Abstract
A zoonotic A/sw/H1avN1 1C.2.2 influenza virus infection was detected in a German child that presented with influenza-like illness, including high fever. There was a history of close contact with pigs 3 days before symptom onset. The child recovered within 3 days. No other transmissions were observed. Serological investigations of the virus isolate revealed cross-reactions with ferret antisera against influenza A(H1N1)pdm09 virus, indicating a closer antigenic relationship with A(H1N1)pdm09 than with the former seasonal H1N1 viruses.
Keywords: Germany, zoonotic infections, viral infections, influenza, influenza virus, sentinel surveillance, epidemiology, laboratory
During routine surveillance at the National Influenza Centre in Germany in June 2020, a nasal swab was conspicuous because qPCR for the influenza A virus matrix protein (MP) and N1 neuraminidase (NA) genes were positive, whereas the haemagglutinin (HA) qPCR gave no results. The sample underwent whole genome sequencing and results pointed to a zoonotic influenza virus originating from swine. Here we describe the clinical features of the infection as well as the results of antigenic and genetic characterisation of this zoonotic influenza virus.
Description of the case and setting
The diagnostic sample originated from a 2.5-year-old child who lived on a farm, had regular contact with pigs, most recently 3 days before symptom onset, and was not vaccinated against influenza. The child had influenza-like illness over 3 days, displaying fever up to 40 °C, a sore throat, rhinorrhoea, headaches, myalgias, some fussiness and one episode of emesis, and slept a lot. Afterwards, they recovered quickly and fully. The child was not treated with antiviral drugs. No other family member, including the child’s 5-month-old sibling, showed any symptoms, although some of them had been in close contact with the pigs. Four weeks later, 15 pigs of all age groups held at the farm and six family members were swabbed. All nasal swabs were negative, indicating absence of further virus circulation at this location. Four family members tested positive for rhinoviruses, but not the child who had had influenza. Because these swabs were qPCR-negative, virus isolation was not attempted from the pigs’ swabs.
The pig herd of the farm has 600 fattening pigs. Every 4 weeks, 120 new pigs (ca 30 kg, 8–9-weeks-old) are introduced from another farm in Germany. The pig farm is situated outside of the village and no one except the farmer, his family and the veterinarian have access to it. The pig feed is generated by the farm from its own harvest. The pigs are not vaccinated against influenza. Two weeks before the child was infected, a new batch of pigs arrived at the farm. At that time, some pigs were displaying a cough, for which they were treated with antibiotics. Thus, the infection was most probably introduced to the herd via the new batch of pigs.
Antigenic characterisation
Virus isolation from the child’s nasal swab was successful in MDCK-SIAT cells and embryonated hens’ eggs. The virus was termed influenza A/Hessen/47/2020 (HES/2020). Antigenic characterisation showed that cross-reactivity was highest with swine hyperimmune serum directed against influenza A/sw/H1avN1 virus (Table 1) [1]. Further investigations using ferret antisera demonstrated cross-reactivity with the wildtype and vaccine influenza A(H1N1)pdm09 viruses, but not with the previous seasonal influenza A(H1N1) viruses (i.e. those circulating before 2009).
Table 1. Cross-reactivity of HES/2020 and other influenza A(H1N1) viruses investigated by haemagglutination inhibition using turkey erythrocytes, Germany, June 2020.
