Abstract
Objectives
To determine aetiology of illness among children and adults presenting during outbreak of severe respiratory illness in Southern Province, Sri Lanka, in 2018.
Design
Prospective, cross-sectional study.
Setting
1600-bed, public, tertiary care hospital in Southern Province, Sri Lanka.
Participants
410 consecutive patients, including 371 children and 39 adults, who were admitted with suspected viral pneumonia (passive surveillance) or who met case definition for acute respiratory illness (active surveillance) in May to June 2018.
Results
We found that cocirculation of influenza A (22.6% of cases), respiratory syncytial virus (27.8%) and adenovirus (AdV) (30.7%; type B3) was responsible for the outbreak. Mortality was noted in 4.5% of paediatric cases identified during active surveillance. Virus type and viral coinfection were not significantly associated with mortality.
Conclusions
This is the first report of intense cocirculation of multiple respiratory viruses as a cause of an outbreak of severe acute respiratory illness in Sri Lanka, and the first time that AdV has been documented as a cause of a respiratory outbreak in the country. Our results emphasise the need for continued vigilance in surveying for known and emerging respiratory viruses in the tropics.
Keywords: respiratory infections, public health, epidemiology, public health
Strengths and limitations of this study.
In the midst of an outbreak, we implemented active surveillance for acute respiratory illness and rapidly set up and validated laboratory testing for respiratory pathogens in a lower-resource setting.
The compilation of data from both passive and active surveillance methods provides a comprehensive description of this outbreak.
Data collected through active and passive surveillance were inherently different, with less clinical and outcome data collected through passive surveillance.
Different molecular testing protocols were used in conducting testing of samples collected through active and passive surveillance.
Introduction
Important knowledge gaps exist in the epidemiology of respiratory viral infections in the tropics and subtropics. Current surveillance systems for respiratory viruses target the identification of novel viruses, such as the recently emerged SARS-CoV-2, given their potential for widespread devastation.1 However, the circulation and disease burden of more familiar viruses such as influenza and respiratory syncytial virus (RSV) has only recently received attention in the tropics.2 3 Elucidating seasonal variation in the prevalence of common respiratory viral infections is important locally and regionally for preventative and control activities, and detecting shifts in local transmission patterns or severe clinical manifestations also has global significance.4
In Sri Lanka, data regarding respiratory virus seasonality are limited and largely confined to influenza.5–7 As per the Sri Lankan Ministry of Health, influenza occurs in two peaks annually, with the larger peak occurring in May to July and the second peak occurring in November to January, with most infections due to influenza A.7–9 National epidemiological data indicated 3646 hospital admissions and 35 deaths due to influenza (0.96% of influenza admissions) in 2017, with approximately one-fifth of cases occurring in patients<5 years of age.10 Anecdotally, increased numbers of hospitalisations and deaths due to influenza are noted during peak periods, although robust data are lacking. Data regarding the burden and seasonality of other respiratory viruses in Sri Lanka are limited.5
In May to June 2018, clinicians noted an increase in case numbers of children and adults presenting with acute respiratory infection in Southern Province, Sri Lanka. Cases were noted to be of high severity in children, and many of the severe cases were referred to Teaching Hospital Karapitiya (THK), the largest tertiary referral centre in the Southern Province and one of only four hospitals in the country with a paediatric intensive care unit (PICU). The onset of cases corresponded with the expected first peak of the bimodal influenza season for the province.5 9 11 However, rapid diagnostic testing by immunochromatographic test (ICT) was negative for influenza A and B in the majority of initial cases, raising suspicion for another causative pathogen. Consequently, we supplemented ongoing laboratory-based, passive surveillance at THK with active surveillance to determine the aetiology of acute respiratory illness during this suspected outbreak.
Methods
Setting
THK is a public, 1600-bed hospital with 10 adult (≥12 years) medical wards, 3 paediatric wards, 4 adult ICUs and 1 PICU. All care is provided free of charge to patients. THK serves as the primary tertiary referral centre for the Southern Province, with the majority of admissions being self-referrals from patients in the surrounding Galle district.
Laboratory-based passive surveillance
During the period of the suspected outbreak (May to June 2018), THK had ongoing laboratory-based, passive surveillance for influenza. Clinician-identified cases suspected to be viral pneumonia were referred to the virology laboratory, where a rapid ICT for influenza A and B was performed on nasopharyngeal (NP) samples using the SD BIOLINE Influenza Ag test (Abbott, Abbott Park, Illinois). At the time of the outbreak, THK did not have capacity to perform molecular testing for respiratory viruses on site. When the ICT was negative for influenza, an aliquot of the sample was sent to the national reference laboratory, Medical Research Institute (MRI), in the capital, Colombo (approximately 60 miles away), for additional aetiological testing.NP samples were generally collected 24–72 hours after admission, placed in viral transport media (VTM; Sigma Virocult Duo, Medical Wire & Equipment, Corsham, England), separated into aliquots, and frozen at −80°C before shipment to MRI where the following viral targets were identified using multiplex polymerase chain reaction (PCR) (Altona Diagnostics, Hamburg, Germany and National Institute of Virology, Pune, India): influenza A, B, human adenovirus (AdV) and respiratory syncytial virus A and B (RSVA and RSVB). The time required to ship samples to MRI delayed reporting of results to THK clinicians, sometimes after cases had expired or were discharged.
