Abstract
Clinical genome and exome sequencing can diagnose pediatric patients with complex conditions that often require follow-up care with multiple specialties. The American Academy of Pediatrics emphasizes the role of the medical home and the primary care pediatrician in coordinating care for patients who need multidisciplinary support. In addition, the electronic health record (EHR) with embedded clinical decision support is recognized as an important component in providing care in this setting. We interviewed 6 clinicians to assess their experience caring for patients with complex and rare genetic findings and hear their opinions about how the EHR currently supports this role. Using these results, we designed a candidate EHR clinical decision support application mock-up and conducted formative exploratory user testing with 26 pediatric primary care providers to capture opinions on its utility in practice with respect to a specific clinical scenario. Our results indicate agreement that the functionality represented by the mock-up would effectively assist with care and warrants further development.
Keywords: decision support systems, clinical genetic testing, electronic health records
INTRODUCTION
Due to steadily lowering costs, clinical genome and exome sequencing (CGES) is used increasingly in pediatric settings. Although most tests are ordered by pediatric specialty physicians,1 pediatric primary care physicians (PCPs) provide routine management and care coordination for tested children.2,3 CGES frequently identifies complicated and rare genetic diagnoses, possibly with complex or long-term care requirements across multiple providers.2,4 CGES can also result in secondary findings unrelated to the initial indication for testing,5,6 which can require follow-up by primary care clinicians and specialists who did not originally order the test.
The American Academy of Pediatrics recommends a medical home approach to care in these situations where care is required from multiple specialists, and emphasizes the role of the PCP in coordinating the care of children who are seen by multiple specialty providers, including genetics.7,8 A medical home approach has been shown to improve outcomes for children, such as lower hospitalization rates.9 Among the recommendations for implementing the medical home is the use of information systems and tools to support care-coordination activities. One such approach is to integrate clinical decision support (CDS) software into the electronic health record (EHR). In the context of CGES, pediatricians face unique challenges navigating ethical, legal, and social consideration (eg, related to risk alleles for adult-onset disease).10 Health screening and intervention workflows are often markedly different in pediatric settings, which affects CDS design. As children mature from a state of complete dependence on their caregivers into adolescence, with increasing levels of autonomy, the educational materials that pediatricians provide must be tailored to their developmental level and must always consider the changing perspective of their caregivers during the process of child development. However, when appropriately prepared,11 pediatricians also have the opportunity to markedly improve health outcomes for children in their care through diligent screening and early intervention in disease processes.
EHR-based CDS is recognized as an important tool for improving many aspects of clinical care, including genomic testing.12–14 The Clinical Sequencing Exploratory Research (CSER) consortium, together with the Electronic Medical Records and Genomics network, found that hospitals are already incorporating genomic data into their EHRs and identified CDS for medically actionable results as a high priority among respondents.15 The network, which has focused on CDS for pharmacogenomic variants, has identified multiple barriers to implementing CDS for genomic test results.16 Barriers to implementing CDS are often organizational, related to communication, staffing, and approval.17 These organizational barriers have been addressed by developing evidence of effectiveness for other types of CDS,18 for example, in the domains of medication interaction alerts,19 chronic disease management,20 and preventive health care reminders.21 It is likely that part of the challenge at present is that physicians and other organizational decision-makers do not understand the value of CDS for genomic test results.
Given (1) the expectation that PCPs will provide medical homes for all children under their care regardless of complexity, (2) the increasing availability of CGES, and (3) the lack of appropriate CDS embedded in EHRs to help pediatricians understand and appropriately act on CGES results, we initiated a pilot project to guide CDS implementation efforts for CGES results in primary care pediatric settings. We investigated PCP perspectives on genomic testing, proposed a candidate design for a CGES CDS tool based on these perspectives, and evaluated the candidate design with PCPs.
