Abstract
Objective
Our objective is to create a source of synthetic electronic health records that is readily available; suited to industrial, innovation, research, and educational uses; and free of legal, privacy, security, and intellectual property restrictions.
Materials and Methods
We developed Synthea, an open-source software package that simulates the lifespans of synthetic patients, modeling the 10 most frequent reasons for primary care encounters and the 10 chronic conditions with the highest morbidity in the United States.
Results
Synthea adheres to a previously developed conceptual framework, scales via open-source deployment on the Internet, and may be extended with additional disease and treatment modules developed by its user community. One million synthetic patient records are now freely available online, encoded in standard formats (eg, Health Level-7 [HL7] Fast Healthcare Interoperability Resources [FHIR] and Consolidated-Clinical Document Architecture), and accessible through an HL7 FHIR application program interface.
Discussion
Health care lags other industries in information technology, data exchange, and interoperability. The lack of freely distributable health records has long hindered innovation in health care. Approaches and tools are available to inexpensively generate synthetic health records at scale without accidental disclosure risk, lowering current barriers to entry for promising early-stage developments. By engaging a growing community of users, the synthetic data generated will become increasingly comprehensive, detailed, and realistic over time.
Conclusion
Synthetic patients can be simulated with models of disease progression and corresponding standards of care to produce risk-free realistic synthetic health care records at scale.
Keywords: electronic health records, computer simulation, patient-specific modeling, clinical pathways, RS-EHR
BACKGROUND AND SIGNIFICANCE
Health care lags other industries in information technology, data exchange, and interoperability. To close these gaps, developers require access to large repositories of high-quality health datasets for a range of secondary uses that have no clinical or medical implications, including software development, testing, and clinical training.1–4 However, access to real electronic health record (EHR) data is hindered by legal, privacy, security, and intellectual property restrictions. Where real datasets are unavailable, developers have frequently turned to anonymized health records even where there is no need for real data. Anonymized EHRs are bought and sold by a range of government,5,6 commercial corporate, insurance,7 and clinical groups.7–10 The use of real patient records, even those that have been anonymized, comes with issues of privacy, confidentiality, and consent. For instance, there is an especially high risk of harm from public disclosure and identification of individuals resulting from the release or use of anonymized health records, and multiple examples of re-identification of these records have already been observed and publicized.5,11–15 Backlash from these breaches reduces the number of anonymized datasets available for research and development both directly and indirectly, by ad hoc and perfunctory legal remedies that place unrealistic burdens on users to safeguard data.16–18 As a result, a minefield of legal concerns and policy frameworks effectively prevents research and learning.19 To circumvent these challenges, an international research collaboration involving the MITRE Corporation and the HIKER Group (spanning Massey University, New Zealand; the University of Montana, USA; and Macquarie University, Australia) has developed an approach, method, and open-source system for generating realistic synthetic EHRs (RS-EHRs).
Synthetic data generation
A variety of synthetic data generation (SDG) methods have been developed across a wide range of domains, and these approaches described in the literature exhibit a number of limitations.20
First, the collective knowledge of SDG methods has not been well synthesized. Within the health care domain, many approaches to SDG are focused on investigation of pathophysiology, such as synthesis of gene expression21 or neuronal structure data.22 Some SDG projects within health care are either too specific or too general in scope to produce RS-EHRs across a useful range of patient types and clinical conditions.23,24 The most promising published attempts to generate high-quality RS-EHRs are limited in their reliance on real patient records, scope, or quality.24–27 Buczak et al. developed the Synthetic Electronic Medical Records Generator (EMERGE) as a methodology for creating EHRs. Choi et al.27 proposed the medical Generative Adversarial Network (medGAN) to generate realistic synthetic EHRs. While EMERGE and medGAN claim to address privacy concerns through synthesized census-related patient demographic data, real patient records were used in the methods. In some cases, the methods are quite close to anonymization; for example, EMERGE derived the care patterns for populating the synthetic patient record from a real EHR. Thus, any synthetic records developed using the EMERGE methodology are potentially susceptible to re-identification, as are other methods that rely on anonymization.25 MedGAN is quite promising in its use of a real EHR only to check whether or not the generated EHRs are realistic.
Second, there are few systematic methods for assessing the quality and realism of synthetic health record data. Specifically, validation of claims of success and methodologies in SDG is often superficial and limited to overall structural appearance or general statistical comparison, or focused on the speed and number of records created; meanwhile, few studies attempt to assess the validity of the generated data.20 The only complete realism validation method for SDG documented at present was developed by one of the authors, and includes relevant examples drawn from the domain of midwifery.20
Third, the majority of SDG approaches reported in the literature are not described in sufficient detail to replicate the experiments reported, preventing future projects from advancing these efforts.20 Thorough documentation enables repeatability and reusability.28,29 Synthetic data derived from methods without complete documentation cannot be validated, reducing the utility of such methods for the wider scientific community.30–32 A well-documented but incorrect method is therefore preferable to any other method where the documentation itself is incomplete.33 Nonetheless, multiple researchers have reported that the documentation of SDG methods is almost always incomplete, insufficient in detail, and replete with omissions.20,32
There are systems that generate synthetic EHRs that have not been described in peer-reviewed literature. Kartoun24 presents pregenerated datasets of synthetic EHR with an insufficient explanation of how the datasets were generated. These datasets exhibit several inconsistencies between health problems, age, and gender (eg, a randomly sampled male patient reportedly became pregnant and experienced spontaneous abortion at the age of 66 years). MDClone26 builds synthetic data based on analysis of existing cohorts, anonymizes the data, and produces statistically similar populations without reusing any of the original data points. This approach is limited in that it is not open source, not freely available, and requires access to real patient data as an input. Additionally, there is PatientGen, proprietary software developed by the Michigan Health Information Network, which generates statistically based synthetic patients based upon regional demographics, prevalent health incidents, and health states. PatientGen34 tests health information systems without exposing patients to potential data privacy risks. PatientGen is currently not open source and not freely available.