Active surveillance
To augment ongoing passive surveillance, a local outbreak response team was quickly assembled to establish active surveillance including implementation of a multiplex, respiratory viral, real-time polymerase chain reaction (RT-PCR) platform at the affiliated Faculty of Medicine, University of Ruhuna (Galle, Sri Lanka). We conducted active surveillance concurrently with the existing passive surveillance programme to improve case identification and collect additional clinical information. Active surveillance of all paediatric wards and the PICU at THK was conducted from 28 May 2018 through 21 June 2018 by trained MBBS-qualified research assistants. Cases were identified as patients with a history of fever (subjective or observed in hospital) plus one or more of the following symptoms of respiratory illness: cough, difficulty breathing, shortness of breath, and hypoxia or tachypnea (as noted in the medical record by the treating physician). In children<5 years, lower chest wall indrawing/retractions, central cyanosis, inability to drink/breastfeed, vomiting everything, altered consciousness and convulsions were also considered symptoms of respiratory illness as per WHO Integrated Management of Childhood Illness guidelines.12 Patients were enrolled within 24–72 hours after admission, except on the first day of active surveillance when all patients meeting the case definition were enrolled.
For active surveillance cases, NP samples were placed in VTM, separated into aliquots, and frozen at −20°C before being tested for influenza A, AdV and RSV with RT-PCR using a Mic qPCR Cycler (Bio Molecular Systems, Queensland, Australia). Primers and probes were sourced from IDTdna (Coraville, Iowa, USA). Probes were labelled at the 5′ end with carboxyfluorescein (FAM), with an internal ZEN quencher and a 3′ Iowa Black Dark FQ Quencher. A week-long training of local laboratory technical staff and quality assurance/control was directed by an expert in molecular diagnostics (BP Nicholson) to set up the RT-PCR platform. The platform was established, validated and processing samples within 2 weeks.
Reporting of viral test results
A subset of case samples was tested on both the passive and active surveillance PCR platforms and results were compared to assess concordance. A positive virus identification from either platform was considered a positive result and reported to clinicians caring for case subjects. If different viral targets were identified between platforms, all positive viral identifications were reported.
Sequencing of AdV isolates
Sequencing of identified AdV isolates was performed for a convenience subset of isolates identified through passive surveillance (whole virus sequencing) and active surveillance (hexon gene fragment sequencing). Whole virus sequencing was performed at Hong Kong University (Peiris Lab, Hong Kong). Nucleic acid was extracted and quantified, and the hexon gene was amplified using consensus primers and Sanger sequenced for virus subtype identification.13 14 Subsequently, primers were designed based on reference genomes of the virus subtype for performing long PCRs with overlapping to amplify the complete virus genome. Long PCRs were performed using LA Taq DNA polymerase Hot-Start version (Takara, Nojihigashi, Japan) and products sequenced with Illumina NovaSeq 6000 platform (Illumina, San Diego, California). Sequencing libraries were prepared using the NextEra XT library preparation kit (Illumina) and paired-end reads with length of 150 nucleotides were generated. Sequencing raw read data were mapped to a reference genome using BWA and data processed with SAMtools.15 16 AdV full genomes were generated by taking the consensus of the mapped sequencing read data with a minimal sequencing read coverage of 100 times at each nucleotide position.
For hexon gene sequencing, a section of the AdV hexon gene was amplified using primers previously described.13 The PCR products were confirmed via gel electrophoresis, and successful amplicons were purified through agarose gel filtration to remove unincorporated primers, nucleotides and salts. The purified fragments were sequenced using bidirectional Sanger sequencing reactions by Eton Bioscience, Research Triangle Park, North Carolina, USA. The chromatogram files were aligned using SeqMan Pro, Lasergene 16 Package (DNASTAR, Madison, Wisconsin, USA). All primer sequences were trimmed from the contiguous sequences.