METHODS
We explored the current practice of genomic medicine by conducting a preliminary round of semistructured interviews with 6 clinicians. Based on the broad challenges that emerged from these interviews, we designed a CDS tool prototype and collected data on the usability and utility of the design from a separate sample of 26 PCPs through a design walk-through of the tool.22,23
Study setting
Our study was conducted between April 1, 2015, and December 31, 2015, as part of the CSER consortium’s portfolio of research in the Pediatric Research Consortium affiliated with The Children’s Hospital of Philadelphia (CHOP). This network of 31 pediatric practices spans urban, suburban, and rural regions of Pennsylvania and New Jersey and includes both academic and nonacademic practices. Prior work has shown that the demographic characteristics of children who receive care at these practices are representative of the region.24 Although CGES is not yet widespread in our network, it is increasingly available. Much of our PCPs’ experience to date has been with genetic tests for specific conditions. This network of practices had been using EpicCare (Epic Systems Inc., Verona, WI, USA) as their EHR for over 10 years at the time of this study.
Semistructured interviews
The semistructured interviews with 6 clinicians focused on their experience and attitudes toward genomic testing in general and CGES tests specifically, barriers to the utilization of genomic test results in patient care, educational and informational needs, and EHR functionality relevant to CGES testing. The clinicians represented multiple perspectives on genomic testing and primary care workflow as follows: clinical site leadership, EHR expertise, care coordination for medically complex children, medical education (2 clinicians), and genetic domain expertise.22 The interviews were audio recorded, and 1 author (EK) took written notes of salient themes during the interviews. Two authors (JP and DK) reviewed the audio recordings and independently generated themes related to the challenges of accessing and using genomic test results. Themes were discussed among these 3 authors and refined until there was 100% agreement.
Development of low-fidelity mock-ups
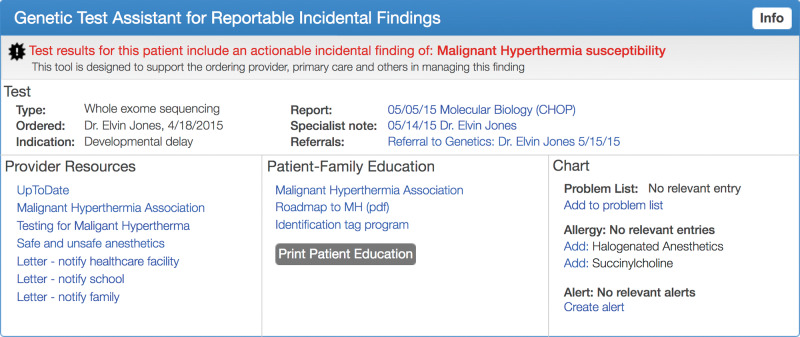
Based on the results of these interviews, we created a set of interactive mock-ups (Axure, San Diego, CA, USA) to represent the clinical scenario of a child presenting to a PCP with a medically actionable incidental finding of malignant hyperthermia susceptibility due to a pathogenic RYR1 variant.22,25 This variant is among the reportable incidental findings recommended by the American College of Medical Genetics and Genomics.5 We developed the mock-up in consultation with a pediatric expert on malignant hyperthermia. Using a set of 15 separate mock-up screens, we presented a potential clinical workflow including (1) notification of an actionable result, (2) review of specialist notes, (3) access to clinician and patient education content, and (4) assistance with PCP documentation activities (see Figure 1 and Supplementary Appendix A).
Figure 1.
EHR integrated intervention interactive mock-up, including a provider alert, direct linkage to results report, specialist note, education resources for clinicians, patients and families, note templates, problem list, and other documentation resources.
Design walk-through
We evaluated the mock-up by performing a formative user test based on modified design walk-through method26,27 with pediatric PCPs recruited from 5 practices in CHOP’s research network of 31 practices. Practice sites were selected to include both teaching (urban) and nonteaching (suburban) practices. Practice managers received a letter inviting them to participate in the study, and when the invitation was accepted, a 1-hour user test session was scheduled. The practice manager invited PCPs from the practice to the user test session.
