Abstract
Lack of access to high-frequency, high-volume patient-derived data, such as mechanical ventilator waveform data, has limited the secondary use of these data for research, quality improvement, and decision support. Existing methods for collecting these data are obtrusive, require high levels of technical expertise, and are often cost-prohibitive, limiting their use and scalability for research applications. We describe here the development of an unobtrusive, open-source, scalable, and user-friendly architecture for collecting, transmitting, and storing mechanical ventilator waveform data that is generalizable to other patient care devices. The system implements a software framework that automates and enforces end-to-end data collection and transmission. A web-based data management application facilitates nontechnical end users’ abilities to manage data acquisition devices, mitigates data loss and misattribution, and automates data storage. Using this integrated system, we have been able to collect ventilator waveform data from >450 patients as part of an ongoing clinical study.
Keywords: respiration, artificial, translational medical research, ventilators, mechanical, intensive care units, monitoring, physiologic, patient ventilator asynchrony
INTRODUCTION
The intensive care unit (ICU) is a data-rich, information-poor environment with limited access to high-volume patient data for secondary use. Waveform data streaming from physiologic monitoring devices are used by clinicians for diagnosis, prognostication, and assessing response to treatment, but are not routinely instantiated in the electronic health record (EHR).1 Vendor-derived hardware and software do not always allow collection of raw data and can cost thousands of dollars,2,3 making it potentially cost-prohibitive to deploy for a large, multipatient research study.4 Lack of access to these data sources is a major barrier to the transformation of ICU data into actionable information for translational research, quality improvement, and clinical decision support.1
These limitations are well illustrated by the use case of mechanical ventilation (MV). Ventilators are computerized devices that regulate the pressure, flow, and delivery of supplemental oxygen to support patients with acute respiratory failure. Ventilators display streaming waveform data representing pressure, flow, and volume (Figure 1A) that are used by clinicians to diagnose and manage patients in real time, but these data are not routinely captured.4–6 Lack of data access has hindered the study of MV and pathologic patient-ventilator interactions (PVIs), and previous studies of PVIs may have been limited, in part, to small sample sizes and isolated periods of data collection resulting from the lack of passive, unobtrusive, scalable, and easy-to-use informatics infrastructure.2,7–13 Improved informatics infrastructure would enhance the study of PVI and serve as a generalizable model for the development of systems to capture and store data streams from other critical care patient monitoring devices. Passive data-collection mechanisms would be particularly useful, given that they would allow researchers to enroll patients in a study, set up data collection, and then allow the collection mechanism to continue operating autonomously until data collection ends. For researchers located at institutions that lack the will or the funds necessary to implement an expansive data-collection infrastructure, access to research-focused architectures for streaming waveform data collection would be highly valuable. With this in mind, we set out to develop an automated, scalable, and intuitive physical and software infrastructure to unobtrusively acquire, wirelessly transmit, and store large volumes of streaming mechanical ventilator data.
Figure 1.
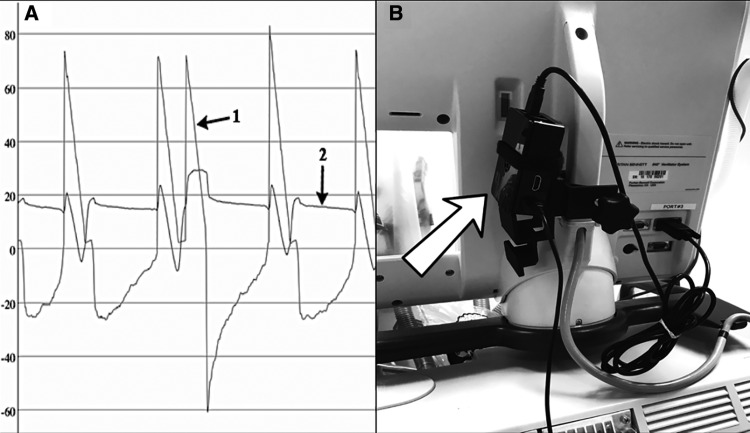
(A) Flow-time (arrow 1) and pressure-time (arrow 2) waveforms captured from the ventilator of a patient with severe acute respiratory distress syndrome. These waveforms are displayed in real time on ventilator user interfaces for diagnosis and management, but are not routinely captured for secondary use or decision support. (B) Image of the RPi attached to a ventilator. The arrow is pointing to the RPi, which is positioned behind the ventilator’s monitor screen. The RPi is attached behind the monitor to avoid influencing patient care or provider behavior while collecting data.