Patient and public involvement
It was not appropriate or possible to involve patients or the public in the design, or conduct, or reporting, or dissemination plans of our research
Results
In total, 410 individual patients were identified during passive and active surveillance, of whom 371 (90.5%) were children and 388 (94.6%) underwent testing for virus identification. A total of 87 patients were represented in both passive and active surveillance. The median age at presentation was 1.3 (IQR 0.7–3.3) years and 325 (79.3%) were less than 5 years of age. The median duration of illness at hospital admission was 5 (IQR 3–11) days, and the median length of hospitalisation was 5 (IQR 4–8) days.
Passive surveillance
NP swabs from 205 paediatric and 12 adult (≥18 years) cases were collected after referral to the THK Infection Control team, and 120 (55.3%) were positive for a respiratory virus (table 1). Among paediatric patients, positive viral identification was as follows: 41 (20.0% of all tested children) for influenza A, 9 (4.4%) for influenza B, 38 (18.5%) for AdV and 54 (26.3%) for RSV. All but one RSV-positive sample underwent testing for further identification as RSVA (40 19.5%) or RSVB (14, 6.8%). Coinfection was detected in 21 (10.2%) of children. Of adults, two (16.7% of all tested) were positive for influenza A. No adult samples were positive for influenza B, AdV, RSVA or RSVB.
Table 1.
Results from PCR for cases identified during 2018 respiratory viral outbreak in Southern Province, Sri Lanka*
| Viral target | Passive surveillance n=231 | Active surveillance n=179 | Overall n=410 | ||||||
| <2 years n=136 | ≥2 and <5 years n=41 | ≥5 years n=40 | <2 years n=107 | ≥2 and <5 years n=41 | ≥5 years n=29 | <2 years n=243 | ≥2 and <5 years n=82 | ≥5 years n=69 | |
| MonoInfection | 63 (46.3) | 20 (48.8) | 16 (40.0) | 67 (62.6) | 25 (61.0) | 18 (62.1) | 130 (53.5) | 45 (54.9) | 34 (49.3) |
| Influenza A | 13 (9.6) | 8 (19.5) | 12 (30.0) | 10 (9.3) | 12 (29.3) | 6 (20.7) | 23 (9.5) | 20 (24.4) | 18 (26.1) |
| Influenza B† | 3 (2.2) | 1 (2.4) | 3 (7.5) | 0 | 0 | 0 | 3 (1.2) | 1 (1.2) | 3 (4.3) |
| AdV | 20 (14.7) | 4 (9.8) | 1 (2.5) | 31 (29.0) | 11 (26.8) | 10 (34.5) | 51 (21.0) | 15 (18.3) | 11 (15.9) |
| RSV | 27 (19.8) | 7 (17.1) | 0 | 26 (24.3) | 2 (4.9) | 2 (6.9) | 53 (21.8) | 9 (11.0) | 2 (2.9) |
| CoInfection | 14 (10.3) | 5 (12.2) | 2 (5.0) | 21 (19.6) | 6 (14.6) | 5 (17.2) | 35 (14.4) | 11 (13.4) | 7 (10.1) |
| Influenza A+AdV | 1 (0.7) | 0 | 0 | 6 (5.6) | 4 (9.8) | 3 (10.3) | 7 (2.9) | 4 (4.9) | 3 (4.3) |
| Influenza A+RSV | 4 (2.9) | 2 (4.9) | 1 (2.5) | 2 (1.9) | 1 (2.4) | 0 | 6 (2.5) | 3 (3.6) | 1 (1.4) |
| AdV+RSV | 6 (4.4) | 3 (7.3) | 0 | 12 (11.2) | 1 (2.4) | 1 (3.4) | 18 (7.4) | 4 (4.9) | 1 (1.4) |
| Other | 3 (2.2) | 0 | 1 (2.5) | 1 (0.9) | 0 | 1 (3.4) | 4 (1.6) | 0 | 2 (2.9) |
| Negative | 55 (40.4) | 13 (31.7) | 16 (40.0) | 19 (17.8) | 10 (24.4) | 6 (20.7) | 74 (30.4) | 23 (28.0) | 22 (31.9) |
| Not tested | 4 (2.9) | 3 (7.3) | 6 (15.0) | 0 | 0 | 0 | 4 (1.6) | 3 (3.6) | 6 (8.7) |
| Total positives | 77 (56.6) | 25 (61.0) | 18 (45.0) | 88 (82.2) | 31 (75.6) | 23 (79.3) | 165 (67.9) | 56 (68.3) | 41 (59.4) |
Frequency (%) are listed in the table by age group.
*Age missing for 16 patients.
†Testing only performed at Medical Research Institute, the national reference laboratory.
AdV, adenovirus; RSV, respiratory syncytial virus.
Active surveillance
NP swabs were obtained from 166 paediatric and 11 adult patients meeting the case definition for acute respiratory infection, and 142 (80.2%) were positive for a respiratory virus. Positive viral identification among children was as follows: 43 (25.9% of all children tested) for influenza A, 76 (45.8%) for AdV and 49 (29.5%) for RSV. Among adults, seven tested positive for a respiratory virus, with two influenza A, four AdV and one AdV and influenza A coinfection.
