Abstract
Objective
Large electronic health record (EHR) datasets are increasingly used to facilitate research on growth, but measurement and recording errors can lead to biased results. We developed and tested an automated method for identifying implausible values in pediatric EHR growth data.
Materials and Methods
Using deidentified data from 46 primary care sites, we developed an algorithm to identify weight and height values that should be excluded from analysis, including implausible values and values that were recorded repeatedly without remeasurement. The foundation of the algorithm is a comparison of each measurement, expressed as a standard deviation score, with a weighted moving average of a child’s other measurements. We evaluated the performance of the algorithm by (1) comparing its results with the judgment of physician reviewers for a stratified random selection of 400 measurements and (2) evaluating its accuracy in a dataset with simulated errors.
Results
Of 2 000 595 growth measurements from 280 610 patients 1 to 21 years old, 3.8% of weight and 4.5% of height values were identified as implausible or excluded for other reasons. The proportion excluded varied widely by primary care site. The automated method had a sensitivity of 97% (95% confidence interval [CI], 94–99%) and a specificity of 90% (95% CI, 85–94%) for identifying implausible values compared to physician judgment, and identified 95% (weight) and 98% (height) of simulated errors.
Discussion and Conclusion
This automated, flexible, and validated method for preparing large datasets will facilitate the use of pediatric EHR growth datasets for research.
Keywords: growth, data quality, research methodology
BACKGROUND AND SIGNIFICANCE
Secondary analyses of data in electronic health records (EHRs) from clinical encounters are increasingly common.1,2 Although clinically obtained data have limitations, they permit evaluation of larger and more diverse populations than is feasible in prospective research-specific evaluations, and therefore represent an invaluable resource for a learning health care system.3 Methods for automated validation and preparation of EHR data are needed to produce unbiased analyses efficiently and reproducibly.
In pediatrics, evaluation of growth is fundamental, and many pediatric research studies include some aspect of growth as an outcome or other variable. The clinical growth measurements obtained in day-to-day care are susceptible to error beyond the imprecision inherent in any anthropometric measurement.4 Some errors result from minor problems with measurement technique. While these errors can be important in certain analyses, they are often small and generally impossible to detect after measurements are recorded. Larger measurement technique errors can result in values that are biologically implausible and can cause problems for many analyses. Implausible values can also result from various types of recording errors including leaving out, adding, or swapping digits; unit errors (eg, measuring weight in pounds but recording it in kilograms); recording weight as height, or vice versa; recording the measurement for a different child; or other causes.
Lawman et al.5 recently published a review of methods for identifying biologically implausible values in large growth datasets and identified 11 different methods. Use of these methods on a single dataset resulted in rates of severe obesity ranging from 7.2% to 8.6%, a relative variation of almost 20%. One approach is to identify likely implausible values and eliminate them from further analysis (also referred to as data cleaning) by excluding those outside of a predetermined plausible range. The range for this trimming process can be based on absolute measurements, percentiles from a published growth reference, or limits based on the study dataset.1,6,7 Trimming is simple to perform and will identify very large errors. However, this approach also excludes extreme values that are not in error (true outliers) and includes values that are within the cutoffs but incorrect for an individual (erroneous inliers).8 Freedman et al.9 recently demonstrated that most of the values flagged as implausible by cutoffs recommended by the World Health Organization are likely true outliers.
Deviation from an expected growth trajectory can also be used to identify implausible values. Three methods identified by Lawman defined implausibility at least in part as due to the change in growth of individuals; all of them involved measurements taken at defined intervals.10–12 Yang and Hutcheon described using conditional percentiles as a strategy to identify implausible values that can be used in growth data with variable intervals. None of these trajectory-based methods have been validated. Inspection of growth measurements for each individual patient by researchers is another commonly used method.13 While flexible, this method depends on the expertise of the inspecting individual, is not reproducible, and is prohibitively time-consuming for large datasets that can include millions of data points. Unfortunately, many publications do not describe the method, if any, used to identify and exclude implausible values.
OBJECTIVE
We describe the development and validation of an automated method for cleaning longitudinal pediatric growth data from EHRs. Earlier versions of the automated method have been used to clean data for 2 prior studies,14,15 but details of the method have not previously been published. We compared the results of a revised version of this automated method with cleaning performed by experienced pediatricians and evaluated its performance when identifying simulated errors.
METHODS
Population
The study included patients 1 to 21 years of age from the Children’s Hospital of Philadelphia Pediatric Research Consortium and the American Academy of Pediatrics’ electronic Pediatric Research in Office Settings network.16 Through this network, Health Insurance Portability and Accountability Act–limited EHR datasets17 from pediatric offices throughout the United States have been combined to facilitate research. The deidentified dataset used for this study included 2 million measurements from 280 610 patients at 47 primary care practice sites from 2009 to 2011. Data for infants younger than 1 year of age were not available in this dataset.