| Antiserum | Ferret antiseraa | Swine hyperimmune serab | ||||||
|---|---|---|---|---|---|---|---|---|
| Virus | Brisbane/2/2018 A(H1N1)pdm09 |
Michigan/45/2015 A(H1N1)pdm09 |
California/7/2009 A(H1N1)pdm09 |
Brisbane/59/2007 seasonal H1N1 |
PR/8/1934 H1N1 34 |
2688/2010 A(H1N1)pdm09 |
12653/2010 A/sw/H1pdmN2 |
Re230/1992 A/sw/H1avN1 |
| HES/2020c | 1,280 | 1,280 | 640 | < 10 | < 10 | 160 | < 10 | 2,560 |
| Brisbane/2/2018 A(H1N1)pdm09 |
10,240 | 5,120 | 2,560 | < 10 | < 10 | 5,120 | 320 | 640 |
| Michigan/45/2015 A(H1N1)pdm09 |
320 | 640 | 320 | < 10 | < 10 | 640 | 80 | 160 |
| California/7/2009 A(H1N1)pdm09 |
80 | 160 | 320 | < 10 | < 10 | 1,280 | 160 | 160 |
| Brisbane/59/2007 Seasonal H1N1 |
< 10 | < 10 | < 1:10 | 80 | < 10 | < 10 | < 10 | < 10 |
| PR/8/1934 H1N1 of 1930s |
< 10 | < 10 | < 1:10 | < 10 | 1,280 | 160 | 80 | 80 |
| Finistere/2899/1982 A/sw/H1avN1 | 320 | 40 | 80 | < 10 | < 10 | 80 | < 10 | 640 |
| Greven/2889/2004 /A/sw/H1avN1 | < 10 | < 10 | < 10 | < 10 | < 10 | < 10 | < 10 | 320 |
| Heinsberg/8905/2009 A/sw/H1avN1 | 10 | < 10 | < 10 | < 10 | < 10 | < 10 | < 10 | 320 |
| 2688/2010 A(H1N1)pdm09d |
80 | 80 | 320 | < 10 | < 10 | 5,120 | 1,280 | 80 |
| 12653/2010 A/sw/H1pdmN2d |
< 10 | < 10 | < 10 | < 10 | < 10 | 160 | 5,120 | 80 |
| Re230 /1992 A/sw/H1avN1d |
< 10 | < 10 | < 10 | < 10 | < 10 | 160 | 640 | 5,120 |
a Post-infection sera of ferrets.
b Hyperimmune sera of pigs were established according to [1].
c Zoonotic influenza A/sw/H1avN1 virus (A/Hessen/47/2020) described in this study.
d For genetic analysis of these viruses see also [18,29,30]; antisera against influenza A/sw/H1pdmN2 viruses cross-react minimally, or not at all, with A(H1N1)pdm09 and swine H1avN1 viruses because the antigenic distance is larger between them [29].
The Table shows reciprocal haemagglutination inhibition titres.
Blood samples from 14 of 15 pigs were found to be seropositive against the infecting virus (HES/2020). In haemagglutination inhibition (HI) tests against HES/2020, titres ranged from 1:10 to 1:160. All pig sera were negative against influenza A(H1N1)pdm09 virus (A/Brisbane/2/2018).
Sequence analysis showed that the majority of HA antigenic sites were conserved between influenza A/sw/H1avN1 and A(H1N1)pdm09 viruses (Table 2) [2]. In accordance with International Health Regulations, the case was reported to World Health Organization (WHO) via the Early Warning and Response System (EWRS) [3] and the virus was provided to the WHO Collaborating Centre London for further characterisation [4].
Table 2. Comparison of amino acids in the antigenic sites of the haemagglutinin molecule of HES/2020 vs influenza A(H1N1) viruses, Germany, June 2020.