This paper introduces Synthea, an approach, method, and system for RS-EHR generation that advances previous works in several ways. First, our approach guarantees fully synthetic output by accepting only publicly available information and health statistics as inputs. Second, our method generates data based on models of clinical workflow and disease progression that can be easily inspected, modified, and refined, facilitating transparency and continuous improvement. Third, our method includes a temporal model that covers a patient’s entire lifetime instead of focusing on one health problem or disease. Fourth, our method achieves scalability by facilitating collaboration among experts from a range of clinical and technical backgrounds.
Conceptual framework for synthetic EHR generation
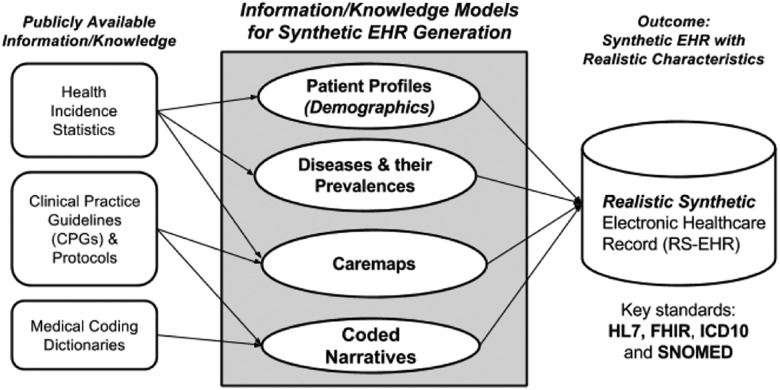
The framework for the synthetic data generation process utilized by Synthea is based on the use of PADARSER, the Publicly Available Data Approach to the Realistic Synthetic EHR.35 The PADARSER framework, unlike EMERGE25 and medGAN,27 assumes that access to the real EHR is impossible or undesirable, relying instead on publicly available datasets to populate the synthetic EHR. Figure 1 presents the PADARSER framework.
Figure 1.
PADARSER as the conceptual framework for Synthea.
PADARSER: (1) emphasizes the use of publicly available health statistics, (2) assumes that access to the real EHR is impossible, (3) makes use of clinical guidelines or protocols in the form of care maps, and (4) employs methods that guarantee inherent realistic properties in the resulting synthetic EHR, making them sufficient enough to replace real records for secondary uses that require realistic but not real EHRs. Privacy preservation is the central aspect of PADARSER, hence public data gathered from aggregate health incident statistics, clinical practice guidelines (CPGs), and medical coding dictionaries are extrapolated and injected into the generation process and applied to the synthetic patient. Care maps are developed from clinician input and CPGs, and a temporal model for each patient is configured using state-transition machines, as demonstrated by both content modelling for synthetic E-Health records20 and Synthea. Realism is improved with regionally prevalent aggregate data sets, clinician input, and CPGs. The resulting RS-EHR has sufficient properties of realism to replace the need for the real EHR in many secondary uses, especially at the aggregated population level, while avoiding hazards associated with the re-identification of anonymized records.
Synthea uses the PADARSER framework in a top-down approach that generates a skeletal synthetic EHR with coded entries in the Health Level-7 (HL7) Fast Healthcare Interoperability Resources (FHIR) standard format for the entire lifetime of the synthetic patient. Meanwhile, the GRiSER (Generating the Realistic Synthetic EHR) method20,35 takes a bottom-up approach by generating robustly detailed synthetic EHR entries for health problems associated with midwifery using the content modelling for synthetic E-Health records web-based application.20 Eventually, ongoing work from the top-down and bottom-up approaches will converge into a complete and comprehensive RS-EHR generation system.
OBJECTIVES
In support of PULSE@MassChallenge, a no-equity global digital health startup incubator designed to connect entrepreneurs with industry, academia, and resources,36 MITRE agreed to provide synthetic health care records for residents of a virtual Commonwealth of Massachusetts in a health information exchange. The data and exchange would be open and available to all participants, eliminating the issues and risks related to using real or deidentified data. To accomplish this goal, the team created an open-source synthetic health simulation, Synthea, that generates patients, simulates their entire life, and outputs their EHRs. Those records were then made publicly available for industry, innovators, research, and education free of legal, privacy, security, financial, and intellectual property restrictions. When the project started, the focus was limited to the progression of type 2 diabetes and related treatments. As we progressed, we quickly expanded the scope to include cardiovascular disease and other common ailments. The goal of the software is to produce increasingly realistic data that are suitable for innovation, development, education, and other nonclinical secondary uses where access to real data is not mandatory and realistic synthetic data will suffice. Therefore, it is important to note the limitation of synthetic data in the realm of clinical research – where synthetic data are not appropriate, nuanced, or suitably rich for biomedicine, genetics, or pharmaceutical discovery. Synthetic data may be useful for training or education in those scenarios, but real clinical discovery requires real data.