Software
The method was initially developed using Stata (Statacorp LP, College Station, TX, USA) with the published code tested in version 13.1 (Supplementary File S1). The method was translated into R version 3.1.3 (R Foundation for Statistical Computing, Vienna, Austria) (Supplementary File S2) to facilitate its use in a specific computing environment. The translation into R was facilitated by detailed English instructions (Supplementary File S3).18–22
Description of method
The expected size of a child and the expected variability in the sizes of children in a population vary with age. Therefore, pediatric growth is most commonly evaluated using a growth reference that allows determination of a child’s percentile compared to other children of the same age and sex. For this study, we used the Centers for Disease Control and Prevention (CDC) growth reference, which is recommended for use in children >2 years of age in the United States.23,24 When determining the appropriate reference, we assumed that measurements were recumbent for children <2 years and standing height for those >2 years. Since most measurements were for children >2 years, we refer to all linear growth measurements as “height” measurements.
In research settings, pediatric growth is often described using z-scores, which represent the distance, in standard deviations, of a value from the reference median.23 Z-scores can be adjusted for skewness, which is particularly important in evaluation of weight, for which the distribution is considerably skewed. Accounting for skewness in z-scores can have very large effects on extreme z-score values, so modifications have been developed that allow some correction for skewness but better represent absolute differences in attained growth.20,25 One modification, the standard deviation (SD) score,20 was used for this study in order to better identify errors at the extremes of attained growth. As an example, for a 16-year-old male with a change in weight of 150 kg to 200 kg, the z-score has a relatively small increase from 3.7 to 4.5, while the SD score increases from 5.9 to 9.2.
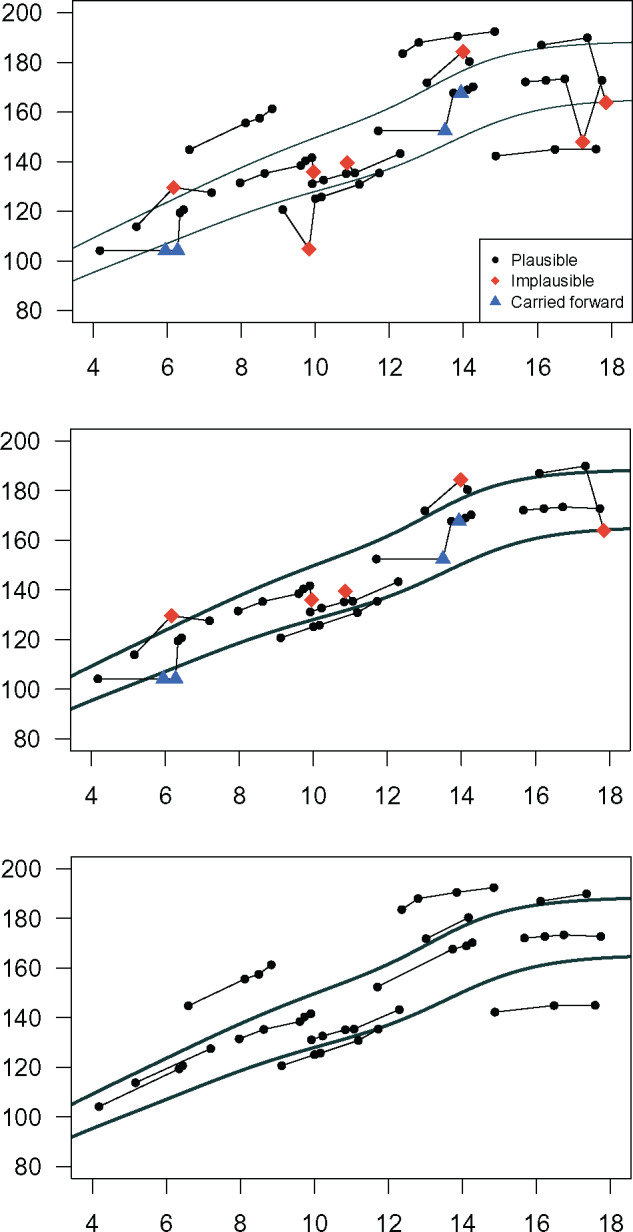
A child’s weight or height z-score or SD score can and often does change over time, but repeated measures of weight (or height) within each child are typically highly correlated.26 This correlation serves as the foundation for our method: SD scores of recorded values of weight or height are compared with a weighted moving average for the child to identify values with an unusually large deviation from the expected value. To estimate a moving average with data points at variable intervals from the point of interest, we used inverse-distance weighting, a method often used for data in physical space.27 This weighted average is a mean exponentially weighted by the inverse of the distance from the value of interest raised to a power, p. Higher values of p increase the relative influence of measures closer to the point of interest. To apply this method to growth data, distance in age (measured in number of days) was used rather than distance in space (Figure 1 ).
Figure 1.
The formula used for calculating exponentially weighted moving averages of standard deviation scores.
The cutpoints for the deviation between recorded and expected SD scores that led to designation as implausible were refined iteratively with a goal to maximize performance of the algorithm compared to the judgment of 1 author (CD) with clinical and research experience in evaluating pediatric growth. The cutpoints vary based on context and can be found in the attached code and instructions. The cutpoints were chosen with a general preference to include values if there was a substantial chance that they were representative of a child’s true growth. Through iterative trials, P = 1.5 was found to best balance the relative influence of measurements at short and long intervals when comparing performance of the algorithm to CD’s judgment. Performance also improved when 5 was added to all age differences to avoid excessive influence by visits separated by only 1 or 2 days.