METHODS
We developed our architecture to support a large clinical study of patient-ventilator interactions. After extensive review by health system clinical engineering, IT security, and IT network operations, this system and our methodology were approved by the University of California, Davis (UCD) institutional review board (protocol no. 647002). All subjects or their surrogates provided informed consent per the requirement of the study protocol (see Supplementary Material for additional methods). Our system requirements were that it should: (1) enable passive, continuous, and automated data collection from multiple mechanical ventilators simultaneously; (2) be nearly undetectable by providers so as not to influence patient care14,15; (3) ensure temporally accurate data to preserve data linkage between collected ventilator waveform data (VWD) and EHR data16–18; (4) include automated archival storage; and (5) ensure ease of use by nontechnical end users for both data-acquisition hardware and data-management software.
We chose Raspberry PiTM (RPi) microcomputers for data collection. The RPi is a small, powerful Linux-based computer that, with appropriate interface drivers, can easily attach to a ventilator and rapidly begin collecting VWD. The RPi is small enough that it can be connected discreetly, without affecting provider behavior or interfering with patient care (Figure 1B). We achieved temporal accuracy in VWD by using the hospital’s Network Time Protocol (NTP) servers to ensure that all VWD time stamps were accurate to within 5 seconds of the actual time.19
Since all VWD are written into files on the RPi, archiving the data requires moving the files wirelessly from the RPi to a central location. We first experimented with command line file-transfer functions and open-source graphical user interface (GUI) file-transfer tools on a study laptop, but found this process frequently resulted in lost or misattributed VWD and required a high level of user training. Because linkage of data to the correct patient is of critical importance, we pursued an automated solution and created a clinical data supervisor web application (CSA) to handle persistent archival transfer and storage. The CSA automates transfer of VWD from multiple RPi’s to a temporary central file-storage server. The CSA also incorporates a GUI-based, single human checkpoint that allows study personnel to link the transferred files to the appropriate study subject’s unique study identifier. Finally, we created a database storage plug-in for the CSA that operates after study personnel have ensured file attribution and automatically populates newly aggregated VWD into a relational database containing other EHR-derived data. This architecture and workflow enables VWD collection from multiple devices operating simultaneously across the hospital. All steps are fully automated in VWD acquisition, except for a single data-quality assurance step, and meet the functional goals of reducing human error in physical acquisition, data misattribution, and linkage.
RESULTS
Data collection
The RPi’s only need 2 additional hardware components for collecting VWD: an RS-232 DB-9–to–USB null modem adapter and a power cable (see Supplementary Material for additional details on hardware selection and ventilator communication settings). We wrote specifically designed software in PythonTM for capturing VWD that only requires connecting the serial port of the ventilator to the RPi’s USB port using the DB-9–to–USB adapter and the input of the power supply to the RPi. Once connected, our software automatically begins passive collection of VWD from the ventilator. In order for it to remain unobtrusive, we attach each RPi behind the ventilator monitor, where it operates unseen, to avoid influencing patient care (Figure 2).
Figure 2.
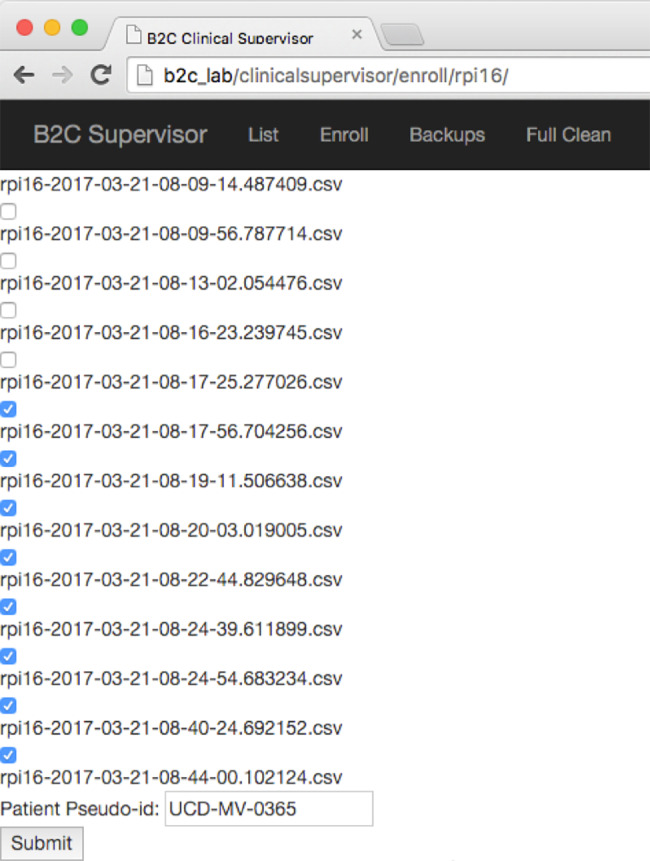
Clinical data supervisor web application user interface. Using the Enroll function, study personnel can list files residing on a specified RPi computer, select files of interest for backup, append selected file names with a unique study identifier (patient pseudo-id), and delete selected files from the device. Renamed files are then automatically uploaded to a study-specific relational database for subsequent computation by waveform analysis software and linkage with EHR-derived data. The user interface was designed to be simple, intuitive, and fast to allow rapid data attribution and file aggregation by study personnel with minimal computer skills.