MATERIALS AND METHODS
We developed an open-source synthetic health simulation called Synthea that simulates synthetic patients from cradle to grave. As listed in Table 1, our simulation includes models for the top 10 reasons patients visit their primary care physicians and the top 10 chronic conditions responsible for years of life lost, as indicated by the Global Burden of Disease data for the United States.37,38
Table 1.
Diseases and treatments modeled within Synthea
| Top 10 reasons patients visit primary care providers | Top 10 years of life lost (as of 2013) | |
|---|---|---|
| 1 | Routine infant/child health check | Ischemic heart disease |
| 2 | Essential hypertension | Lung cancer |
| 3 | Diabetes mellitus | Alzheimer’s disease |
| 4 | Normal pregnancy | Chronic obstructive pulmonary disease |
| 5 | Respiratory infections (pharyngitis, bronchitis, sinusitis) | Cerebrovascular disease |
| 6 | General adult medical examination | Road injuries |
| 7 | Disorders of lipoid metabolism | Self-harm |
| 8 | Ear infection (otitis media) | Diabetes mellitus |
| 9 | Asthma | Colorectal cancer |
| 10 | Urinary tract infection | Drug use disorders (limited to opioids) |
Synthea was developed with numerous data sources collected on the internet, including US Census Bureau demographics, Centers for Disease Control and Prevention prevalence and incidence rates, and National Institutes of Health reports. The source code and disease models include annotations and citations for all data, statistics, and treatments. These models of diseases and treatments interact appropriately with the health record. For example, procedures and diagnoses correspond with patient-physician encounters, labs are recorded when they are completed, and patients’ records are annotated when they die. When data were publicly available, treatments were modeled on published care maps, otherwise they were constructed by the authors based on publicly available documentation.
RESULTS
The Synthea software design adheres to the PADARSER conceptual framework while being architected to scale to the web and allow additional disease and treatment modules to be added by the community. A million patient records are freely available on the Internet using standard formats (HL7 FHIR and Consolidated-Clinical Document Architecture) and using standard application program interfaces (HL7 FHIR) at https://syntheticmass.mitre.org/fhir/metadata.
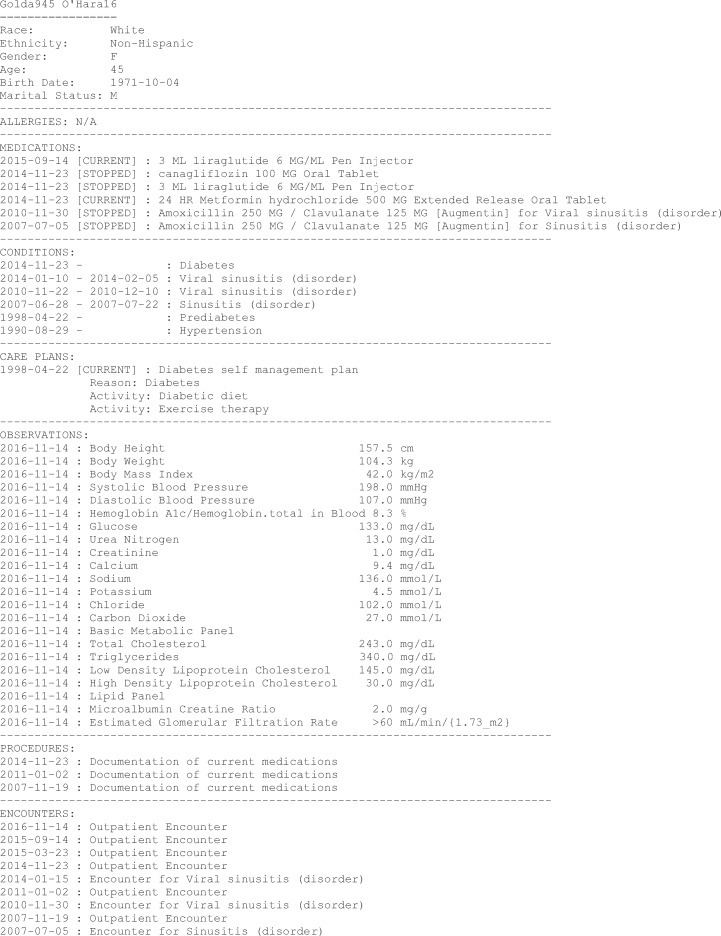
An abridged sample synthetic patient record without longitudinal data (eg, historical vital signs) is displayed in Listing 1, using plain text rather than FHIR JavaScript Object Notation (JSON) for readability.
Listing 1.
Sample synthetic patient data (abridged).