The median SD score for the children in this study changed across ages, as has been demonstrated in US and other populations.14,28,29 For example, the median weight SD score for males in this population increased from −0.01 at 2 years to 0.24 at 3 years. The variation in mean with age led to larger deviations between SD scores for measurements taken at certain age intervals, with a corresponding increase in the deviation in SD scores and an increased likelihood of excluding measurements for implausibility over these intervals. To minimize variation in performance of the algorithm based on age interval, we recentered the SD scores by subtracting the median SD score for weight and height at each age from the calculated SD score. A sensitivity analysis that performed recentering by site, rather than for the entire study sample combined, produced results that differed for only 0.04% of measurements.
One challenge in using exponentially weighted moving averages is that individual values with large errors also distort the moving average of surrounding values, even if those values are without error. We used several methods in combination to confirm that values with a large deviation were implausible. The primary method required determining 3 moving averages for each measurement: 1 using the method described above, 1 excluding the preceding value if one was present, and 1 excluding the following value if one was present. The deviation of the measurement’s SD score from all 3 moving averages had to be beyond a cutoff in order for the measurement to be designated implausible. Additionally, the algorithm excluded only 1 value at a time per child, even if multiple values met the criteria for exclusion. After the value with the most extreme deviation was excluded, the method was reapplied to the remaining values without the impact of the excluded value to determine if additional exclusions were appropriate. Finally, we ensured that a potentially implausible value deviated in the same direction from the values before and after it, and that adjusting the weight or height by a small amount (5% and 1 cm, respectively) did not bring the deviation of the measurement’s SD score below the cutoff for implausibility. If any of these criteria were not met, the value was not deemed implausible.
Several types of errors interfered with the performance of this method and were handled separately:
Unit errors and switches. English-to-metric conversion errors (unit errors) and recording weight as height or vice versa (switches) can produce large errors in recorded growth measurements. However, suspected unit errors and switches can, in fact, result from other errors (such as a missed or added digit) or be compounded by additional errors. To balance the desire to use measurements that provide information about growth with the potential for introducing error, we developed conservative criteria for choosing which potential unit errors and switches could be corrected. Specifically, the deviation between the value corrected for unit error and the moving average had to be more than −0.3 and less than 0.3, corresponding to ±0.5 SDs from the mean deviation in the study dataset. First and last measurements were treated with special care, because the moving averages for these measurements represent extrapolations rather than interpolations. We excluded, rather than correcting, likely unit errors and switches that were the first or last measurement for a subject because of the increased uncertainty regarding the expected value.
Very extreme values. Values with a z-score or SD score less than −25 or more than 25, representing 25 or more standard deviations from the mean, were universally found to be implausible as true measurements in this dataset. Excluding these measurements without the use of moving averages simplified the remaining steps.
Carried forward. Some measurement values were found to be identical over time for the same child, indicating that these values were likely carried forward from the initial value rather than remeasured. This can happen because an EHR requires entry of a measurement when a clinician does not believe remeasurement is clinically necessary. Because of the imprecision inherent in all growth measurement, relatively few independent measurements documented with appropriate precision should be identical to the prior measurement even if the child’s size has not changed.4 The proportion of children with identical sequential measurements varied widely across primary care practice sites. Inspection of the site with the lowest proportion revealed that even at that site, most values that were identical to the prior value were unlikely to be the result of an independent measurement. Therefore, all values identical to the prior value were excluded, because they were likely to have been carried forward.
Duplicates. Some subjects had more than 1 value for height or weight on the same day. This could result from measurements being taken in more than 1 office (eg, a primary care and subspecialty visit on the same day), remeasuring in the same office because of concern over possible error, or other reasons. For analysis, we wanted to include a maximum of 1 value for each child and parameter per day. Generally, the duplicate with the lowest value of deviation between the recorded value and moving average was retained for analysis. If there was too little information to determine an appropriate value to retain, such as if the duplicate values were not similar to each other and were the only values for that child and parameter, no values were retained for analysis.
Additionally, several types of implausible values were not fully addressed by the moving average method and were addressed specifically after excluding most implausible values:
Height absolute differences. Height monotonically increases in childhood, with exceedingly rare exceptions (such as a vertebral compression fracture), whereas weight decreases relatively often because of illness, intentional weight loss, or other reasons. Any decrease in height >3 cm in sequential measurements was considered beyond the bounds of acceptable measurement error. Height velocity percentiles were used as a starting point to identify additional pairs of values with implausible growth patterns (values in Supplementary File S4).21,22 Once these patterns were identified, differences in moving averages were calculated to determine which of the measurements in the implausible pair was more likely to be erroneous.
Single measurements and pairs. When only 1 or 2 measurements were available for a child and parameter, different criteria were used to identify measurements as implausible, primarily comparing the SD score to the available SD scores for the other parameter (eg, height if evaluating weight) for the same child. Cutoffs for single measurements and pairs were defined with the goal of making the proportion of implausible extreme values for these children similar to the proportion for children with ≥3 measurements.
