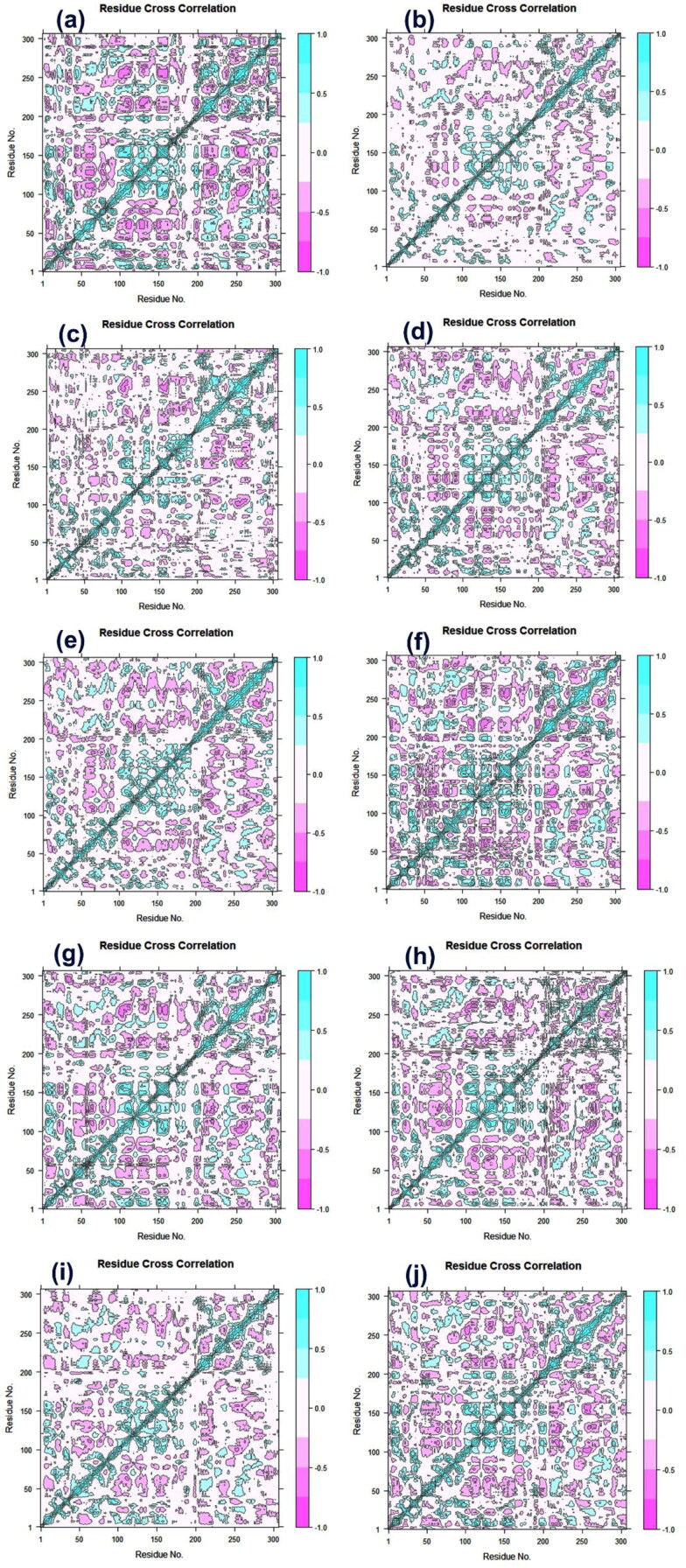
Furthermore, to calculate the conformational dynamics changes instituted in SARS-CoV-2 Mpro structure in each complex due to inhibitory activity of docked ligands, DCCM analysis was computed based on the positions of Cα atoms. Figure 7 shows high correlated motions which varies from the light blue to cyan color (+1) while anti-correlated motions are in the range from light purple to red brick color (−1). Analysis of residue cross correlation suggested a considerable correlated motions and dynamic changes in all the systems by comparison to N7 inhibitor (Figures 10 and S17). The calculated results established that screened drugs and N3 inhibitors significantly induced the restricted conformation changes in the viral protease structure during 100 ns MD simulation (Figure 10).
Figure 10.
Dynamic cross-correlation matrix analysis for SARS-CoV-2 Mpro complexed with screened FDA approved drugs, i.e. (a) R428, (b) Etoposide, (c) MK-3207, (d) CEP-11981, (e) Teniposide, (f) Orvepitant, (g) VS-5584, (h) UK-432097, (i) Tadalafil, and (j) Setileuton. Herein, cyan color explains positive correlation and cyan red signifies the negative correlation in the movement of residues during the course of 100 ns simulation interval.
Based on the combinatorial computational analysis, including structure-based virtual screening, molecular docking, binding free energy calculations, MD simulation and post-MD simulation analysis for the screened FDA drugs with viral protease, suggested the drugs, R428, Teniposide, VS-5584, and Setileuton with comparatively higher stability and affinity with SARS-CoV-2 Mpro against N3 inhibitor via strong intermolecular interactions formation as well disturbing the conformation of viral protease active pocket.