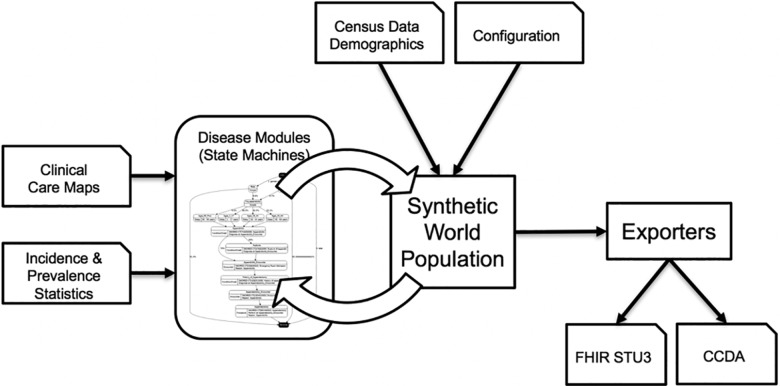
The high-level software architecture of Synthea is illustrated in Figure 2. Clinical care maps and statistics are used to construct models of disease progression and treatment in a Generic Module Framework that encodes these models as state transition machines in an open and documented JSON format.39 Census data and configuration options seed the synthetic world. Each module computes state transitions (if any) for every person at every timestep in the synthetic world. Timesteps are configurable, and default to 7 days. Synthea has logic to handle events that occur within the timestep (eg, it should take much less than an hour to complete a routine medical encounter). Each state or transition in a module can trigger condition onsets, encounters, medication prescriptions, and other clinical events.
Figure 2.
Synthea software architecture.
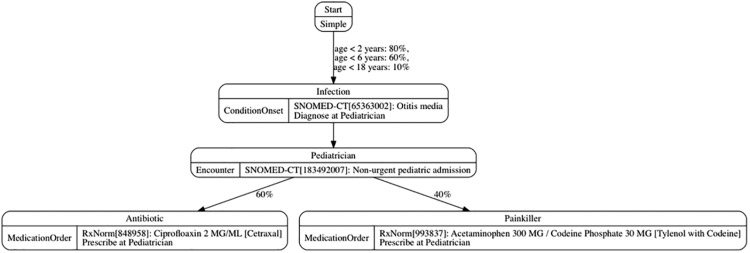
Figure 3 illustrates a simplified example generic module of childhood ear infections. In this example, children get ear infections at different rates based on their age, are then diagnosed at an encounter, and are prescribed either an antibiotic or a painkiller. This example demonstrates several different types of states and transitions.
Figure 3.
Simplified example of a Synthea module.
Listing 2 shows the corresponding JSON definitions for the “Infection” and “Pediatrician” states illustrated in Figure 3. Each state has a name (eg, “Infection”) with a type. Depending on the state type, other attributes (eg, medical terminology codes to use in a diagnosis) or transitions to other states will be defined.
Listing 2.
Partial JSON representation of a Synthea module.
Synthea currently supports 2 categories of states: control and clinical. The control states manage starting, terminating, and delaying the progression through a module state machine. Control states are also used to conditionally control module flow and to read/write attributes on the patient. Clinical states introduce encounters, symptoms, conditions, medications, observations, and care plans to the EHR.
Control states
Synthea currently supports 7 types of control states: Initial, Terminal, Simple, Guard, Delay, SetAttribute, and Counter.
The Initial state is the starting point of a module.
A Terminal state is where the module ends. Some modules do not terminate (eg, simple respiratory diseases, to which people are susceptible their entire lives). All modules always stop and exit when a patient dies.
Simple states are used to control the flow or increase the readability of a module.
Guard states filter out patients if they do not match specific criteria (eg, demographic filters based on age or gender).
Delay states pause the execution of the module until the specified amount of time in the simulation has passed (eg, wait 3–4 weeks for symptoms to emerge).
SetAttribute states set a named attribute/value pair to a patient. These attributes can later be accessed by Guard states, Clinical states, or transitions. For example, several modules can set an attribute regarding prescription opioids (related to a traumatic injury, for instance). This attribute may be later be referenced and used by other modules, such as the opioid addiction module.
Counter states increase or decrease the value of a named attribute on a patient. These attributes can later be accessed by Guard states, Clinical states, or transitions. For example, this can be used to count the number of chemotherapy treatments administered to a patient.
Clinical states
Synthea currently supports 11 types of clinical states: Encounter, ConditionOnset, ConditionEnd, MedicationOrder, MedicationEnd, CarePlanStart, CarePlanEnd, Procedure, Observation, Symptom, and Death. The details of clinical states often include medical terminology codes.
Encounter states represent a point where a patient receives health care services, such as visiting a primary care physician or going to the emergency room. Encounters can either process immediately or wait for the patient’s next regularly scheduled wellness exam.
ConditionOnset and ConditionEnd states represent the onset and end of a condition such as a disease. The condition will not be diagnosed and present on the patient’s record until a target encounter has occurred.
MedicationOrder and MedicationEnd states indicate the initial prescribing and ending of medication.
CarePlanStart and CarePlanEnd states indicate the starting and ending of a planned delivery of care. For example, the care plan associated with a hip replacement might include physical therapy and stretching exercises.