Error load. Some children had a substantial proportion of values deemed implausible. For example, some had growth measurements that were not correlated with each other and could have represented EHR test patients inadvertently included in the research dataset. For these, all values for 1 or both parameters were excluded depending on the total number of measurements and the proportion that were deemed implausible.
Evaluation of true outliers and erroneous inliers
In the absence of a universally accepted definition of an outlier, we considered 2 definitions of outliers based on the cutoffs in Calle et al.6 (moderate outliers, z-score less than −3 or more than 3, corresponding to percentiles of 0.1 and 99.9) and extreme outliers19 (z-score less than −5 or more than 5). For each set of limits, we determined the proportion of measurements in the original dataset with a CDC z-score beyond these limits that were deemed plausible by the automated method. To evaluate true inliers, we determined the proportion of measurements deemed implausible (not including duplicate or carried forward measurements) with a CDC z-score ≥ −3 and ≤ 3.
Validation
We used 2 methods to evaluate the performance of the automated method: (1) comparison with physician judgment and (2) identification of simulated errors.
Comparison with physician judgment: We compared the results of the method with growth chart review by 2 authors who were not involved in the development or refinement of the method (AF and RW). Both reviewers are experienced pediatricians and clinical researchers who assess growth on a regular basis. We randomly selected 400 primary validation measurements, stratified by growth parameter and sex, with 100 meeting each the following 4 criteria: implausible with |SD score|>3, implausible with |SD score|≤3, plausible with |SD score|>3, and plausible with |SD score|≤3. Stratification by SD score allowed us to specifically evaluate the accuracy of the method with respect to true outliers and erroneous inliers. We selected an additional 118 values to evaluate special error types: duplicates, unit errors, and switches.
The reviewers were provided with plotted growth curves for all patients who had a value selected for validation. The selected value was not marked. Unit errors and switches were presented after correction. Each reviewer independently marked all measurements that he deemed implausible. Discordance between the reviewers was resolved by consensus after their initial review. For values for which the reviewers and the automated method were discordant, the reviewers were asked to classify the discordant value in 1 of 3 ways: as a measurement that they and other reasonable pediatricians (1) would include, (2) would exclude, or (3) might either include or exclude. During this final stage, the growth data were presented in a new random order with new randomly selected identifiers to prevent the reviewers’ initial answers from influencing their classification answers.
The primary outcome for this analysis was the sensitivity and specificity of the automated method for identifying implausible values using reviewer consensus as the gold standard in the 400 primary validation measurements.
We evaluated for differences in accuracy by age group (1 to <3, 3 to <12, and 12 to 21 years) and time interval from the preceding measurement (1 to <30 days, 30 to <365 days, and ≥365 days) using a chi-squared test. We also replicated Yang’s conditional percentile method in our dataset and evaluated its sensitivity and specificity using the physician judgment gold standard, restricted to second and subsequent measurements, because the Yang method is not designed to evaluate the first measurement for a patient.30
Simulated data. Two statistician authors (MR and RL) who were not involved in the development or refinement of the automated method simulated errors in a version of the dataset for which all data for patients with any identified implausible values had been excluded. The error simulation followed a prespecified protocol (details provided in Supplementary File S5). Multiple error types were introduced, and measurements could be affected by >1 error. The automated method was then used to identify the simulated errors. We determined the proportion of simulated errors correctly identified by the automated method and the proportion of measurements deemed implausible by the automated method that represented simulated errors. For a randomly selected sample of 100 height and weight measurements, 2 pediatricians (AF and RW) and a registered dietitian (JL) manually reviewed the manipulated growth patterns to determine whether the introduced errors would be considered implausible by expert reviewers. Each reviewer was asked to review 100 patients for both height and weight (some with multiple introduced errors). Fifty patients for each measurement type were evaluated by 2 reviewers, and the remaining 50 by a single reviewer. This approach reduced the burden on the physician reviewers while maximizing the number of measurements that would be reviewed twice.
Ethics
The study was approved by the American Academy of Pediatrics Institutional Review Board. The Children’s Hospital of Philadelphia Institutional Review Board deemed the study not human subjects research and exempt from review.
RESULTS
We evaluated 1 355 717 weight and 644 878 height measurements for 280 610 patients ages 1 to 21 years, with 92% of the measurements from children 4 to 18 years (Table 1). Overall, 3.8% of weight values and 4.5% of height values were excluded. Notably, the proportion of values excluded, and the most common reasons for exclusion, varied substantially by site (Table 2).
Table 1.
Demographics, overall and by primary care practice site
| Overall | Site minimum | Site maximum | |
|---|---|---|---|
| Number of patients | 280 610 | 432 | 17 933 |
| Number of visits per patient | |||
| Median | 6 | 4 | 12 |
| Maximum | 172 | 23 | 172 |
| Age (years) | |||
| Median | 8.9 | 4.9 | 10.6 |
| Sex | |||
| % Male | 49 | 43 | 57 |
| Racea | |||
| % Asian | 2 | 0 | 9 |
| % Black | 21 | 0 | 96 |
| % Native American | 0.3 | 0 | 3 |
| % White | 51 | 0 | 89 |
| % Multiple | 0.4 | 0 | 6 |
| % Explicit unknown | 3 | 0 | 9 |
| Ethnicity | |||
| % Hispanic | 4 | 0 | 41 |
aDocumentation of race varied by site; some patients had more than 1 category selected.
