Abstract
SARS-CoV-2 IgG screening of 1,000 antenatal serum samples in the Oxford area, United Kingdom, between 14 April and 15 June 2020, yielded a 5.3% seroprevalence, mirroring contemporaneous regional data. Among the 53 positive samples, 39 showed in vitro neutralisation activity, correlating with IgG titre (Pearson’s correlation p<0.0001). While SARS-CoV-2 seroprevalence in pregnancy cohorts could potentially inform population surveillance, clinical correlates of infection and immunity in pregnancy, and antenatal epidemiology evolution over time need further study.
Keywords: antenatal, pregnancy, COVID-19, antibody, IgG, serology, epidemiology, prevalence, ELISA, neutralisation
During the first peak of the coronavirus disease (COVID-19) pandemic in the United Kingdom (UK) between April and June 2020, we set out to generate a benchmark estimate of antenatal severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) seroprevalence in a population of pregnant women in the Oxford area. We also evaluated an IgG enzyme-linked immunosorbent assay (ELISA) head-to-head with a pseudotyped virus neutralisation test in this group, and investigated the extent to which routine antenatal testing for SARS-CoV-2 IgG antibody could inform population surveillance efforts.
Samples and processing pipeline
We analysed antenatal serum samples that had been taken in the Oxford area, during the first trimester of pregnancy (typically at 8–12 weeks’ gestation), from women aged 17 to 48 years (median: 32 years; interquartile range (IQR): 28–35) over a period spanning 9 weeks between 14 April and 15 June 2020. This coincided with the first peak of the national and local COVID-19 pandemic (Figure 1A). Samples were collected after routine clinical laboratory tests had been completed (Figure S1). We used primary sample identifiers to obtain year of birth, self-reported ethnicity, Oxfordshire postcode district (first three or four digits), and index of multiple deprivation (IMD) from the electronic patient records (EPR). We excluded samples for which no EPR record was available, and those with a missing postcode, generating a final dataset of 1,000 consecutive samples with supporting clinical metadata. To track laboratory processing, samples were assigned a new unique barcode identifier. Aliquots were kept at 4 °C throughout and prepared for laboratory assays using a Janus liquid handler (PerkinElmer, Waltham, Massachusetts, United States) (Figure S1). For comparing results from our cohort of pregnant women, to a regional (South-East England) seroprevalence estimate during a similar period, we used data from the Office for National Statistics (ONS), which had overseen a large population serosurvey [1].
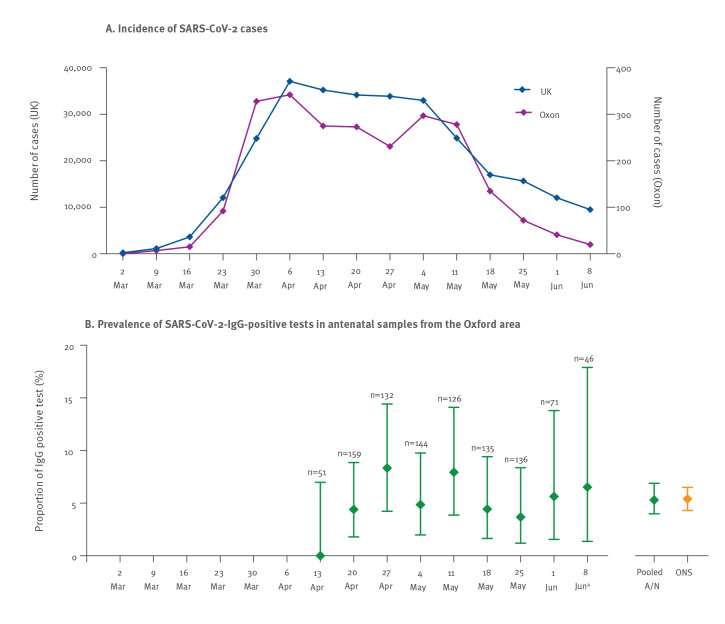
Figure 1.
(A) Incidence of SARS-CoV-2 cases in Oxfordshire and the United Kingdom during the first peak of the COVID-19 pandemic, shown in parallel with (B) prevalence of SARS-CoV-2-IgG-positive antenatal samples from the Oxford area, presented by week, March–June 2020
A/N: antenatal; COVID-19: coronavirus disease; ONS: Office of National Statistics data; Oxon: Oxfordshire; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
(A) Incidence of SARS-CoV-2 cases, based on nose/throat swab reverse-transcription PCR testing; purple line shows diagnosed cases in Oxfordshire; blue line shows diagnosed cases in England (data from public sources [22,23]). (B) SARS-CoV-2 IgG prevalence in antenatal serum samples is shown in green, from the week commencing 13 April (bars show 95% confidence intervals; number at the top of each bar indicates number sampled during this week). Final two bars show pooled data for the Oxford area A/N sample collection (green), compared with data for South-East England collected by the ONS, for which SARS-CoV-2 IgG prevalence is shown for the period 26 April–8 June, measured using the same ELISA (orange) [1].
a Data for the last 2 weeks of our serosurvey are pooled due to small numbers tested in the final week, where testing was only conducted on 1 day (15 June).