4. Conclusion
This study was aimed to repurpose the FDA approved drugs as potential inhibitor of SARS-CoV-2 Mpro using structure-based virtual screening approach and validation by MD simulation. Initially, 1051 compounds were obtained from structure-based virtual screening of Druglib database and only top 10 drug molecules were selected as potential ligands of SARS-CoV-2 Mpro for further intermolecular and complex stability. The selected drugs showed significant interactions with the catalytic (His41 and Cys145) and other substrate-binding residues in docked complexes as well as during 100 ns MD simulation. Thereof, based on collective observations, R428, Teniposide, VS-5584, and Setileuton drugs were suggested to hold the potential to significantly occupy the active pocket of SARS-CoV-2 Mpro and can be used as its inhibitors. Hence, computational screening and validation of top 10 drugs recorded in this study can be further evaluated against in vitro SARS-CoV-2 Mpro inhibition and viral infection for the drug formulation against SARS-CoV-2 infection.
Supplementary Material
Acknowledgments
Authors are highly thankful to Dr. Amaresh Kumar Sahoo, Indian Institute of Information Technology, Prayagraj, India for providing his kind support in binding free energy calculation using Prime MMGBSA module of Schrodinger Suite 2019.2.
Funding Statement
This stu
dy was supported by The Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia, under grant no. FP-3-42.
Disclosure statement
The authors declare no competing interests.
References
- Bharadwaj, S., Lee, K. E., Dwivedi, V. D., & Kang, S. G. (2020). Computational insights into tetracyclines as inhibitors against SARS-CoV-2 M-pro via combinatorial molecular simulation calculations. Life Sciences, 257, 118080. 10.1016/j.lfs.2020.118080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj, S., Lee, K. E., Dwivedi, V. D., Yadava, U., Nees, M., & Kang, S. G. (2020). Density functional theory and molecular dynamics simulation support Ganoderma lucidum triterpenoids as broad range antagonist of matrix metalloproteinases. Journal of Molecular Liquids, 311, 113322. 10.1016/j.molliq.2020.113322 [DOI] [Google Scholar]
- Bharadwaj, S., Lee, K. E., Dwivedi, V. D., Yadava, U., Panwar, A., Lucas, S. J., Pandey, A., & Kang, S. G. (2019). Discovery of Ganoderma lucidum triterpenoids as potential inhibitors against Dengue virus NS2B-NS3 protease. Scientific Reports, 9(1), 19059. 10.1038/s41598-019-55723-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj, S., Rao, A. K., Dwivedi, V. D., Mishra, S. K., & Yadava, U. (2020). Structure-based screening and validation of bioactive compounds as Zika virus Methyltransferase (MTase) inhibitors through first-principle density functional theory, classical molecular simulation and QM/MM affinity estimation. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1747545 [DOI] [PubMed] [Google Scholar]
- Bowers, K. J., Chow, D. E., Xu, H., Dror, R. O., Eastwood, M. P., Gregersen, B. A., Sacerdoti, F. D.Shaw, D. E., Shan, Y., Salmon, J. K., Moraes, M. A., Kolossvary, I., Klepeis, J. L. (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters. Paper presented at the SC’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. IEEE. [Google Scholar]
- Canducci, F., Debiaggi, M., Sampaolo, M., Marinozzi, M. C., Berrè, S., Terulla, C., Gargantini, G., Cambieri, P., Romero, E., & Clementi, M. (2008). Two‐year prospective study of single infections and co‐infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. Journal of Medical Virology, 80(4), 716–723. 10.1002/jmv.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. K., Lo, S. C., Wang, Y. S., & Hou, M. H. (2016). Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discovery Today, 21(4), 562–572. 10.1016/j.drudis.2015.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Liu, Q. Y., & Guo, D. Y. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92(4), 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, P., Rolain, J. M., Lagier, J. C., Brouqui, P., & Raoult, D. (2020). Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. International Journal of Antimicrobial Agents, 55(4), 105932. 10.1016/j.ijantimicag.2020. 105932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J., Li, F., & Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology, 17(3), 181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot, R. J., Baker, S. C., Baric, R. S., Brown, C. S., Drosten, C., Enjuanes, L., Fouchier, R. A. M., Galiano, M., Gorbalenya, A. E., Memish, Z. A., Perlman, S., Poon, L. L. M., Snijder, E. J., Stephens, G. M., Woo, P. C. Y., Zaki, A. M., Zambon, M., & Ziebuhr, J. (2013). Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the coronavirus study group. Journal of Virology, 87(14), 7790–7792. 10.1128/JVI.01244-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L. Y., Hu, S. S., & Gao, J. J. (2020). Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discoveries & Therapeutics, 14(1), 58–60. 10.5582/ddt.2020.01012 [DOI] [PubMed] [Google Scholar]