Table 2.
Characteristics of excluded height and weight measurements, overall and by primary care site
| Type of excluded measurement | Weight |
Height |
||||
|---|---|---|---|---|---|---|
| Overall (%) | N | Site minimum %–maximum % | Overall, % (N) | N | Site minimum %–maximum % | |
| Total | 1 355 717 | 644 878 | ||||
| Included | 96.2 | 1 302 850 | 83.8–99.7 | 95.5 | 615 391 | 71.9–99.4 |
| Carried forward | 3.5 | 46 673 | 0.2–13.2 | 3.5 | 22 352 | 0.4–16.2 |
| Duplicate | 0.2 | 3031 | 0–13.9 | 0.4 | 2736 | 0–12.5 |
| Extreme | 0.02 | 262 | 0–0.1 | 0.04 | 246 | 0–0.3 |
| Primary moving average method | 0.09 | 1210 | 0–0.5 | 0.3 | 1603 | 0.02–2.3 |
| Absolute | N/A | 0.2 | 1505 | 0–2.3 | ||
| Singles/pairs | 0.006 | 87 | 0–0.04 | 0.01 | 86 | 0–0.05 |
| Error load | 0.01 | 136 | 0–0.1 | 0.02 | 148 | 0–0.8 |
| Corrected switch | 0.001 | 20 | 0–0.04 | 0.003 | 20 | 0–0.05 |
| Corrected unit error | 0.01 | 160 | 0–0.1 | 0.01 | 63 | 0–0.07 |
The results for the Stata and R code were discordant for 382 measurements (0.2%). We evaluated a selection of discordant points and found the discrepancies to be related to instability of floating point arithmetic.
True outliers and erroneous inliers
Most moderate outliers were not excluded; 90% (weight [19 194/21 303]) and 63% (height [4413/7060]) of moderate outliers, and 46% (weight [772/1668]) and 17% (height [311/1826]) of extreme outliers were not deemed implausible (Figures 2 and 3 ). Of all measurements excluded due to implausibility, 50% (weight [849/1695]) and 59% (height [2117/3558]) were inliers.
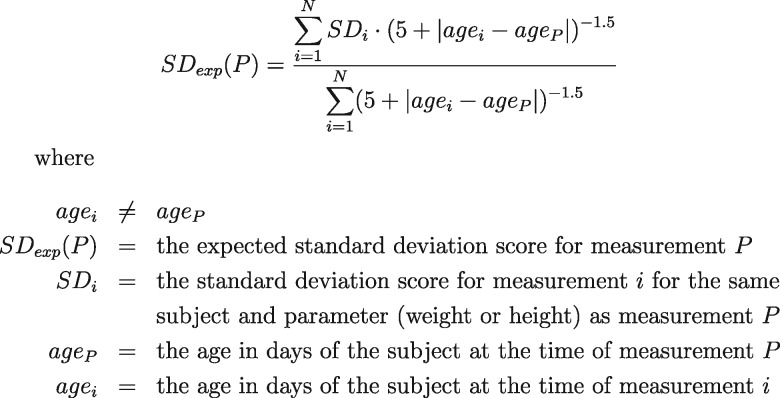
Figure 2.
Twelve individual male weight trajectories were selected as exemplars and plotted against Centers for Disease Control 5th and 95th percentiles (A) without cleaning, (B) cleaned by removing height values with |SD|>3, and (C) cleaned using the automated method. The legend indicates the status of individual values as determined by the automated method.
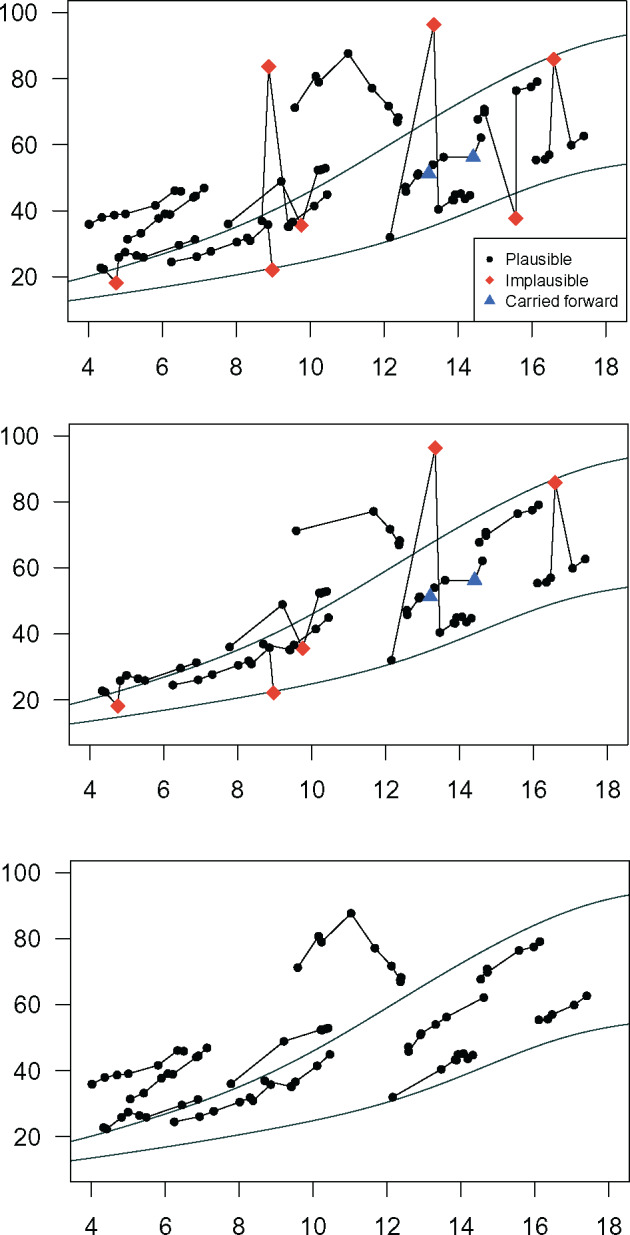
Figure 3.