The design walk-through was performed in small group sessions (1 session at each participating site). At the start of the session, the PCP study subjects completed a short survey that included questions about demographics (gender, age, role, clinical training), Likert scale ratings of attitudes toward genomic testing, and relevant clinical training. The facilitators then gave a short slide presentation on CGES and presented the malignant hyperthermia scenario. The facilitator then stepped though the 15 mock-up screens. Following the scenario-based walkthrough, PCPs completed additional survey questions, including Likert scale ratings for tool features that addressed each challenge identified in the preliminary interviews, and a set of questions based on the Technology Acceptance Model (TAM),28 which has previously been applied to health care information technology, including primary care and pediatrics.29–31 The TAM survey included 14 questions from 3 dimensions: 10 perceived usefulness, 3 perceived ease of use, and 1 intent to use (see Supplementary Appendix B). Survey responses were entered into a REDCap database,32 then exported and analyzed with the statistical program R version 3.30,33 adding the “psych,” “likert,” and “ggplot” modules to analyze survey responses and generate figures. Demographic characteristics of study participants were compared with those of the larger CHOP network of PCPs and tested for significant differences using a chi-square goodness-of-fit test with a critical value of 0.05. We calculated Cronbach’s alpha to assess internal consistency within the TAM dimensions of perceived ease of use and perceived usefulness.
RESULTS
Semistructured interviews
Six PCPs participated in the preliminary round of semistructured interviews. PCPs were enthusiastic about the subject and readily shared their descriptions of real experiences and ideas on how to address the challenges they described. Based on an analysis of the interviews, we identified 6 major challenges to utilizing genomic test results in the care of patients: (1) failure of notification of test results; (2) confusing genomic test reports; (3) difficulty providing care coordination for complex cases; (4) lack of communication with genetics specialists; (5) lack of educational materials for providers, patients, and families; and (6) lack of consistency in clinical documentation. Table 1 presents illustrative quotes from interviewees for each challenge.
Table 1.
Interviews with PCPs revealed challenges to access and use of genomic test results
| Challenge | Provider Quote |
|---|---|
| Notification | “I keep checking [the EHR] to see if there are [genomic test] results, no notification whatsoever!” |
| Confusing genomic test reports | “[The genomic test report contains] all kinds of information I do not understand.” |
| Care coordination | “[The genomic test report has a] list of a million things to do… [There were] referrals to eight specialties!” |
| Communication | “As far as I know, there is no option to speak to a genetic counselor.” |
| Education | “[Genetics] has jumped so far from when I was in medical school.” |
| Documentation | “It has not yet been decided… who is responsible for the problem list.” |
Design walk-through
In our second activity, 26 PCPs from 5 practices affiliated with CHOP participated in the design walk-through.
Demographics
PCP participants are described in Table 2. Mean age was 49.7 (range 29–63). Mean years in practice was 17.4 (range 1–38). With the exception of the proportion of clinicians in teaching practices, these characteristics are not significantly different from the overall network. In addition, 10 subjects (38%) responded that they had no patients with genomic testing in the prior year, 14 (54%) had between 1 and 5 patients, and 2 (8%) had 6–10 patients.
Table 2.