Procedure states indicate that a medical procedure is performed on the patient, such as an appendectomy, colonoscopy, or ultrasound.
Observation states represent the capturing and reporting of patient vital signs and other diagnostic or clinical information.
Symptom states indicate the level of severity of symptoms that a patient is currently experiencing. If symptoms are sufficiently severe, then the patient will schedule an encounter to seek treatment.
The Death state represents the death of the patient. The Death state may process immediately or provide a time representing the remaining life expectancy given the patient’s condition. The Death state may indicate cause of death, which will be represented in the patient’s record as a death certificate.
Transitions
Synthea currently supports 4 types of transitions: direct, distributed, conditional, and complex.
Direct transitions are the simplest; they transition directly to the indicated state.
Distributed transitions will transition to one of several possible states based on the configured distribution. Distribution values are from 0.0 to 1.0, such that a value of 0.55 would indicate a 55% chance of transitioning to the corresponding state.
Conditional transitions will transition to one of several possible states based on conditional logic. A conditional transition consists of an array of condition/transition pairs that are tested in the order they are defined. The first condition that evaluates to true will result in a transition to its corresponding transition state. Conditional logic supports And, Or, Not, At Least, At Most operations upon gender, race, socioeconomic status, and any other patient attribute. Conditional logic can also access the medical record to look at observations, conditions, medications, and care plans.
Complex transitions are a combination of direct, distributed, and conditional transitions. A complex transition consists of an array of condition/transition pairs that are tested in the order they are defined. The first condition that evaluates to true will result in a transition based on its corresponding direct transition or distributions. If the condition/transition pair defines a direct transition, it will transition directly to that named state. If the condition/transition pair defines distributions, it will then transition to one of these according to the same rules as the distributed transition.
Validation of Synthea
An initial validation was performed on the quantities and qualities being produced by the Synthea generator, in particular the type 2 diabetes (T2D) modules. T2D is represented in Synthea by 2 modules: one for disease progression, including patient symptoms and complications such as nephropathy and retinopathy, and one for standards of care, including medications such as insulin and procedures such as dialysis. The interactions between these modules are expected to produce realistic output at the patient level, such that each medical record reflects authentic standards of care, and at the population level, such that prevalence of T2D and complications are close to reality. As real-world statistical information does not always exist regarding the progression of T2D and complications through various stages, in many cases the probability that a patient will transition from one stage to the next is an estimate intended to produce realistic aggregate results.
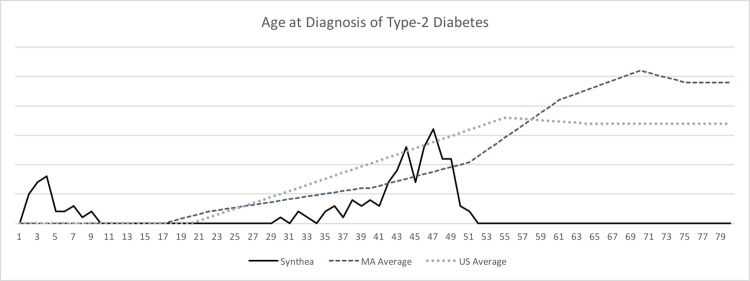
The methodology for this validation involved using synthetic EHRs generated by the T2D modules, contrasting and comparing statistical and treatment properties with publicly available statistics. Some variance with real-world statistics was expected in these first results, as the input data, constraints, algorithms, and methods of Synthea until this point had remained unverified. Figure 4 presents a graph comparing the age at diagnosis for Synthea synthetic patients with the Massachusetts state and US national averages.
Figure 4.
Graph of age at diagnosis of type 2 diabetes.
While Synthea roughly approximated age at diagnosis curves, it incorrectly generated 20% of patients in the infant age group (ages 2–5) and failed to note diabetes onset after age 52 years. Further results indicated that Synthea incorrectly generated prevalence of T2D by racial group. Comorbid conditions such as neuropathy and amputations were generated for nearly every synthetic T2D patient, meaning that overall, a Synthea patient was 4000 times more likely to undergo a diabetes-related amputation than the national or state average. Synthea patients were 110 times more likely to suffer kidney failure and end-stage renal disease, surviving between 2 and 3 times longer than similar real patients. A summary of these generated prevalence statistics is captured in the “Initial Test Run” column of Table 2.
Table 2.