Eleven individual male height trajectories were selected as exemplars and plotted against Centers for Disease Control 5th and 95th percentiles (A) without cleaning, (B) cleaned by removing weight values with |SD|>3, and (C) cleaned using the automated method. The legend indicates status of individual values as determined by the automated method.
Validation
Comparison with physician judgment. The automated method had a sensitivity of 97% (95% CI, 94–99%) and a specificity of 90% (95% CI, 85–94%) for identifying implausible values compared to physician judgment (Table 3). Agreement between the automated method and physician judgment (κ = 0.87, 95% CI, 0.83–0.91) was similar to the agreement between the 2 reviewers (κ = 0.79, 95% CI, 0.74–0.84). The automated method and physician reviewers were discordant for 27/400 measurements in the primary analysis. Most of the 27 discordant values (81%, n = 22) were included by the physician reviewers and excluded by the automated method. Seven of those 22 measurements were extreme outliers that were classified as “definitely exclude” during the second review by both physicians, indicating that the measurements were likely missed during the initial review. If these likely missed measurements had been deemed implausible by the physician reviewers, specificity would improve to 93%.
Table 3.
Comparison of automated method for identification of implausible measurements with physician judgment for a stratified random selection of measurements
| Primary analysis | ||||
|---|---|---|---|---|
| Type of measurement | Inlier/outlier | Automated method results | Concordant (%) | n/Total |
| Weight | Inlier | Plausible | 98 | 49/50 |
| Implausible | 96 | 48/50 | ||
| Outlier | Plausible | 100 | 50/50 | |
| Implausible | 84 | 42/50 | ||
| Height | Inlier | Plausible | 98 | 49/50 |
| Implausible | 82 | 41/50 | ||
| Outlier | Plausible | 94 | 47/50 | |
| Implausible | 94 | 48/50 | ||
| Overall | 93 | 373/400 | ||
| Special error types | ||||
| Duplicates | 100 | 40/40 | ||
| Corrected unit errors | 95 | 38/40 | ||
| Corrected switchesa | 89 | 34/38 | ||
aOnly 38 of the 40 corrected switches were selected for validation because of sex-stratified selection.
Accuracy was not associated with age (P = .2) or age interval (P = .7). The Yang method had higher specificity (98%) but much lower sensitivity (50%) than our method.
Simulated errors. The automated method correctly identified 95% (weight) and 98% (height) of simulated errors. Of the values the automated method deemed implausible, 98% (weight) and 97% (height) were simulated errors. For a sample of 109 weight and 123 height values for which the algorithm failed to identify a simulated error, at least 1 physician reviewer also indicated that the measurement may have been plausible for 71% of weight and 60% of height measurements.
DISCUSSION
We demonstrated the validity of an automated method for preparing pediatric EHR growth data for analysis that addresses many of the weaknesses of previously used methods. Specifically, it is able to distinguish erroneous and true values for both outliers and measurements in the typical range. It is also able to handle flexible age intervals that no validated methods have previously addressed. Unlike manual individual inspection, the method is feasible to use on very large datasets and is reproducible.
The flexibility of this method is crucial when evaluating retrospective EHR datasets collected from very large networks.31,32 Expected variability in attained growth changes with age, and expected variability in growth velocity changes with both age and the interval between measurements.22 Restricting analyses to children with the same intervals between measurements, such as children who attend all well-child visits as scheduled, would create a high potential for bias. Our algorithm demonstrated validity across ages and age intervals. Because many analyses focus on children at the extremes of size, such as children with obesity or failure to thrive, the ability to avoid excluding true outliers is also particularly valuable. The ability to exclude erroneous inliers may improve power in analysis of growth velocity by eliminating falsely elevated variance. The most common error types varied widely by site, highlighting the need for a method that can identify multiple types of errors.