Combined surveillance data of paediatric cases
The epidemic curve of cases among paediatric patients admitted to THK for the duration of the suspected outbreak is depicted in figure 1. In the case of duplicates (ie, cases that were represented in both passive and active screening), only the active case result was included. Of a total of 371 paediatric NP samples collected through active and passive surveillance, ≥1 viral target was detected in 253 (68.2%) patients: 84 (22.6%) were positive for influenza A, 9 (2.4%) for influenza B, 114 (30.7%) for AdV and 103 (27.8%) for RSV. Of 87 cases for whom samples were tested on both PCR platforms, there was >90% agreement in results. Coinfections (defined as >1 viral identification on either platform) were detected in 53 (12.9%) of cases.
Figure 1.
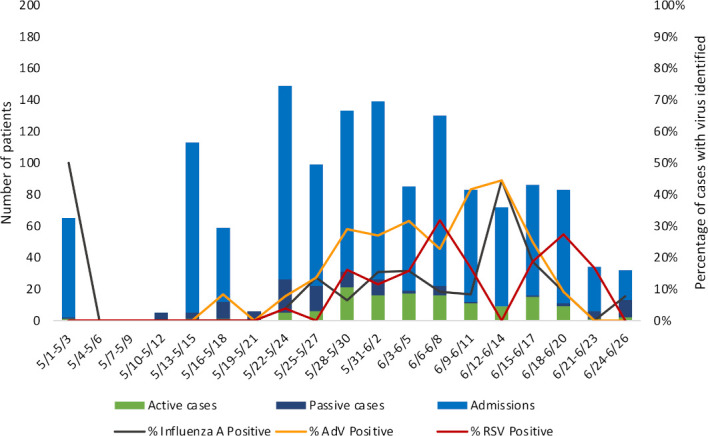
Epidemic curve of 2018 respiratory viral outbreak among paediatric patients in Southern Province, Sri Lanka. Paediatric (age<18 years) cases identified during both passive and active surveillance are depicted. AdV, adenovirus; RSV, respiratory syncytial virus.
Outcomes in paediatric patients
Paediatric cases identified through active surveillance were followed during hospitalisation for the occurrence of the following severe outcomes: repeat admissions for the same illness, ICU admission/transfer, use of supplemental oxygen (O2), use of mechanical ventilation (MV) and death (table 2). Data collected from passive cases were incomplete, thus are not reported. The median (IQR) duration of index hospitalisation was 5 (4–8) days. A total of 17 fatal paediatric cases who had samples sent for testing were observed (active=8, passive=9), with 3 testing positive for influenza A, 4 for AdV, 3 for RSV, 1 for influenza A and AdV, and 6 testing negative. Overall, among paediatric cases identified during active surveillance, eight (4.5%) died: two had influenza A identified, two had AdV, one had RSV, one had influenza A+AdV and two had negative testing results. Cases with RSV detection were more likely to have any severe outcome (table 2) compared with cases with AdV or influenza A; however, this finding was not statistically significant and cases with each of these viruses developed severe outcomes. A minority (9, 5.0%) of paediatric cases captured during active surveillance were immunocompromised due to a congenital syndrome or other chronic illness, but these cases represented more than one-third (37.5%) of fatal cases identified by active surveillance. Cases in which ≥1 virus was identified were as likely as cases in which no virus was identified to develop adverse clinical outcomes.
Table 2.
Outcomes of cases during 2018 respiratory viral outbreak in Southern Province, Sri Lanka, delineated by type of virus detected
| Viral detection result | Number with test result | Outcome | |||||
| Repeat admission* | ICU | O2 | MV | Death | Any severe outcome† | ||
| Influenza A | 26 | 3 (11.5) | 3 (11.5) | 5 (19.2) | 3 (11.5) | 2 (7.7) | 6 (23.1) |
| AdV | 48 | 7 (14.6) | 5 (10.4) | 14 (29.2) | 3 (6.2) | 2 (4.2) | 19 (39.6) |
| RSV | 30 | 2 (6.7) | 2 (6.7) | 14 (46.7) | 1 (3.3) | 1 (3.3) | 15 (50.0) |
| Influenza A+AdV | 12 | 1 (8.3) | 1 (8.3) | 2 (16.7) | 1 (8.3) | 1 (8.3) | 3 (25.0) |
| Influenza A+RSV | 3 | 1 (33.3) | 0 | 0 | 0 | 0 | 1 (33.3) |
| AdV+RSV | 14 | 2 (14.3) | 2 (14.3) | 5 (35.7) | 0 | 0 | 6 (42.8) |
| Other coInfection | 2 | 1 (50.0) | 0 | 0 | 0 | 0 | 1 (50.0) |
| Negative | 31 | 2 (6.4) | 6 (19.4) | 9 (29.0) | 4 (12.9) | 2 (6.4) | 9 (29.0) |
| Total | 166 | 19 (11.4) | 19 (11.4) | 49 (29.5) | 12 (7.2) | 8 (4.8) | 60 (36.1) |
Only cases and outcomes of paediatric patients collected during active surveillance are documented. Frequency and percentage of cases with the indicated viral test result are documented.