Model prevalence statistics
| Prevalence of condition/medication | Real prevalence | Initial test run | Synthea rerun A | Synthea rerun B | Synthea rerun C |
|---|---|---|---|---|---|
| Prediabetes | 38.00 | 28.18 | 33.14 | 33.64 | 33.78 |
| Diabetes | 8.80 | 9.63 | 6.79 | 6.86 | 6.60 |
| Diabetes by race, black | 12.80 | 9.56 | 11.90 | 9.66 | 10.66 |
| Diabetes by race, Hispanic | 14.20 | 10.02 | 11.47 | 11.99 | 11.60 |
| Diabetes by race, Asian | 16.00 | 9.87 | 13.45 | 12.44 | 13.12 |
| Diabetes by race, white | 6.50 | 9.60 | 5.14 | 5.43 | 5.04 |
| Hypertension | 29.60 | 25.36 | 31.40 | 30.77 | 30.75 |
| Hypertension given diabetes | 59.40 | 68.80 | 50.20 | 53.80 | 53.00 |
| Diabetic retinopathy (DR) | 40.30 | 98.00 | 32.90 | 30.00 | 30.50 |
| Proliferative DR | 7.00 | 86.80 | 5.60 | 4.90 | 5.60 |
| Diabetic macular edema | 11.00 | 88.00 | 7.60 | 6.60 | 8.40 |
| Blindness among diabetic patients | 1.00 | 88.10 | 0.30 | 0.20 | 0.10 |
| Diabetic neuropathy | 50.00 | 98.00 | 42.10 | 38.40 | 42.60 |
| Amputations | 1.00 | 80.20 | 1.40 | 1.40 | 0.80 |
| Diabetic nephropathy | 34.50 | 98.00 | 34.80 | 34.70 | 30.80 |
| Diabetic microalbuminuria | 28.80 | 92.20 | 8.70 | 10.20 | 8.70 |
| Diabetic end-stage renal disease | 0.78 | 82.00 | 0.00 | 0.00 | 0.00 |
| Oral medication | 63.30 | 98.00 | 78.30 | 80.10 | 80.60 |
| Insulin | 30.80 | 0.00 | 39.90 | 40.10 | 35.70 |
| Living diabetics generated (n) | 1000 | 1000 | 1000 | 1000 |
The diabetes disease progression module within Synthea was subsequently modified, primarily by altering the transition probabilities between progressive stages of the 3 main hallmarks of the disease: neuropathy, nephropathy, and retinopathy. Subsequent runs of the simulation produced individual patient records with realistic disease progression and treatments. However, due to the variability of the simulation, replicating population-level statistics proved challenging. For example, the probability of a diabetic patient developing mild eye damage is determined by a “roll of the dice” recurring at each timestep. Once mild eye damage occurs, nonproliferative retinopathy can develop based on a second probability distribution per timestep, which can develop into proliferative retinopathy based on a third probability distribution, eventually leading to macular edema and blindness – each with its own probability distribution per timestep. In general, each run of Synthea is different due to the probabilistic nature of the simulation – the dice rolls come out slightly different each time. As a probability-based patient simulation, Synthea does not predetermine what will happen to each patient in advance, so population-level statistics approach real prevalence based on the probabilities used rather than exactly mirroring those prevalences. This differs from an alternative approach of prepopulating a set of patients exactly matching a set of statistics, and then postprocessing them to fill in the details of disease progression and treatment after the fact. To illustrate the population-level variability in Synthea, the prevalence of diabetic conditions and medications across several simulation runs is summarized in the “rerun” columns of Table 2.
Table 2 shows that disease prevalences in the “reruns” are often underestimated, most notably with retinopathy, neuropathy, and diabetic microalbuminuria. Conversely, medications are often overestimated, probably due to exact prescribing by doctors according to the standard of care and complete medication adherence by patients. Variability between runs is the result of randomness inherent in the probabilistic technique.
In conclusion, the initial validation of the T2D module showed that several population-level statistical issues with regard to prevalence needed to be addressed, and these did not represent an insurmountable challenge to generating realistic synthetic patient records. The flexibility of the approach and of Synthea allows results to be tuned with publicly available statistics for any given global population or infirmity. The significance of the outcome of this validation is that the Synthea generator provides a solid first step toward realizing the goal of generating a dataset of patients that can be representative of the 7 million Massachusetts health care consumers. Modules based on a number of the most prevalent conditions and diseases are being developed and, with input from clinicians and other research groups, are being iteratively tuned to greater degrees of accuracy.
Future work
Synthea has been established as an open-source community project. Synthea-generated EHRs can be used for a variety of secondary uses, and Synthea itself is available for modification, experimentation, or incorporation into other projects. We are aware of several academic researchers performing validation on the data, as well as analytics, in their student projects. We are also aware of several health IT vendors in the FHIR community using the data for development, testing, and public demonstrations of their FHIR-based apps. We hope to add more disease modules with collaboration within the health IT and clinical communities. Our aim is to scale horizontally by positioning the clinical community to iteratively contribute care maps and knowledge (as generic modules) to help produce increasingly realistic patient data for their medical specialties and therapeutic areas. Hence one of our immediate future efforts is to develop a care map authoring tool that would be publicly available as a web application to facilitate the creation of a shared library of care maps that can be used with Synthea or other projects. Other future work areas are summarized in Table 3.
Table 3.