This method was designed for use in research, but also has potential as a quality improvement or clinical tool. The wide variation in the types of identified errors by site speaks to the potential of targeted approaches to reduce measurement and recording errors. The high variability across primary care practice sites also supports the principle that aggregation of data across sites to create large samples for analysis should always consider site-level sources of measurement error. A modified method could also be used to identify an expected range for measurements of a specific child and improve the accuracy of point-of-care EHR alerts indicating that a child’s measurement might be inaccurate.
Adapting this method for use with other types of clinical EHR data may be possible, but presents special challenges. Most clinical measurements are not as highly correlated as growth data and can vary, in either direction, much more quickly. For example, applying it to vital sign data would be inappropriate, given the wide variation in vital signs that can occur in a short time period. Certain laboratory measurements might be more suited to evaluation by this method, although clinical laboratory measurements are generally performed and recorded using much higher standards for quality control than growth measurements.
Despite efforts to err on the side of not excluding borderline measurements, the method was more likely than the physician reviewers to deem measurements implausible. The wide variation in error type frequency noted among the large number of evaluated sites was unexpected. Notwithstanding the large number of evaluated sites, it is possible that additional types of errors not handled well by this method could be relatively common in other datasets.33 The method is not yet validated for infants or for head circumference and will likely require modification to adequately address newborn weight loss and other differences in infant growth; this work is ongoing. Finally, the automated method cannot identify measurements with small errors that still have the potential to cause bias. Nevertheless, we argue that this method represents a substantial methodological advance, allowing researchers to more effectively use the increasing amount of available growth data in EHRs.
CONCLUSION
We developed an automated method for preparing large quantities of longitudinal pediatric EHR height and weight data for analysis that is valid, flexible, and reproducible. This method has wide potential for use in retrospective analyses of pediatric growth data, regardless of whether growth is used as an outcome, exposure of interest, or potential confounder.
Funding
This project is supported by the Health Resources and Services Administration (HRSA) of the US Department of Health and Human Services (HHS) under grant number R40MC24943 and title “Primary Care Drug Therapeutics CER in a Pediatric EHR Network,” number UB5MC20286 and title “Pediatric Primary Care EHR Network for CER,” and number UA6MC15585 and title “National Research Network to Improve Child Health Care.” Funding was also provided by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) under the Best Pharmaceuticals for Children Act. This information, content, and conclusions are those of the authors and should not be construed as the official position or policy of, nor should any endorsements be inferred by, HRSA, HHS, NICHD, or the US government.
Competing Interests
The authors have no competing interests to declare.
Contributors
CD developed and refined the algorithm, contributed to the design and analysis of the algorithm validation, and drafted the manuscript. MR planned the simulated error validation and created the simulated error dataset, and revised the manuscript for important intellectual content. ARL contributed to the design and analysis of the algorithm validation and revised the manuscript for important intellectual content. AGF acquired the data, contributed to the design and analysis of the algorithm validation, and revised the manuscript for important intellectual content. RCW acquired the data, contributed to the design and analysis of the algorithm validation, and revised the manuscript for important intellectual content. RWG refined the algorithm, contributed to the design and analysis of the algorithm validation, and revised the manuscript for important intellectual content.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank Janeen Leon for her assistance with simulated error validation.
References
- 1. Smith N, Coleman KJ, Lawrence JM. et al. Body weight and height data in electronic medical records of children. Int J Pediatr Obes. 2010;53:237–42. [DOI] [PubMed] [Google Scholar]
- 2. Koebnick C, Coleman KJ, Black MH. et al. Cohort profile: the KPSC Children’s Health Study, a population-based study of 920 000 children and adolescents in southern California. Int J Epidemiol. 2012;413:627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Institute of Medicine. The Learning Healthcare System: Workshop Summary. Washington, DC: National Academies Press; 2007. [PubMed] [Google Scholar]
- 4. Roche AF. Growth, Maturation, and Body Composition: The Fels Longitudinal Study 1929–1991. New York: Cambridge University Press; 1992. [Google Scholar]
- 5. Lawman HG, Ogden CL, Hassink S, Mallya G, Vander Veur S, Foster GD. Comparing methods for identifying biologically implausible values in height, weight, and body mass index among youth. Am J Epidemiol. 2015;1824:359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calle EE, Thun MJ, Petrelli JM, Rodriguez C, Heath CW. Body-mass index and mortality in a prospective cohort of U.S. adults. N Engl J Med. 1999;34115:1097–105. [DOI] [PubMed] [Google Scholar]
- 7. Spencer EA, Appleby PN, Davey GK, Key TJ. Validity of self-reported height and weight in 4808 EPIC-Oxford participants. Public Health Nutr. 2002;54:561–65. [DOI] [PubMed] [Google Scholar]
- 8. Winkler W. Problems with Inliers. Census Bur Res Rep Ser RR9805. 1998. www.census.gov/srd/papers/pdf/rr9805.pdf. Accessed November 11, 2011.