PCP demographics and comparison to CHOP network
| Category | Characteristic | No. of providers (%) | CHOP network (%) | P value |
|---|---|---|---|---|
| Gender | Female | 22 (85) | 80 | 0.556 |
| Role | Physician | 23 (88) | 83 | 0.458 |
| Nurse practitioner | 3 (12) | 17 | ||
| Primary care practice type | Teaching | 16 (62) | 36 | 0.006 |
| Nonteaching | 10 (38) | 64 |
Genetics topics
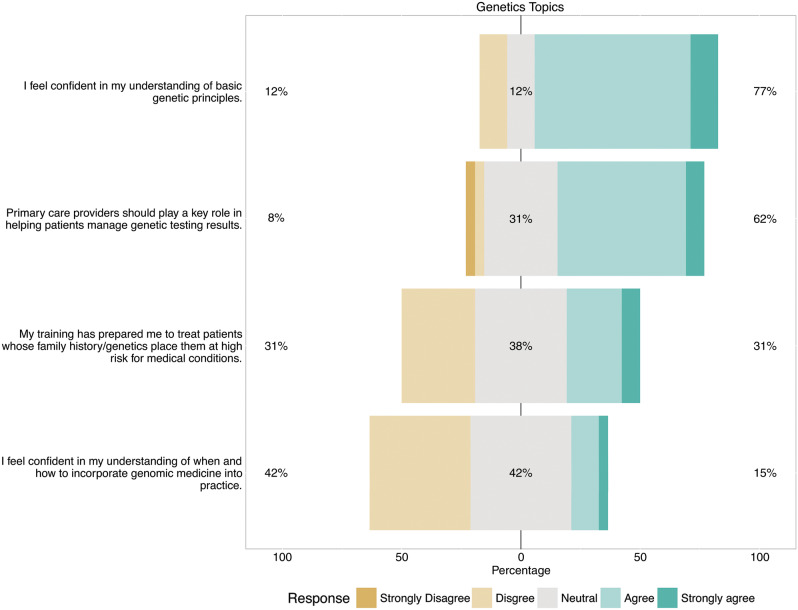
Seventy-seven percent of subjects indicated that they were confident in their understanding of basic genetic principles and 62% agreed that PCPs should help patients manage genomic testing results. By contrast, 31% agreed that their training was sufficient and 15% were confident in their ability to incorporate genomic medicine into practice (Figure 2).
Figure 2.
Bar chart summarizing Likert responses to items regarding clinicians’ perspectives on their role in genomic testing. Each bar is centered on the response of “neutral.” Bars extending to the right indicate favorable responses (up to 100%) and those extending to the left indicate unfavorable responses (up to 100%). The percentages reported at the left and right of each bar indicate the exact percentages of unfavorable vs favorable responses, respectively (excluding neutral responses).
Tool functionality
Seven functions that addressed the challenges identified in the semistructured interviews were illustrated in the prototype mock-up. The vast majority of subjects indicated that all 7 functions were important: patient/family education content (100%), clinician education content (100%), EHR notifications (96%), problem list entry (92%), letter templates (92%), link to report (92%), and specialist note (92%). All subjects agreed that the tool would be useful, even for very few patients, and all responded that they would use the tool if it were available.
TAM survey
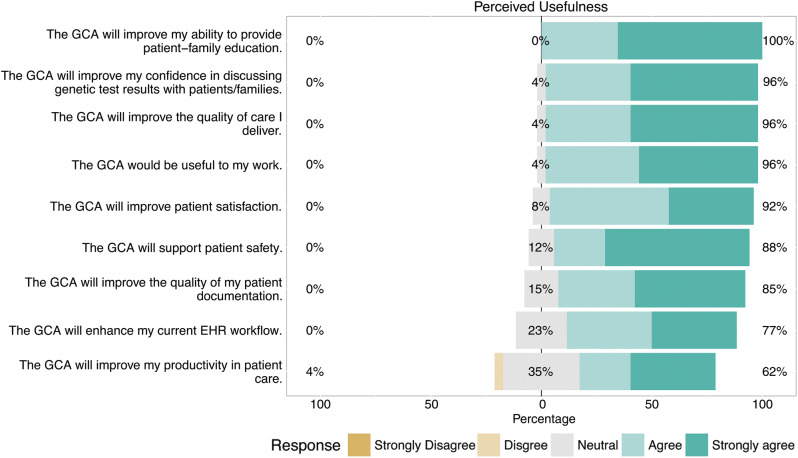
We performed Cronbach’s alpha test on the 2 TAM dimensions and both results, perceived usefulness (0.89) and perceived ease of use (0.88), exceeded the accepted range of 0.70 for internal consistency and reliability. Agreement on the 10 perceived usefulness questions ranged from 100% to 62% (Figure 3). Agreement for the 3 perceived ease of use questions were as follows: 85% agreement that the tool was clear and understandable, 73% agreement that the tool was easy to use, but only 50% agreement that the tool could be used without any training.