| Amino acid in the antigenic sitea | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site Sa | Site Sb | ||||||||||||||||
| Virus | HA clade/genotype | 124 | 125 | 155 | 157 | 159 | 160 | 162 | 163 | 164 | 153 | 156 | 185 | 189 | 190 | 193 | 195 |
| HES/2020 | 1C.2.2 | P | N | G | S | P | K | R | N | S | K | N | D | Q | T | Q | N |
| swDUEL/2012 | 1C.2.2 | P | N | G | S | P | K | R | K | S | K | N | D | Q | T | Q | N |
| swLUED/2013 | 1C.2.1 | P | N | G | S | P | K | S | T | S | K | N | D | Q | T | Q | N |
| NL/2016 | 1C.2.1 | P | N | E | S | P | K | S | T | S | K | N | D | Q | T | Q | N |
| swSHA/2013 | 1C.2.3/G1 | P | N | G | S | P | K | S | K | S | K | N | D | Q | T | Q | N |
| swHEN/2018 | 1C.2.3/G4 | P | N | G | S | P | K | S | K | S | K | N | D | Q | T | Q | N |
| swSHA/2014 | 1C.2.3/G5 | P | N | G | S | P | K | S | K | S | K | N | D | Q | T | Q | N |
| swANH/2015 | 1C.2.3/G6 | P | N | G | S | P | K | S | K | S | K | N | D | Q | T | Q | N |
| GU-MA/2019 | pdm09 | P | N | G | S | P | K | N | Q | T | K | N | I | E | S | Q | A |
| MICH/2015 | pdm09 | P | N | G | S | P | K | N | Q | S | K | N | T | Q | S | Q | A |
| Site Ca1 | Site Ca2 | Site Cb | |||||||||||||||
| Virus | HA clade/genotype | 166 | 170 | 204 | 237 | 135 | 137 | 140 | 142 | 221 | 222 | 70 | 71 | 73 | 74 | 75 | 115 |
| HES/2020 | 1C.2.2 | T | G | S | G | A | S | G | N | R | E | L | L | A | N | S | E |
| swDUEL/2012 | 1C.2.2 | T | G | S | G | A | S | G | N | R | E | L | L | A | N | S | E |
| swLUED/2013 | 1C.2.1 | T | G | S | G | A | S | G | K | R | E | L | I | A | N | S | E |
| NL/2016 | 1C.2.1 | T | G | S | G | A | S | G | K | R | E | L | I | A | N | S | E |
| swSHA/2013 | 1C.2.3/G1 | T | G | S | G | A | S | G | N | R | G | L | L | A | N | S | E |
| swHEN/2018 | 1C.2.3/G4 | T | G | T | G | S | S | G | N | R | E | L | L | A | N | S | E |
| swSHA/2014 | 1C.2.3/G5 | T | G | S | G | S | S | G | N | R | E | L | L | A | N | S | E |
| swANH/2015 | 1C.2.3/G6 | T | G | S | G | A | S | G | N | R | E | L | L | A | N | S | E |
| GU-MA/2019 | pdm09 | I | G | S | G | A | P | G | K | R | D | L | S | A | R | S | E |
| MICH/2015 | pdm09 | I | G | S | G | A | P | G | K | R | D | L | S | A | S | S | E |
HA: haemagglutinin.
a H1 numbering without signal sequence.
Virus names from top to bottom: A/Hessen/47/2020, A/swine/Duelmen/15075/2012, A/swine/Luedinghausen/18391/2013, A/Netherlands/3315/2016, A/swine/Shandong/39/2013, A/swine/Henan/SN13/2018, A/swine/Shandong/S113/2014, A/swine/Anhui/1227/2015, A/Guangdong-Maonan/SWL1536/2019, A/Michigan/45/2015.
Shaded cells: amino acid differences relative to HES/2020; presentation of antigenic sites adapted from [2].
Genetic characterisation
The genetic classification of HES/2020 is F (polymerase basic protein 2, PB2), G (polymerase basic protein 1, PB1), I (polymerase acidic protein, PA), 1C.2.2 (HA), F (nucleoprotein, NP), 1F (NA), F (MP), 1E (nonstructural proteins, NS) [5,6]. It is unrelated to the recently reported G4 reassortant EA(H1N1) viruses circulating in China [2]. Sequences were submitted to GISAID and the accession numbers were as follows: PB2: EPI1757436, PB1: EPI1757437, PA: EPI1757435, HA: EPI1757439, NP: EPI1757432, NA: EPI1757438, MP: EPI1757434 and NS: EPI1757433. Blast analysis and phylogenetic analysis demonstrated that the segments of HES/2020 are closely related to those of different viruses: HA (Figure) and NA to influenza A/swine/Germany/Ellerbrock-IDT14696/2012 (swELLE/2012, H1N1, HA-1C.2.2) and A/swine/Duelmen/15075/2012 (swDUEL/2012, H1N1, HA-1C.2.2); MP, NP, NS and PB1 to A/swine/Luedinghausen/18391/2013 (swLUED/2013, H1N1, HA-1C.2.1) and to zoonotic A/Netherlands/3315/2016 (NL/2016, H1N1, HA-1C.2.1) [7]; PA and PA-X to A/swine/Belgium/Heist-op-den-Berg-363/2012 (swHEIST/2012, H1N1, HA-1C.2.1); and PB2 to A/swine/Belgium/Oostkamp-26/2012 (swOOST/2012, H1N2, HA-1B.1.2.1). The genetic composition of HES/2020 indicates several intra- and inter-clade reassortments.