Upon connecting to a ventilator, our software performs the essential task of connecting to an NTP server. After connecting to NTP, a time stamp is written at the top of each file so that we can later feed-forward the time for each recorded observation; data collection does not begin if an NTP server cannot be contacted. The feed-forward rate was based on the 50 Hz fixed sampling rate of the Puritan Bennet Model 840TM (PB-840) ventilators (Covidien, Dublin, Republic of Ireland) used in this study. We initially attempted to collect data by using the RPi system clock to attach a time stamp to each observation; however, this method yielded computational race conditions, resulting in data corruption. As a result, we opted for the less computationally complex system of feed-forward time-stamping.
PB-840 VWD records air flow, pressure, and “BS” (breath start) and “BE” (breath end) tokens to describe when a breath has started and ended. Sequential files ≤2 h in length are stored locally on the RPi and are backed up automatically every hour to minimize data loss in the event of hardware failure. Ventilator system configuration and alarm statuses can also be collected, but require ventilator setup that is exclusive to collecting VWD.20 In consequence, we chose to only collect VWD and not configuration settings and alarms for this study.
Data aggregation
Management of each RPi, including data aggregation, is performed by our CSA, which intentionally introduces a single human checkpoint into the data-aggregation workflow to ensure proper data attribution. The CSA provides a GUI for the following operations: (1) List all VWD files on the RPi’s; (2) Transfer selected VWD files from the RPi to an intermediate storage location for further processing; (3) Backup VWD files to prevent catastrophic data loss; (4) Append file names with each study subject’s unique identifier; and (5) Delete VWD files on the RPi’s once they have been renamed and transferred to another location.
Ensuring proper data attribution is a major challenge in the ICU, since an RPi can be switched to a new patient’s ventilator before being cleaned of a previous patient’s VWD. To solve this problem, new files are made every 2 h with file names that include date/time stamps corresponding to the time of file creation. The CSA GUI allows research personnel to efficiently select files on a given RPi that belong to a specific study subject (Figure 2), cross-referencing file date/time stamps with the time spent on the ventilator as described in the EHR and study records. Any files that cannot be unambiguously attributed to a study subject are easily deleted to prevent data misattribution.
Data storage
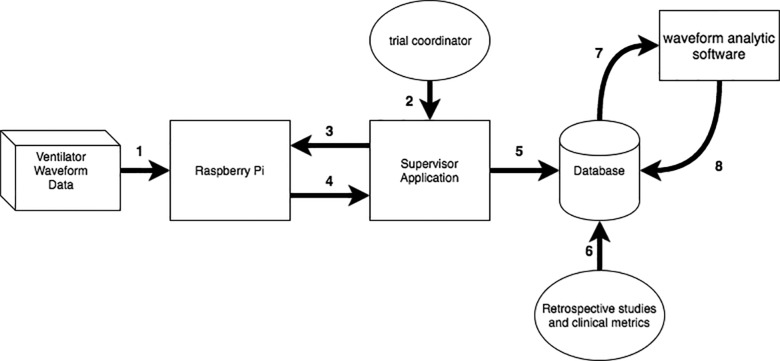
To automate data storage, we created a plug-in for the CSA that automatically uploads VWD into a database after transferred files are properly attributed. Using temporal metadata encoded in the VWD, we can then analyze the VWD with specialized computing algorithms that can detect adverse patient-ventilator interactions21–24 and combine the resulting information with EHR data sources to create a more complete picture of patient state (Figure 3). Without professional clinical research coordinators, we have used these methods to collect a dataset with approximately 48 million breaths from >450 patients, as illustrated in Table 1.