*Repeat admission=any prior hospitalisation(s) for the same illness.
†Any severe outcome=repeat admission, ICU, O2, MV or death.
AdV, adenovirus; ICU, intensive care unit admission; MV, requiring mechanical ventilation (which in some cases was provided outside ICUs due to space constraints); O2, requiring any form of supplemental oxygen; RSV, respiratory syncytial virus.
Genotyping and phylogenetic analyses
Of cases that were positive for AdV, a convenience sample of nine cases (two from passive surveillance; seven from active surveillance) was selected for sequencing. Table 3 indicates the clinical characteristics of cases that were selected for sequencing. Through analysis of the short hexon gene fragment, it was possible to assign all samples as species B type 3. The sequences were strongly monophyletic with 100% bootstrap support. Sequences were compared with known B3 strains in the Indo-Pacific region, and there was >99% similarity. Compared sequences included sequences from India (GenBank Accession No KF268210.1), Malaysia (KU145043.1, KU145041.1, KU145036.1), Taiwan (EF486497.1, KC570876.1), Hong Kong (KY511691.1), Korea (KY320276.1), China (KR090803.1), Japan (AB900154.1, AB900151.1) and the Philippines (AB742370.1, AB742369.1).
Table 3.
Clinical characteristics of 10 patients with type 3 adenovirus detection
| ID | Surveillance type | Date* | Coinfection (influenza/ RSV test result) | Age | O2/MV | Outcome | Hospitalisation duration |
| 1 | Passive | May 12 | Negative | 6 months | Yes, intubated | Not available | Not available |
| 2 | Passive | May 14 | Negative | 3.6 years | Missing | Not available | Not available |
| 3 | Passive | May 23 | Negative | 10 months | Missing | Died | Not available |
| 4 | Active | May 23 | RSV | 3 weeks | No | Discharged | 8 days |
| 5 | Active | May 29 | Negative | 3.5 years | 02 | Discharged | 11 days |
| 6 | Active | May 24 | Negative | 12 years | No | Discharged | 13 days |
| 7 | Active | May 24 | Negative | 1.2 years | No | Discharged | 13 days |
| 8 | Active | May 25 | Negative | 4 years | 02 | Discharged | 11 days |
| 9 | Active | May 29 | Influenza A | 2.5 years | No | Not available | Not available |
| 10 | Active | June 18 | Influenza A | 1 year | No | Discharged | 13 days |
All dates listed are for 2018.
*Date of sample receipt for cases obtained through passive surveillance; date of admission for cases obtained through active surveillance.
MV, requiring mechanical ventilation; O2, requiring any form of supplemental oxygen; RSV, respiratory syncytial virus.
Discussion
We describe an outbreak of respiratory viral illness that differed from previously reported outbreaks in Sri Lanka in the following important ways: (1) greater severity among previously healthy children, (2) cocirculation of multiple respiratory viruses and (3) identification of AdV as a cause of severe illness in children and adults.
This severe outbreak overwhelmed the already overburdened healthcare infrastructure and alarmed the general public. The demand for ICU beds, respiratory support equipment and physician attention quickly exceeded capacity, and delays in coordination among healthcare, public health and government officials initially hindered outbreak response efforts. Moreover, most of the fatalities occurred in children who were previously healthy with all requisite vaccinations, which was particularly disturbing to parents.
The influenza A and RSV infections noted during this outbreak were of greater severity than generally seen, for reasons that are not completely clear. National epidemiological data indicate that 0.96% of influenza admissions die during hospitalisation; in contrast, 7.7% of paediatric influenza A patients identified during active surveillance succumbed to their illness during the current outbreak.10 Robust data regarding morbidity and mortality associated with RSV do not exist in Sri Lanka, but a systematic review and meta-analysis of studies from 32 countries noted 6.21 deaths per 1000 children among medically attended cases; 3.3% of children with RSV identified during active surveillance died during this outbreak.17 As with any outbreak, incomplete data regarding actual case counts may show higher severity than in reality. However, anecdotally, clinicians noted higher severity of respiratory illness during this outbreak than during prior peak influenza seasons. Outbreaks such as this one highlight the need for evaluating the value and cost-effectiveness of influenza vaccination in Sri Lanka, where the vaccine is not available through the public healthcare sector.