Figure.
Phylogenetic analysis of the haemagglutinin gene (1,695 bp) of influenza A viruses
The phylogenetic analyses of the other coding sequences (NA, MP, NP, NS, NS1, PA, PA-X, PB1, PB1-F2, PB2) are shown in Supplementary Figures S1–S10. Virus genomes were analysed by whole genome sequencing and were phylogenetically evaluated with Mega7 (neighbour-joining method, midpoint rooted, bootstrap test with 1,000 replicates, Kimura 2-parameter method, partial deletion (site coverage cut-off: 5%). Sixty-one influenza A viruses were characterised: 1A.3.3.2/H1N1pdm09 (light blue), 1B.1.2.1 (black), 1C.1 (grey), 1C.2 including reassorted A(H1N2)-viruses (orange), 1C.2.1 (green), 1C.2.2 including zoonotic A/Hessen/47/2020 (red, italics, framed in black) and 1C.2.3 including genotypes G1/G4/G5/G6 (blue) [2]. Framed items: closely related viruses that are identified by BLAST analysis of each segment (data not shown) and used as reference viruses for further analysis: swELLE/2012 and swDUEL/2012 for HA and NA, zoonotic NL/2016 and swLUED/2013 for MP, NP, NS and PB1, swHEIST/2012 for PA and swOOST/2012 for PB2.
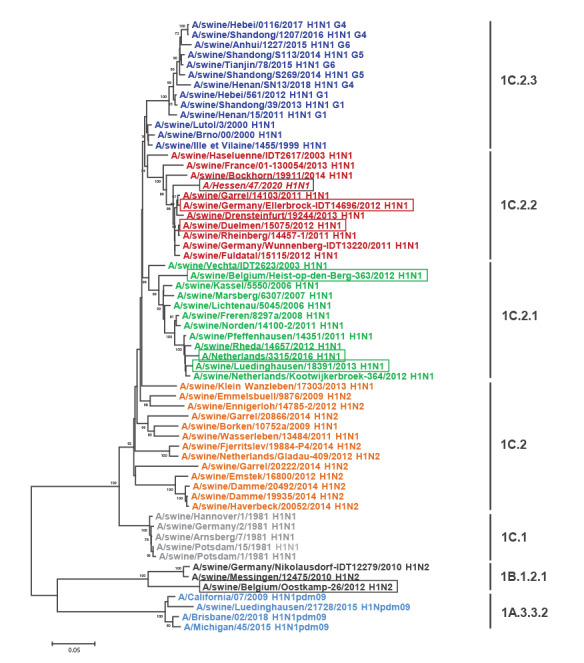
Nucleotide sequence variation was highest over the usually well conserved NP and PA-X coding sequences (Twelve coding sequences were analysed: HA, NA, M1, M2, NP, NS1, NEP, PA, PA-X, PB1, PB1-F2, PB2 with a length of 1,701, 1,410, 759, 294, 1,497, 693, 366, 2,151, 759, 2,274, 273, 2,280 nt, respectively). They displayed nucleotide identities of 95% each, whereas all other coding sequences displayed nucleotide sequence identity > 95% relative to the reference sequence. Reference sequences were swDUEL/2012 for HA and NA, swLUED/2013 for MP, NP, NS and PB1, swHEIST/2012 for PA and swOOST/2012 for PB2). Amino acid (AA) sequence variation was highest over the regulator proteins of the host innate immune response, NS1, PA-X and PB1-F2 (identities of 95%, 94% and 95%, respectively) [8,9]. Variant calling for HES/2020 and another zoonotic virus, NL/2016 [7], relative to the reference viruses, demonstrated that the number of substitutions common to both HES/2020 and another zoonotic virus, NL/2016, was highest for the PB1-F2 protein (four of five substitutions) Table 3). In contrast to NL/2016, PB1-F2 of HES/2020 is full-length at 90 AA. Phylogenetic analyses of MP, NP, NS, NS1, PB1 and PB1-F2 demonstrated that the two zoonotic viruses are closely related (Figure, Supplementary Figures S1–S10). To detect substitutions with potential functional relevance in the HES/2020 genome, the FluSurver online tool was employed (https://flusurver.bii.a-star.edu.sg/), identifying substitutions in the HA receptor binding domain (D222E) [10], NP (K48Q;R98K;R99K [11], R351K;V353I;Q357K [12]) and PB2 (D701N) [13] (Supplementary Table S1). The substitutions NP-Q357K, PA-X-R57K, PA-R57K, PA-T639A are present in both zoonotic viruses and in both analyses (FluSurver and the genetic comparison in Table 3).