Figure 3.
Bar chart summarizing Likert responses to items on the TAM regarding perceived usefulness of the genomic care assistant. Each bar is centered on the response of “neutral.” Bars extending to the right indicate favorable responses (up to 100%) and those extending to the left indicate unfavorable responses (up to 100%). The percentages reported at the left and right of each bar indicate the exact percentages of unfavorable vs favorable responses, respectively (excluding neutral responses).
There were no statistically significant associations between survey responses and clinician demographics.
DISCUSSION
We found that the majority of PCPs were already caring for children who receive genetic and genomic testing. Similarly, a majority of PCPs felt that they should play a role in managing genetic and genomic test results. However, the clinicians we interviewed reported gaps in their ability to provide care for these patients due to challenges such as understanding the test results and coordinating care related to the results. All clinicians indicated that access to educational materials related to test results was important. Our CDS tool mock-up was positively received, with a high degree of agreement that such a tool would be usable and would aid in the care of patients with genomic testing. The only exception was that half of the clinicians indicated on the TAM survey that they would need training to use the CDS features, but it is possible they misinterpreted “training” in this context to mean “genetics medical education.”
Our results aligned well with recent literature documenting that CDS for genomic test results is a high priority for successful implementation of genomic medicine.15 Also, the specific importance placed on educational materials has been observed in studies of genomic CDS in oncology and medication reconciliation.34,35 These studies found a similar desire by clinicians for relevant educational materials that support the interpretation of results in the context of patient care.
Localization of CDS
The CDS design mock-up we developed and validated in our study provides guidance for future implementation efforts at institutions that seek to promote the role of PCPs as the patient-centered medical home for children who have CGES results. However, specific local workflows must always be considered in any implementation of health information technology.36 Future implementation efforts must examine these workflows at a granular level to ensure that they meet the needs of individual institutions. Fortunately, toolkits such as the SMART platform provide significant flexibility in adapting design prototypes to local workflow requirements.34,37
Future directions
To meet the care coordination goals of the medical home, primary and specialty care providers need a unified view of the clinical implications of genomic test results. Evaluation of the perspectives and combined information needs of primary care and specialty providers is warranted. Additionally, new interoperable knowledge bases will be required to provide both CDS and references to educational materials. This need has been confirmed and studied in other areas of genomic decision support.38,39 We used low-cost user-centered design methods to formulate a preliminary picture of the need and an initial direction for possible solutions for genomic CDS in pediatric primary care, but the specific knowledge base requirements of clinicians should be explored further. The degree to which we found that clinicians have already been engaged in genomic medicine, coupled with the apparent gap in access to basic informational resources, indicates that further study and progress toward real solutions is warranted.
Limitations
Our study was limited to 1 institution, and clinicians from teaching practices (those involved in the education of residents and medical students) were overrepresented in our sample. However, there were no associations between the demographic characteristics of clinicians in our sample and the survey responses, making it more likely that results would be similar at other institutions. Also, given the fundamental difficulties providers reported regarding notification of genomic test results and finding results in the EHR, it is possible that the positive response to the CDS mock-up was overstated, given the current challenges faced by PCPs. Additionally, pediatric settings that have significantly different workflows related to genetic evaluation may have different requirements for genomic decision support than those identified in our study. Finally, as noted previously, there may have been a misinterpretation of the term “training” in the context of the standardized TAM survey to mean “genetics medical education.”
CONCLUSIONS
Pediatric primary care physicians are already managing children who have had genomic testing completed, and they feel they have an important role in clinical management based on those results. Access to educational materials was identified as the most important feature of the proposed tool. Clinicians reported that our CDS prototype’s features were both usable and useful in the management of children who have had CGES tests performed.
Funding
This work was supported by National Human Genome Research Institute grant number UO1HG006546.