Influenza A, RSV and AdV were all implicated in this outbreak of severe respiratory illness, with each contributing to approximately one-third of cases. Cocirculating viruses during the expected peak of influenza in the region may have impacted outbreak severity and duration, as was seen in the early weeks of the 2009 H1N1 influenza pandemic.18 19 Further studies into the drivers of viral cocirculation, as well as implications for severe disease and potential for outbreaks, are greatly needed in the tropics and subtropics. Viral coinfection was identified in 12.9% of individual cases. In comparison, approximately 20% of children with community-acquired pneumonia were noted to have viral coinfection in the USA, and 7.9% of children<5 years presenting to the outpatient setting of this hospital with influenza-like illness were documented to have viral coinfection in 2013–2015.5 20 Individuals with coinfection did not have more severe outcomes in this study, in contrast to other reports; further investigation is needed to distinguish coinfection from codetection in this and other outbreak settings.21
One likely contributing factor to the severity of illness seen was the emergence of human AdV—the most prevalent virus identified among outbreak cases—which had not previously been recognised as a cause of clinically significant illness in Sri Lanka. Our findings add to the growing number of reports of AdV as an emerging infectious disease in children and adults in Asia, and underscore the importance of surveillance programmes for respiratory viruses.22–27 Our findings are consistent with studies that indicate that over 80% of AdV infections occur in children<4 years due to a lack of humoral immunity.28 In addition, while severe infection generally occurs in immunocompromised patients, with AdV dissemination or severe respiratory failure developing in 10%–30% of cases, fatalities have been described in healthy children and adults.28
AdV-3, which is among the most common serotypes globally, was the culprit AdV strain in this outbreak and has been documented as a cause of respiratory illness and outbreaks in Asia.22 29–31 While generally associated with milder disease, AdV can cause fatal pneumonia in children, including in healthy children.32–36 In addition, AdV-3 has been associated with epidemics in healthy adult military recruits.37 Increased severity of adenoviral infections can be seen with recombinant AdVs or novel genotypic variants; sequencing of the hexon gene did not reveal such recombination in our study, though we did not perform restriction enzyme analysis to identify potentially novel strains.38 39 Currently, an enteric-coated vaccine exists for AdV types 4 and 7 among military recruits, and given the prevalence of AdV-3, there are efforts underway to develop a vaccine that covers type 3.28 40 Such a vaccine may prove useful if further outbreaks occur in this setting in the future. The occurrence of severe illness due to AdV also highlights the need for further investigation of therapeutics such as cidofovir and brincidofovir.28
Several limitations must be noted. There were some differences in the information resulting from active and passive surveillance, with AdV and RSV infections as well as all coinfections being more prevalent among active surveillance cases than passive surveillance cases. Such differences may be due to variations in sample collection or testing and from a more specific case definition being used in active surveillance. Reassuringly, there was >90% agreement in results among samples tested on both PCR platforms. We believe that the compilation of data from both passive and active surveillance methods is a strength in this report, as these efforts were carried out in the midst of an ongoing epidemic to better delineate aetiology and outcome of illness. As passive surveillance preceded active surveillance, more cases were identified through the former mechanism and inherently fewer illness and outcome data were collected. In addition, we may have missed some viral coinfections, since we could only test for a limited panel of viruses given resources available during the outbreak.
In conclusion, our results highlight the need for a better understanding of the epidemiology of respiratory viruses in the tropics, specifically for the following: (1) patterns of coinfection and cocirculation of influenza, AdV and RSV and (2) severe cases of AdV infection/coinfection, which are almost certainly under-recognised.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the clinical staff at Teaching Hospital Karapitiya, particularly in the Departments of Microbiology and Pediatrics. In addition, the authors thank the laboratory staff, especially Nishantha Abayawaradana Gunasekara, at the Department of Microbiology, Faculty of Medicine, University of Ruhuna, Galle, Sri Lanka; at the Medical Research Institute, Colombo, Sri Lanka; and at Hong Kong University School of Public Health, Hong Kong.
Footnotes
Contributors: SV: design of study, data collection, data analysis, manuscript preparation, manuscript editing and review. GW: data collection, generation of laboratory testing data, manuscript preparation, manuscript editing and review. ND, JJ, BP and RK: data collection, generation of laboratory testing data, manuscript editing and review. CH, VD and CKB: design of study, data collection, manuscript editing and review. TS: data analysis, manuscript preparation, manuscript editing and review. SA: data collection, manuscript editing and review. BPN, JSMP, GCG and SG: generation of laboratory testing data, manuscript editing and review. AN: design of study, generation of laboratory testing data, manuscript editing and review. CWW: design of study, data analysis, manuscript editing and review. LGT: design of study, data analysis, manuscript preparation, manuscript editing and review.