Figure 3.
Schematic of our data acquisition and management infrastructure. (1) Raw ventilator waveform data are sent to the RPi via serial connection. (2) Study investigator visits supervisor application, (3) chooses RPi, and selects files to extract or delete. (4) Files are extracted from the RPi and labeled with a unique patient identifier. (5) All data are then transferred using SQL and stored in a database. (6) Electronic health record and other clinical metadata are entered into the database. (7) VWD are processed using waveform analysis software to generate breath-by-breath physiologic metadata and detect relevant clinical events. (8) Time-stamped data derived from VWD processing are uploaded into the database for subsequent research and quality improvement studies.
Table 1.
Summary statistics describing the data collected during the course of our study to date
| Pathophysiology | Patients | Breaths | Hours |
|---|---|---|---|
| ARDS | 129 | 21 153 573 | 15 474 |
| COPD | 72 | 8 069 244 | 3649 |
| Other | 266 | 18 768 135 | 14 788 |
| Total | 467 | 47 990 952 | 33 911 |
Abbreviations: ARDS, acute respiratory distress syndrome; COPD, chronic obstructive pulmonary disease.
Scalability and adding new devices
Provisioning additional RPi devices enables simultaneous VWD collection from multiple study subjects and replacement of faulty devices. The lack of availability of study personnel with advanced computing expertise may limit the ability to instantiate software onto newly purchased RPi’s, limiting the scalability of our VWD collection architecture. To address this issue, we developed a managed, partially automated workflow to initialize new RPi’s by creating images (files containing the RPi operating system and necessary custom data-acquisition programs) that store preconfigured details of our software that can be copied to the RPi’s microSD card in a single step, resulting in a fully functional RPi without the need to install multiple software components (see Supplementary Material). After flashing the microSD card, VWD can be collected, provided a public NTP server is accessible. Extra configuration details, like changing default user passwords, can be managed with Ansible, an IT automation platform.25 This workflow substantially decreases the number of steps necessary for RPi deployment and may allow nontechnical users to assist more experienced personnel in provisioning.
Security
As a prerequisite to receiving approval of our study methods from the UCD institutional review board, the security of the proposed system was thoroughly analyzed by the UCD clinical engineering, IT networking, and security teams. In collaboration with clinical engineering, we determined that the PB-840 can perform read-only operations via the RS-232 port, preventing remote operation of the PB-840 via the RPi’s.20 With the cooperation of the clinical networking group, we were able to link the RPi’s to a private, monitored, firewalled, encrypted partition of the UCD Medical Center network that is closed to non-whitelisted devices and unable to communicate with other partitions. Additional security steps were taken to ensure that the RPi’s could not communicate with the CSA directly, and that the CSA could perform actions on the RPi’s limited only to transferring, deleting, and listing VWD files. All VWD transferred on the network is encrypted by the secure copy protocol. All VWD stored on the RPi’s is non–protected health information, so resting encryption was not required. For additional details on security, including hardening of the RPi’s and CSA, see the Supplementary Material.
DISCUSSION
Rapid advances in patient-monitoring devices, wireless communication, and computational capacity are leading to the generation of large volumes of patient-generated data in clinical environments. Limited access to reliable electronic data from medical devices represents a formidable challenge to the secondary use of these data for research, quality improvement, and decision support.5 Researchers in particular face problems including lack of institutional infrastructure, limited funding, and a dearth of technical expertise required to collect electronic data.4
To address these issues in the context of mechanical ventilation research, we developed an inexpensive and easy-to-use system that fully automates the end-to-end workflow for data acquisition, wireless communication, and storage, except for a single intentional human checkpoint to ensure proper data attribution. Our system enables temporally accurate waveform data collection and database storage with an unobtrusive physical presence, to avoid influencing patient care or introducing observer bias, and requires limited training by study personnel.14,15 Furthermore, instructions are provided to enable researchers to use our platform at other institutions (see Supplementary Materials).