Table 3. Non-synonymous substitutions in the coding sequences of HES/2020 relative to closely related swine influenza viruses and common substitutions with the zoonotic NL/2016 virus, Germany, June 2020.
| CDS | Substitutions of HES/2020 relative to reference sequencesa | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HA1b | T14Ac | G53K | V57L | I80V | K163N | I214T | M227I | H253Y | V265I | T267M | D269N | H271R | K278M | K302E | Q311H | |
| HA2b | Q353H | Q365R | D399G | S451A | N473D | D474E | ||||||||||
| NA | M15L | A76V | A79E | S82P | L140M | D210S | V211I | K220R | A232V | E311D | V338I | T340I | S369N | V389I | T396I | N398D |
| M1 | G30S | |||||||||||||||
| M2 | T28I | F48S | ||||||||||||||
| NP | S16G | K105V | Q357K | V363I | A423T | R452K | S482N | N498S | ||||||||
| NS1 | K44R | S48N | R67C | A86T | R88H | M98I | V111L | I123V | Y165S | A191T | N209I | T215I | ||||
| NEP | K18R | T52S | L55H | |||||||||||||
| PA | H24Y | R57K | I66S | R104K | I184L | K204R | E206D | E252G | I268L | K269R | L335I | H346N | M374V | G388S | T639A | V712M |
| PA-X | H24Y | R57K | I66S | R104K | I184L | R199K | N204D | K206T | S207L | E209G | T212I | I216T | S219F | P224L | K252E | |
| PB1 | I69V | I111M | K213N | K571R | V632I | G636E | V640I | A648S | I682V | S741A | ||||||
| PB1-F2 | T39M | S63F | K73R | stop80W | K81R | |||||||||||
| PB2 | S12L | I255V | A351T | K353R | R389K | C409R | M473V | A598T | D611E | |||||||
AA: amino acid; CDS: coding sequences; HES/2020: influenza A/sw/H1avN1 (A/Hessen/47/2020).
a Reference sequences were as follows: swDUEL/2012 for HA and NA, swLUED/2013 for MP, NP, NS and PB1, swHEIST/2012 for PA and swOOST/2012 for PB2.
b H1 numbering without signal sequence.
c AA substitution within the HA signal sequence.
Consistent AA substitutions that occur in both zoonotic viruses HES/2020 and NL/2016 virus are labelled in bold; AA substitutions that differ from the reference viruses and between zoonotic HES/2020 and zoonotic NL/2016 virus are labelled in bold and italics and only the AA of HES/2020 is displayed; the change tag to tgg at codon 80 revealed an extension of HES/2020 PB1-F2 to 90 AA and is shown in italics.
Resistance characterisation
While HES/2020 does not exhibit NA or PA mutations conferring resistance against neuraminidase inhibitors or baloxavir marboxil, its M2 sequence contains the AA substitutions L26I, V27A and S31N, all of which are associated with adamantane resistance (amantadine and rimantadine). Phenotypic susceptibility testing against oseltamivir, peramivir and zanamivir confirmed that HES/2020 was sensitive to all neuraminidase inhibitors authorised in Europe.