Funding: The authors acknowledge funding from the National Institutes of Health Fogarty Training Grant (D43 TW009337), Thrasher Research Foundation Early Career Award (grant number N/A), National Institutes of Allergy and Infectious Diseases (K23AI125677), and the Duke Hubert-Yeargan Center for Global Health (grant number N/A).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication: Not required.
Ethics approval: Ethical approval for this study was obtained from the Ethical Review Committee of the Faculty of Medicine, University of Ruhuna (Sri Lanka) and from the Duke University Institutional Review Board (USA). All research activities conformed to the principles embodied in the Declaration of Helsinki.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: No data are available.
References
- 1.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA 2020;323:1239–42. 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Reeves RM, Wang X, et al. Global patterns in monthly activity of influenza virus, respiratory syncytial virus, parainfluenza virus, and metapneumovirus: a systematic analysis. Lancet Glob Health 2019;7:e1031–45. 10.1016/S2214-109X(19)30264-5 [DOI] [PubMed] [Google Scholar]
- 3.Caini S, Andrade W, Badur S, et al. Temporal patterns of influenza A and B in tropical and temperate countries: what are the lessons for influenza vaccination? PLoS One 2016;11:e0152310. 10.1371/journal.pone.0152310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Research needs for the battle against respiratory viruses (brave), 2013. Available: http://www.who.int/influenza/patient_care/clinical/BRaVe_Research_Agenda_2013.pdf?ua=1 [Accessed 5 Mar 2018].
- 5.Shapiro D, Bodinayake CK, Nagahawatte A, et al. Burden and seasonality of viral acute respiratory tract infections among outpatients in southern Sri Lanka. Am J Trop Med Hyg 2017;97:88–96. 10.4269/ajtmh.17-0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perera KVHKK, Chan KH, Ma E, et al. Influenza virus infections among a sample of hospital Attendees in Ragama, Sri Lanka. Ceylon Med J 2010;55:40–4. 10.4038/cmj.v55i2.1987 [DOI] [PubMed] [Google Scholar]
- 7.Rafeek RAM, Divarathna MVM, Noordeen F. History and current trends in influenza virus infections with special reference to Sri Lanka. Virusdisease 2017;28:225–32. 10.1007/s13337-017-0390-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epidemiology Unit SLMoH Health education presentation on seasonal infuenza 2015, 2015. [Google Scholar]
- 9.Tillekeratne LG, Bodinayake CK, Nagahawatte A, et al. An under-recognized influenza epidemic identified by rapid influenza testing, southern Sri Lanka, 2013. Am J Trop Med Hyg 2015;92:1023–9. 10.4269/ajtmh.14-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ministry of Health NaIM Annual health statistics 2017, 2019. [Google Scholar]
- 11.Tillekeratne LG, Bodinayake CK, Simmons R, et al. Respiratory viral infection: an underappreciated cause of acute febrile illness admissions in southern Sri Lanka. Am J Trop Med Hyg 2019;100:672–80. 10.4269/ajtmh.18-0699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Revised WHO Classification and treatment of pneumonia in children at health facilities: evidence summaries. Geneva, 2014. [PubMed] [Google Scholar]
- 13.Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol 2006;151:1587–602. 10.1007/s00705-005-0722-7 [DOI] [PubMed] [Google Scholar]
- 14.Wolffs PFG, Bruggeman CA, van Well GTJ, et al. Replacing traditional diagnostics of fecal viral pathogens by a comprehensive panel of real-time PCRs. J Clin Microbiol 2011;49:1926–31. 10.1128/JCM.01925-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011;27:2987–93. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein RT, Bont LJ, Zar H, et al. Respiratory syncytial virus hospitalization and mortality: systematic review and meta-analysis. Pediatr Pulmonol 2017;52:556–69. 10.1002/ppul.23570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratnamohan VM, Taylor J, Zeng F, et al. Pandemic clinical case definitions are non-specific: multiple respiratory viruses circulating in the early phases of the 2009 influenza pandemic in New South Wales, Australia. Virol J 2014;11:113. 10.1186/1743-422X-11-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascalis H, Temmam S, Turpin M, et al. Intense co-circulation of non-influenza respiratory viruses during the first wave of pandemic influenza pH1N1/2009: a cohort study in reunion island. PLoS One 2012;7:e44755. 10.1371/journal.pone.0044755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain S, Williams DJ, Arnold SR, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med 2015;372:835–45. 10.1056/NEJMoa1405870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resch B, Puchas C, Resch E, et al. Epidemiology of respiratory syncytial virus-related hospitalizations and the influence of viral coinfections in southern Austria in a 7-year period. Pediatr Infect Dis J 2020;39:12–16. 10.1097/INF.0000000000002494 [DOI] [PubMed] [Google Scholar]