Areas of future work
| Area of future work | Description |
|---|---|
| Model validation | Comprehensive and rigorous module and clinical validation |
| Medication adherence | Adherence rates for prescriptions: medications, care plans (eg, physical exercise) – currently patients always take their medicine |
| Drug-drug interactions | Prescriptions are applied on a module-by-module basis, and these are not adequately deconflicted by any centralized process |
| Physiological data | Modeling organ systems and functions to produce heart rate monitoring streams, etc. |
| Imaging data | Procedures such as X-rays and ultrasounds occur and are recorded, but no synthetic imagery is actually produced |
| Clinical notes | Highly detailed physician notes are not produced |
| Clinician quality | Varying quality of clinicians – currently all clinicians act perfectly according to the modeled standard of care |
| System capacity | Currently there is unlimited care, so restricting access to clinicians, hospital beds, etc. |
| Claims data | Claims and other financial data in addition to the health record |
DISCUSSION
The lack of risk-free health records has been a longstanding issue in health care that has hindered innovation and raised the barrier of entry into the industry – which lags other industries in information technology, data exchange, and interoperability. Approaches and tools are available to generate risk-free synthetic data. Synthea establishes an open-source project for the health IT and clinical community to reuse, experiment with, and generate synthetic data. The quality of synthetic data will improve over time and become increasingly realistic with community contributions. The synthetic data are of suitable quality for a variety of nonclinical secondary uses for >20 clinical conditions. The synthetic data are not suitable or appropriate for research into diseases not covered by the project or research focused on clinical discovery.
CONCLUSION
Synthetic patients can be simulated with models for disease progression and corresponding standards of care to produce risk-free health care records at large scale. Thus, Synthetic electronic health care records for synthetic patients can be generated from publicly available health statistics and clinical guidelines or standards of care from which models of disease progression could be based.
Source code is available online at https://synthetichealth.github.io/synthea and RS-EHR records are available online via a FHIR API at https://syntheticmass.mitre.org/fhir.
FUNDING
The research reported in this publication was supported by the MITRE Innovation Program. Approved for public release; distribution unlimited. Case number 16-2025.
COMPETING INTERESTS
None.
CONTRIBUTORS
Synthea was initially proposed by MK. Software development was completed by JW, AQ, CM, DH, and CD. Conceptual framework and review were provided by KD, TG, and SMcL. Data validation was performed by SMcL. Clinical input and review was provided by JN. All authors contributed to the writing and final approval of this manuscript.
References
- 1. Moniz L., Buczak A. L., Hung L., Babin S., Dorko M., Lombardo J.. Construction and Validation of Synthetic Electronic Medical Records. Online J Public Health Inform. 2009;11: ojphi.v1i1.2720. http://doi.org/10.5210/ojphi.v1i1.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vinzamuri B, Reddy C. Cox Regression with Correlation Based Regularization for Electronic Health Records. Wayne State University; 2013. http://dmkd.cs.vt.edu/papers/ICDM13.pdf [Google Scholar]
- 3. Weiss J, Page D. Forest-based point process for event prediction from electronic health records.European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases . University of Wisconsin; 2013. http://www.ecmlpkdd2013.org/wp-content/uploads/2013/07/128.pdf [Google Scholar]
- 4. Braunstein M. From EHR to Healthcare App Platform. Information Week: Healthcare. 2014. http://www.informationweek.com/healthcare/electronic-health-records/from-ehr-to-healthcare-appplatform/a/d-id/1316263. Accessed July 25, 2017. [Google Scholar]
- 5. Sweeney L, Abu A, Winn J. Identifying Participants in the Personal Genome Project by Name. Harvard University: Data Privacy Lab; 2013. http://dataprivacylab.org/projects/pgp/1021-1.pdf. Accessed July 25, 2017. [Google Scholar]
- 6. Hoeksma J. The NHS’ care.data scheme: What are the risks to privacy? Brit Med J. 2014;348: g1547. [DOI] [PubMed] [Google Scholar]
- 7. Tanner A. This little-known firm is getting rich off your medical data. Fortune. 2016. http://fortune.com/2016/02/09/ims-health-privacy-medical-data/. Accessed July 25, 2017. [Google Scholar]
- 8. Frenkel J. Doctors selling medical records. Herald Sun. May 25, 2005. [Google Scholar]
- 9. Peel D. Personal health data is for sale. Health Privacy Summit. Slides and talking points; 2012. http://goo.gl/bprN1B. Accessed July 25, 2017. [Google Scholar]
- 10. Tate R, Beloff N, Al-Radwan B, et al. Exploiting the potential of large databases of electronic health records for research using rapid search algorithms and an intuitive query interface. J Am Inform Assoc. 2014;212:292–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ross Anderson. Under threat: patient confidentiality and NHS computing. Drugs and Alcohol Today. 2006;64:13–17. [Google Scholar]
- 12.Broken Promises of Privacy: Responding to the Surprising Failure of Anonymization (August 13, 2009). UCLA Law Rev.2010;57:1701, U of Colorado Law Legal Studies Research Paper No. 9–12. Available at SSRN: https://ssrn.com/abstract=1450006.