ELISA for detection of IgG to SARS-CoV-2 spike protein
Samples were tested using a new 384-well ELISA established at the University of Oxford, which detects IgG to trimeric SARS-CoV-2 spike protein, with a sensitivity of 99.1% (95% confidence interval (CI): 97.8–99.7) and specificity 99.0% (95% CI: 98.1–99.5) as recently described [2,3]. The threshold for positivity in this assay is 8.0×106 standard units.
Pseudotyped virus neutralisation assays
SARS-CoV-2 pseudotyped microneutralisation (pMN) assays were undertaken using methods previously described [4,5]. Briefly, a lentivirus particle was constructed to display the full SARS-CoV-2 spike protein. Infectivity was determined by incubating the pseudovirus particle together with twofold serial dilutions of test sera with HEK-293T-ACE2-plasmid-transfected cells, generating a luciferase read out in relative light units (RLU) after incubation at 37 °C for 72 hours. The laboratory work was undertaken blinded to the results of the serology assay and to the location of positive controls on the plates.
Ethical statement
This work was approved by the South Central Research Ethics Committee (Ref: 08/H0606/139).
Prevalence and distribution of IgG in antenatal population
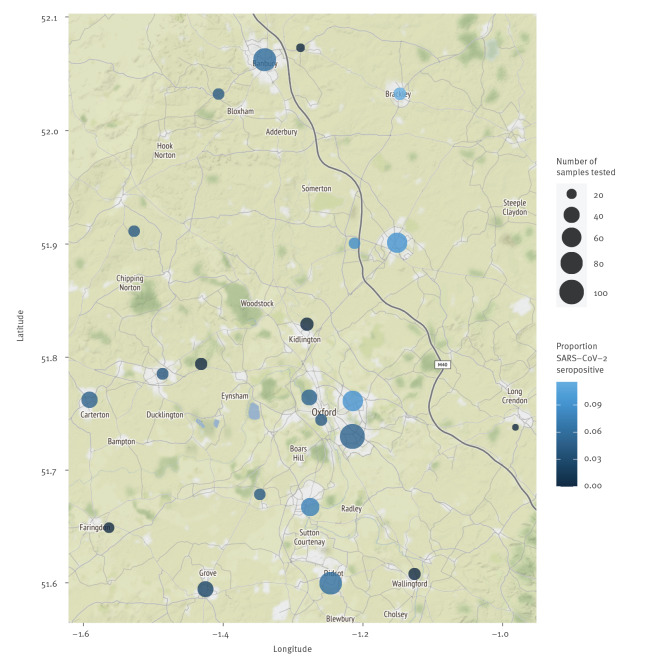
The overall prevalence of SARS-CoV-2 anti-spike IgG in this antenatal cohort was 53/1,000 (5.3%; 95% CI: 4.0–6.9%), which closely mirrors ONS population surveillance data for South-East England in a similar time period (26 April–8 June), at 5.4% (95%CI: 4.3–6.5%); (Figure 1). There was no observed change in prevalence by week surveyed (Figure 1B) and IgG status was not associated with maternal age (p = 0.6), self-reported ethnicity (p = 1.0), or IMD score (p = 1.0); (Figure S2). Sampling density was highest in urban areas (clustered around Oxford city centre), but there were no obvious geographical hotspots, with seropositive samples originating from across the region (Figure 2).
Figure 2.
Map showing the location and prevalence of SARS-CoV-2 IgG-positive tests in antenatal women, Oxford area, South-East England, United Kingdom, March–June 2020 (n = 1,000 antenatal women testeda)
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
a Data are shown for postcodes in which ≥ 10 serum samples were collected.