- Du Toit, A. (2020). Outbreak of a novel coronavirus. Nature Reviews Microbiology, 18(3), 123–123. 10.1038/s41579-020-0332-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi, V. D., Bharadwaj, S., Afroz, S., Khan, N., Ansari, M. A., Yadava, U., Tripathi, R. C., Mishra, S. K., Kang, S. G. (2020). Anti-dengue infectivity evaluation of bioflavonoid from Azadirachta indica by dengue virus serine protease inhibition. Journal of Biomolecular Structure & Dynamics, 1–4. 10.1080/07391102.2020.1734485 [DOI] [PubMed] [Google Scholar]
- Dwivedi, V. D., Tripathi, I. P., Bharadwaj, S., Kaushik, A. C., & Mishra, S. K. (2016). Identification of new potent inhibitors of dengue virus NS3 protease from traditional Chinese medicine database. Virusdisease, 27(3), 220–225. 10.1007/s13337-016-0328-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Sahin, A. T., & Yelekci, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure & Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier, R. A. M., Kuiken, T., Schutten, M., van Amerongen, G., van Doornum, G. J. J., van den Hoogen, B. G., Peiris, M., Lim, W., Stöhr, K., & Osterhaus, A. D. M. E. (2003). Aetiology: Koch’s postulates fulfilled for SARS virus. Nature, 423(6937), 240–240. 10.1038/423240a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giwa, A., & Desai, A. (2020). Novel coronavirus COVID-19: An overview for emergency clinicians. Emergency Medicine Practice, 22(2), 1–21. [PubMed] [Google Scholar]
- Grant, B. J., Rodrigues, A. P., ElSawy, K. M., McCammon, J. A., & Caves, L. S. (2006). Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics, 22(21), 2695–2696. 10.1093/bioinformatics/btl461 [DOI] [PubMed] [Google Scholar]
- Hing, E., Cherry, D. K., & Woodwell, D. A. J. A. D. (2006). National ambulatory medical care survey: 2004 summary. Advance Data, 374(374), 1–33. [PubMed] [Google Scholar]
- Jevšnik, M., Uršič, T., Žigon, N., Lusa, L., Krivec, U., & Petrovec, M. (2012). Coronavirus infections in hospitalized pediatric patients with acute respiratory tract disease. BMC Infectious Diseases, 12(1), 365. 10.1186/1471-2334-12-365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Kim, J.-M., Chung, Y.-S., Jo, H. J., Lee, N.-J., Kim, M. S., Woo, S. H., Park, S., Kim, J. W., Kim, H. M., & Han, M.-G. (2020). Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health and Research Perspectives, 11(1), 3–7. 10.24171/j.phrp.2020.11.1.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirtipal, N., Bharadwaj, S., & Kang, S. G. (2020). From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 85, 104502. 10.1016/j.meegid.2020.104502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt, K., & Cohen, J. (2020). Race to find COVID-19 treatments accelerates. Science, 367(6485), 1412–1413. 10.1126/science.367.6485.1412 [DOI] [PubMed] [Google Scholar]
- Labbé, C. M., Rey, J., Lagorce, D., Vavruša, M., Becot, J., Sperandio, O., Villoutreix, B. O., Tufféry, P., & Miteva, M. A. (2015). MTiOpenScreen: A web server for structure-based virtual screening. Nucleic Acids Research, 43(W1), W448–W454. 10.1093/nar/gkv306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. Z. (2020). Drug treatment options for the 2019-new coronavirus (2019-nCoV). Bioscience Trends, 14(1), 69–71. 10.5582/bst.2020.01020 [DOI] [PubMed] [Google Scholar]
- Mena-Ulecia, K., Tiznado, W., & Caballero, J. (2015). Study of the differential activity of thrombin inhibitors using docking, QSAR, molecular dynamics, and MM-GBSA. PLoS One, 10(11), e0142774. 10.1371/journal.pone.0142774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Team, R. C. (2013). R: A language and environment for statistical computing. Team, R. C. [Google Scholar]
- Trott, O., & Olson, A. J. (2010). Software news and update auto dock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret, A., Dina, J., Gouarin, S., Petitjean, J., Corbet, S., & Freymuth, F. (2006). Detection of the new human coronavirus HKU1: A report of 6 cases. Clinical Infectious Diseases, 42(5), 634–639. 10.1086/500136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek, L., Pyrc, K., Jebbink, M. F., Vermeulen-Oost, W., Berkhout, R. J. M., Wolthers, K. C., Wertheim-van Dillen, P. M. E., Kaandorp, J., Spaargaren, J., & Berkhout, B. (2004). Identification of a new human coronavirus. Nature Medicine, 10(4), 368–373. 10.1038/nm1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10, 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Yang, M., Ding, Y., Liu, Y., Lou, Z., Zhou, Z., Sun, L., Mo, L., Ye, S., Pang, H., Gao, G. F., Anand, K., Bartlam, M., Hilgenfeld, R., & Rao, Z. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences of the United States of America, 100(23), 13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q., Li, S., Xue, F., Zou, Y., Chen, C., Bartlam, M., & Rao, Z. (2008). Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. Journal of Virology, 82(17), 8647–8655. 10.1128/JVI.00298-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W., China Novel Coronavirus Investigating and Research Team. (2020). A novel coronavirus from patients with Pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.