Several efforts to develop waveform data-acquisition infrastructure have been described, but these examples are limited in several important ways, including the need for a human to be actively involved at multiple points in data collection, the intrusiveness of monitoring devices that could influence provider behavior or adversely affect patient care, or the use of high-cost and proprietary products.2,7–13 In an important proof-of-concept study, Howard demonstrated the ability to wirelessly gather data from PB-840 ventilators using a laptop computer and ad hoc Bluetooth and WiFi local area networks.26 OpenICETM is another software platform aimed at demonstrating the feasibility of an open-source waveform data-acquisition architecture.12 A key difference between OpenICE and our system is that our platform uses minimal network bandwidth and allows for passive data collection, making it ideal for conducting a longitudinal, low-maintenance research study on existing clinical and research networks. Our system extends these previous efforts by demonstrating the value of automating the end-to-end data-acquisition, transmission, and storage workflow, incorporating the use of enterprise wireless networking to eliminate geographic limitations on data collection, and restricting human-software interaction to a single workflow checkpoint. Finally, our results validate the generalizability and utility of an open-source, integrated architecture model deployed in a large, geographically and technologically complex health system.
Our system serves as a use case for the development of automated, wireless research-focused data-acquisition systems in complex health care environments. While our architecture is currently limited to use with mechanical ventilators that provide open data access through an RS-232 serial port or similar interface, such as the PB-840 ventilator, the framework can be generalized to other patient-monitoring devices that allow similar data access. Due to variance in network topology and security across institutions, there may be technical issues that must be addressed before implementation. For example, our devices currently transfer data on the hospital clinical wireless network, but this might not be possible in cases where wireless signal availability is poor or institutional barriers prohibit research devices on facility networks. In the latter case, a separate freestanding wireless local area network may be required for implementation.26 Finally, our design is meant to be operated by users with minimal technical training, but end-to-end system setup remains complex and needs to be performed with assistance from IT professionals with systems administration experience.
In summary, we have developed a novel platform for acquiring waveform data from mechanical ventilators using unobtrusive, low-cost microcomputers, and demonstrated the ability to automate the process of data acquisition, wireless transmission, and database storage of VWD. Our system offers research teams a new avenue for accessing VWD data and serves as a generalizable model for the ongoing development of systems to acquire, transmit, and store other patient data streams for secondary use.
FUNDING
This work was generously supported by the National Heart, Lung, and Blood Institute Emergency Medicine K12 Clinical Research Training Program, grant number K12 HL108964; the Center for Information Technology Research in the Interest of Society, grant number 2014-227; the UC Davis Clinical and Translational Science Center, grant number UL1 RR024146; and the National Center for Advancing Translational Sciences and National Institutes of Health, grant number UL1 TR001860. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CONTRIBUTORS
JYA, GBR, JPD, NRA, and BTK all conceptualized and designed this study. ECG, MKL, and JN contributed to successful implementation of study components and assisted with manuscript review. All authors were significantly involved in all stages of the study and approved the final version of the manuscript.
COMPETING INTERESTS
The authors declare no conflicts of interest related to this work.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful for the dedicated support of the nursing, respiratory care, information technology, and clinical engineering staff at UC Davis Medical Center who helped make this project possible.
REFERENCES
- 1. Halpern NA. Innovative designs for the smart ICU: Part 1: from initial thoughts to occupancy. Chest. 2014;1452:399–403. [DOI] [PubMed] [Google Scholar]