Quantification and interpretation of viral neutralisation
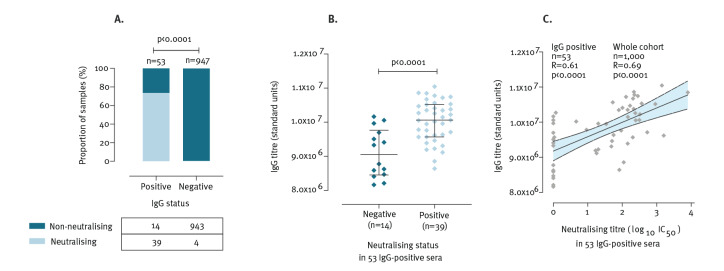
Overall, neutralising activity was detected in 43/1,000 (4.3%) samples. Among these, 39 were IgG-positive and four IgG-negative. Thus, 39 of 53 IgG-positive sera had evidence of neutralising activity in vitro (Figure 3A). Among the IgG-positive samples, those with neutralising activity had significantly higher quantitative IgG titres than those that were non-neutralising, p < 0.0001 (Figure 3B), in keeping with previous reports [6,7]. Among the 53 IgG-positive samples, half maximal inhibitory concentration (IC50) in pMN was correlated with IgG concentration; for each log10(IC50) increase in neutralising activity, IgG titres were 410,000 (95%CI: 260,000–560,000) units higher (p < 0.0001, Figure 3C).
Figure 3.
Relationship between SARS-CoV-2 IgG (standard units by ELISA) and neutralisation (based on pseudovirus microneutralisation assay) in serum samples from antenatal women in the Oxford area, United Kingdom, (n = 1,000 women tested)
CI: confidence interval; ELISA: enzyme-linked immunosorbent assay; IC50: half maximal inhibitory concentration; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
(A) Concordance between IgG status (i.e. negative or positive; threshold for positivity set at 8.0×106 standard units) and neutralising activity of serum samples (n = 1,000); p-value by Fisher’s Exact Test; (B) IgG titres in samples meeting the threshold for IgG-positivity (n = 53), grouped according to neutralisation status; lines indicate median and interquartile range; p-value by Mann–Whitney U test. Four IgG-negative samples in which neutralising activity was detected are shown in Figure S3; (C) Relationship between quantitative ELISA read-out and neutralisation expressed as log10 IC50 in IgG-positive samples (n = 53). Shaded area shows 95% CI. R and p-value by Pearson’s correlation are presented both the 53 datapoints shown, and for the whole sample set of 1,000 sera (among which those with no measurable neutralising activity were assigned as zero).
Neutralising activity suggests that the anti-spike antibodies measured by the pMN assay offer protection against infection and/or disease in the majority of samples designated seropositive by ELISA, but further clinical correlation is required.
Interestingly, 4/947 (0.4%) SARS-CoV-2 IgG-negative samples had neutralising activity, of which three had IgG titres close to the positive threshold (Figure S3). This result highlights the stringent threshold set for IgG-positivity (to assure assay specificity), suggesting that in some cases sufficient IgG titres may be present to mediate protective immunity even when the sample is reported as seronegative. In the lowest-titre IgG-negative sample with neutralising activity (Figure S3), it is possible that cross-reactive antibodies (not detected on a specific anti-spike IgG ELISA) could be responsible for neutralisation, with corresponding protection against infection or disease; this hypothesis warrants further investigation.
Discussion
SARS-CoV-2 antibody testing has been scaled up at pace in many countries throughout 2020 to develop insights into the epidemiology of infection and inform interventions for infection prevention and control [2,8-11]. In the UK, serum samples are collected as part of routine antenatal care, providing an opportunity for serosurveillance with fewer logistical and resource implications than other population groups [12]. There is interest in the extent to which pregnant women might represent a ‘sentinel group’, providing an accessible snapshot of SARS-CoV-2 seroprevalence in the general population. However, pregnant women represent a small part of the overall 18–40 year-old population, and although our study results suggest that seroprevalence in this group is congruent with population estimates, this observation should be extrapolated with caution and may change over time. Antenatal incidence may decline compared with other population groups, as pregnant women behave cautiously, with close observance of social distancing and other measures to reduce exposure [13]. Samples taken at 8 to 12 weeks’ gestation, when pregnant women may not yet have fully established behaviours to increase their protection, might be more relevant as a sentinel group for the wider population than serology performed at delivery, but the observation we make in the first epidemic wave may not hold true in future outbreaks.
Our sample collection represents local population density, with urban areas relatively over-represented, but – given the low seroprevalence – we were underpowered to identify associations between seropositivity and other maternal characteristics. The IMD of the region sampled is not representative of the UK, as Oxfordshire ranks 142/151 of upper tier local authority districts, putting it in the 10% least deprived areas of the UK. Other studies of SARS-CoV-2 IgG seroprevalence in pregnancy have generated higher point estimates than in our setting, for example seroprevalence was 8% in a study in France [14], 10% in Italy [15], and 14% in Spain [16]; differences may reflect timing of sample collection and local population epidemiology, as well as different performance characteristics of the assays used to measure antibodies.