Discussion
This is the sixth zoonotic swine influenza virus infection in humans investigated at the German National Influenza Centre (in 2007: A/sw/H1avN1 and A/sw/H3N2 in Lower Saxony, in 2010: A/sw/H1avN1 in Lower Saxony, in 2011: A/sw/H1huN2 and A/sw/H1avN1 in Lower Saxony) [14]. Of the five previously reported cases, two occurred in children and one in an immunocompromised adult; influenza A/sw/H1avN1 infections were the most common [14]. All previous German cases were detected in Lower Saxony, the federal state with the second largest pig population in Germany. The case described here is the first from a region with a low density of pig holdings, i.e. Hesse.
The genetic diversity of influenza A viruses in the European pig population is increasing [15-17]. A/sw/H1avN1 are the predominant swine influenza viruses in Germany [18]. Among them, the two most prevalent lineages are H1avN1 1C.2.2 and H1avN1 1C.2.1. Other swine influenza viruses include H1huN2 and H3N2 viruses as well as H1pdmN1 and H1pdmN2 viruses [15-18]. An increasing number of reassortments between these viruses augment the diversity of influenza virus populations in swine.
Swine influenza viruses acquired adamantane resistance in the late 1980s [19]. The influenza A(H1N1)pdm09 virus contains the MP gene from A/sw/H1avN1 viruses which confers adamantane resistance via the M2-S31N mutation in MP gene 2 [20]. This mutation was common in all seasonal influenza A viruses circulating globally during the last years [21]. In addition to S31N, HES/2020 contains the M2 AA substitutions L26I, V27A which are also associated with adamantane resistance. The M2-L26I and M2-V27A mutations can be found sporadically in influenza A viruses [21].
Swine influenza viruses have acquired some resistance genes against human myxovirus resistance protein MxA during their evolution in pigs, facilitating their transmission to humans [12]. Pig-to-human influenza virus transmissions are not rare, especially in close contact settings such as agricultural fairs [22], and sporadic zoonotic transmission of swine influenza A(H1N1) virus has been reported [23,24]. The farm child was the only member of his family who was infected, although some of the other family members had also been exposed. The infection of a child is not surprising. Because of their limited exposure history, young children display a narrower (if any) immune response to influenza virus than adults [25].
Our serology investigations indicate some level of cross-reactivity between influenza A(H1N1)pdm09 virus and A/sw/H1avN1 viruses in ferrets. This is in line with previous findings that influenza A(H1N1)pdm09 infection induces broadly neutralising (not strain-specific) antibodies [26]. Antibodies against influenza A/sw/H1avN1 viruses in the human population are rare [27,28]. On the other hand, sera of human volunteers collected 3–7 weeks after vaccination with the annual 2017/18 vaccine all reflected antibodies against influenza A/sw/H1avN1 virus at varying microneutralisation titres and none was negative [15]. Although the family members of the zoonotic case had not been vaccinated, they may have been exposed to human and swine influenza A viruses before, potentially resulting in pre-existing immunity which might impair transmission of influenza A/sw/H1avN1 influenza virus.
However, the rising genetic diversity among swine influenza viruses, involving antigenic drift and shift, may increase divergence from influenza A/sw/H1avN1 viruses in the future. In particular, swine reassortant viruses may quickly acquire antigenic changes, and this is where substantial zoonotic potential may arise.
Acknowledgements
We thank Prof Dr Timm Harder and Prof Dr Martin Beer, Friedrich-Loeffler-Institute, Greifswald - Isle of Riems, Germany for establishment of ferret antisera and Mareen Adam, Heike Fischer, Susi Hafemann, Ute Hopf-Guevara, Carmen Karstädt-Schulze, Katja-Irena Madaj, Jeanette Milde, Bettina Mischke, Christine Spingies, Anneliese Schindel, Kathrin Seidel, Nathalie Tollard, Robert Koch Institute, Berlin, Germany for technical assistance.
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: RD and WH designed the study, RD, MW, DYO, SD wrote the manuscript, RD, MW, BB, MH-K, CG, RV, AMH, KG, AT, SA, JR, SD, SB, TW contributed to the investigations, all authors read and edited the manuscript.