- 9. Freedman DS, Lawman HG, Skinner AC, McGuire LC, Allison DB, Ogden CL. Validity of the WHO cutoffs for biologically implausible values of weight, height, and BMI in children and adolescents in NHANES from 1999 through 2012. Am J Clin Nutr. 2015;1025:1000–06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lawman HG, Mallya G, Veur SV. et al. Trends in relative weight over 1 year in low-income urban youth. Obesity. 2015;232:436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim J, Must A, Fitzmaurice GM. et al. Incidence and remission rates of overweight among children aged 5 to 13 years in a district-wide school surveillance system. Am J Public Health. 2005;959:1588–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sturm R, Datar A. Body mass index in elementary school children, metropolitan area food prices and food outlet density. Public Health. 2005;11912:1059–68. [DOI] [PubMed] [Google Scholar]
- 13. Saari A, Harju S, Mäkitie O, Saha M-T, Dunkel L, Sankilampi U. Systematic growth monitoring for the early detection of celiac disease in children. JAMA Pediatr. 2015;1693:e1525. [DOI] [PubMed] [Google Scholar]
- 14. Daymont C, Neal A, Prosnitz A, Cohen MS. Growth in children with congenital heart disease. Pediatrics. 2013;1311:e236–42. [DOI] [PubMed] [Google Scholar]
- 15. Gerber JS, Bryan M, Ross RK. et al. Antibiotic exposure during the first 6 months of life and weight gain during childhood. JAMA. 2016;31512:1258–65. [DOI] [PubMed] [Google Scholar]
- 16. Fiks AG, Grundmeier RW, Margolis B. et al. Comparative effectiveness research using the electronic medical record: an emerging area of investigation in pediatric primary care. J Pediatr. 2012;1605:719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. National Institutes of Health. How Can Covered Entities Use and Disclose Protected Health Information for Research and Comply with the Privacy Rule? 2007. http://privacyruleandresearch.nih.gov/pr_08.asp. Accessed August 4, 2015.
- 18. CDC Percentile Data Files with LMS Values. August 2009. http://www.cdc.gov/growthcharts/percentile_data_files.htm. Accessed November 14, 2014.
- 19. Vidmar S, Carlin J, Hesketh K. Standardizing anthropometric measures in children and adolescents with new functions for egen. Stata J. 2004;41:50–55. [Google Scholar]
- 20. Cut-Offs to Define Outliers in the 2000 CDC Growth Charts. Atlanta, GA: National Center for Chronic Disease Prevention and Health Promotion, Centers for Disease Control and Prevention; 2014. http://www.cdc.gov/nccdphp/dnpa/growthcharts/resources/BIV-cutoffs.pdf. Accessed July 28, 2014. [Google Scholar]
- 21. Tanner JM, Davies PS. Clinical longitudinal standards for height and height velocity for North American children. J Pediatr. 1985;1073:317–29. [DOI] [PubMed] [Google Scholar]
- 22. WHO Multicentre Growth Reference Study Group. WHO Child Growth Standards: Growth Velocity Based on Weight, Length and Head Circumference: Methods and Development. Geneva, Switzerland: World Health Organization; 2009. [Google Scholar]
- 23. Kuczmarski RJ, Ogden CL, Guo SS. et al. 2000 CDC Growth Charts for the United States: methods and development. National Center for Health Statistics. Vital Health Stat. 2002;11246:1–190. [PubMed] [Google Scholar]
- 24. Grummer-Strawn LM, Reinold C, Krebs NF. Use of World Health Organization and CDC growth charts for children aged 0–59 months in the United States. MMWR Recomm Rep. 2010;59(RR-9):1–15. [PubMed] [Google Scholar]
- 25. WHO Multicentre Growth Reference Study Group. WHO Child Growth Standards Length/Height-for-Age, Weight-for-Age, Weight-for-Length, Weight-for-Height and Body Mass Index-for-Age: Methods and Development. Geneva, Switzerland: World Health Organization; 2006. [Google Scholar]
- 26. Cole TJ. Conditional reference charts to assess weight gain in British infants. Arch Dis Child. 1995;731:8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shepard D. A Two-dimensional Interpolation Function for Irregularly-spaced Data. New York, USA: ACM Press; 1968:517–24. [Google Scholar]
- 28. Cole TJ, Williams AF, Wright CM. Revised birth centiles for weight, length and head circumference in the UK-WHO growth charts. Ann Hum Biol. 2011;381:7–11. [DOI] [PubMed] [Google Scholar]
- 29. Juliusson PB, Roelants M, Hoppenbrouwers K, Hauspie R, Bjerknes R. Growth of Belgian and Norwegian children compared to the WHO growth standards: prevalence below –2 and >2 SD and the effect of breastfeeding. Arch Child. 2010;9610:916–21. [DOI] [PubMed] [Google Scholar]
- 30. Yang S, Hutcheon JA. Identifying outliers and implausible values in growth trajectory data. Ann Epidemiol. 2016;261:77–80.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;214:578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fiks AG, Grundmeier RW, Steffes J. et al. Comparative effectiveness research through a collaborative electronic reporting consortium. Pediatrics. 2015;361:e215–24. [DOI] [PubMed] [Google Scholar]
- 33. Wu S, Miller T, James M. et al. Negation’s not solved: generalizability versus optimizability in clinical natural language processing. PLoS One. 2014;911:e112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.