Competing interests
The authors have no competing interests to declare.
Contributors
JP initiated the study, led the team in study design, performed data collection, and drafted and revised the manuscript. He is the guarantor. DK selected methods, recruited and designed data collection surveys, performed data collection, analyzed survey data, and drafted and revised the manuscript. EK performed literature searches and data collection, and revised the manuscript. JM performed statistical analysis and revised the manuscript. BB advised on methods, designed data collection surveys, performed literature review, and revised the manuscript. RG designed data collection surveys, recruited subjects, performed data collection, and revised the manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors want to thank the network of primary care clinicians and their patients and families for contributing to this project and clinical research facilitated through the Pediatric Research Consortium at The Children’s Hospital of Philadelphia. We want to thank Ian Krantz and Nancy Spinner and the CHOP CSER project team for their support and critical review of the design and initial results of this study.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
References
- 1. Tarini BA, Zikmund-Fisher BJ, Saal HM et al. . Primary care providers’ initial evaluation of children with global developmental delay: a clinical vignette study. J Pediatrics. 2015;1676:1404–08.e1. [DOI] [PubMed] [Google Scholar]
- 2. Schaefer GB, Larson IA, Bolick J et al. . What is the role of clinical genetics in the patient-centered medical home?: A commentary from the Medical Home Workgroup of the Heartland Regional Genetics and Newborn Screening Collaborative. Genet Med. 2016;185:440–42. [DOI] [PubMed] [Google Scholar]
- 3. Scott J, Trotter T. Primary care and genetics and genomics. Pediatrics. 2013;132(Suppl 3):S231–37. [DOI] [PubMed] [Google Scholar]
- 4. Yang Y, Muzny DM, Xia F et al. . Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;31218:1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Solomon BD, Hadley DW, Pineda-Alvarez DE et al. . Incidental Medical Information in Whole-Exome Sequencing. Pediatrics. 2012;1296:e1605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Green RC, Berg JS, Grody WW et al. . ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;157:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Medical Home Initiatives for Children With Special Needs Project Advisory Committee. The Medical Home. Pediatrics. 2002;1101:184–86.12093969 [Google Scholar]
- 8. AAP Genetics in Primary Care Institute [Internet]. https://geneticsinprimarycare.aap.org/. Accessed October 1, 2016.
- 9. Cooley WC, McAllister JW, Sherrieb K et al. . Improved outcomes associated with medical home implementation in pediatric primary care. Pediatrics. 2009;1241:358–64. [DOI] [PubMed] [Google Scholar]
- 10. Clayton EW, McCullough LB, Biesecker LG et al. . Addressing the ethical challenges in genetic testing and sequencing of children. Am J Bioeth. 2014;142015:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vassy JL, Korf BR, Green RC. How to know when physicians are ready for genomic medicine. Sci Transl Med. 2015;7287:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Welch BM, Eilbeck K, Fiol G Del et al. . Technical desiderata for the integration of genomic data with clinical decision support. J Biomed Inform. 2014;51:3–7. [DOI] [PubMed] [Google Scholar]
- 13. Masys DR, Jarvik GP, Abernethy NF et al. . Technical desiderata for the integration of genomic data into Electronic Health Records. J Biomed Inform. 2012;453:419–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Overby CL, Kohane I, Kannry JL et al. . Opportunities for genomic clinical decision support interventions. Genet Med. 2013;1510:817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shirts BH, Salama JS, Aronson SJ et al. . CSER and eMERGE: Current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc. 2015;226:1231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCarty CA, Chisholm RL, Chute CG et al. . The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;41:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herr TM, Bielinski SJ, Bottinger E et al. . Practical considerations in genomic decision support: The eMERGE experience. J Pathol Inform. 2015;6:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shojania KG, Jennings A, Mayhew A et al. . The effects of on-screen, point of care computer reminders on processes and outcomes of care. Cochrane Database of Systematic Rev. 2009. CD001096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Awdishu L, Coates CR, Lyddane A et al. . The impact of real-time alerting on appropriate prescribing in kidney disease: a cluster randomized controlled trial. J Am Med Inform Assoc. 2016;233:609–16. [DOI] [PubMed] [Google Scholar]