- 2. Beitler JR, Sands SA, Loring SH, et al. Quantifying unintended exposure to high tidal volumes from breath stacking dyssynchrony in ARDS: the BREATHE criteria. Intensive Care Med. 2016;429:1427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murias G, Montanya J, Chacón E, et al. Automatic detection of ventilatory modes during invasive mechanical ventilation. Critical Care. 2013;17:2. BioMed Central. 2016;20(1):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raghupathi W, Raghupathi V. Big data analytics in healthcare: promise and potential. Health Inform Sci Syst. 2014;2:1. BioMed Central. 2014;2(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Auffray C, Balling R, Barroso I, et al. Making sense of big data in health research: towards an EU action plan. Genome Med. 2016;8:1. BioMed Central. 2016;8(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murdoch TB, Detsky AS. The Inevitable Application of Big Data to Health Care. JAMA. 2013;30913:1351–52. [DOI] [PubMed] [Google Scholar]
- 7. Oliveira S, Portela CF, Santos MF. Pervasive universal gateway for medical devices. In: Recent Advances in Electrical Engineering and Education Technologies (SCI 2014). 2014;205–10. [Google Scholar]
- 8. Mulqueeny Q, Ceriana P, Carlucci A, Fanfulla F, Delmastro M, Nava S. Automatic detection of ineffective triggering and double triggering during mechanical ventilation. Intensive Care Med. 2007;3311:2014–18. [DOI] [PubMed] [Google Scholar]
- 9. Chanques G, Kress JP, Pohlman A, et al. Impact of ventilator adjustment and sedation-analgesia practices on severe asynchrony in patients ventilated in assist-control mode. Critical Care Med. 2013;419:2177–87. [DOI] [PubMed] [Google Scholar]
- 10. Szlavecz A, Chiew YS, Redmond D, et al. The Clinical Utilisation of Respiratory Elastance Software (CURE Soft): a bedside software for real-time respiratory mechanics monitoring and mechanical ventilation management. BioMed Engineering 2014;13:1. BioMed Central. 2014;13(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pohlman MC, McCallister KE, Schweickert WD, et al. Excessive tidal volume from breath stacking during lung-protective ventilation for acute lung injury. Critical Care Med. 2008;3611:3019–23. [DOI] [PubMed] [Google Scholar]
- 12. Plourde J, Arney D, Goldman JM. OpenICE: An Open, Interoperable Platform for Medical Cyber-physical Systems. IEEE; 2014: 221–21. [Google Scholar]
- 13. Blanch L, Villagra A, Sales B, et al. Asynchronies during mechanical ventilation are associated with mortality. Intensive Care Med. 2015;414:633–41. [DOI] [PubMed] [Google Scholar]
- 14. Chen LF, Vander Weg MW, Hofmann DA, Reisinger HS. The Hawthorne effect in infection prevention and epidemiology. Infect Control Hospital Epidemiol. 2015;3612:1444–50. [DOI] [PubMed] [Google Scholar]
- 15. McCambridge J, Witton J, Elbourne DR. Systematic review of the Hawthorne effect: new concepts are needed to study research participation effects. J Clin Epidemiol. 2014;673:267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacobs I, Nadkarni V, Bahr J, et al. Cardiac arrest and cardiopulmonary resuscitation outcome reports: update and simplification of the Utstein templates for resuscitation registries. Resuscitation. 2004;633:233–49. [DOI] [PubMed] [Google Scholar]
- 17. Holman CDJ, Bass JA, Rosman DL, et al. A decade of data linkage in Western Australia: strategic design, applications and benefits of the WA data linkage system. Aust Health Rev. 2008;324:766–77. [DOI] [PubMed] [Google Scholar]
- 18. Ludvigsson JF, Otterblad-Olausson P, Pettersson BU, Ekbom A. The Swedish personal identity number: possibilities and pitfalls in healthcare and medical research. Eur J Epidemiol. 2009;2411:659–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mills D, Burbank J, Kasch W. Network Time Protocol Version 4: Protocol and Algorithms Specification. RFC Editor; 2010. [Google Scholar]
- 20. Covidien. Operator's and Technical Reference Manual: Puritan Bennett 800 Series Ventilator System. Covidien; 2011. www.medtronic.com/content/dam/covidien/library/us/en/product/acute-care-ventilation/PB840_Technical_Reference_Manual_EN_10067720D00.pdf. Accessed July 29, 2017. [Google Scholar]
- 21. Adams J, Lieng M, Kuhn B, et al. Automated mechanical ventilator waveform analysis of patient-ventilator asynchrony. Chest. 2015;1484:175A. [Google Scholar]
- 22. Mulqueeny Q, Ceriana P, Carlucci A, Fanfulla F, Delmastro M, Nava S. Automatic detection of ineffective triggering and double triggering during mechanical ventilation. Intensive Care Med. 2007;3311:2014–18. [DOI] [PubMed] [Google Scholar]
- 23. Chiew YS, Pretty CG, Beatson A, et al. Automated Logging of Inspiratory and Expiratory Non-synchronized breathing (ALIEN) for Mechanical Ventilation. IEEE; 2015: 5315–8. [DOI] [PubMed] [Google Scholar]
- 24. Blanch L, Sales B, Montanya J, et al. Validation of the Better Care® system to detect ineffective efforts during expiration in mechanically ventilated patients: a pilot study. Intensive Care Med. 2012;385:772–80. [DOI] [PubMed] [Google Scholar]
- 25. Heap M. Advanced Ansible. Berkeley, CA: Apress; 2016:137–57. [Google Scholar]
- 26. Howard WR. Wireless on-demand and networking of Puritan Bennett 840 ventilators for direct data capture. Respir Care. 2007;5211:1530–41. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.