References
- 1.Dürrwald R, Krumbholz A, Baumgarte S, Schlegel M, Vahlenkamp TW, Selbitz H-J, et al. Swine influenza A vaccines, pandemic (H1N1) 2009 virus, and cross-reactivity. Emerg Infect Dis. 2010;16(6):1029-30. 10.3201/eid1606.100138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun H, Xiao Y, Liu J, Wang D, Li F, Wang C, et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci USA. 2020;117(29):17204-10. 10.1073/pnas.1921186117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO). Influenza at the human-animal interface. Summary and assessment, from 9 May to 10 July 2020. Geneva: WHO; 2020. Available from: https://www.who.int/influenza/human_animal_interface/Influenza_Summary_IRA_HA_interface_10_07_2020.pdf?ua=1
- 4.World Health Organization (WHO). Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness. Geneva: WHO; 2020. Available from: https://www.who.int/influenza/vaccines/virus/202009_zoonotic_vaccinevirusupdate.pdf?ua=1
- 5.Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, et al. A phylogeny-based global nomenclature system and automated annotation tool for h1 hemagglutinin genes from swine influenza A viruses. MSphere. 2016;1(6):e00275-16. 10.1128/mSphere.00275-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu G, Rowley T, Garten R, Donis RO. FluGenome: a web tool for genotyping influenza A virus. Nucleic Acids Res. 2007;35(Web Server issue):W275-9. 10.1093/nar/gkm365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraaij PLA, Wildschut ED, Houmes RJ, Swaan CM, Hoebe CJ, de Jonge HCC, et al. Severe acute respiratory infection caused by swine influenza virus in a child necessitating extracorporeal membrane oxygenation (ECMO), the Netherlands, October 2016. Euro Surveill. 2016;21(48):30416. 10.2807/1560-7917.ES.2016.21.48.30416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zell R, Krumbholz A, Eitner A, Krieg R, Halbhuber KJ, Wutzler P. Prevalence of PB1-F2 of influenza A viruses. J Gen Virol. 2007;88(Pt 2):536-46. 10.1099/vir.0.82378-0 [DOI] [PubMed] [Google Scholar]
- 9.Xu G, Zhang X, Liu Q, Bing G, Hu Z, Sun H, et al. PA-X protein contributes to virulence of triple-reassortant H1N2 influenza virus by suppressing early immune responses in swine. Virology. 2017;508:45-53. 10.1016/j.virol.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 10.Wedde M, Wählisch S, Wolff T, Schweiger B. Predominance of HA-222D/G polymorphism in influenza A(H1N1)pdm09 viruses associated with fatal and severe outcomes recently circulating in Germany. PLoS One. 2013;8(2):e57059. 10.1371/journal.pone.0057059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dornfeld D, Petric PP, Hassan E, Zell R, Schwemmle M. Eurasian Avian-Like Swine Influenza A Viruses Escape Human MxA Restriction through Distinct Mutations in Their Nucleoprotein. J Virol. 2019;93(2):e00997-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mänz B, Dornfeld D, Götz V, Zell R, Zimmermann P, Haller O, et al. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 2013;9(3):e1003279. 10.1371/journal.ppat.1003279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459(7249):931-9. 10.1038/nature08157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buda S, Haas W, Baillot A, Beyrer K, Monazahian M, Pulz M, et al. Humane Fälle mit Infektion durch Schweineinfluenzaviren. [Human cases of infection with swine influenzaviruses]. Epid Bulletin. 2011;39:357-9. German. Available from: https://www.rki.de/DE/Content/Infekt/EpidBull/Archiv/2011/Ausgaben/39_11.pdf?__blob=publicationFile [Google Scholar]
- 15.Henritzi D, Petric PP, Lewis NS, Graaf A, Pessia A, Starick E, et al. Surveillance of European domestic pig populations identifies an emerging reservoir of potentially zoonotic swine influenza A viruses. Cell Host Microbe. 2020;28(4):614-627.e6. 10.1016/j.chom.2020.07.006 [DOI] [PubMed] [Google Scholar]