- 20. Roshanov PS, Misra S, Gerstein HC et al. . Computerized clinical decision support systems for chronic disease management: a decision-maker–researcher partnership systematic review .Implement Sci. 2011;6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fiks AG, Grundmeier RW, Biggs LM et al. . Impact of clinical alerts within an electronic health record on routine childhood immunization in an urban pediatric population. Pediatrics. 2007;1204:707–14. [DOI] [PubMed] [Google Scholar]
- 22. Poon EG, Wang SJ, Gandhi TK et al. . Design and implementation of a comprehensive outpatient Results Manager. J Biomed Inform. 2003;36(1–2):80–91. [DOI] [PubMed] [Google Scholar]
- 23. Johnson CM, Johnson TR, Zhang J. A user-centered framework for redesigning health care interfaces. J Biomed Inform. 2005;38:75–87. [DOI] [PubMed] [Google Scholar]
- 24. Feemster KA, Li Y, Grundmeier R et al. . Validation of a pediatric primary care network in a US metropolitan region as a community-based infectious disease surveillance system. Interdiscip Perspect Infect Dis. 2011;2011. doi:10.1155/2011/219859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rose AF, Schnipper JL, Park ER et al. . Using qualitative studies to improve the usability of an EMR. J Biomed Inform. 2005;381:51–60. [DOI] [PubMed] [Google Scholar]
- 26. Nielsen J. Usability Engineering. 1993;443:362. [Google Scholar]
- 27. Kushniruk AW, Patel VL. Cognitive and usability engineering methods for the evaluation of clinical information systems. J Biomed Inform. 2004;371:56–76. [DOI] [PubMed] [Google Scholar]
- 28. Davis FD. User acceptance of information technology: system characteristics, user perceptions and behavioral impacts. Int J ManMachine Stud. 1993;383:475–87. [Google Scholar]
- 29. Holden RJ, Karsh BT. The Technology Acceptance Model: its past and its future in health. J Biomed Inform. 2010;431:159–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Schaik P, Flynn D, Van Wersch A et al. . The acceptance of a computerised decision-support system in primary care: a preliminary investigation. Behav Inf Technol. 2004;235:321–26. [Google Scholar]
- 31. Chismar WG, Wiley-Patton S. Test of the technology acceptance model for the internet in pediatrics. Proc AMIA Symp. 2002;155–59. PMC2244480. [PMC free article] [PubMed] [Google Scholar]
- 32. Harris PA, Taylor R, Thielke R, Payne J et al. . Research electronic data capture (REDCap): A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;422:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 34. Warner JL, Rioth MJ, Kohane IS et al. . SMART precision cancer medicine: a FHIR-based app to provide genomic information at the point of care. J Am Med Inform Assoc. 2016;234:701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nishimura AA, Shirts BH, Tarczy-Hornoch P, Physician et al. . perspectives of CYP2C19 and clopidogrel drug-gene interaction active clinical decision support alerts. Int J Med Inform. 2016;86:117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shiffman RN. Best Practices for Implementation of Clinical Decision Support. Springer: Clinical Decision Support Systems; 2016: 99–109. [Google Scholar]
- 37. Mandl KD, Mandel JC, Kreda DA et al. . The SMART Platform: early experience enabling substitutable applications for electronic health records. J Am Med Inform Assoc. 2012;194:597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoffman JM, Dunnenberger HM, Freimuth RR et al. . Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC). J Am Med Inform Assoc. 2016;234:796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Overby CL, Tarczy-Hornoch P, Veenstra DL et al. . Feasibility of incorporating genomic knowledge into electronic medical records for pharmacogenomic clinical decision support. BMC Bioinformatics. 2010;11 (Suppl 9):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.