Mapping the exposure of pregnant women to SARS-CoV-2 infection is important, as they represent a potentially vulnerable group [17,18]; more evidence is required to determine whether pregnancy is a risk factor either for acquisition of infection or for severe maternal disease [19,20], and to identify adverse fetal or neonatal outcomes. More focus is also needed for potentially high-risk groups (for example according to ancestry, deprivation, maternal age), and to understand the impact of changes in behaviour (and thus exposure) during pregnancy in different populations over time. Prospective surveillance through programmes such as INTERCOVID have been established to address these questions [21].
In conclusion, our study suggests that routine surveillance of SARS-CoV-2 antibody in sera collected early in pregnancy may be a useful tool for monitoring wider population IgG prevalence, but scrutiny will be required to track this observation over time.
Acknowledgements
We are grateful to the biochemistry and microbiology laboratory staff at Oxford University Hospitals NHS Foundation Trust for the handling and curation of the samples, and to all those supporting maternal and newborn health in our region during the pandemic.
Funding
This work was supported by NIHR Oxford Biomedical Research Centre. Funding for laboratory work was provided by the von Opel Foundation award to SG. PCM is funded by a Wellcome intermediate fellowship (ref. 110110/Z/15/Z). DWE is a Robertson Foundation Fellow. SFL is a Wellcome Trust Clinical Research Training Fellow (ref. 102176/B/13/Z). KKC is a Medical Research Foundation PhD student (ref. MRF-145-0004-TPG-AVISO). BDM is supported by the Kennedy Trust for Rheumatology Research and by the SGC, a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute [OGI-055], Innovative Medicines Initiative (EU/EFPIA) [EUbOPEN grant no. 875510], Janssen, Merck KGaA, Darmstadt, Germany, MSD, Novartis Pharma AG, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome. PK is funded by Wellcome Trust (ref. WT109965MA) and an NIHR Senior Fellowship.
Supplementary Data
Conflict of interest: DWE declares lecture fees from Gilead outside the submitted work.
Authors’ contributions: Sheila F Lumley: Sample collection, writing primary manuscript; David W Eyre: Concept and study design, data analysis, writing primary manuscript; Anna L McNaughton: Writing primary manuscript, laboratory work; Alison Howarth, Sarah Hoosdally, Stephanie B Hatch, James Kavanagh, Kevin K Chau, Stuart Cox, Kate Dingle, Justine Rudkin, Kathryn Auckland, Daniel Ebner: Laboratory work; Louise O Downs, Laura Dunn, Anita Justice, Susan Wareing, Monique I Andersson: Sample collection; Alexander Fyfe, Jai Bolton, Robert Paton: Data analysis; Alexander J Mentzer: Concept and study design; Katie Jeffery: Concept and study design, sample collection; Tim James: Sample collection, laboratory work, supervision; Tim E A Peto: Concept and study design, supervision; Brian D Marsden: Informatics; Gavin Screaton, Richard J Cornall: Supervision; Paul Klenerman: Concept and study design, supervision, reviewing and editing manuscript; David Stuart: Supervision, laboratory work; Derrick W Crook: Concept and study design, supervision; Nicole Stoesser: Concept and study design, sample collection, data analysis, reviewing and editing manuscript; Stephen H Kennedy: Concept and study design, ethics; Craig Thompson: Concept and study design, laboratory work, data analysis, reviewing and editing manuscript; Sunetra Gupta: Concept and study design, funding, supervision, project management; Philippa C Matthews: Concept and study design, project management, sample collection, data analysis, supervision, writing primary manuscript.
References
- 1.Statistical bulletin: Coronavirus (COVID-19) Infection Survey pilot: England, 25 June 2020. Office for National Statistics, in partnership with IQVIA, Oxford University and UK Biocentre. Available from: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/coronaviruscovid19infectionsurveypilot/latest
- 2.Emmenegger M, De Cecco E, Lamparter D, Jacquat RPB, Ebner D, Schneider MM, et al. Early plateau of SARS-CoV-2 seroprevalence identified by tripartite immunoassay in a large population. medRxiv. 2020; 2020.05.31.20118554.