- 16.Zell R, Groth M, Krumbholz A, Lange J, Philipps A, Dürrwald R. Displacement of the Gent/1999 human-like swine H1N2 influenza A virus lineage by novel H1N2 reassortants in Germany. Arch Virol. 2020;165(1):55-67. 10.1007/s00705-019-04457-w [DOI] [PubMed] [Google Scholar]
- 17.Zell R, Groth M, Krumbholz A, Lange J, Philipps A, Dürrwald R. Novel reassortant swine H3N2 influenza A viruses in Germany. Sci Rep. 2020;10(1):14296. 10.1038/s41598-020-71275-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zell R, Groth M, Krumhbolz A, Lange J, Philipps A, Dürrwald R. Cocirculation of swine H1N1 influenza A virus lineages in Germany. Viruses. 2020;12(7):762. 10.3390/v12070762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krumbholz A, Schmidtke M, Bergmann S, Motzke S, Bauer K, Stech J, et al. High prevalence of amantadine resistance among circulating European porcine influenza A viruses. J Gen Virol. 2009;90(Pt 4):900-8. 10.1099/vir.2008.007260-0 [DOI] [PubMed] [Google Scholar]
- 20.Hurt AC. The epidemiology and spread of drug resistant human influenza viruses. Curr Opin Virol. 2014;8:22-9. 10.1016/j.coviro.2014.04.009 [DOI] [PubMed] [Google Scholar]
- 21.Dong G, Peng C, Luo J, Wang C, Han L, Wu B, et al. Adamantane-resistant influenza a viruses in the world (1902-2013): frequency and distribution of M2 gene mutations. PLoS One. 2015;10(3):e0119115. 10.1371/journal.pone.0119115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowman AS, Walia RR, Nolting JM, Vincent AL, Killian ML, Zentkovich MM, et al. Influenza A(H3N2) virus in swine at agricultural fairs and transmission to humans, Michigan and Ohio, USA, 2016. Emerg Infect Dis. 2017;23(9):1551-5. 10.3201/eid2309.170847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Reeth K, Nicoll A. A human case of swine influenza virus infection in Europe--implications for human health and research. Euro Surveill. 2009;14(7):19124. [PubMed] [Google Scholar]
- 24.Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis. 2007;44(8):1084-8. 10.1086/512813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krammer F. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol. 2019;19(6):383-97. 10.1038/s41577-019-0143-6 [DOI] [PubMed] [Google Scholar]
- 26.Wrammert J, Koutsonanos D, Li G-M, Edupuganti S, Sui J, Morrissey M, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208(1):181-93. 10.1084/jem.20101352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoschler K, Thompson C, Casas I, Ellis J, Galiano M, Andrews N, et al. Population susceptibility to North American and Eurasian swine influenza viruses in England, at three time points between 2004 and 2011. Euro Surveill. 2013;18(36):20578. 10.2807/1560-7917.ES2013.18.36.20578 [DOI] [PubMed] [Google Scholar]
- 28.Vandoorn E, Leroux-Roels I, Leroux-Roels G, Parys A, Vincent A, Van Reeth K. Detection of H1 swine influenza A virus antibodies in human serum samples by age group. Emerg Infect Dis. 2020;26(9):2118-28. 10.3201/eid2609.191796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange J, Groth M, Schlegel M, Krumbholz A, Wieczorek K, Ulrich R, et al. Reassortants of the pandemic (H1N1) 2009 virus and establishment of a novel porcine H1N2 influenza virus, lineage in Germany. Vet Microbiol. 2013;167(3-4):345-56. 10.1016/j.vetmic.2013.09.024 [DOI] [PubMed] [Google Scholar]
- 30.Fu Y, Dürrwald R, Meng F, Tong J, Wu N-H, Su A, et al. Infection studies in pigs and porcine airway epithelial cells reveal an evolution of A(H1N1)pdm09 influenza A viruses toward lower virulence. J Infect Dis. 2019;219(10):1596-604. 10.1093/infdis/jiy719 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