- 22.Chang S-Y, Lee C-N, Lin P-H, et al. A community-derived outbreak of adenovirus type 3 in children in Taiwan between 2004 and 2005. J Med Virol 2008;80:102–12. 10.1002/jmv.21045 [DOI] [PubMed] [Google Scholar]
- 23.Ng O-T, Thoon KC, Chua HY, et al. Severe pediatric adenovirus 7 disease in Singapore linked to recent outbreaks across Asia. Emerg Infect Dis 2015;21:1192–6. 10.3201/eid2107.141443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Zhou W, Zhao Y, et al. Molecular typing and epidemiology profiles of human adenovirus infection among paediatric patients with severe acute respiratory infection in China. PLoS One 2015;10:e0123234. 10.1371/journal.pone.0123234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajkumar V, Chiang CSM, Low JM, et al. Risk factors for severe adenovirus infection in children during an outbreak in Singapore. Ann Acad Med Singap 2015;44:50–9. [PubMed] [Google Scholar]
- 26.Lai C-Y, Lee C-J, Lu C-Y, et al. Adenovirus serotype 3 and 7 infection with acute respiratory failure in children in Taiwan, 2010-2011. PLoS One 2013;8:e53614. 10.1371/journal.pone.0053614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie L, Yu X-F, Sun Z, et al. Two adenovirus serotype 3 outbreaks associated with febrile respiratory disease and pharyngoconjunctival fever in children under 15 years of age in Hangzhou, China, during 2011. J Clin Microbiol 2012;50:1879–88. 10.1128/JCM.06523-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch JP, Kajon AE. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin Respir Crit Care Med 2016;37:586–602. 10.1055/s-0036-1584923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen M, Zhu Z, Huang F, et al. Adenoviruses associated with acute respiratory diseases reported in Beijing from 2011 to 2013. PLoS One 2015;10:e0121375. 10.1371/journal.pone.0121375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi EH, Kim HS, Park KH, et al. Genetic heterogeneity of the hexon gene of adenovirus type 3 over a 9-year period in Korea. J Med Virol 2006;78:379–83. 10.1002/jmv.20550 [DOI] [PubMed] [Google Scholar]
- 31.Lin Y-C, Lu P-L, Lin K-H, et al. Molecular epidemiology and phylogenetic analysis of human adenovirus caused an outbreak in Taiwan during 2011. PLoS One 2015;10:e0127377. 10.1371/journal.pone.0127377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coleman KK, Wong CC, Jayakumar J, et al. Adenoviral infections in Singapore: should new antiviral therapies and vaccines be adopted? J Infect Dis 2019;9:2178 10.1093/infdis/jiz489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wo Y, Lu Q-B, Huang D-D, et al. Epidemical features of HAdV-3 and HAdV-7 in pediatric pneumonia in Chongqing, China. Arch Virol 2015;160:633–8. 10.1007/s00705-014-2308-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebelo-de-Andrade H, Pereira C, Gíria M, et al. Outbreak of acute respiratory infection among infants in Lisbon, Portugal, caused by human adenovirus serotype 3 and a new 7/3 recombinant strain. J Clin Microbiol 2010;48:1391–6. 10.1128/JCM.02019-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y-J, Hong J-Y, Lee H-J, et al. Genome type analysis of adenovirus types 3 and 7 isolated during successive outbreaks of lower respiratory tract infections in children. J Clin Microbiol 2003;41:4594–9. 10.1128/JCM.41.10.4594-4599.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong JY, Lee HJ, Piedra PA, et al. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin Infect Dis 2001;32:1423–9. 10.1086/320146 [DOI] [PubMed] [Google Scholar]
- 37.Ryan MAK, Gray GC, Smith B, et al. Large epidemic of respiratory illness due to adenovirus types 7 and 3 in healthy young adults. Clin Infect Dis 2002;34:577–82. 10.1086/338471 [DOI] [PubMed] [Google Scholar]
- 38.Halstead DC, Gray GC, Meyer KS, et al. Recombinant adenovirus type 3 and type 14 isolated from a fatal case of pneumonia. Rev Med Microbiol 2010;21:28–30. 10.1097/MRM.0b013e3283393523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebeck MG, McCarthy TA, Capuano AW, et al. Emergent US adenovirus 3 strains associated with an epidemic and serious disease. J Clin Virol 2009;46:331–6. 10.1016/j.jcv.2009.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T, Zhou Z, Tian X, et al. A recombinant trivalent vaccine candidate against human adenovirus types 3, 7, and 55. Vaccine 2018;36:2199–206. 10.1016/j.vaccine.2018.02.050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