- 13. El Emam K, Jonker E, Arbuckle L, Malin B. A systematic review of re-identification attacks on health data. PLOS One. 2011; 6(12):e28071 10.1371/journal.pone.0028071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gymrek M, McGuire A, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science. 2013;v339:6117. [DOI] [PubMed] [Google Scholar]
- 15. McLachlan S, Dube K, Gallagher T. Using the CareMap with health incidents statistics for generating the realistic synthetic electronic healthcare record. 2016 IEEE International Conference on Healthcare Informatics (ICHI), 2016, (pp. 439–48). Chicago, IL. doi: 10.1109/ICHI.2016.83. [Google Scholar]
- 16. Dunlevy S. Encrypted private medical records released by Department of Health are vulnerable. News Limited. 2016. http://goo.gl/5OPAO3. Accessed July 25, 2017. [Google Scholar]
- 17. Middleton K. Millions of Australians caught in data breach. The Saturday Paper: Monthly. 2016. https://www.thesaturdaypaper.com.au/news/politics/2016/10/08/millions-australians-caught-health-records-breach/14758452003833. Accessed July 25, 2017. [Google Scholar]
- 18. Rollins A. Medicare data breach prompts law change. Australian Med Online J. 2016. https://ama.com.au/ausmed/medicare-data-breach-prompts-law-change. Accessed July 25, 2017. [Google Scholar]
- 19. Kosseim P, Brady M. Policy by procrastination: Secondary use of electronic health records for health research purposes. McGill JL Health. 2008;2:5. [Google Scholar]
- 20. McLachlan S. Realism in Synthetic Data Generation. MPhil Thesis Computer Science and Information Technology, Massey University: New Zealand; 2016. [Google Scholar]
- 21. Van den Bulcke T, Van Leemput K, Naudts B, et al. SynTReN: a generator of synthetic gene expression data for design and analysis of structure learning algorithms. BMC Bioinformatics. 2006;7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ascoli GA, Krichmar JL, Scorcioni R, Nasuto SJ, Senft SL. Computer generation and quantitative morphometric analysis of virtual neurons. Anat Embryol. 2001;204:283–301. [DOI] [PubMed] [Google Scholar]
- 23. Mwogi TS, Biondich PG, Grannis SJ. An evaluation of two methods for generating synthetic HL7 segments reflecting real-world health information exchange transactions. AMIA Annual Symp Proc. 2014;2014:1855–63. [PMC free article] [PubMed] [Google Scholar]
- 24. Uri Kartoun. 2016. A Methodology to Generate Virtual Patient Repositories. arXiv:1608.00570 [cs.CY] (2016). https://arxiv.org/pdf/1608.00570.pdf [Google Scholar]
- 25. Buczak AL, Babin S, Moniz L. Data-driven approach for creating synthetic electronic medical records. BMC Med Inform Decis Mak. 2010;10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. MDClone. Introducing a New Clinical Data Paradigm. 2016. https://www.mdclone.com. Accessed July 25, 2017. [Google Scholar]
- 27. Choi E, Biswal S, Malin B, Duke J, Stewart WF, Sun J. Generating Multi-label Discrete Electronic Health Records using Generative Adversarial Networks. CoRR abs/1703.06490. 2017. [Google Scholar]
- 28. Crawford S, Stucki L. Peer review and the changing research record. J Am Soc Inf Sci. 1990;41:223–28. [Google Scholar]
- 29. Creswell JW. Research Design: Qualitative, Quantitative, and Mixed Method Approaches. Thousand Oaks, CA: Sage Publications; 2003. Print. [Google Scholar]
- 30. Birkin M, Turner A, Wu B. Proc. Second Int. Conf. e-Social Science, Manchester, UK, A Synthetic Demographic Model of the UK Population: Methods, Progress and Problems. National Centre for e-Social Science; 2006. See http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.143.8415. Accessed July 25, 2017. [Google Scholar]
- 31. Collins H. Changing Order: Replication and Induction in Scientific Practice. University of Chicago Press; 1985. Print. http://press.uchicago.edu/ucp/books/book/chicago/C/bo3623576.html. [Google Scholar]
- 32.Stodden V. Data Sharing in Social Science Repositories: Facilitating Reproducible Computational Research. 2010. https://web.stanford.edu/∼vcs/papers/nips2010Stodden12062010.pdf. Accessed July 25, 2017. [Google Scholar]
- 33. Bolon-Canedo V, Sanchez-Marono N, Alonso-Betanzos A. A review of feature selection methods on synthetic data. Knowledge Inform Syst. 2013;341. [Google Scholar]
- 34. Michigan Health Information Network Shared Services (MiHIN). PatientGen (n.d.). https://mihin.org/services/patientgen/. Accessed July 25, 2017.
- 35. Dube K., Gallagher T.. Approach and Method for Generating Realistic Synthetic Electronic Healthcare Records for Secondary Use In: Gibbons J., MacCaull W. eds. Foundations of Health Information Engineering and Systems . FHIES 2013. Lecture Notes in Computer Science, vol8315 Berlin, Heidelberg: Springer; 2014. 10.1007/978-3-642-53956-5_6. Accessed July 25, 2017. [DOI] [Google Scholar]
- 36. PULSE@MassChallenge http://masschallenge.org/programs/pulse. Accessed July 25, 2017.
- 37. Global Burden of Disease, United States Years of Life Lost. http://ghdx.healthdata.org/global-burden-disease-study-2013-gbd-2013-data-downloads. Accessed November 2016 (no longer available). http://www.healthdata.org/united-states. Accessed July 25, 2017.
- 38. U.S. Census Bureau, Statistical Abstract of the United States: 2001 (121st edition) Washington, DC 2001. Library of Congress Card No. 4-18089. Table No. 159. p. 109.
- 39. Synthea Generic Module Framework. https://github.com/synthetichealth/synthea/wiki/Generic-Module-Framework. Accessed July 25, 2017.