- 3.National SARS-CoV-2 Serology Assay Evaluation Group Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis. 2020;S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson C, Grayson N, Paton R, Lourenço J, Penman B, Lee LN, et al. Neutralising antibodies to SARS coronavirus 2 in Scottish blood donors - a pilot study of the value of serology to determine population exposure. Infectious Diseases (except HIV/AIDS). medRxiv; 2020. doi: 10.1101/2020.04.13.20060467 [DOI]
- 5.Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):680-6. 10.1080/22221751.2020.1743767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep Med. 2020;1(3):100040. 10.1016/j.xcrm.2020.100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterhoff D, Glück V, Vogel M, Schuster P, Schütz A, Neubert P, et al. A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization. Infection. 2020. 10.1007/s15010-020-01503-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sood N, Simon P, Ebner P, Eichner D, Reynolds J, Bendavid E, et al. Seroprevalence of SARS-CoV-2-Specific Antibodies Among Adults in Los Angeles County, California, on April 10-11, 2020. JAMA. 2020;323(23):2425-7. 10.1001/jama.2020.8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Percivalle E, Cambiè G, Cassaniti I, Nepita EV, Maserati R, Ferrari A, et al. Prevalence of SARS-CoV-2 specific neutralising antibodies in blood donors from the Lodi Red Zone in Lombardy, Italy, as at 06 April 2020. Euro Surveill. 2020;25(24):2001031. 10.2807/1560-7917.ES.2020.25.24.2001031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyre DW, Lumley SF, O’Donnell D, Campbell M, Sims E, Lawson E, et al. Oxford University Hospitals Staff Testing Group Differential occupational risks to healthcare workers from SARS-CoV-2 observed during a prospective observational study. eLife. 2020;9:e60675. 10.7554/eLife.60675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, et al. Cochrane COVID-19 Diagnostic Test Accuracy Group Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;6:CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buekens P, Alger J, Bréart G, Cafferata ML, Harville E, Tomasso G. A call for action for COVID-19 surveillance and research during pregnancy. Lancet Glob Health. 2020;8(7):e877-8. 10.1016/S2214-109X(20)30206-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett GA, Milne SJ, Hehir MP, Lindow SW, O’connell MP. Health anxiety and behavioural changes of pregnant women during the COVID-19 pandemic. Eur J Obstet Gynecol Reprod Biol. 2020;249:96-7. 10.1016/j.ejogrb.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattern J, Vauloup-Fellous C, Zakaria H, Benachi A, Carrara J, Letourneau A, et al. Post lockdown COVID-19 seroprevalence and circulation at the time of delivery, France. Infectious Diseases (except HIV/AIDS). medRxiv; 2020. doi: 10.1101/2020.07.14.20153304 [DOI] [PMC free article] [PubMed]
- 15.Cosma S, Borella F, Carosso A, Sciarrone A, Cusato J, Corcione S, et al. The "scar" of a pandemic: Cumulative incidence of COVID-19 during the first trimester of pregnancy. J Med Virol. 2020;jmv.26267. 10.1002/jmv.26267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crovetto F, Crispi F, Llurba E, Figueras F, Gómez-Roig MD, Gratacós E. Seroprevalence and presentation of SARS-CoV-2 in pregnancy. Lancet. 2020;396(10250):530-1. 10.1016/S0140-6736(20)31714-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galang RR, Chang K, Strid P, Snead MC, Woodworth KR, House LD, et al. Severe Coronavirus Infections in Pregnancy: A Systematic Review. Obstet Gynecol. 2020;136(2):262-72. 10.1097/AOG.0000000000004011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen J, Vignaux O, Jacquemard F. Covid-19 in pregnant women: General data from a French National Survey. Eur J Obstet Gynecol Reprod Biol. 2020;251:267-8. 10.1016/j.ejogrb.2020.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maleki Dana P, Kolahdooz F, Sadoughi F, Moazzami B, Chaichian S, Asemi Z. COVID-19 and pregnancy: a review of current knowledge. Infez Med. 2020;28(suppl 1):46-51. [PubMed] [Google Scholar]
- 20.Maraschini A, Corsi E, Salvatore MA, Donati S. Coronavirus and birth in Italy: results of a national population-based cohort study. Obstetrics and Gynecology. medRxiv; 2020. doi: 10.1101/2020.06.11.20128652 [DOI] [PubMed]
- 21.INTERCOVID • INTERGROWTH-21st [Accessed 1 Jul 2020]. Available from: https://intergrowth21.tghn.org/intercovid/
- 22.Coronavirus (COVID-19) in the UK. [Accessed 16 Sep 2020]. Available from: https://coronavirus.data.gov.uk/cases
- 23.World Health Organization (WHO). WHO Coronavirus Disease (COVID-19) Dashboard. Geneva: WHO. [Accessed 16 Sep 2020]. Available from: https://covid19.who.int/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.